Arrhythmias are common clinical problems. Atrial fibrillation (AF) affects 2–5 million Americans, is a common cause of cardiovascular morbidity including hospitalization, heart failure, and stroke, and is associated with increased mortality [1]. Sudden cardiac death (SCD) due to very rapid ventricular arrhythmias (ventricular tachycardia, VT, and ventricular fibrillation, VF) is the commonest cause of death among American adults, accounting for 250,000–500,000 cases annually, 10–20% of all adult death [2]. This review will outline the challenges and opportunities presented in applying contemporary genetic analyses to understanding variability in drug response in this area. Relevant clinical features are first discussed, followed by a description of challenges in the field and current status.
Atrial fibrillation
Most patients with AF have concomitant structural abnormalities of the heart such as left ventricular hypertrophy. In a minority, AF occurs in the absence of such abnormalities, and is termed “lone AF”. A family history of AF is common in affected individuals, and is even more common in those with lone AF [3,4]. Other studies associate indices of oxidant stress [5] or renin-angiotensin activation [6–8] with the arrhythmia. One setting in which AF is especially common is in patients who have undergone cardiac surgery, where the incidence can be as high as 25% in the week after the procedure. In this setting, inflammatory pathways have been implicated [9,10].
Clinical presentations in AF are highly heterogeneous. AF results in loss of coordinated atrial activity, and irregular and often rapid ventricular response rates due to conduction of atrial fibrillatory activity through the atrioventricular node. Therapies are thus designed to prevent the arrhythmia and maintain normal atrial activity (“rhythm control”) or to slow conduction through the AV node, thereby slowing the ventricular response rate (“rate control”). It is intuitively obvious that a strategy to prevent the arrhythmia and maintain normal cardiac electrical activity should be superior to one designed to ignore the arrhythmia but treat its symptoms; nevertheless, randomized trials comparing rhythm control to rate control have shown, if anything, slight superiority of the rate control strategy [11]. One reasonable explanation for this apparent paradox is that the drugs used for rhythm control are not uniformly effective, and do carry serious cardiac and non-cardiac risks of their own.
In patients who are highly symptomatic with the arrhythmia, a rate control strategy is often ineffective. In such individuals non-pharmacologic, catheter-based ablative therapies are now being developed to identify specific regions within the atria – notably extensions of the left atrial myocardium into pulmonary veins – that drive fibrillatory activity. These approaches are meeting with increasing, although currently incomplete, success [12,13].
The highly variable clinical presentations, including variable and unpredictable responses to drugs and other therapies, uncertain etiologies, variable associations with concomitant heart disease, and familial aggregation, notably in lone AF, all point to a potential role for genetic variants in the etiology of the arrhythmia as well as in the variable response to drugs. Candidate pathways for modulating such variability include the ion channels and associated proteins that determine electrical activity, genes associated with structural heart disease, inflammatory or oxidant stress pathways, and developmental pathways determining atrial architecture.
Patients with AF are at increased risk for thrombus formation and thromboembolism. AF is common in patients with stroke, and thromboembolism is the presumed mechanism. Randomized clinical trials have shown that therapy with the vitamin K antagonist anticoagulant warfarin can reduce stroke incidence 60–80% with an acceptable risk of bleeding [14]. Warfarin is a cumbersome drug to use, since it requires intensive monitoring of its pharmacologic endpoint (extent of anticoagulant assessed as the “International Normalized Ratio,” or INR) and doses vary widely among individuals. At least 50% of this dose variability is now attributed to genetic variants in the coding region of CYP2C9 (which bio-inactivates the drug) and in the promoter region of VKORC1 (encoding the pharmacologic target) [15,16]. A large randomized multi-center trial will start in the spring of 2009 to examine the contribution of pre-prescription genotyping to improving warfarin therapy [17].
Sudden cardiac death
In most cases of SCD, there is underlying coronary artery disease. VF may occur during an acute myocardial infarction [18]; as well, myocardial infarction may lead to myocardial scarring and increased susceptibility to VF weeks, months, and years later [19]. Efforts to prevent SCD have been directed almost exclusively at the latter group, in whom the extent and severity of underlying structural heart disease predicts increasing risk, thereby allowing evaluation of preventive therapies. By contrast, predicting who will develop VF during an acute myocardial infarction, and deploying preventive antiarrhythmic therapies, has proven much more problematic; the best antiarrhythmic in this setting is an effort to reduce myocardial ischemia (e.g. mechanical or pharmacological thrombolysis). Pharmacologic therapies have been relatively unsuccessful in reducing SCD in such patients (and occasionally increase risk [20]). Accordingly, placement of implantable cardioverter-defibrillator devices (ICDs) is the approach of choice for patients judged to be at high risk for SCD due to underlying heart disease [21].
In addition to those patients with advanced coronary or other heart disease, there is a smaller group of patients, often younger, who are at high risk because of inherited heart diseases, such as hypertrophic cardiomyopathy or the congenital long QT syndromes (LQTS) [2]. Linkage analysis in large kindreds has led to identification of disease genes, with the subsequent recognition that mutations in multiple genes can culminate in very similar phenotypes, and penetrance can be highly variable [22]. Thus, advances in identifying genetic causes of these diseases in probands with obvious phenotypes (like electrocardiographic abnormalities, left ventricular hypertrophy, or SCD) has led to the identification of family members who are mutation carriers and yet who may have very little clinical phenotype. In patients with certain subtypes of LQTS, treatment with beta-blockers, a relatively benign pharmacologic therapy, appears to be effective in reducing SCD risk [23]. Otherwise, in patients who have “monogenic” arrhythmia syndromes, ICDs are the only preventive therapy deployed. Given the intrusive nature of this treatment, and the relatively young age of many affected mutation carriers, efforts are underway to understand modifiers of the arrhythmia phenotype (accounting for SCD in some individuals and no clinical phenotype in others) and to thereby stratify a risk for SCD.
A number of studies discussed below implicate a family history of SCD as a risk factor for SCD. Therefore, risk may include a genetic component, and identifying that component could direct improved prophylactic therapies. Candidate pathways for modulating such risk include those pathways implicated in the rarer monogenic diseases (contractile proteins, cell-cell adhesion proteins, ion channel proteins). As well, experiments in animals suggest that release of membrane lipids during ischemia may be especially arrhythmogenic [24]; thus, another candidate pathway for mediating SCD risk is variability in the acute electrophysiological response to a coronary thrombus. Identification of risk pathways for SCD could lead to development of new antiarrhythmic drugs.
The long QT syndrome and the problem of drug-induced torsades de pointes
The cardiac cycle is characterized by cardiac depolarization, contraction, and then a recovery process mediated by return of the action potential to its baseline state, repolarization. The congenital LQTS [23] is characterized by a prolonged repolarization phase, and a morphologically distinctive polymorphic ventricular tachycardia, termed torsades de pointes (TdP); TdP is the cause of death in most cases of the congenital LQTS. Therapy with certain antiarrhythmic and other drugs can also prolong the QT interval, and has been associated with TdP [25]. The incidence of this drug-induced long QT syndrome (diLQTS) is as high as 5% with certain antiarrhythmic drugs, and it can also occur during therapy with drugs not used in therapy of cardiovascular disease. These include some antipsychotics, antibiotics, and antihistamines; however, this is not a class effect and the risk for each drug must be evaluated individually. Clinical risk factors for the drug-induced arrhythmia include underling slow heart rates, hypokalemia, female gender, recent conversion from AF, and structural heart disease such as left ventricular hypertrophy [25]. However, predicting the arrhythmia in an individual still is not possible.
One common form of the congenital LQTS arises from loss of function mutations in KCNH2 (also termed HERG), whose expression results in a potassium current important in cardiac repolarization, IKr. Virtually all drugs that cause diLQTS are IKr blockers, and this has led to routine pre-clinical screening for KCNH2 block by new drugs in development. However, the relationship between potency of IKr block and downstream risk of TdP remains uncertain [25]. As described below, efforts are underway to identify genomic contributors to diLQTS risk.
Challenges in arrhythmia genetics and pharmacogenetics
With this background, the opportunities and challenges in applying contemporary genomic approaches to clinical arrhythmia problems can be enunciated.
Case ascertainment
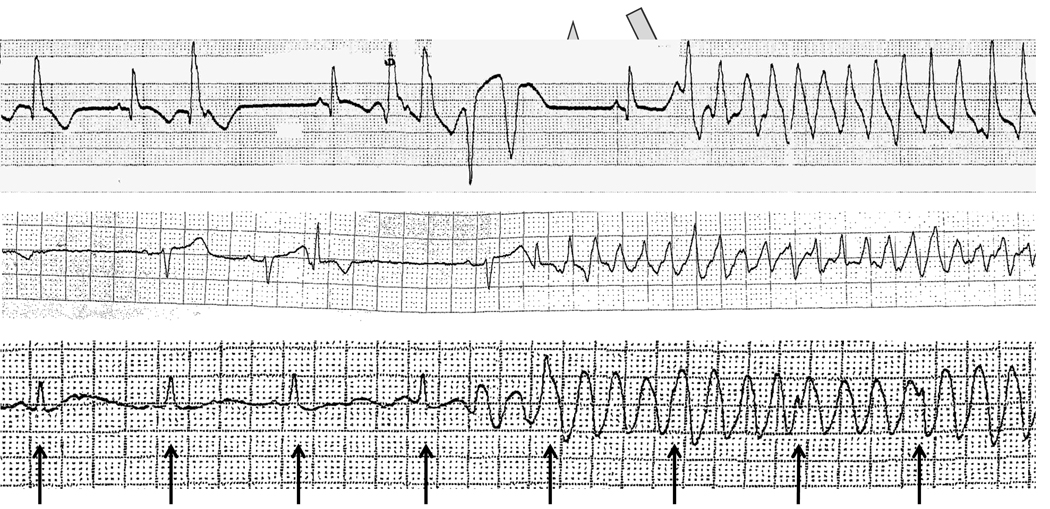
Challenges include making a precise diagnosis, understanding the clinical subtype if necessary, and identifying clinical covariates. For example, Figure 1 illustrates three patients referred for evaluation of TdP. The top rhythm shows typical features of diLQTS: very clear QT prolongation and a pause in cardiac rhythm prior to the development of the arrhythmia. The second shows no QT prolongation, and was observed in a patient with acute myocardial ischemia, another setting in which polymorphic ventricular tachycardia (but without QT prolongation) can be seen; this arrhythmia is clinically and mechanistically distinct from diLQTS. The third appears to be a polymorphic ventricular tachycardia, but is actually a recording artifact superimposed on a regular rhythm indicated by the arrows. Including either of the latter cases in a series of TdP to investigate underlying genetic causes would be an obvious experimental mistake.
Figure 1.
Cardiac rhythm recordings in 3 patients referred for evaluation of torsades de pointes. The top recording shows typical features of diLQTS: very clear QT prolongation (arrow) and a pause in cardiac rhythm (star) prior to the development of the arrhythmia. The second shows no QT prolongation, and was observed in a patient with acute myocardial ischemia, another setting in which polymorphic ventricular tachycardia (but without QT prolongation) can be seen; this arrhythmia is clinically and mechanistically distinct from diLQTS. The third appears to be a polymorphic ventricular tachycardia, but is actually a recording artifact superimposed on a regular rhythm (indicated by the arrows).
Clinical covariates
A second challenge in the broad area is identifying clinical covariates. For example, in virtually all case series of diLQTS, there is a two-threefold higher increase in prevalence in women [26]. The mechanisms for this gender-specific risk are not well understood, but failing to correct for gender would also be an experimental mistake.
Definition of endpoints, and the use of surrogate endpoints
The most appropriate endpoint for any study of arrhythmias is either symptoms or a cardiovascular event such as syncope or death. However, these may be difficult to quantify (in the case of symptoms) or relatively uncommon (in the case of “hard” endpoints). Thus, surrogate endpoints linked to the final phenotypes have been deployed in this field. For example, in studies of diLQTS risk, it is common use the extent of drug related prolongation of the QT interval as a marker of risk, rather than the arrhythmia itself. The fibrillatory rate during AF [27] may be a reflection of differing underlying atrial electrophysiologic derangements, and thus may be a surrogate for understanding AF genetics. The surrogate approach has the virtue that larger numbers of subjects, with a readily acquired and quantified endpoint for pharmacologic therapy, can be accrued. On the other hand, the relationship between the surrogate and the true endpoint, like SCD, diLQTS or AF may be non-linear or occasionally non-existent. In the 1980s, suppression of isolated premature ventricular contractions (PVCs) was taken as a surrogate for reducing SCD risk until the Cardiac Arrhythmia Suppression Trial showed increased SCD rates in patients receiving drugs that potently suppressed PVCs [20].
Identifying candidate genes for arrhythmia phenotypes
The simplest approach is one which considers obvious candidate genes, such as those known to encode ion channels, their ancillary subunits, elements of physiologic systems that modulate cardiac electrical activity (adrenergic, oxidant, inflammatory signaling), or genes whose protein products modulate electrical activity in other ways (e.g. calcium control genes).
Unbiased approaches are also being used to identify loci associated with increased arrhythmia prevalence as described below. A variation on this approach is to use variability in electrocardiographic parameters, which are readily recorded in large numbers of subjects, as surrogates for arrhythmia susceptibility. A genome-wide association study (GWAS) analyzing variability in the distribution of QT interval durations in normal subjects identified a locus on chromosome 1 near a gene encoding an ancillary subunit of the neuronal nitric oxide synthase gene (NOS1AP) [28]. The encoded protein, termed CAPON, is expressed in heart and modulates cardiac ion channel function through mechanisms that remain to be determined [29].
Another interesting unbiased approach to identifying genes modulating diLQTS risk has been high-throughput screening in sensitized embryonic zebrafish. Heart rate is readily recorded in these animals, and exposure to IKr blockers results in predictable heart rate slowing [30]. Screening large numbers of mutagenized zebrafish for excessive heart rate slowing to low dose blockers, or resistance to heart slowing with high dose blockers, identified multiple new genes modulating this drug response phenotype, including elements of the heg-san-vtn pathway [31].
Accumulating case series
AF is a common arrhythmia and does not usually cause death. Thus, accruing very large numbers of well-characterized patients is feasible, and has been important starting point for clinical and genetic studies. Further, AF is sufficiently prevalent within a population that large population-based studies, such as Framingham or the Rotterdam Heart study [32], can, over time accrue sufficiently large numbers of patients with incident AF to enable studies of clinical and genetic predictors of the arrhythmia.
By contrast, SCD represents a greater challenge in case ascertainment. One approach is to examine SCD in large population based studies, such as Framingham, the Rotterdam Heart Study [33], the Atherosclerosis Risk In Communities (ARIC) Study, or the Cardiovascular Health Study (CHS) [34]. These efforts undertake extensive phenotyping of patients at entry and during followup, and thus may provide new information on clinical and genetic risk predictors of the event. The Paris prospective study recruited 7,000 middle aged men in the late 1960s, followed them for a quarter century, and identified family history as a risk factor for SCD [35]. Another approach is to accrue SCD cases as they occur (through emergency medical services or coronary care units) and to analyze the problem in case-control fashion. Two studies have taken this approach and both suggest family history is a risk factor for SCD. One, in Seattle, studied cases identified by emergency services with out of hospital SCD [36]. The second, in the Netherlands, identified a cohort of 330 patients developing within 90 minutes of the onset of their first acute myocardial infarction [37]. The control subjects were 372 developing their first myocardial infarction, but not displaying VF. The most powerful predictor of SCD was a family history of SCD, with an odds ratio of 2.72 (95% CI 1.84 to 4.03).
For much rarer events – such as diLQTS – such large population based studies are not suitable. Here, databases accrued during drug development (by industry) or by networks of investigators brought together for specific case and control identification and curation represent alternate approaches. The Trans-Atlantic Network of Excellence in “Preventing SCD” represents such an example (http://www.allianceagainstscd.org/definition.php). Investigators in this network have accrued over 200 cases of diLQTS, and controls exposed to culprit drugs and not developing marked QT prolongation. These cohorts represent an important starting point for studies evaluating clinical as well as genetic risk factors for the arrhythmia [38].
Electronic medical records (EMR) linked to large collections of DNA may represent another mechanism for accruing cases and controls for clinical or genetic studies of risk. At Vanderbilt, we have created such a DNA resource, termed BioVU, that links de-identified medical records to DNA samples obtained during routine clinical care [39]. Rapid accrual of large numbers of cases, and very extensive phenotypic information contained in electronic medical records represent advantages of this approach. For common, readily ascertained phenotypes, such as AF and accompanying heart disease, using such an EMR approach seems feasible [40], although its utility for more complex phenotypes, such as diLQTS, remains to be evaluated. As of February 2009, the Vanderbilt resource includes 1.7 million deidentified electronic records, and over 50,000 are associated with DNA samples. The widespread use of electronic medical records may allow generation of much larger collections of genetic studies. In addition, integrating DNA research with the electronic medical records is a first step to incorporating DNA markers of risk of disease or of variable drug responses into clinical practice.
Current status of AF genetics and pharmacogenetics
There are considerable data that support the idea that AF includes a genetic component. To date, linkage analyses in large kindreds have identified four distinct genetic loci for the arrhythmia [41–44]. At one of these, a mutation has been identified in NUP155, encoding a nuclear pore protein not previously implicated in the arrhythmia [45], opening a new pathway to understanding arrhythmia susceptibility and perhaps new drug development.
In addition, smaller kindreds have been analyzed by screening coding regions of candidate genes for non-synonymous variation. Using the latter approach, mutations in ion channel genes have been associated with AF [46–51]. In these cases, the strength of the evidence relies on the fact that mutations in plausible candidate genes were identified and that they associate with the arrhythmia across families, although a formal lod score is usually absent. In addition, a common in vitro electrophysiologic finding is increased outward current, a change predicted to shorten atrial action potentials which in experimental animals predisposes to AF. Increased outward current is seen with “gain of function” mutations in potassium channel genes, loss of function in cardiac calcium channel genes [52], and with mutations in the atrial natriuretic peptide gene (NPPA) which have also been reported in familial AF [53].
A loss of function mutation in an atrial potassium channel gene, KCNA5, has been described in one kindred with familial AF [54]. The predicted effect of this mutation is to prolong atrial action potential, indicating there are multiple electrophysiologic derangements in mutant potassium channels that can predispose to AF. