Abstract
Occupational exposure to formaldehyde has been linked to nasopharyngeal carcinoma. To date, mechanistic explanations for this association have primarily focused on formaldehyde-induced cytotoxicity, regenerative hyperplasia and DNA damage. However, recent studies broaden the potential mechanisms as it is now well established that formaldehyde dehydrogenase, identical to S-nitrosoglutathione reductase, is an important mediator of cGMP-independent nitric oxide signaling pathways. We have previously described mechanisms by which formaldehyde can influence nitrosothiol homeostasis thereby leading to changes in pulmonary physiology. Considering evidences that nitrosothiols govern the Epstein-Barr virus infection cycle, and that the virus is strongly implicated in the etiology of nasopharyngeal carcinoma, studies are needed to examine the potential for formaldehyde to reactivate the Epstein-Barr virus as well as additively or synergistically interact with the virus to potentiate epithelial cell transformation.
Introduction
The International Agency for Research on Cancer (IARC) has classified formaldehyde as a human carcinogen based, in part, on epidemiological evidence that formaldehyde increases the risk of nasopharyngeal carcinoma (NPC) [1-4]. This cancer exhibits remarkable geographical distribution that is posited to result from environmental factors, host genetic factors (i.e. race), and genetic variation in the gammaherpesvirus Epstein-Barr virus (EBV) that is widely believed to play a role in the etiology of NPC [5-9]. Mechanisms posited to explain formaldehyde carcinogenicity have invoked evidence for cytotoxicity, compensatory cell proliferation, and genotoxicity in animal bioassays [4,10-13]. However, mathematical models of the human respiratory passages indicate that the highest doses of formaldehyde are predicted to be above the hard palate, with relatively less vapor reaching the nasopharynx [14]. Histopathological samples from workers exposed to formaldehyde exhibit only mild nasal tissue pathology [4,15], suggesting that tissue damage might not occur in more distal regions. Thus, available data do not paint a coherent mechanism for formaldehyde to increase the risk of NPC, and moreover the association continues to be disputed [16-18].
Few articles published on formaldehyde toxicity over the past decade have acknowledged that the class III alcohol dehydrogenase (ADH3), also termed formaldehyde dehydrogenase and S-nitrosoglutathione (GSNO) reductase, is a key mediator of the nitric oxide (NO)-related post-translational protein modification known as S-nitrosylation (Figure 1a) [19-24]. The S-nitrosylation of protein cysteine residues by NO and GSNO regulates a broad spectrum of cellular proteins and functions [25-36]. Dysregulation of nitrosothiol homeostasis has been implicated in diseases of the central nervous system, cardiovascular system, and lung [37-43]. We have previously described how ADH3 and nitrosothiols influence pulmonary physiology, and how formaldehyde might influence nitrosothiol homeostasis [24,44,45]. Considering that nitrosothiols are implicated in the regulation of EBV and other viruses [28,29,32,46], herein we describe mechanisms by which formaldehyde might influence the reactivation of the EBV and additionally interact with EBV latency programs to potentiate epithelial cell transformation.
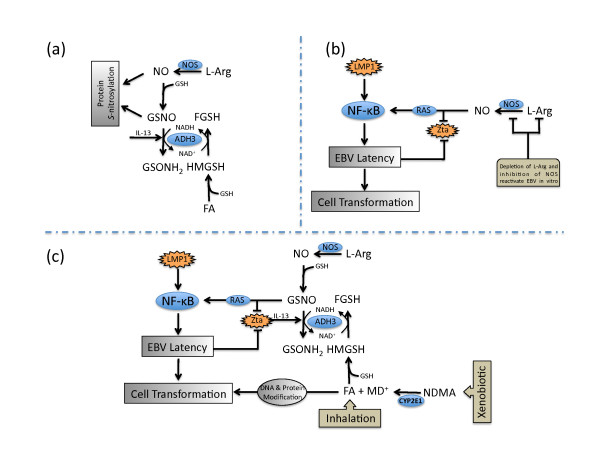
Figure 1.
Simplified hypothetical model for formaldehyde interaction with EBV. a) Alcohol dehydrogenase 3 (ADH3) catalyzes the oxidation of GSH-conjugated formaldehyde (S-hydroxymethylglutathione, HMGSH) to S-formylglutathione (FGSH), and the reduction of GSH-conjugated NO (S-nitrosoglutathione, GSNO) to glutathione sulfinamide (GSONH2). Both NO and GSNO regulate protein function by S-nitrosylating cysteine residues, albeit with differential specificity. Excessive formaldehyde can accelerate GSNO reduction via NAD+/NADH cofactor recycling leading to decreased cellular nitrosothiols. Interleukin-13 has recently been shown to increase ADH3 expression. b) EBV latency programs that contain LMP1 promote latency, in part, through activation of NF-κB pathways. NO appears to be required for latency in programs that lack LMP1, as NOS inhibition can reactivate the EBV in vitro. Similar to LMP1, this process likely involves activation of NF-κB. This NO-mediated regulation of EBV is cGMP-independent and involves S-nitrosylation of RAS by NO and/or GSNO. In addition, binding of the early lytic transcription factor, Zta, to DNA is inhibited by redox and nitrosative modification of cysteine residues. c) Nitrosothiols influence the EBV life cycle. Increased ADH3 expression or excessive formaldehyde exposure from xenobiotic metabolism (e.g. N-nitrosodimethylamine, NDMA) or exogenous sources may accelerate GSNO reduction thereby promoting EBV reactivation. Zta may also increase IL-13, which may in turn increase ADH3 expression. In addition to EBV reactivation, formaldehyde mediated protein modification and DNA reactivity can interact with established EBV latency programs to promote epithelial cell transformation.
Discussion
(a) The EBV Life Cycle and NPC
The World Health Organization has periodically classified NPC based on histology, yet all NPC appears to be variants of squamous cell carcinoma [5]. Though NPC risk varies geographically, nearly all cases and types of NPC demonstrate EBV-positive cells and thus EBV is strongly implicated in the etiology of NPC [5,7,8]. Over 90% of the population is EBV positive, yet EBV is rarely detected in normal human epithelial biopsies, and when it is detected, it is almost always in carcinomas and in the latent phase [6,8,47]. The EBV exhibits three latency programs [48]. Latency III is the least restrictive and involves the expression of six nuclear antigens (EBNA1-6), three latent membrane proteins (LMP1, LMP2A, and LMP2B), and untranslated nuclear RNAs. Latency I is the most restrictive and expresses LMP2A and EBNA1 and is associated with memory B-cells. The Latency II program expresses EBNA1 and the three LMPs, and is the program associated with NPC [48]. These latency programs and other factors compete to repress and promote the activity of the EBV lytic transcription factor Zta [47]. Importantly, both EBV reactivation and EBV latency programs are thought to contribute to the etiology of NPC [6,8,49-52]. The former process may increase the risk of infection to target epithelial cells, whereas the latter may increase the risk of cell transformation. The potential influence of formaldehyde on these processes is described in the sections that follow.
(b) Nitrosothiols Influence the EBV Life Cycle
In 1968, the EBV was reported to be reactivated in cells cultured in media deficient in the nonessential amino acid and NO precursor L-arginine [53]. Over 25 years later, NO synthase (NOS) inhibitors were shown to reactivate EBV in human lymphocytes exhibiting Type I latency but not Type III [29]. This specificity was posited to relate to the fact that LMP1 (not present in Type I latency) and NO converge on similar signaling pathways such as NF-κB [29,34]. NO also prevented B-cell apoptosis, which is consistent with evidence that LMP1 promotes uncontrolled B-cell proliferation [29,48]. This suppression of the lytic phase and apoptosis by NO was reportedly mediated through cGMP-independent signaling pathways, and likely involves nitrosothiol modification of p21 Ras [29,34,54]. NO also suppresses the lytic phase of EBV in epithelial cell lines; and moreover, the established induction of EBV reactivation by TPA (12-O-tetradecanoylphorbol-13 acetate) coincided with decreased levels of NOS [55]. It is also noteworthy that NO inhibition increased the expression of the early lytic transcription factor Zta in both lymphocytes and epithelial cells [29,55]. The binding of this viral transcription factor to DNA is reported to be inhibited by both redox and nitrosative mechanisms that involve critical cysteine residues [28]. Interestingly, Zta is reported to induce the expression of interleukin-13, which may in turn up-regulate ADH3 [56,57]. Although further studies are needed to examine these responses together in individual cell types and in in vivo systems, the data suggest that nitrosothiol homeostasis plays a dual role in promoting the latent phase and inhibiting the lytic phase of the EBV in lymphocytes and epithelial cells expressing certain latency programs (Figure 1b).
To date, little is known about what conditions might lead to altered nitrosothiol homeostasis in vivo. It is becoming increasing clear, however, that environmental exposures to allergens can elevate ADH3 expression in airways thereby decreasing nitrosothiol levels sufficiently to cause patent changes in pulmonary physiology [41,57,58]. Moreover, evidence suggests that exposure to relatively high levels of formaldehyde vapor can lead to immune responses, elevated ADH3, and increased GSNO breakdown in rodent airways [45,59,60]. Additionally, formaldehyde can accelerate GSNO reduction several fold in cell and cell-free systems through NAD+/NADH cofactor recycling on ADH3, as well as decrease cellular nitrosothiol levels in vitro (Figure 1a)) [61,62]. Together, these data suggest that formaldehyde can accelerate GSNO reduction, which might promote EBV reactivation in cells lining the airways (Figure 1c). This reactivation, in turn, could lead to increased viral shedding in nasopharyngeal lymphocytes and subsequent infection of basal epithelial cells [8]. It is notable, though circumstantial, that the relative risk of NPC among occupationally exposed workers is reported to be stronger among EBV seropositive individuals [2].
(c) EBV Latency and Cell Transformation
Several mechanisms are implicated in EBV-mediated cell transformation. EBNA1, EBNA2, and LMP1 have all been shown to induce oxidative stress, genomic instability, or DNA repair inhibition [63-66]. In fact, LMP1 is posited to be an important oncogene and a key modulator in the pathogenesis of NPC through activation of NF-κB signaling pathways related to cell cycle control, apoptosis and transformation [51]. LMP1 acts as a ligand-independent CD40 receptor that functions, in part, by preventing the ubiquitination of TNF-receptor associated factors [50]. Other EBV proteins manipulate the fate of host proteins by accelerating or decelerating their degradation through the ubiquitin-proteosome system, while simultaneously decelerating their own ubiquitination and degradation [50,67]. These myriad effects imparted by EBV latency programs likely perturb many cellular functions in subtle ways that can increase spontaneous transformation or otherwise potentiate cells for transformation. As noted previously, the etiology of NPC involves host, viral and environmental factors. Considering that the functions of many proteins may be modulated by S-nitrosylation, it is conceivable that excess formaldehyde exposure may further alter protein functions in EBV-positive cells. Additionally, formaldehyde itself can deplete GSH levels and can react with proteins and peptides [68-70]. Recent genomics analyses in yeast cells clearly indicate that oxidative stress pathways are induced by formaldehyde exposure [70]. These analyses also identified protein fate (processing and degradation, folding and stabilization, and ubiquitination pathways) as a major category of altered gene expression; formaldehyde induced 14-19% of genes involved in ubiquitination processes, whereas methanol (which is oxidized to formaldehyde) induced only 1% of these genes [70]. While this simple eukaryotic system may not directly inform in vivo mammalian tissue responses, there is little reason to believe that it cannot provide sentinel information as to how formaldehyde interacts with cellular macromolecules. These effects on protein function may partially explain the synergistic effect formaldehyde has on genetic damage in mammalian cells [71,72]. Moreover, such effects on proteins may explain the association between NPC and consumption of salted fish containing nitrosamines like N-nitrosodimethylamine, as the cytochrome P450-mediated oxidation of these compounds can generate formaldehyde (Figure 1c) [5,7,73-76].
Conclusion
NPC is a multifactorial disease thought to involve host, viral, and environmental factors. Evidence for an association between NPC and occupational exposure to formaldehyde remains debated, despite its classification as a human carcinogen [1-4,16-18]. Data described herein indicate that NO, acting through cGMP-independent pathways, can influence the EBV life cycle. As such, excessive environmental exposure to formaldehyde might perturb nitrosothiol homeostasis and potentiate EBV reactivation - thereby increasing the chance of basal epithelial cell infection. Data also suggest that protein stability and degradation are significantly affected by formaldehyde exposure; such changes might arise from direct formaldehyde modification of proteins, redox changes, or altered protein nitrosylation. Thus, concomitant with multiple perturbations caused by EBV latency programs, formaldehyde might additionally act additively or synergistically with EBV to promote epithelial cell transformation. Future investigations on these mechanisms may shed additional light as to the plausibility of epidemiological associations between formaldehyde exposure and NPC. Finally, the mechanisms presented herein might also be operative for other diseases associated with formaldehyde exposure that have etiologies involving EBV, such as lymphoproliferative disorders.
List of abbreviations
ADH3: class III alcohol dehydrogenase; CYP2E1: cytochrome P450 2E1; EBNA: Epstein-Barr nuclear antigens; EBV: Epstein-Barr virus; FA: formaldehyde; FSH: S-formylglutathione; GSH: glutathione; GSNO: S-nitrosoglutathione; GSONH2: glutathione sulfinamide; GSSG: oxidized glutathione; HMGSH: S-hydroxymethylglutathione; IARC: International Agency for Research on Cancer; L-arg: L-arginine; LMP: latent membrane protein; MD+: methyldiazonium ion; NAD: Nicotinamide adenine dinucleotide; NF-κB: nuclear factor kappa-light-chain-enhancer of activated B cells; NDMA: N-nitrosodimethylamine; NO: nitric oxide; NOS: nitric oxide synthase; NPC: nasopharyngeal carcinoma; TNF: tumor necrosis factor; TPA: 12-O-tetradecanoylphorbol-13 acetate
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CT drafted the manuscript and constructed Figure 1. RG collaborated on all analytical and editorial decisions. Both authors read and approved the final manuscript.
Contributor Information
Chad M Thompson, Email: cthompson@toxstrategies.com.
Roland C Grafström, Email: Roland.Grafstrom@ki.se.
Acknowledgements
Portions of this work were supported by grants from the Swedish Cancer and Allergy Fund, the Swedish Fund for Research without Animal Experiments, and Karolinska Institutet.
References
- Vaughan TL, Stewart PA, Teschke K, Lynch CF, Swanson GM, Lyon JL, Berwick M. Occupational exposure to formaldehyde and wood dust and nasopharyngeal carcinoma. Occup Environ Med. 2000;57:376–384. doi: 10.1136/oem.57.6.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildesheim A, Dosemeci M, Chan CC, Chen CJ, Cheng YJ, Hsu MM, Chen IH, Mittl BF, Sun B, Levine PH. Occupational exposure to wood, formaldehyde, and solvents and risk of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2001;10:1145–1153. [PubMed] [Google Scholar]
- Hauptmann M, Lubin JH, Stewart PA, Hayes RB, Blair A. Mortality from solid cancers among workers in formaldehyde industries. Am J Epidemiol. 2004;159:1117–1130. doi: 10.1093/aje/kwh174. [DOI] [PubMed] [Google Scholar]
- IARC. Formaldehyde, 2-Butoxyethanol and 1-tert-Butoxypropan-2-ol. Lyon: IARC; 2006. [PMC free article] [PubMed] [Google Scholar]
- Barnes L, Kapadia SB. The biology and pathology of selected skull base tumors. J Neurooncol. 1994;20:213–240. doi: 10.1007/BF01053041. [DOI] [PubMed] [Google Scholar]
- Lo KW, Huang DP. Genetic and epigenetic changes in nasopharyngeal carcinoma. Semin Cancer Biol. 2002;12:451–462. doi: 10.1016/s1044579x02000883. [DOI] [PubMed] [Google Scholar]
- Chan AT, Teo PM, Johnson PJ. Nasopharyngeal carcinoma. Ann Oncol. 2002;13:1007–1015. doi: 10.1093/annonc/mdf179. [DOI] [PubMed] [Google Scholar]
- Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol. 2002;12:431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- Nguyen-Van D, Ernberg I, Phan-Thi Phi P, Tran-Thi C, Hu L. Epstein-Barr virus genetic variation in Vietnamese patients with nasopharyngeal carcinoma: full-length analysis of LMP1. Virus Genes. 2008;37:273–281. doi: 10.1007/s11262-008-0262-9. [DOI] [PubMed] [Google Scholar]
- Whalan JE, DeVoney D, Thompson CM, White P, Vandenberg JJ. Proposed Cancer Mode of Action for Formaldehyde Based on EPA Cancer Guidelines. Annual Meeting of the Society of Toxicology: 2006; San Diego, CA. 2006.
