The budding yeast Saccharomyces cerevisiae is widely used as a model organism for studying many biological processes and systems, particularly for understanding the cell cycle. The yeast peptide pheromone α-factor (WHWLQLKPGQPMY) activates the mating pathway in MATa cells, arresting the cell cycle in the G1 phase. This arrest is used as a tool for synchronizing yeast cultures and for investigating signaling events involved in the pathway and other processes related to morphogenesis and transcription. Herein we report the synthesis and activity of an α-factor analogue designed to control the on/off state of the mating signal. Incorporating a photocleavable residue at a flexible site in the peptide allowed UV degradation in situ to mimic the natural protease degradation of this signaling peptide. The control over the α-factor signal achieved serves as proof of concept for use of this analogue as a tool. It also and allows access to previously impractical experiments that will yield highly time-resolved information about the yeast cell cycle and mating pathway.
The binding of the α-factor to its G-protein-coupled receptor Ste2p in haploid MATa yeast cells initiates a MAP kinase cascade that results in cell-cycle arrest, cell morphogenesis (to extend a cell-wall projection toward the source of the peptide), and mating-specific genes.[1,2] The peptide binds to the Ste2p receptor in a specific conformation, resulting in the termini being buried in the receptor with the central flexible region exposed.[1,3] Under normal circumstances, the Bar1 protease is excreted extracellularly to degrade excess peptide and to control the level of α-factor signaling.[4] Generally, α-factor-arrest experiments are performed in a liquid medium and require washing of cells in fresh buffer solution or medium to remove the α-factor peptide and allow release into the cell cycle.[5] However, precise temporal control of release is difficult to achieve in a multistep washing procedure and results in the loss of information about the initiation of the cell cycle. This also means that experiments involving arrest and release in formats other than liquid medium cannot be performed. An analogue of the α-factor, which could be degraded in a controlled manner in situ, would be useful for a better control of the release and more-rapid analysis of the physiological consequences of dropping below the pheromone-signaling threshold for the mating response. This will open the door to previously impractical experiments, such as microscopically studying the cytoskeletal changes that are the result of the change from pheromone-associated polarity to bud-site-selection polarity, or gene-expression analysis of the first few minutes after removal of the mating signal to obtain information about the switch that turns on cell-cycle-initiation factors such as cyclins and ultimately results in the decision to leave G1 arrest.
Although caging of bioactive molecules for subsequent activation by using UV light is a common strategy,[6–11] the use of photocleavable functionality for deactivation of peptides and proteins[12] is relatively unexplored. Backbone cleavage through 2-nitrophenyl amino acid derivatives provides an interesting way to specifically turn off a biological signal of interest. We show herein a successful proof of concept for applying this strategy to control the activity of a signaling peptide.
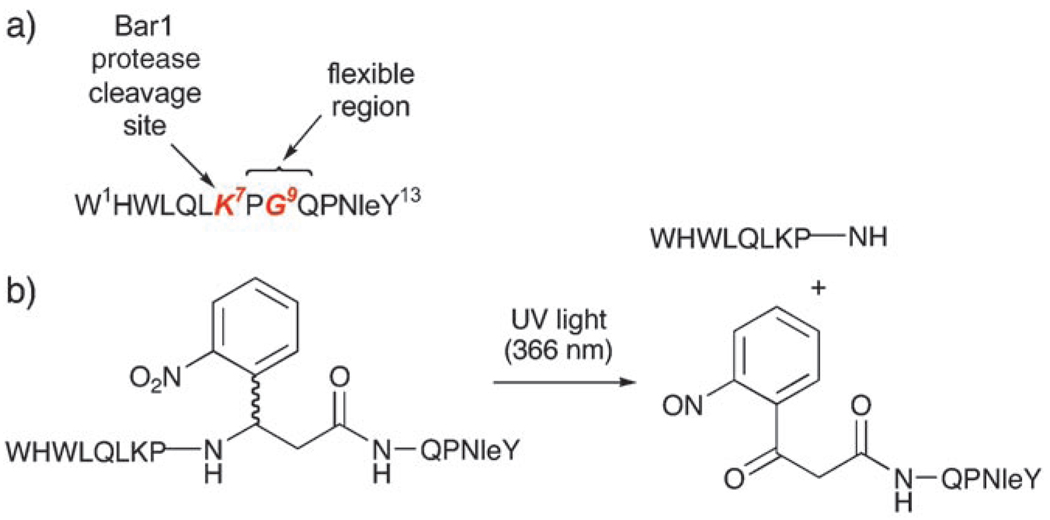
Substituting a position in the flexible region of the α-factor with an amino acid analogue that can be artificially cleaved should mimic Bar1 protease degradation at Lys-7 and results in “turning off” of the α-factor signal (Scheme 1a). As the structure of this peptide is very important to its activity, selection of the mutation site was important. Extensive work by Naider and Becker has established the importance of particular residues to the activity, giving us a good basis on which to design the analogue.[1] For example, alanine scanning of this peptide with both l- and d-Ala showed that l-Ala at position 9 (Scheme 1a) disrupted the binding of the peptide to the receptor and reduced the activity. Notably, however, at this position, d-Ala gave increased binding and a fivefold increase in activity. This suggested that, depending on the structural constraints imposed by the linkage, bulkier amino acids may be tolerated at position 9, for example, cleavable 2-nitrophenyl amino acid derivatives such as the commercially available (2-nitro)-β-phenylalanine ([d/l]-(β)2-Npa; Scheme 1b).
