Abstract
Purpose:
The DNA replication licensing machinery is integral to the control of proliferation, differentiation and maintenance of genomic stability in human cells. We have analyzed replication licensing factors (RLFs), together with DNA ploidy status, to investigate their role in progression of penile squamous cell carcinoma (PeScc) and to assess their utility as novel prognostic tools.
Experimental Design:
In a cohort of 141 patients, we linked protein expression profiles of the standard proliferation marker Ki67 and the RLFs Mcm2 and geminin to clinicopathological variables, ploidy status and clinical outcome.
Results:
Increased Ki67, Mcm2 and geminin levels were each significantly associated with arrested tumor differentiation (p<0.0001) and aneuploidy (p≤0.01). Accelerated cell cycle progression was linked to increasing tumor size, stage and depth of invasion. Aneuploid tumors significantly correlated with tumor grade (p<0.0001). Biomarker expression and DNA ploidy status were significant predictors of locoregional disease progression (Mcm2 [p=0.02], geminin [p=0.02], Ki67 [p=0.03] and aneuploidy [p=0.03]) in univariate analysis. Importantly, aneuploidy was a strong independent prognosticator for overall survival (HR 4.19, 95% CI 1.17-14.95, p=0.03). Used in conjunction with conventional pathological information, multiparameter analysis of these variables can stratify patients into low or high risk groups for disease progression (Harrell's c-index=0.88).
Conclusions:
Our findings suggest that RLFs and tumor aneuploidy may be used as an adjunct to conventional prognostic indicators, identifying men at high risk of disease progression. Our results also identify the DNA replication initiation pathway as a potentially attractive therapeutic target in PeScc.
Keywords: DNA replication licensing, geminin, aneuploidy, prognosis, penile carcinoma
INTRODUCTION
Penile carcinoma is a rare cancer in the Western world with an incidence of 0.1 to 0.9 per 100,000 males. However, there is notable geographic variation with much higher incidences in Africa, Asia and South America (1). Patients with squamous cell carcinoma (PeScc) represent the largest subgroup (98% of all cases) and typically present with primary lesions on the glans, foreskin or shaft of the penis (2). Locoregional lymph node status is currently the most powerful prognostic indicator identified in penile cancer. Early, radical inguinal lymphadenectomy has been demonstrated to convey a distinct survival benefit (3-6), but these surgical techniques are limited by their associated high rates of morbidity and mortality (7) Furthermore, controversy exists over patient selection for both radical surgical and chemotherapeutic interventions. Treatment selection is currently based on the patient's age and health status and clinical lymph node stage (cN), together with conventional prognostic factors including histological grade, TMN stage and depth of invasion (8, 9). However, a proportion of men will be incorrectly staged using these algorithms and potentially inappropriately treated (10). Therefore, it is imperative to isolate novel biomarkers in patients with poor prognostic characteristics in order to identify high-risk patients and facilitate treatment stratification.
Tumors acquire a growth advantage over normal tissues through a variety of mechanisms including acquisition of aneuploidy and dysregulation of the mechanisms that control cellular proliferation. The DNA replication licensing pathway has emerged as a powerful downstream mechanism for controlling the proliferative state of cells and ensures that DNA is replicated once and only once per cell cycle, thus maintaining genomic stability (11, 12). During late mitosis and early G1 phase, the replication licensing factors (RLFs) ORC, Cdc6, Cdt1 and Mcm2-7 assemble into pre-replicative complexes (pre-RCs), which render replication origins “licensed” for DNA synthesis. During S phase, Cdc7 kinase and CDKs induce a conformational change in the pre-RC, resulting in recruitment of additional initiator proteins that collectively promote DNA unwinding and recruitment of DNA polymerases (13, 14). During S-G2-M phases the presence of the licensing repressor protein geminin prevents inappropriate re-initiation events at origins that have already been activated (15, 16). Recent studies suggest that dysregulation of replication licensing in early tumorigenesis may arise as a consequence of oncogene-induced cell proliferation, which can cause either under- or over-replication of chromosomal DNA and therefore contribute to the development of aneuploidy commonly seen during multistep tumor progression to an aggressive cancer phenotype (17).
Mcm2-7 (MCM) are expressed throughout the cell cycle (G1-S-G2-M), but are tightly downregulated during exit into out-of-cycle quiescent (G0), differentiated or senescent states (11, 12, 14, 18-21). Thus the MCM proteins represent novel biomarkers of growth and have been confirmed as powerful markers for cancer detection and prognostication in a wide range of tumor types (11, 17, 22). Moreover, expression profiling of MCM together with Ki67 (standard proliferation marker) and geminin (biomarker of S-G2-M progression) allows cells in out-of-cycle states to be distinguished from those residing in-cycle, and can assign cells to G1 and S-G2-M phase (23, 24). Mcm2–7 protein expression also identifies noncycling cells with proliferative potential. The Mcm2/Ki67 ratio therefore defines the proportion of cells that are licensed to proliferate. Consequently, the higher the Mcm2/Ki67 ratio, the greater the proportion of cells that reside in a licensed noncycling state (11, 23-27). Because Ki67 is present throughout the cell cycle in proliferating cells, the ‘geminin/Ki67’ ratio may be used as an indicator of the relative length of G1 phase and the rate of cell cycle progression (11, 23-27). Similarly, the ‘Ki67-geminin’ labeling index can be used to identify the numbers of cells transiting G1 phase (27-29). This information is valuable for determining the cell cycle kinetics of dynamic tumor cell populations and has been shown to be of prognostic significance (11, 23-27).
Complex signaling pathways interlinked with redundant growth regulatory mechanisms contribute to the diverse and heterogeneous effects of oncogenic mutations observed in diverse tumor types (30). Attempts to formulate improved biomarkers for cancer detection and progression alongside the development of novel chemotherapeutic agents against these new molecular targets have met with limited success to date (31). Targeting the DNA replication licensing pathway, which acts as an integration point for upstream mitogenic signaling pathways, is an attractive alternative approach to the identification of new prognostic and predictive markers (11). In light of the biological, prognostic and therapeutic implications of these cell cycle regulators in tumorigenesis, we have investigated their role in the progression of penile carcinoma. We have used multiparameter analysis of Mcm2, geminin and Ki67 to study the cell cycle kinetics of this tumor type in vivo and how deregulation of the replication licensing pathway is linked to acquisition of aneuploidy and clinical outcome. Our findings provide new insights into the biological mechanisms involved in tumor progression of penile carcinoma and how these novel biomarkers of growth might be exploited to predict the in vivo behavior of this rare tumor type.