- Kerns WD, Pavkov KL, Donofrio DJ, Gralla EJ, Swenberg JA. Carcinogenicity of formaldehyde in rats and mice after long-term inhalation exposure. Cancer Res. 1983;43:4382–4392. [PubMed] [Google Scholar]
- Monticello TM, Miller FJ, Morgan KT. Regional increases in rat nasal epithelial cell proliferation following acute and subchronic inhalation of formaldehyde. Toxicol Appl Pharmacol. 1991;111:409–421. doi: 10.1016/0041-008x(91)90246-b. [DOI] [PubMed] [Google Scholar]
- Woutersen RA, van Garderen-Hoetmer A, Bruijntjes JP, Zwart A, Feron VJ. Nasal tumours in rats after severe injury to the nasal mucosa and prolonged exposure to 10 ppm formaldehyde. J Appl Toxicol. 1989;9:39–46. doi: 10.1002/jat.2550090108. [DOI] [PubMed] [Google Scholar]
- Kimbell JS, Subramaniam RP, Gross EA, Schlosser PM, Morgan KT. Dosimetry modeling of inhaled formaldehyde: comparisons of local flux predictions in the rat, monkey, and human nasal passages. Toxicol Sci. 2001;64:100–110. doi: 10.1093/toxsci/64.1.100. [DOI] [PubMed] [Google Scholar]
- Holmstrom M, Wilhelmsson B, Hellquist H, Rosen G. Histological changes in the nasal mucosa in persons occupationally exposed to formaldehyde alone and in combination with wood dust. Acta Otolaryngol. 1989;107:120–129. doi: 10.3109/00016488909127488. [DOI] [PubMed] [Google Scholar]
- Marsh GM, Youk AO, Buchanich JM, Erdal S, Esmen NA. Work in the metal industry and nasopharyngeal cancer mortality among formaldehyde-exposed workers. Regul Toxicol Pharmacol. 2007;48:308–319. doi: 10.1016/j.yrtph.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Marsh GM, Youk AO, Morfeld P. Mis-specified and non-robust mortality risk models for nasopharyngeal cancer in the National Cancer Institute formaldehyde worker cohort study. Regul Toxicol Pharmacol. 2007;47:59–67. doi: 10.1016/j.yrtph.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Bosetti C, McLaughlin JK, Tarone RE, Pira E, La Vecchia C. Formaldehyde and cancer risk: a quantitative review of cohort studies through 2006. Ann Oncol. 2008;19:29–43. doi: 10.1093/annonc/mdm202. [DOI] [PubMed] [Google Scholar]
- Hoog JO, Stromberg P, Hedberg JJ, Griffiths WJ. The mammalian alcohol dehydrogenases interact in several metabolic pathways. Chem Biol Interact. 2003;144:175–181. doi: 10.1016/s0009-2797(02)00225-9. [DOI] [PubMed] [Google Scholar]
- Jensen DE, Belka GK, Du Bois GC. S-Nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochem J. 1998;331:659–668. doi: 10.1042/bj3310659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivusalo M, Baumann M, Uotila L. Evidence for the identity of glutathione-dependent formaldehyde dehydrogenase and class III alcohol dehydrogenase. FEES Lett. 1989;257:105–109. doi: 10.1016/0014-5793(89)81797-1. [DOI] [PubMed] [Google Scholar]
- Staab CA, Hellgren M, Hoog JO. Medium- and short-chain dehydrogenase/reductase gene and protein families: Dual functions of alcohol dehydrogenase 3: implications with focus on formaldehyde dehydrogenase and S-nitrosoglutathione reductase activities. Cell Mol Life Sci. 2008;65:3950–3960. doi: 10.1007/s00018-008-8592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uotila L, Koivusalo M. Formaldehyde dehydrogenase from human liver. Purification, properties, and evidence for the formation of glutathione thiol esters by the enzyme. J Biol Chem. 1974;249:7653–7663. [PubMed] [Google Scholar]
- Thompson CM, Sonawane B, Grafstrom RC. The ontogeny, distribution, and regulation of alcohol dehydrogenase 3: implications for pulmonary physiology. Drug Metab Dispos. 2009;37:1565–1571. doi: 10.1124/dmd.109.027904. [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Kokkola T, Savinainen JR, Monkkonen KS, Retamal MD, Laitinen JT. S-nitrosothiols modulate G protein-coupled receptor signaling in a reversible and highly receptor-specific manner. BMC Cell Biol. 2005;6:21. doi: 10.1186/1471-2121-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CD, Benhar M, Keys JR, Rockman HA. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- Wang P, Day L, Dheekollu J, Lieberman PM. A redox-sensitive cysteine in Zta is required for Epstein-Barr virus lytic cycle DNA replication. J Virol. 2005;79:13298–13309. doi: 10.1128/JVI.79.21.13298-13309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick JB, Asano K, Izumi K, Kieff E, Stamler JS. Nitric oxide produced by human B lymphocytes inhibits apoptosis and Epstein-Barr virus reactivation. Cell. 1994;79:1137–1146. doi: 10.1016/0092-8674(94)90005-1. [DOI] [PubMed] [Google Scholar]
- Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- Schonhoff CM, Daou MC, Jones SN, Schiffer CA, Ross AH. Nitric oxide-mediated inhibition of Hdm2-p53 binding. Biochemistry. 2002;41:13570–13574. doi: 10.1021/bi026262q. [DOI] [PubMed] [Google Scholar]
- Marshall HE, Hess DT, Stamler JS. S-nitrosylation: physiological regulation of NF-kappaB. Proc Natl Acad Sci USA. 2004;101:8841–8842. doi: 10.1073/pnas.0403034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti M, Persichini T, Venturini G, Ascenzi P. S-nitrosylation of viral proteins: molecular bases for antiviral effect of nitric oxide. IUBMB Life. 1999;48:25–31. doi: 10.1080/713803459. [DOI] [PubMed] [Google Scholar]
- Marshall HE, Merchant K, Stamler JS. Nitrosation and oxidation in the regulation of gene expression. FASEBJ. 2000;14:1889–1900. doi: 10.1096/fj.00.011rev. [DOI] [PubMed] [Google Scholar]
- Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- Schonhoff CM, Matsuoka M, Tummala H, Johnson MA, Estevez AG, Wu R, Kamaid A, Ricart KC, Hashimoto Y, Gaston B. S-nitrosothiol depletion in amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2006;103:2404–2409. doi: 10.1073/pnas.0507243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannu R, Singh I. Pharmacological strategies for the regulation of inducible nitric oxide synthase: neurodegenerative versus neuroprotective mechanisms. Neurochem Int. 2006;49:170–182. doi: 10.1016/j.neuint.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Lima B, Lam GK, Xie L, Diesen DL, Villamizar N, Nienaber J, Messina E, Bowles D, Kontos CD, Hare JM. Endogenous S-nitrosothiols protect against myocardial injury. Proc Natl Acad Sci USA. 2009;106:6297–302. doi: 10.1073/pnas.0901043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston B, Singel D, Doctor A, Stamler JS. S-nitrosothiol signaling in respiratory biology. Am J Respir Crit Care Med. 2006;173:1186–1193. doi: 10.1164/rccm.200510-1584PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson EM, Gaston B. SNOR and wheeze: the asthma enzyme? Trends Mol Med. 2005;11:481–484. doi: 10.1016/j.molmed.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Wu H, Romieu I, Sienra-Monge JJ, Estela Del Rio-Navarro B, Anderson DM, Jenchura CA, Li H, Ramirez-Aguilar M, Del Carmen Lara-Sanchez I, London SJ. Genetic variation in S-nitrosoglutathione reductase (GSNOR) and childhood asthma. J Allergy Clin Immunol. 2007;120:322–328. doi: 10.1016/j.jaci.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PE, Ryckman KK, Williams SM, Patel N, Summar ML, Sheller JR. Genetic variants of GSNOR and ADRB2 influence response to albuterol in African-American children with severe asthma. Pediatr Pulmonol. 2009;44:649–654. doi: 10.1002/ppul.21033. [DOI] [PubMed] [Google Scholar]
- Thompson CM, Grafstrom RC. Mechanistic considerations for formaldehyde-induced bronchoconstriction involving S-nitrosoglutathione reductase. J Toxicol Environ Health A. 2008;71:244–248. doi: 10.1080/15287390701598259. [DOI] [PubMed] [Google Scholar]
- Thompson CM, Subramaniam RP, Grafstrom RC. Mechanistic and dose considerations for supporting adverse pulmonary physiology in response to formaldehyde. Toxicol Appl Pharmacol. 2008;233:355–359. doi: 10.1016/j.taap.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Mannick JB, Stamler JS, Teng E, Simpson N, Lawrence J, Jordan J, Finberg RW. Nitric oxide modulates HIV-1 replication. J Acquir Immune Defic Syndr. 1999;22:1–9. doi: 10.1097/00042560-199909010-00001. [DOI] [PubMed] [Google Scholar]
- Flint SJ, Enquist LW, Racaniello VR, Skalka AM. Molecular Biology. Third. I. Washington, DC: ASM Press; 2009. Principles of Virology. [Google Scholar]
- Dantuma NP, Masucci MG. The ubiquitin/proteasome system in Epstein-Barr virus latency and associated malignancies. Semin Cancer Biol. 2003;13:69–76. doi: 10.1016/s1044-579x(02)00101-3. [DOI] [PubMed] [Google Scholar]
- Dolcetti R, Masucci MG. Epstein-Barr virus: induction and control of cell transformation. J Cell Physiol. 2003;196:207–218. doi: 10.1002/jcp.10263. [DOI] [PubMed] [Google Scholar]
- Masucci MG. Epstein-Barr virus oncogenesis and the ubiquitin-proteasome system. Oncogene. 2004;23:2107–2115. doi: 10.1038/sj.onc.1207372. [DOI] [PubMed] [Google Scholar]
- Zheng H, Li LL, Hu DS, Deng XY, Cao Y. Role of Epstein-Barr virus encoded latent membrane protein 1 in the carcinogenesis of nasopharyngeal carcinoma. Cell Mol Immunol. 2007;4:185–196. [PubMed] [Google Scholar]
- Schulz TF, Cordes S. Is the Epstein-Barr virus EBNA-1 protein an oncogen? Proc Natl Acad Sci USA. 2009;106:2091–2092. doi: 10.1073/pnas.0812575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle W, Henle G. Effect of arginine-deflcient media on the herpes-type virus associated with cultured Burkitt tumor cells. J Virol. 1968;2:182–191. doi: 10.1128/jvi.2.3.182-191.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander HM, Ogiste JS, Pearce SF, Levi R, Novogrodsky A. Nitric oxide-stimulated guanine nucleotide exchange on p21ras. J Biol Chem. 1995;270:7017–7020. doi: 10.1074/jbc.270.13.7017. [DOI] [PubMed] [Google Scholar]
- Gao X, Tajima M, Sairenji T. Nitric oxide down-regulates Epstein-Barr virus reactivation in epithelial cell lines. Virology. 1999;258:375–381. doi: 10.1006/viro.1999.9748. [DOI] [PubMed] [Google Scholar]
- Tsai SC, Lin SJ, Chen PW, Luo WY, Yeh TH, Wang HW, Chen CJ, Tsai CH. EBV Zta protein induces the expression of interleukin-13, promoting the proliferation of EBV-infected B cells and lymphoblastoid cell lines. Blood. 2009;114:109–118. doi: 10.1182/blood-2008-12-193375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que LG, Yang Z, Stamler JS, Lugogo NL, Kraft M. S-Nitrosoglutathione Reductase -- An Important Regulator in Human Asthma. Am J Respir Crit Care Med. 2009;180:226–231. doi: 10.1164/rccm.200901-0158OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que LG, Liu L, Yan Y, Whitehead GS, Gavett SH, Schwartz DA, Stamler JS. Protection from experimental asthma by an endogenous bronchodilator. Science. 2005;308:1618–1621. doi: 10.1126/science.1108228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WW, Kim EM, Lee EH, Yun HJ, Ju HR, Jeong MJ, Hwang KW, Sul D, Kang HS. Formaldehyde exposure induces airway inflammation by increasing eosinophil infiltrations through the regulation of reactive oxygen species production. Environmental Toxicology and Pharmacology. 2007;24:174–182. doi: 10.1016/j.etap.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Yi C, Ke K, Xiaohua L, Xu Y. Up-regulation of GSNO reductase in mice lungs by formaldehyde inhalation. The 1st International Conference on Bioinformatics and Biomedical Engineering. Wuhan, China. 2007. pp. 294–297.
- Staab CA, Alander J, Brandt M, Lengqvist J, Morgenstern R, Grafstrom RC, Hoog JO. Reduction of S-nitrosoglutathione by alcohol dehydrogenase 3 is facilitated by substrate alcohols via direct cofactor recycling and leads to GSH-controlled formation of glutathione transferase inhibitors. Biochem J. 2008;413:493–504. doi: 10.1042/BJ20071666. [DOI] [PubMed] [Google Scholar]
- Staab CA, Alander J, Morgenstern R, Grafstrom RC, Hoog JO. The Janus face of alcohol dehydrogenase 3. Chem Biol Interact. 2009;178:29–35. doi: 10.1016/j.cbi.2008.10.050. [DOI] [PubMed] [Google Scholar]
- Gruhne B, Sompallae R, Marescotti D, Kamranvar SA, Gastaldello S, Masucci MG. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc Natl Acad Sci USA. 2009;106:2313–2318. doi: 10.1073/pnas.0810619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan SH, Tai CC, Lin CS, Hsu WB, Chou SF, Lai CC, Chen JY, Tien HF, Lee FY, Wang WB. Epstein-Barr virus nuclear antigen 2 disrupts mitotic checkpoint and causes chromosomal instability. Carcinogenesis. 2009;30:366–375. doi: 10.1093/carcin/bgn291. [DOI] [PubMed] [Google Scholar]
- Chen YR, Liu MT, Chang YT, Wu CC, Hu CY, Chen JY. Epstein-Barr virus latent membrane protein 1 represses DNA repair through the PI3K/Akt/FOXO3a pathway in human epithelial cells. J Virol. 2008;82:8124–8137. doi: 10.1128/JVI.00430-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerimele F, Battle T, Lynch R, Frank DA, Murad E, Cohen C, Macaron N, Sixbey J, Smith K, Watnick RS. Reactive oxygen signaling and MARK activation distinguish Epstein-Barr Virus (EBV)-positive versus EBV-negative Burkitt's lymphoma. Proc Natl Acad Sci USA. 2005;102:175–179. doi: 10.1073/pnas.0408381102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci USA. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstrom RC, Jernelov MI, Dypbukt JM, Sundquvist K, Atzori L, Zheng X, Mohr U, Adler KB, Dungworth DI, Harris CC. Correlations between in vitro and in vivo investigations in inhalation toxicology. Washington, DC: ILSI Press; 1996. Aldehyde toxicity and thiol redox state in cell cultures from human aerodigestive tract; pp. 319–336. [Google Scholar]
- Metz B, Kersten GF, Hoogerhout P, Brugghe HF, Timmermans HA, de Jong A, Meiring H, ten Hove J, Hennink WE, Crommelin DJ. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J Biol Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- Yasokawa D, Murata S, Iwahashi Y, Kitagawa E, Nakagawa R, Hashido T, Iwahashi H. Toxicity of Methanol and Formaldehyde Towards Saccharomyces cerevisiae as Assessed by DNA Microarray Analysis. Appl Biochem Biotechnol. 2009. in press . [DOI] [PubMed]
- Grafstrom RC, Curren RD, Yang LL, Harris CC. Genotoxicity of formaldehyde in cultured human bronchial fibroblasts. Science. 1985;228:89–91. doi: 10.1126/science.3975633. [DOI] [PubMed] [Google Scholar]
- Grafstrom RC, Fornace AJ Jr, Autrup H, Lechner JF, Harris CC. Formaldehyde damage to DNA and inhibition of DNA repair in human bronchial cells. Science. 1983;220:216–218. doi: 10.1126/science.6828890. [DOI] [PubMed] [Google Scholar]
- Liteplo RG, Meek ME, Windle W, International Programme on Chemical Safety. N-Nitrosodimethylamine. Geneva: World Health Organization; 2002. [Google Scholar]
- Godoy W, Albano RM, Moraes EG, Pinho PR, Nunes RA, Saito EH, Higa C, Filho IM, Kruel CD, Schirmer CC. CYP2A6/2A7 and CYP2E1 expression in human oesophageal mucosa: regional and inter-individual variation in expression and relevance to nitrosamine metabolism. Carcinogenesis. 2002;23:611–616. doi: 10.1093/carcin/23.4.611. [DOI] [PubMed] [Google Scholar]
- Sulc M, Kubickova B, Maslova V, Hodek P. Rabbit liver microsomal system: study of interaction with two model N-nitrosamines and their metabolism. Gen Physiol Biophys. 2004;23:423–433. [PubMed] [Google Scholar]
- Hou DF, Wang SL, He ZM, Yang F, Chen ZC. Expression of CYP2E1 in human nasopharynx and its metabolic effect in vitro. Mol Cell Biochem. 2007;298:93–100. doi: 10.1007/s11010-006-9356-7. [DOI] [PubMed] [Google Scholar]