Scheme 1.
a) Residues of interest in the α-factor sequence; b) UV cleavage of peptide 2.
With these design features in mind, we synthesized the photocleavable peptide 2 (see Table 1) and tested its activity to induce the mating response. In both 1 and 2, the native Met12 was mutated to Norleucine (Nle), a common substitution that is reported to improve the stability of the peptide to oxidative damage.[1] Peptide 1 served as a “native” α-factor control. Peptide 2 was synthesized with racemic d/l-(2-nitro)-β-phenylalanine to provide a means for selective cleavage of the backbone, giving two diastereomeric products that were separated by reverse-phase HPLC into 2a and 2b (see the Supporting Information for characterization of all peptides). The rate of UV-induced cleavage for 2 (as shown in Scheme 1b) was determined at 366 nm, 5 mWcm−2 (5 µm, t1/2≈8 min; see the Supporting Information). Although cleavage may be faster at shorter wavelengths (e.g. 237 nm), 366 nm is a safer wavelength for avoiding DNA damage in cells, so all characterization of UV treatment in biological assays was determined for 366-nm UV light.
Table 1.
Peptides synthesized in this study. (The bold text highlights the variation between the peptides.)
| Peptide | Sequence |
|---|---|
| 1 | WHWLQLKPGQPNleY |
| 2 | WHWLQLKP-[d/l-(β)2-Npa]-QPNleY |
| 2a | WHWLQLKP-[d-(β)2-Npa]-QPNleY |
| 2b | WHWLQLKP-[l-(β)2-Npa]-QPNleY |
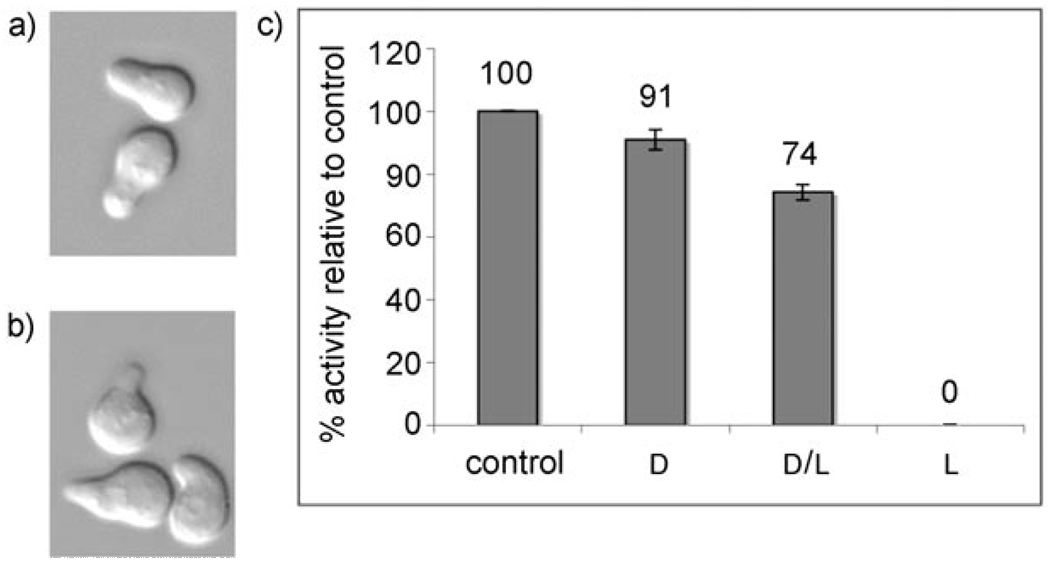
Both diastereomers of 2 were tested for G1 arrest by using “shmoo” activity (to detect morphological changes resulting from activation of the mating pathway) and halo (disc diffusion, to determine the arrest-related growth inhibition) assays (see Figure 1 and the Supporting Information for images of arrest halos). Only one product was active, for which NOESY NMR spectroscopy supported the structural assignment as peptide 2a, the diastereomer containing the d isomer of (2-nitro)-β-phenylalanine (see the Supporting Information and Table 1). Identifying compound 2a, as the active diastereomer is consistent with previous data for bulky d- and l-amino acid substitutions at position 9 of this peptide, where l-amino acids tended to decrease activity, whereas d-amino acids improved activity.[1] The activity of 2a in the halo assay was approximately 90% of the control peptide 1, which is remarkably high activity for such a drastic change in backbone structure. As shown by Naider and Becker,[1] amino acid substitutions throughout this peptide affect the affinity for the receptor and show that it is normally necessary to increase the rigidity of the backbone in a favorable conformation for receptor binding to increase the activity. It is surprising that a β-amino acid with the backbone flexibility provided by the extra methylene group still results in a peptide with satisfactory activity.
Figure 1.
Shmooing of A cells induced by a) control peptide 1 (5 mm) compared with b) d-(β)2-Npa 2a (10 µm); c) Halo assay for relative growth-arrest activity. All activity seen in the diastereomeric peptide mixture resulted from the d diastereomer.
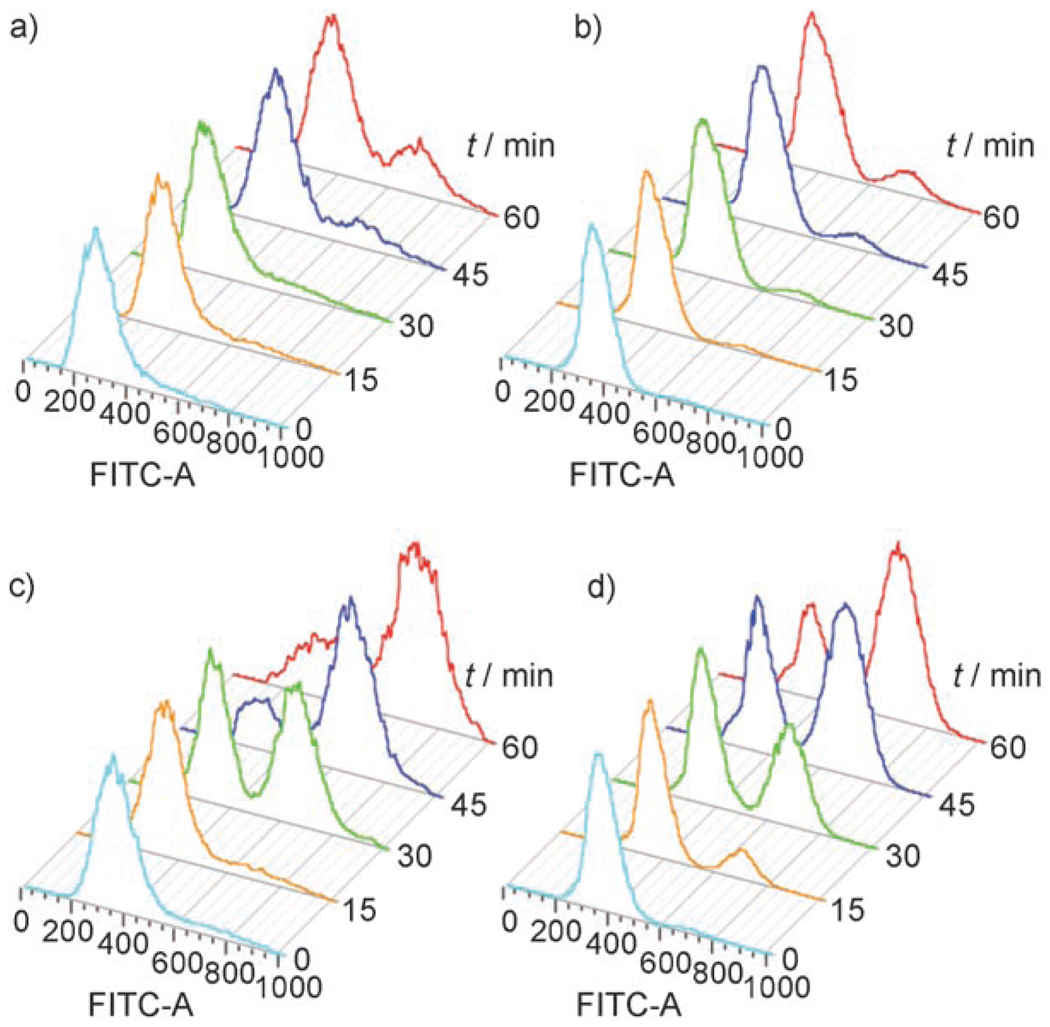
The kinetics of release from G1 arrest for control α-factor 1 (after peptide washout by using standard conditions) and photocleavable peptide 2a (after UV release) were characterized by using the α-factor/nocodozole trap assay (which discriminates with high resolution between cells that have and have not left G1 arrest) and analyzed by flow cytometry to determine the DNA content (fluorescence increases depending on the extent of DNA replication, indicating the progression of the cell cycle). Three hours of treatment at 25°C with 10 µm photocleavable analogue 2 was sufficient to obtain complete G1 arrest of a yeast culture (optical density at 600 nm (OD)600=0.1). After treating this culture for 5 min with UV light (20 mWcm−2), the peptide was 75% degraded (as shown by the HPLC analysis of the supernatant medium, see the Supporting Information) and the synchronized cells were released into the cell cycle with kinetics comparable to those seen after washing out the control α-factor 1 (cells treated with 5 µm 1 to achieve G1 arrest; Figure 2).
Figure 2.
Flow cytometry analysis of the release from G1 arrest by a G1/G2 trap assay. The DNA content was determined as 1N (G1 trapped) or 2N (G2 trapped; SYTOX green staining). a) 1, no release; b) 2a, no release; c) 1, washout; d) 2a, UV (5 min, 366 nm, 20 mWcm−2). FTIC-A=absorption of SYTOX green (fluorescein-based DNA stain).
This strategy for degradation in situ opens up exciting possibilities for G1 arrest and mating experiments in microscopy formats on solid media that were not possible before and should lead to important, novel information about yeast mating and the cell cycle. For example, the rates of oscillations in expression of various genes between different phases of the cell cycle have been extensively characterized by using real-time PCR and microarray experiments.[13–16] However, the earliest times at which it has been possible to measure G1-arrest recovery-related gene induction have been after the first five to seven minutes—the typical time necessary for centrifugation and resuspension in pheromone-free medium. By this time, the cells have already made the decision to leave G1, and CLN3/SWI4 expression levels are beginning to rise accordingly.[17] These genes are known to turn on the expression of CLN1 and CLN2, cyclins that act with the cyclin-dependent kinase Cdc28p to drive the G1 to S transition. Owing to the practical constraints, it has not been possible to look for genes that change before that time frame, and questions remain about what defines the switch that is ultimately responsible for this process.[18, 19]
Further to studying the normal cell-cycle recovery from G1 arrest, another intriguing possibility would be to cleave this peptide at 237-nm UV light to simultaneously induce damage and release cells from G1. This may provide new information about the G1 checkpoint mechanisms that delay entry into the cell cycle after DNA damage.[20] Another interesting application could involve the development of designer signaling mechanisms in mammalian cells by using chimeric receptors[21] that would respond to α-factors[22] (with photocleavable control of the signal) but stimulate pathways that are more relevant to other organisms.
In conclusion, we have shown that this peptide has sufficient activity to arrest cells in G1 at reasonable doses and that it can be specifically degraded by using relatively brief exposure to UV light at a wavelength that is “safe” for most cells. Its activity is related to the conformation of the peptide around its central loop region and agrees with literature data for other “constrained” versions of α-factors containing bulkier d-amino acids at position 9, even though this peptide contains a β-amino acid. We are currently investigating ways to use this cleavable α-factor to address the biological questions detailed above by using genome arrays and physiological and biochemical studies.
Experimental Section
Yeast strains and media: The strain used in this study was SKY809 (W303 background; MATa, ura3-1, can1-100, leu2-3,112 GAL+). Yeast cells were grown in standard rich YPD (1% yeast extract, 2% peptone, 2% dextrose) or a 2X synthetic media lacking tryptophan (2X-Trp) at 25 or 30°C. When necessary, cells were washed in standard rich YEP (1 % yeast extract, 2 % peptone).
Peptide synthesis and purification: Peptides were synthesized manually on a 0.1 mmol scale by using tert-butoxycarbonyl (Boc) solid-phase peptide synthesis, cleaved with anhydrous HF (p-cresol as scavenger), and purified by using reverse-phase HPLC (Grace Vydac C18 column, 10 mm ID × 250 mmL; solvent A: 0.1% aqueous TFA; solvent B: 0.1% TFA in acetonitrile). Peptides were analyzed by using an Agilent 1100 XCT ion trap LC/MS (C18 column, 2.1 mm ID × 50 mmL; gradient 5–65% B at 4% min−1).
Halo disc diffusion assay[23]: Peptide dissolved in dimethylsulfoxide (DMSO; 15 µL total volume) was applied to sterile small paper discs (6.5 mm). wild-type (WT) W303 MATa yeast was grown overnight to saturation in YPD (10 µL), diluted into warm 0.5% agar in 2X-Trp medium (4 mL, dissolved by heating in the microwave and equilibrated in a water bath to 45°C), and poured onto the surface of YPD plates. After cooling briefly to set the agar, the peptide discs were laid on the surface and the plates incubated at 30°C overnight. Halos were visualized by digital photograph, the diameters measured and normalized to the paper disc diameters.
G1 arrest assay with an α-factor/nocodazole trap[20]: Yeast strain SKY809 was grown to saturation (OD600≈1.8) overnight at room temperature in 2X-Trp medium. The culture was diluted in a 1:10 ratio and grown to the log phase at 30°C for 3 h (final OD600≈0.6). This log phase culture was diluted to OD600=0.1 in 6-well plates (2.5 mL total volume) and treated with peptide 1 or 2 for 3 h at room temperature to arrest the cells in the G1 phase. Peptide 1 was removed by pelleting and washing the cells in YEP, then resuspending them in YPD. Peptide 2b was degraded by treatment with a UVP Blak-Ray UV lamp (5 min, 20 mWcm−2). Before and after UV treatment, 250 µl of the cell suspension was removed, pelleted, and the supernatant then removed and filtered for HPLC analysis (Waters 626 with Empower2 software system). After peptide removal, aliquots (250 µL) were quenched with trap medium (250 µL, 10 µmα-factor, 40 µgmL−1 nocodazole) after 0, 15, 30, 45 and 60 min and incubated for 1.5 h in a 24-well plate on a shaker at room temperature. Trapped cells were filtered in a 96-well filter bottom plate (MultiScreen, Millipore) and washed twice with 70% aqueous EtOH to fix them. After all time-point aliquots were finished being trapped, fixed cells were resuspended in 250 µL RNAse (0.25 µgµL−1 in 50 mm tris(hydroxymethyl)aminomethane hydrochloride (Tris-Cl) buffer solution; pH 7.5) and incubated at 55°C for 2 h, followed by washing twice with Tris-Cl buffer solution (100 µL, 50 mm pH 7.5). Cells were then resuspended in 100 µL pepsin solution (5 µgµL−1 in 4.5 mm HCl) and treated at 37°C for 1 h, washed twice with Tris-Cl buffer solution (as above), taken up in 100 µL Tris-Cl buffer solution (as in wash), and sonicated to deaggregate prior to analysis with flow cytometry. Cell suspensions were transferred to flow cytometry tubes containing 400 µL SYTOX green (Molecular Probes; final concentration 5 µm) and analyzed by using a BD FACSCanto flow cytometer.
Supplementary Material
Footnotes
L.P. gratefully acknowledges the NIH for a Ruth L. Kirschstein Individual Postdoctoral Fellowship (F32). S.J.K. is a Scholar of the Leukemia and Lymphoma Society.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Laurie L. Parker, Department of Biochemistry and Molecular Biology, University of Chicago, 929 E. 57th Street, CIS W201A, Chicago, IL 60637 (USA)
Josh W. Kurutz, Department of Biochemistry and Molecular Biology, University of Chicago, 929 E. 57th Street, CIS W201A, Chicago, IL 60637 (USA)
Stephen B. H. Kent, Department of Biochemistry and Molecular Biology, University of Chicago, 929 E. 57th Street, CIS W201A, Chicago, IL 60637 (USA)
Stephen J. Kron, Department of Molecular Genetics and Cellular Biology, University of Chicago, 924 East 57th Street, Knapp R322, Chicago, IL 60637 (USA).