MATERIALS AND METHODS
Study cohort
From January 1988 to January 2007, 141 patients were diagnosed with carcinoma in situ or invasive squamous cell carcinoma of the penis. All patients had been treated within the North ondon Cancer Network and histological specimens were reviewed by a uro-oncology pathologist at diagnosis. Paraffin wax embedded tissue specimens were retrieved from the pathology archives for all patients and clinical information was sourced from hospital medical records. Local research ethics committee approval for the study was obtained from the joint UCL/UCLH Committees on the Ethics of Human Research. Excised tumors were histologically staged using the revised TMN system criteria 2002 (32). Pathological variables of the primary tumor included: grade, local stage, subtype, extent (unifocal/multifocal), tumor size, depth of invasion and lymphovascular invasion. All pathological parameters were recorded by a specialist uro-oncology pathologist and independently reviewed by a second pathologist. Tumor grade was defined using Broders's classification (33): well differentiated (grade 1), moderately differentiated (grade 2) and poorly differentiated (grade 3). Tumor size was defined as the maximal dimension and depth of invasion measured from adjacent normal epithelium to the deepest invasive point. Lymphovascular invasion was determined microscopically and confirmed using antibodies against endothelial markers CD33 and CD34. Lymph node status (pN) was confirmed following pathological review of inguinal and pelvic lymph node specimens attained through prophylactic or delayed lymphadenectomy. Patients entered into surveillance programs without lymph node surgery were classified as negative after 2 years without disease presentation. Twelve patients with carcinoma in situ were removed from most analyses and 11 patients were lost to follow up. Therefore, 118 patients were included in the long term follow-up survival study. The median follow-up time was 20 months (range 0.8 to 162.4 months). Table 1 summarizes the clinicopathological characteristics of the patients. The mean age of all patients at the time of diagnosis was 62 years (range 27 to 87 years).
Table 1.
Patient characteristics
| Frequency (%) | ||
|---|---|---|
| Age (years) | Mean | 62.7 |
| Range | 27 to 86 | |
| Grade | 1 | 26 (18) |
| 2 | 54 (38) | |
| 3 | 49 (35) | |
| Tumor stage (T) | 1 | 60 (47) |
| 2 | 55 (43) | |
| 3 | 12 (9) | |
| 4 | 2 (1) | |
| Subtype | Standard PeScc | 87 (68) |
| Basaloid | 7 (5) | |
| Warty/Verrucous | 6 (4) | |
| Papillary | 18 (14) | |
| Mixed | 11 (9) | |
| Tumor extent | Unifocal | 101 (78) |
| Multifocal | 28 (22) | |
| Vascular invasion | Negative | 101 (78) |
| Positive | 28 (22) | |
| Size (cm) | ≤2 | 45 (35) |
| >2 | 60 (47) | |
| Unknown | 14 (11) | |
| Depth invasion (mm) | ≤5 | 58 (45) |
| 6-10 | 27 (21) | |
| 11-20 | 17 (13) | |
| >20 | 11 (9) | |
| Unknown | 16 (12) | |
| Nodal stage (N) | 0 | 59 (46) |
| 1 | 12 (9) | |
| 2 | 15 (12) | |
| 3 | 9 (7) | |
| Unknown | 34 (26) | |
| Lymph node metastases | Negative | 59 (46) |
| Positive | 37 (28) | |
| Surveillance | 20 (16) | |
| Unknown | 13 (10) | |
| Distant metastases (M) | Negative | 96 (74) |
| Positive | 16 (12) | |
| Unknown | 17 (14) | |
| Penile cancer death | Alive | 92 (71) |
| Dead | 26 (20) | |
| Unknown | 11 (9) | |
| Survival follow up (months) | Median | 20.1 |
| Range | 1 to 160 |
n=129
Antibodies
Affinity-purified rabbit polyclonal antibody against full-length human geminin was previously generated and validated (14, 34). Ki67 MAb (clone MIB-1) was obtained from DAKO (Glostrup, Denmark) and Mcm2 MAb (clone 46) from BD Transduction Laboratories (Lexington, Kentucky). The specificity of Ki67 and Mcm2 MAb has been extensively studied and validated in previous studies (24, 25, 27).
Immunohistochemistry
Consecutive serial sections were cut from each paraffin embedded tissue block representative of the tumor. Three-micrometer sections were cut onto Superfrost Plus slides (Leica Microsystems, Newcastle upon Tyne, UK), dewaxed in xylene and rehydrated through graded alcohol to water. For antigen retrieval, slides were pressure cooked in 0.1 M citrate buffer (pH 6.0) at 103 kPa for 2.5 minutes. Tissue sections were immunostained using the Bond Polymer Define Detection kit and Bond-X automated system (Leica) according to the manufacturer's instructions. Primary antibodies were applied at the following dilutions: Ki67 (1:70), Mcm2 (1:1000) and geminin (1:500). Coverslips were applied with Pertex mounting medium (CellPath, Newtown, UK). Incubation without the primary antibody was used as a negative control and colonic epithelial sections were used as positive controls.
Protein expression profile analysis
Protein expression analysis was performed by determining the labeling index (LI) of the markers in each tumor, as previously described (23-27, 35). Consecutive serial sections cut from the same formalin-fixed paraffin-embedded tissue block were used to stain for the markers. Slides were evaluated at magnification ×100 to select the advancing edge of the tumor. Selected areas at the advancing edge plus 3-5 adjacent fields perpendicular to the advancing front moving progressively towards the centre of the tumor were image-captured at magnification ×400 with a charge coupled device (CCD) camera and AnalySIS image analysis software (SIS, Munster, Germany). Images were subsequently printed for quantitative analysis, which was undertaken with the observer unaware of the clinicopathological variables. Both positive and negative cells within the field were counted and any stromal or inflammatory cells were excluded. A minimum of 500 cells were counted for each case. The LI was calculated using the following formula: LI = number of positive cells/total number of cells × 100. Normal foreskin and colon specimens were used as external controls. Normal adjacent epithelium acted as an additional internal control. Reassessment of 10 randomly selected cases by an independent assessor showed high levels of agreement.
DNA image cytometry
For each case, one 40 μm section of formalin-fixed, paraffin-embedded tissue, obtained from the same block as that assessed for immunohistochemistry, was used to prepare a suspension of nuclei. Sections were deparaffinised in xylene, rehydrated through decreasing alcohol gradient, and washed twice in PBS. Rehydrated tissue sections were incubated at 37°C in a shaker water bath for 2 hours in the presence of 20 mg/ml bacterial proteinase type XXIV (Sigma). Chilled PBS was added to stop enzymatic digestion and the suspension of nuclei was filtered through a nylon mesh filter and centrifuged at 1500 rpm for 5 minutes. The supernatant was then discarded and the pellet resuspended in 3 ml fresh PBS. A volume of 100 μl of the suspension was cytospun at 1500 rpm for 5 minutes to prepare a monolayer on a Superfrost Plus slide (Visions Biosystems, UK). The density of the nuclear preparation on the slide was checked under a light microscope and an adjusted volume of the suspension was cytospun if correction of the density was required. The monolayer preparations were air-dried and fixed overnight in 4% formaldehyde. After washing in distilled water, slides were incubated in 5 M HCl for 1 hour at room temperature for hydrolysis. Slides were then rinsed in distilled water and incubated in Feulgen-Schiff's solution for 2 hours in the dark. Finally, the slides were washed in running tap water for 10 minutes, dehydrated in increasing alcohol gradient, cleared in Xylene, and coverslipped.
The Fairfield DNA Ploidy System (Fairfield Imaging, Nottingham, UK) was used for image processing, analysis and classification. This consisted of a Zeiss Axioplan microscope equipped with a 40/0.75 objective lens (Zeiss, Jena, Germany), a 546 nm green filter and a black and white high-resolution digital camera (C4742-95; Hamamatsu Photonics K.K., Hamamatsu, Japan) with 1024 × 1024 pixels at 10 bit per pixel. The integrated optical density of each nucleus was calculated based on measurements of optical density and area. Background optical density was measured and corrected for each nucleus. At least 1000 nuclei were scanned for each case and stored in galleries, which were then edited to discard spliced, overlapping and pyknotic nuclei. A minimum of 300 tumor nuclei per case was used by Histogram Draftsman 1.4 (Fairfield Imaging) to create the histograms. Lymphocytes and plasma cells were included as diploid internal controls and sections of high-grade bladder cancer and normal foreskin tissue as external controls for aneuploid and diploid populations respectively.
Ploidy histograms were constructed for 121 cases with nine cases not linked to survival data. Twenty specimens could not be fully processed due to insufficient or poor quality tissue material. Histograms were classified according to the following previously published criteria (36): The tumor was classified as diploid if only one G0/G1 peak (2c) was present, the number of nuclei in the G2 (4c) peak did not exceed 10% of the total number of nuclei and the number of nuclei with a DNA content exceeding 5c did not exceed 1%. A tumor was defined as tetraploid when a peak in the 4c position was present together with a peak in the 8c position or the fraction of nuclei in the 4c region exceeded 10% of the total number of nuclei. A tumor was defined as polyploid when a peak in the 8c position was present together with a peak in the 16c position. The tumor was defined as aneuploid when non-euploid peaks were present or the number of nuclei with a DNA content exceeding 5c/9c, not representing euploid populations, exceeded 1%. The histograms were classified by two independent assessors with a high level of agreement, and without knowledge of the clinicopathological variables. For the purposes of statistical analysis, tetraploid and polyploid tumors were grouped together with aneuploid tumors.
Statistical analysis
Relationships between biomarker expression and other factors were assessed using the Mann-Whitney U, Kruskal-Wallis and Jonckheere-Terpstra tests. Data were summarized as the median value and inter-quartile range of labeling indices observed across the cohort. Multivariable analyses for lymph node status were carried out in three steps using logistic regression: (1) all factors were assessed separately and those with p<0.05 were retained; (2) remaining pathological and biomarker factors were entered into two separate models and backward elimination (BE) applied with p=0.05; (3) remaining factors were entered into a single model and BE applied to produce a final model. Multivariable analyses for overall survival were carried out in a similar fashion using Cox's proportional hazards model. The discriminatory ability of this model was quantified using Harrell's c-index (37), which is analogous to the receiver operating characteristic (ROC) area and gives the probability that two randomly selected patients have concordant predictions and survival times. The c-index takes values between 0.5 (random predictions) to 1 (perfect concordance). Patients were divided into tertile model-based risk groups and labeled as low-, medium- and high-risk for disease progression. Patients with incomplete data were excluded from multivariable analyses. All tests were two-sided and used a significance level of 0.05 with 95% confidence intervals and no allowances were made for multiple hypothesis testing. All analyses were performed using Stata 10 for Windows (StataCorp, College Station, Texas).
RESULTS
RLF expression in normal, dysplastic and malignant penile squamous epithelium
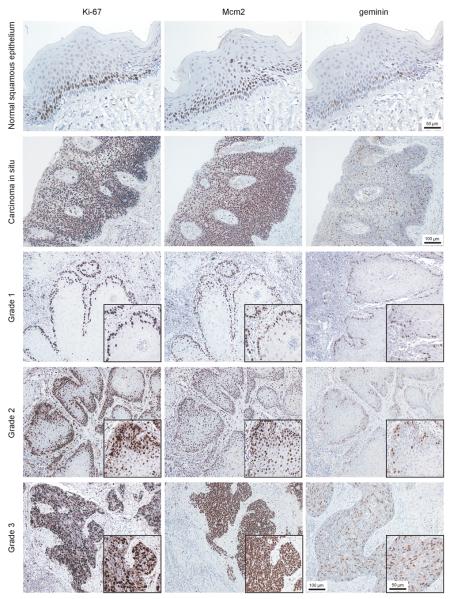
Protein expression profiles for Mcm2, Ki67 and geminin were determined in benign, dysplastic and malignant lesions of the penis (Figure 1) using previously characterized monospecific antibodies against Mcm2, geminin and Ki67 (14, 25, 34). In normal penile squamous epithelium Mcm2, geminin and Ki67 expression was restricted to the basal and suprabasal layers forming the transit amplifying compartment (TAC). Cells in the superficial layers demonstrated a fully differentiated phenotype with flattened morphology and showed low Mcm2, Ki67 and geminin labeling indices (<4%). In striking contrast, dysplastic lesions showed very high replication Mcm2, geminin and Ki67 expression throughout the full thickness of the epithelium, reflecting an expanded proliferative compartment and arrested differentiation (Figure 1). As expected, penile cancers showed high levels of Mcm2, geminin and Ki67 expression, indicative of a hyperproliferative state (Figure 1).
Figure 1.
Photomicrographs of paraffin wax embedded tissue sections of representative normal squamous epithelium, carcinoma in situ and penile squamous cell carcinoma (grades 1 to 3) immunohistochemically stained with antibodies to Ki67, Mcm2 and geminin (magnification x200). Inset demonstrates immunostaining at high magnification (x400).
Relationship between RLF expression, DNA ploidy and clinicopathological characteristics
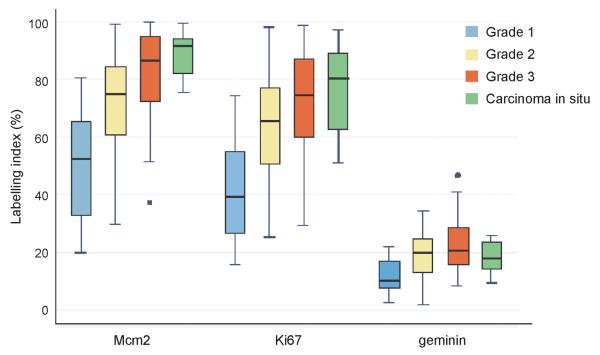
Mcm2, geminin and Ki67 labeling indices were highly significantly associated with tumor grade, with more poorly differentiated tumors showing a higher labeling index (all p<0.0001) (Table 2). Median Mcm2 expression was greater than median Ki67 expression, with both biomarkers mapped over a broad range within each tumor grade. Mcm2 and Ki67 levels were higher than geminin expression in these tumors, reflecting the lower growth fraction identified by geminin, which is only present during S-G2-M (14). There was strong correlation between all biomarkers tested, highlighted by the high concordance between Mcm2 and Ki67 labeling indices (Pearson coefficient ρ=0.87), consistent with their linkage to the cell division cycle. The ‘Ki67-geminin’ score was associated with an increase in tumor grade (p<0.0001), indicative of an increase in the number of cells transiting G1 phase (11, 27). Thus the proportion of tumor cells actively cycling increases with increasing grade. There was little evidence, however, of an increase in the ‘geminin/Ki67’ ratio with increasing grade. This ratio is an indicator of the relative length of G1 phase, and the results suggest that increased recruitment of cells into the cell division cycle was not linked to accelerated cell cycle progression as seen in other tumor types, e.g. epithelial ovarian cancer (25). There was evidence of a trend for decreasing ‘Mcm2/Ki67’ ratio with increasing grade (p=0.09), reflecting a shift in the proportion of nonproliferating cells that are licensed for DNA replication in well differentiated tumors to a population of actively cycling cells in poorly differentiated tumors (25-27, 34).
Table 2.
Relationship between biomarker expression* and tumor differentiation
| Grade 1 (n=26) |
Grade 2 (n=54) |
Grade 3 (n=49) |
p-value† | |
|---|---|---|---|---|
| Mcm2 | 53 (34-65)‡ |
75 (61-84) |
86 (74-95) |
<0.0001 |
| Ki67 | 39 (27-55) |
66 (51-77) |
75 (60-87) |
<0.0001 |
| geminin | 10 (9-17) |
20 (14-24) |
21 (17-29) |
<0.0001 |
| Mcm2/Ki67 § | 1.20 (1.09-1.39) |
1.13 (1.04-1.21) |
1.10 (1.04-1.22) |
0.09 |
| geminin/Ki67 ∥ | 0.30 (0.20-0.34) |
0.30 (0.23-0.35) |
0.30 (0.25-0.38) |
0.22 |
| Ki67 - geminin# | 28 (22-39) |
46 ( 39-54) |
50 (43-61) |
<0.0001 |
n=129
Jonckheere-Terpstra test
Labeling index (expressed as percentages)
Median (inter-quartile range)
Mcm2/Ki67 indicates the ratio of licensed to actively proliferating cells
geminin/Ki67 ratio indicates the relative length of G1 phase
Ki67-geminin represents the percentage of cells that are transiting G1
We observed an association between both high geminin LI and an increase in the ‘geminin/Ki67’ ratio with advanced tumor growth. An increase in these indices is indicative of accelerated cell cycle transit and was linked to increasing tumor stage (geminin p=0.05; geminin/Ki67 p=0.02) and depth of invasion (geminin p=0.02; geminin/Ki67 p=0.03) (Table 3). Furthermore, we noted a significant association between increased biomarker expression and ‘geminin/Ki67’ ratio with increasing tumor size (Mcm2 p<0.01; Ki67 p=0.02; geminin p<0.001; geminin/Ki67 p<0.01) (Table 3). Thus larger tumors contain a greater proportion of cycling cells and this increased growth fraction is also coupled to accelerated cell cycle transit. Interestingly, a strong association was found for increased Mcm2, Ki67 and geminin expression with aggressive tumor subtypes (Table 3). Previous research has shown that verrucous and warty tumors behave in a biologically indolent fashion with low risk for disease progression compared to more aggressive variants, such as papillary and basaloid tumors, which are associated with poorer clinical outcomes (38). We observed that low risk tumors were significantly associated with lower Ki67 and RLF expression, with the lower geminin and ‘Ki67-geminin’ scores indicating smaller numbers of cells transiting S-G2-M and G1 phase, respectively, when compared to aggressive papillary and basaloid tumors. The subtypes associated with poorer clinical outcome therefore displayed a more aggressive cell cycle phenotype.
Table 3.
Relationship between biomarker expression* and clinicopathological variables
| N | Mcm2 | Ki67 | geminin | Mcm2/Ki67** | geminin/Ki67‡‡ | Ki67-geminin†† | ||
|---|---|---|---|---|---|---|---|---|
| Tumor stage | T1 | 60 | 72 (57-91)‡ |
62 (48-76) |
16 (10-23) |
1.11 (1.03-1.24) |
0.27 (0.20-0.35) |
45 (39-53) |
| T2 | 55 | 78 (63-90) |
67 (51-83) |
20 (12-26) |
1.14 (1.06-1.26) |
0.30 (0.25-0.39) |
44 (31-59) |
|
| T3+4 | 14 | 78 (60-83) |
66 (53-76) |
20 (16-25) |
1.14 (1.07-1.24) |
0.33 (0.31-0.36) |
44 (39-47) |
|
|
|
||||||||
| p-value † | 0.39 | 0.72 | 0.05 | 0.38 | 0.02 | 0.59 | ||
|
| ||||||||
|
Lymph node metastases |
Absent | 59 | 74 (59-83) |
63 (47-77) |
18 (11-24) |
1.11 (1.04-1.22) |
0.30 (0.24-0.37) |
45 (29-53) |
| Present | 37 | 82 (67-94) |
70 (60-85) |
23 (17-28) |
1.10 (1.04-1.17) |
0.33 (0.27-0.36) |
50 (39-58) |
|
|
|
||||||||
| p-value § | 0.02 | 0.04 | <0.01 | 0.56 | 0.30 | 0.10 | ||
|
| ||||||||
|
Distant metastases |
Absent | 96 | 75 (59-85) |
65 (50-78) |
18 (11-25) |
1.12 (1.04-1.23) |
0.30 (0.23-0.36) |
45 (34-53) |
| Present | 16 | 82 (72-93) |
69 (60-83) |
23 (15-27) |
1.10 (1.06-1.16) |
0.30 (0.27-0.36) |
54 (39-58) |
|
|
|
||||||||
| p-value § | 0.09 | 0.16 | 0.14 | 0.83 | 0.56 | 0.23 | ||
|
| ||||||||
| Subtype | Standard PeScc |
87 | 74 (61-86) |
60 (50-75) |
18 (13-24) |
1.14 (1.06-1.25) |
0.30 (0.24-0.36) |
44 (31-53) |
| Papillary | 18 | 80 (61-95) |
75 (53-86) |
22 (14-28) |
1.09 (1.03-1.20) |
0.30 (0.25-0.36) |
49 (39-62) |
|
| Basaloid | 7 | 90 (81-99) |
76 (64-87) |
25 (15-28) |
1.14 (1.02-1.27) |
0.26 (0.24-0.38) |
52 (49-54) |
|
| Verrucous + Warty |
6 | 38 (23-51) |
31 (24-45) |
7 (4-12) |
1.22 (1.14-1.43) |
0.26 (0.11-0.36) |
23 (21-35) |
|
| Mixed | 11 | 81 (71-98) |
68 (55-91) |
17 (11-28) |
1.09 (1.04-1.22) |
0.26 (0.16-0.32) |
47 ( 39-61) |
|
|
|
||||||||
| p-value ∥ | <0.01 | <0.01 | 0.03 | 0.59 | 0.70 | <0.01 | ||
|
| ||||||||
| Size | ≤2cm | 45 | 71 (57-78) |
60 (47-69) |
17 ( 9-21) |
1.14 (1.04-1.24) |
0.25 (0.18-0.34) |
44 (33-49) |
| >2cm | 60 | 81 (64-94) |
71 (54-84) |
22 (15-28) |
1.12 (1.06-1.20) |
0.31 (0.27-0.38) |
46 (35-58) |
|
|
|
||||||||
| p-value § | <0.01 | 0.02 | <0.001 | 0.82 | <0.01 | 0.18 | ||
|
| ||||||||
|
Depth of invasion |
≤5mm | 58 | 74 (61-92) |
60 (49-76) |
17 (10-22) |
1.14 (1.04-1.27) |
0.27 (0.20-0.35) |
45 (36-57) |
| 6-10mm | 27 | 76 (58-88) |
70 (46-83) |
20 ( 9-29) |
1.10 (1.07-1.19) |
0.29 (0.24-0.42) |
44 (31-54) |
|
| 10-20mm | 17 | 74 (60-83) |
67 (55-76) |
21 (14-24) |
1.10 (1.00-1.16) |
0.32 (0.29-0.35) |
46 (39-55) |
|
| ≥20mm | 11 | 82 (71-94) |
70 (61-84) |
22 (17-28) |
1.12 (1.06-1.24) |
0.35 (0.28-0.41) |
47 (39-57) |
|
|
|
||||||||
| p-value † | 0.82 | 0.25 | 0.02 | 0.31 | 0.03 | 0.82 | ||
n=129
Labeling index (expressed as percentages)
Median (inter-quartile range)
Jonckheere-Terpstra test
Mann-Whitney test
Kruskal-Wallis test
Mcm2/Ki67 indicates the ratio of licensed to actively proliferating cells
geminin/Ki67 indicates the relative length of G1 phase
Ki67-geminin represents the percentage of cells that are transiting G1
We next sought to investigate the relationship between RLF expression, anaplasia (arrested differentiation) and tumor DNA ploidy in our study cohort. Tumors showing increasing anaplasia also strongly exhibited aneuploidy as determined by image cytometry (p<0.0001) (Supplementary Table 1 and Supplementary Figure 1), suggesting that arrested differentiation and aneuploidy are linked in PeScc. Moreover, the subset of aneuploid tumors was significantly linked to increased Mcm2, geminin and Ki67 expression (p≤0.01), a decreased ‘Mcm2/Ki67’ ratio (p=0.03) and increased ‘Ki67-geminin’ score (p=0.01) (Supplementary Table 2), indicating an increased proportion of actively cycling tumor cells in aneuploid tumors as compared to diploid tumors. Interestingly, further analysis revealed a significant association between aneuploidy and the development of distant metastases (p=0.04).
To investigate the correlation of RLF expression and locoregional disease progression, protein expression profiles for each biomarker were compared with lymph node status (positive/negative) using logistic regression. Ninety-six men with recorded outcomes for these variables were analyzed, with 37 men (38.5%) identified with positive locoregional disease. In univariate analysis, Mcm2 (p=0.02), geminin (p=0.02) and Ki67 (p=0.03) expression were all significant predictors of nodal status. Tumor grade (p=0.02) and stage (p<0.01), presence of vascular invasion (p=0.04) and ploidy status (p=0.03) were also predictive of nodal status, with poorly differentiated tumors (OR 8.76, 95% CI 1.80 to 42.73), high stage disease (OR 4.80, 95% CI 1.81 to 12.74), presence of vascular invasion (OR 2.66, 95% CI 1.04 to 6.75)) and aneuploidy (OR 2.96, 95% CI 1.09 to 8.01) closely linked to advanced disease with regional lymph node involvement. Multivariate analyses were used to construct a final model using these factors. After backwards elimination, biomarker information (Mcm2, Ki67 or geminin LI), vascular invasion and ploidy status were excluded from the final analysis. Tumor grade (p=0.006) and local stage (p=0.003) were independent predictors of positive lymph node disease (Supplementary Table 3). The area under the ROC curve for this model was 0.78 (95% CI 0.69 to 0.87). Although all biomarkers were predictive markers in univariate analysis, no single biomarker was a significant predictor after adjustment for grade and stage. This is due in part to the significant associations between biomarker LI and tumor grade and stage, making it difficult to separate their independent effects.
RLF expression, DNA ploidy, tumor characteristics and overall survival
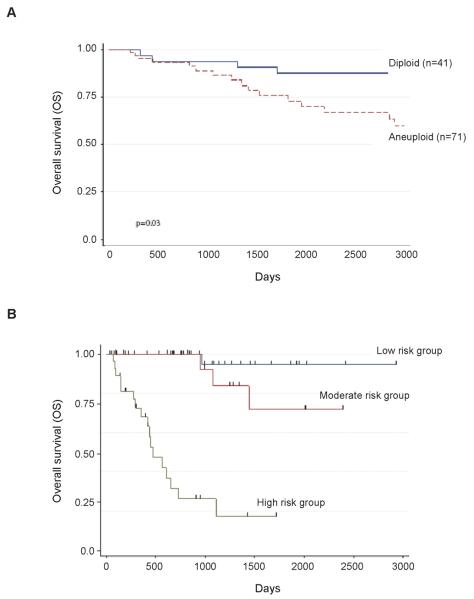
Next, we investigated the survival time for patients with PeScc using Cox's proportional hazards model. The study group for the analysis included 118 men with a recorded survival time. Of these, 26 patients were dead (22%) and 92 alive (78%) at the time of analysis. Univariate analysis showed that Mcm2 (p=0.02) and Ki67 (p=0.04) expression and ‘Ki67-geminin’ score (p=0.03) were all significantly associated with overall survival. Age (p=0.002), tumor stage (p=0.02), depth of invasion (p<0.0001), tumor multifocality (p=0.002), vascular invasion (p=0.04) and ploidy status (p=0.008) were also predictive of overall survival outcomes. Advanced age (HR 1.05 per year, 95% CI 1.02 to 1.1), higher stage tumors (HR 3.89, 95% CI 1.52 to 9.99), multifocal tumors (HR 3.69, 95% CI 1.63 to 8.32), increased depth of invasion (HR 1.045, 95% CI 1.025 to 1.064), presence of vascular invasion (HR per mm 2.23, 95% CI 1.02 to 4.87) and aneuploidy (HR 4.28, 95% CI 1.46 to 12.58) were associated with a significantly shorter overall survival time. It was also noted that lymph node status (negative or positive) was an excellent prognostic factor on univariate analysis (HR 11.07, 95% CI 3.74 to 32.71), in keeping with previous reports (5, 6). Notably, tumor grade failed to predict outcome in this series (p=0.28), emphasizing the limitations with current grading systems. Multivariate analyses were used to construct a final model involving 84 patients (Supplementary Tables 3-7). Backwards elimination resulted in the exclusion of biomarker information (Mcm2, Ki67 or geminin LI), vascular invasion, local tumor stage (pT) and depth of invasion from the final model. However age (p=0.004), lymph node status (p<0.001), tumor multifocality (p=0.002) and aneuploidy (p=0.03) were identified as independent predictors of overall survival (Supplementary Table 8). We quantified the discriminatory ability of this model using Harrell's c-index. In isolation, these predictors have c-index values of: 0.77 (lymph node status), 0.68 (age), 0.65 (aneuploidy) and 0.64 (multifocality). The multivariable model has a c-index of 0.88. Kaplan-Meier curves (Figure 3) demonstrate the survival advantages between patients with diploid versus aneuploid tumors, and between tertile risk groups derived by splitting the patients into equal sized groups based on their predicted risk from the model. Of the 26 deaths analyzed, 24 were cancer specific. We applied the same modeling methodology to cancer-specific survival resulting in the same predictors having univariable associations that were significant at the 5% level. The same biomarker model was then selected (Mcm2 only) but ploidy status was omitted from the pathological model (lymph node metastases, age and extent were selected). The final model omitted Mcm2 (as before) leaving the predictors lymph node metastases, age and extent (for hazard ratios and P-values for this model see Supplementary Table 9).