References
- 1.Naider F, Becker JM. Peptides. 2004;25:1441. doi: 10.1016/j.peptides.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 2.Bardwell L. Peptides. 2004;25:1465. doi: 10.1016/j.peptides.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 3.Lee BK, Khare S, Naider F, Becker JM. J. Biol. Chem. 2001;276:37950. doi: 10.1074/jbc.M103579200. [DOI] [PubMed] [Google Scholar]
- 4.Ciejek E, Thorner J. Cell. 1979;18:623. doi: 10.1016/0092-8674(79)90117-x. [DOI] [PubMed] [Google Scholar]
- 5.Raths SK, Naider F, Becker JM. J. Biol. Chem. 1988;263:17333. [PubMed] [Google Scholar]
- 6.Shigeri Y, Tatsu Y, Yumoto N. Pharmacol. Ther. 2001;91:85. doi: 10.1016/s0163-7258(01)00148-6. [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto M, Solzin J, Helbig A, Hagen V, Ueno S, Kawase O, Maruyama Y, Ogiso M, Godde M, Minakata H, Kaupp UB, Hoshi M, Weyand I. Dev. Biol. 2003;260:314. doi: 10.1016/s0012-1606(03)00236-7. [DOI] [PubMed] [Google Scholar]
- 8.Marriott G, Roy P, Jacobson K. Biophotonics Part A. 2003;360:274. doi: 10.1016/s0076-6879(03)60115-1. [DOI] [PubMed] [Google Scholar]
- 9.Rothman DM, Shults MD, Imperiali B. Trends Cell Biol. 2005;15:502. doi: 10.1016/j.tcb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Toebes M, Coccoris M, Bins A, Rodenko B, Gomez R, Nieuwkoop NJ, van de Kasteele W, Rimmelzwaan GF, Haanen J, Ovaa H, Schumacher TNM. Nat. Med. 2006;12:246. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen A, Rothman DM, Stehn J, Imperiali B, Yaffe MB. Nat. Biotechnol. 2004;22:993. doi: 10.1038/nbt997. [DOI] [PubMed] [Google Scholar]
- 12.England PM, Lester HA, Davidson N, Dougherty DA. Proc. Natl. Acad. Sci. USA. 1997;94:11025. doi: 10.1073/pnas.94.20.11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahler J. Annu. Rev. Genet. 2005;39:69. doi: 10.1146/annurev.genet.39.110304.095808. [DOI] [PubMed] [Google Scholar]
- 14.MacKay VL, Li XH, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, Zhao LP, Morris DR. Mol. Cell. Proteomics. 2004;3:478. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Serikawa KA, Xu XL, MacKay VL, Law GL, Zong O, Zhao LP, Bumgarner R, Morris DR. Mol. Cell. Proteomics. 2003;2:191. doi: 10.1074/mcp.D200002-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Mol. Biol. Cell. 1998;9:3273. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacKay VL, Mai B, Waters L, Breeden LL. Mol. Cell. Biol. 2001;21:4140. doi: 10.1128/MCB.21.13.4140-4148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haase SB, Reed SI. Nature. 1999;401:394. doi: 10.1038/43927. [DOI] [PubMed] [Google Scholar]
- 19.Wittenberg C, Reed SI. Oncogene. 2005;24:2746. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- 20.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Mol. Cell. Biol. 2005;25:8430. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson T. Pharmacol. Ther. 1991;50:425. doi: 10.1016/0163-7258(91)90052-n. [DOI] [PubMed] [Google Scholar]
- 22.Yin DZ, Gavi S, Shumay E, Duell K, Konopka JB, Malbon CC, Wang HY. Biochem. Biophys. Res. Commun. 2005;329:281. doi: 10.1016/j.bbrc.2005.01.130. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman GA, Garrison TR, Dohlman HG. G Protein Pathways Part B. 2002;344:617. doi: 10.1016/s0076-6879(02)44744-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.