Figure 3.
Kaplan-Meier survival curves showing cumulative overall survival across the whole series. (A) Aneuploid tumors demonstrate significantly poorer overall survival (HR 4.19 [1.17 to 14.95]; p=0.03); log rank test p<0.004. (B) Tertile risk groups based on calculated prognostic index (low risk PI < 4.4; intermediate risk 4.4 < PI < 6.2; high risk PI > 6.2) using independent predictive factors for overall survival (Harrell's c-index=0.88; log rank test p<0.001).
DISCUSSION
In this study we have assessed the utility of Mcm2 and geminin, alongside DNA ploidy status, as novel biological predictors of outcome in men with PeScc. Our results demonstrate that Mcm2 and geminin labeling indices and aneuploidy are prognostic indicators and predictors of locoregional metastasis. The inverse relationship between RLF expression and differentiation status recapitulates our findings in the in vitro HL60 monocyte/macrophage differentiation model system (18) and has been noted in several other malignancies (25-28, 34, 35, 39-46). This relationship reflects the mutually antagonistic circuits that control cell proliferation and differentiation in human cells and highlights the potential clinical utility of RLFs for improving current tumor grading systems (33). Our data show that analysis of RLFs and ploidy status in primary biopsy material from PeScc can provide important additional prognostic information and identify those tumors with an aggressive cell cycle phenotype, the latter characterized by an increased growth fraction (i.e. an increase in the numbers of cells traversing G1 [‘Ki67-geminin’ score] and S-G2-M phases [geminin LI]) and accelerated cell cycle transit (geminin/Ki67 LI) (11, 25, 26, 28, 34). Notably, the aggressive tumor cell cycle phenotype was linked to increasing tumor size, stage and depth of invasion and to morphological subtypes associated with an adverse prognosis. The aggressive cell cycle phenotype was also linked to tumor ploidy status, suggesting that dysregulation of the DNA replication licensing pathway and cell cycle machinery is linked to the development of aneuploidy in PeScc. In addition, Mcm2, Ki67 and geminin labeling indices and aneuploidy were identified as significant predictors of locoregional metastasis, and aneuploidy was identified as a predictor of distant metastasis.
We have previously shown in the HL60 monocyte/macrophage differentiation model system that loss of proliferative capacity and cell cycle withdrawal following engagement of the somatic differentiation program is tightly coupled to downregulation of core constituents of the DNA replication licensing machinery including the Mcm2-7 proteins (11, 18). This coupling between loss of proliferative capacity, cell cycle withdrawal, downregulation of the Mcm2-7 helicase complex and differentiation has been observed in anal, bladder, cervical, colonic, esophageal, oral, pancreatic and prostatic epithelia (28, 35, 39, 41, 42, 45-50). In normal stratified squamous penile epithelium, we observed downregulation of RLFs as cells exit the cell cycle and engage the somatic differentiation program. This appears to be a ubiquitous mechanism for lowering the proliferative capacity of cells in stem-transit-differentiating self-renewing tissue systems (11, 14, 18-20). In contrast block to the differentiation program (arrested differentiation) that characterizes dysplastic (preinvasive) lesions is associated with persistent expression of MCM proteins even in surface epithelial layers, indicative of cells failing to withdraw from the cell cycle (11, 14, 18, 20, 45). As previously observed for dysplastic lesions of the cervix, esophagus, bladder, oral and anal mucosa, high-level MCM expression was detected in penile dysplasia (11, 28, 35, 41, 42, 45, 48, 49). Surface sampling of penile lesions followed by immunoexpression analysis for MCM proteins could therefore provide a rapid method for distinguishing benign hyperplastic lesions from dysplasia. This approach has already been exploited in screening for cervical cancer and detection of esophageal, lung, bladder, anal and oral dysplasia (11, 42, 45).
Predictive nomograms for lymph node involvement and cancer specific survival in PeScc using traditional histopathological information alone have been developed, but require prospective validation (51, 52). In our study Mcm2 and Ki67 labeling indices, ‘Ki67-geminin’ score, age, tumor stage, depth of invasion, tumor extent, vascular invasion, and nodal and ploidy status were all identified as predictors of overall survival, with lymph node status, tumor extent and ploidy status identified as independent predictors of overall survival. Interestingly, we have shown that these parameters can be incorporated into a simple prediction model to stratify patients into high-, intermediate- and low-risk groups for disease progression in PeScc. Notably, the multivariable model suggests that low-risk patients are at minimal risk of disease progression compared to men assigned to moderate- or high-risk groups. This is highlighted by overall survival rates at three years of 97%, 93% and 27%, and five-year survival figures of 97%, 72% and 18% for low-, moderate- and high-risk men respectively. This provides a potentially powerful approach for early treatment stratification of patients, with low risk patients assigned to surveillance programs and targeting of high risk patients with radical surgical and adjuvant chemotherapeutic interventions. Further studies in additional patient cohorts are now warranted to confirm the predictive power of this model in the risk stratification of PeScc.
Inhibition of the DNA replication initiation machinery has been shown to provoke a cancer-cell-specific apoptotic response as a result of the loss or impairment of a putative checkpoint for replication-competent origins during tumorigenesis (17, 53). Here we have shown that increasing dysregulation of the DNA replication licensing pathway is linked to emergence of an aggressive cell cycle phenotype that impacts on the in vivo behavior of this tumor type. The DNA replication licensing pathway therefore appears to be also a potentially attractive therapeutic target in PeScc.
STATEMENT OF TRANSLATIONAL RELEVANCE.
Penile cancer is a rare malignancy associated with poor survival outcomes for patients with advanced disease states. Identifying patients who will benefit from aggressive therapeutic strategies remains problematic. The delineation of novel, effective molecular targets capable of delivering powerful, prognostic information as well as defining new therapeutic targets in this clinical setting is challenging. In this study we demonstrate that dysregulation of the DNA replication licensing pathway is intricately linked to tumor progression and aneuploidy. Using a panel of cell cycle biomarkers, we have identified key “proliferation signatures” that reflect aggressive cell cycle phenotypes linked to poorer clinical outcomes. Integration of these biomarkers with conventional clinicopathological parameters in a novel predictive model has potential to facilitate identification of those patients most likely to benefit from radical surgical and chemotherapeutic interventions.
Supplementary Material
Figure 2.
The median (solid black line), interquartile range (boxed), and range (enclosed by lines) of Mcm2, Ki67 and geminin expression are shown according to tumor grade (outlying cases are shown by isolated points). The mean and interquartile range of Mcm2 and Ki67 levels increase with increasing grade compared with geminin.
Abbreviations
- CDK
cyclin dependent kinase
- CIS
carcinoma in situ
- HR
hazard ratio
- LI
labeling index
- MAb
monoclonal antibody
- Mcm2-7
minichromosome maintenance proteins 2-7
- OR
odds ratio
- ORC
origin recognition complex
- PeScc
penile squamous cell carcinoma
- RC
replication complex
- RLF
replication licensing factor
- ROC
receiver operating characteristic
References
- 1.Parkin DM, Muir CS. Cancer Incidence in Five Continents. Comparability and quality of data. IARC Sci Publ. 1992:45–173. [PubMed] [Google Scholar]
- 2.Burgers JK, Badalament RA, Drago JR. Penile cancer. Clinical presentation, diagnosis, and staging. Urol Clin North Am. 1992;19:247–56. [PubMed] [Google Scholar]
- 3.Horenblas S, van TH, Delemarre JF, Moonen LM, Lustig V, van Waardenburg EW. Squamous cell carcinoma of the penis. III. Treatment of regional lymph nodes. J Urol. 1993;149:492–7. doi: 10.1016/s0022-5347(17)36126-8. [DOI] [PubMed] [Google Scholar]
- 4.Kroon BK, Horenblas S, Lont AP, Tanis PJ, Gallee MP, Nieweg OE. Patients with penile carcinoma benefit from immediate resection of clinically occult lymph node metastases. J Urol. 2005;173:816–9. doi: 10.1097/01.ju.0000154565.37397.4d. [DOI] [PubMed] [Google Scholar]
- 5.Ravi R. Correlation between the extent of nodal involvement and survival following groin dissection for carcinoma of the penis. Br J Urol. 1993;72:817–9. doi: 10.1111/j.1464-410x.1993.tb16273.x. [DOI] [PubMed] [Google Scholar]
- 6.Ornellas AA, Kinchin EW, Nobrega BL, Wisnescky A, Koifman N, Quirino R. Surgical treatment of invasive squamous cell carcinoma of the penis: Brazilian National Cancer Institute long-term experience. J Surg Oncol. 2008;97:487–95. doi: 10.1002/jso.20980. [DOI] [PubMed] [Google Scholar]
- 7.Ravi R. Morbidity following groin dissection for penile carcinoma. Br J Urol. 1993;72:941–5. doi: 10.1111/j.1464-410x.1993.tb16304.x. [DOI] [PubMed] [Google Scholar]
- 8.Solsona E, Algaba F, Horenblas S, Pizzocaro G, Windahl T. EAU Guidelines on Penile Cancer. Eur Urol. 2004;46:1–8. doi: 10.1016/j.eururo.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Velazquez EF, Cubilla AL. Penile squamous cell carcinoma: anatomic, pathologic and viral studies in Paraguay (1993-2007) Anal Quant Cytol Histol. 2007;29:185–98. [PubMed] [Google Scholar]
- 10.Hegarty PK, Kayes O, Freeman A, Christopher N, Ralph DJ, Minhas S. A prospective study of 100 cases of penile cancer managed according to European Association of Urology guidelines. BJU Int. 2006;98:526–31. doi: 10.1111/j.1464-410X.2006.06296.x. [DOI] [PubMed] [Google Scholar]
- 11.Williams GH, Stoeber K. Cell cycle markers in clinical oncology. Curr Opin Cell Biol. 2007;19:672–9. doi: 10.1016/j.ceb.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Blow JJ, Hodgson B. Replication licensing--defining the proliferative state? Trends Cell Biol. 2002;12:72–8. doi: 10.1016/s0962-8924(01)02203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–74. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 14.Eward KL, Obermann EC, Shreeram S, et al. DNA replication licensing in somatic and germ cells. J Cell Sci. 2004;117:5875–86. doi: 10.1242/jcs.01503. [DOI] [PubMed] [Google Scholar]
- 15.Wohlschlegel JA, Kutok JL, Weng AP, Dutta A. Expression of geminin as a marker of cell proliferation in normal tissues and malignancies. Am J Pathol. 2002;161:267–73. doi: 10.1016/S0002-9440(10)64178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–86. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blow JJ, Gillespie PJ. Replication licensing and cancer--a fatal entanglement? Nat Rev Cancer. 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barkley LR, Hong HK, Kingsbury SR, James M, Stoeber K, Williams GH. Cdc6 is a rate-limiting factor for proliferative capacity during HL60 cell differentiation. Exp Cell Res. 2007;313:3789–99. doi: 10.1016/j.yexcr.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Kingsbury SR, Loddo M, Fanshawe T, et al. Repression of DNA replication licensing in quiescence is independent of geminin and may define the cell cycle state of progenitor cells. Exp Cell Res. 2005;309:56–67. doi: 10.1016/j.yexcr.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 20.Stoeber K, Tlsty TD, Happerfield L, et al. DNA replication licensing and human cell proliferation. J Cell Sci. 2001;114:2027–41. doi: 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- 21.Stoeber K, Mills AD, Kubota Y, et al. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J. 1998;17:7219–29. doi: 10.1093/emboj/17.24.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldwin P, Laskey R, Coleman N. Translational approaches to improving cervical screening. Nat Rev Cancer. 2003;3:217–26. doi: 10.1038/nrc1010. [DOI] [PubMed] [Google Scholar]
- 23.Kulkarni AA, Kingsbury SR, Tudzarova S, et al. Cdc7 kinase is a predictor of survival and a novel therapeutic target in epithelial ovarian carcinoma. Clin Cancer Res. 2009;15:2417–25. doi: 10.1158/1078-0432.CCR-08-1276. [DOI] [PubMed] [Google Scholar]
- 24.Loddo M, Kingsbury SR, Rashid M, et al. Cell-cycle-phase progression analysis identifies unique phenotypes of major prognostic and predictive significance in breast cancer. Br J Cancer. 2009;100:959–70. doi: 10.1038/sj.bjc.6604924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulkarni AA, Loddo M, Leo E, et al. DNA replication licensing factors and aurora kinases are linked to aneuploidy and clinical outcome in epithelial ovarian carcinoma. Clin Cancer Res. 2007;13:6153–61. doi: 10.1158/1078-0432.CCR-07-0671. [DOI] [PubMed] [Google Scholar]
- 26.Shetty A, Loddo M, Fanshawe T, et al. DNA replication licensing and cell cycle kinetics of normal and neoplastic breast. Br J Cancer. 2005;93:1295–300. doi: 10.1038/sj.bjc.6602829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudderidge TJ, Stoeber K, Loddo M, et al. Mcm2, Geminin, and KI67 define proliferative state and are prognostic markers in renal cell carcinoma. Clin Cancer Res. 2005;11:2510–7. doi: 10.1158/1078-0432.CCR-04-1776. [DOI] [PubMed] [Google Scholar]
- 28.Dudderidge TJ, McCracken SR, Loddo M, et al. Mitogenic growth signalling, DNA replication licensing, and survival are linked in prostate cancer. Br J Cancer. 2007;96:1384–93. doi: 10.1038/sj.bjc.6603718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wharton SB, Maltby E, Jellinek DA, et al. Subtypes of oligodendroglioma defined by 1p,19q deletions, differ in the proportion of apoptotic cells but not in replication-licensed non-proliferating cells. Acta Neuropathol. 2007;113:119–27. doi: 10.1007/s00401-006-0177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bild AH, Potti A, Nevins JR. Linking oncogenic pathways with therapeutic opportunities. Nat Rev Cancer. 2006;6:735–41. doi: 10.1038/nrc1976. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–56. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 32.Sobin LH, Wittekind C. TNM Classification of Malignant Tumours. Wiley; 2002. [Google Scholar]
- 33.Broders AC. Squamous-cell epithelioma of the skin: A study of 256 cases. Ann Surg. 1921;73:141–60. doi: 10.1097/00000658-192102000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wharton SB, Hibberd S, Eward KL, et al. DNA replication licensing and cell cycle kinetics of oligodendroglial tumours. Br J Cancer. 2004;91:262–9. doi: 10.1038/sj.bjc.6601949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng MV, Grossfeld GD, Williams GH, et al. Minichromosome maintenance protein 2 expression in prostate: characterization and association with outcome after therapy for cancer. Clin Cancer Res. 2001;7:2712–8. [PubMed] [Google Scholar]
- 36.Haroske G, Giroud F, Reith A, Bocking A. 1997 ESACP consensus report on diagnostic DNA image cytometry. Part I: basic considerations and recommendations for preparation, measurement and interpretation. European Society for Analytical Cellular Pathology. Anal Cell Pathol. 1998;17:189–200. doi: 10.1155/1998/390837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harell FE. Regression Modeling Strategies. Springer; 2001. [Google Scholar]
- 38.Cubilla AL, Reuter VE, Gregoire L, et al. Basaloid squamous cell carcinoma: a distinctive human papilloma virus-related penile neoplasm: a report of 20 cases. Am J Surg Pathol. 1998;22:755–61. doi: 10.1097/00000478-199806000-00014. [DOI] [PubMed] [Google Scholar]
- 39.Ayaru L, Stoeber K, Webster GJ, et al. Diagnosis of pancreaticobiliary malignancy by detection of minichromosome maintenance protein 5 in bile aspirates. Br J Cancer. 2008;98:1548–54. doi: 10.1038/sj.bjc.6604342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatrath P, Scott IS, Morris LS, et al. Aberrant expression of minichromosome maintenance protein-2 and Ki67 in laryngeal squamous epithelial lesions. Br J Cancer. 2003;89:1048–54. doi: 10.1038/sj.bjc.6601234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freeman A, Morris LS, Mills AD, et al. Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin Cancer Res. 1999;5:2121–32. [PubMed] [Google Scholar]
- 42.Going JJ, Keith WN, Neilson L, Stoeber K, Stuart RC, Williams GH. Aberrant expression of minichromosome maintenance proteins 2 and 5, and Ki-67 in dysplastic squamous oesophageal epithelium and Barrett's mucosa. Gut. 2002;50:373–7. doi: 10.1136/gut.50.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez MA, Pinder SE, Callagy G, et al. Minichromosome maintenance protein 2 is a strong independent prognostic marker in breast cancer. J Clin Oncol. 2003;21:4306–13. doi: 10.1200/JCO.2003.04.121. [DOI] [PubMed] [Google Scholar]
- 44.Obermann EC, Eward KL, Dogan A, et al. DNA replication licensing in peripheral B-cell lymphoma. J Pathol. 2005;205:318–28. doi: 10.1002/path.1695. [DOI] [PubMed] [Google Scholar]
- 45.Williams GH, Romanowski P, Morris L, et al. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci U S A. 1998;95:14932–7. doi: 10.1073/pnas.95.25.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams GH, Swinn R, Prevost AT, et al. Diagnosis of oesophageal cancer by detection of minichromosome maintenance 5 protein in gastric aspirates. Br J Cancer. 2004;91:714–9. doi: 10.1038/sj.bjc.6602028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoeber K, Halsall I, Freeman A, et al. Immunoassay for urothelial cancers that detects DNA replication protein Mcm5 in urine. Lancet. 1999;354:1524–5. doi: 10.1016/S0140-6736(99)04265-8. [DOI] [PubMed] [Google Scholar]
- 48.Stoeber K, Swinn R, Prevost AT, et al. Diagnosis of genito-urinary tract cancer by detection of minichromosome maintenance 5 protein in urine sediments. J Natl Cancer Inst. 2002;94:1071–9. doi: 10.1093/jnci/94.14.1071. [DOI] [PubMed] [Google Scholar]
- 49.Scarpini C, White V, Muralidhar B, et al. Improved screening for anal neoplasia by immunocytochemical detection of minichromosome maintenance proteins. Cancer Epidemiol Biomarkers Prev. 2008;17:2855–64. doi: 10.1158/1055-9965.EPI-08-0288. [DOI] [PubMed] [Google Scholar]
- 50.Torres-Rendon A, Roy S, Craig GT, Speight PM. Expression of Mcm2, geminin and Ki67 in normal oral mucosa, oral epithelial dysplasias and their corresponding squamous-cell carcinomas. Br J Cancer. 2009;100:1128–34. doi: 10.1038/sj.bjc.6604967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ficarra V, Zattoni F, Artibani W, et al. Nomogram predictive of pathological inguinal lymph node involvement in patients with squamous cell carcinoma of the penis. J Urol. 2006;175:1700–4. doi: 10.1016/S0022-5347(05)01003-7. [DOI] [PubMed] [Google Scholar]
- 52.Kattan MW, Ficarra V, Artibani W, et al. Nomogram predictive of cancer specific survival in patients undergoing partial or total amputation for squamous cell carcinoma of the penis. J Urol. 2006;175:2103–8. doi: 10.1016/S0022-5347(06)00313-2. [DOI] [PubMed] [Google Scholar]
- 53.Jackson PK. Stopping replication, at the beginning. Nat Chem Biol. 2008;4:331–2. doi: 10.1038/nchembio0608-331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.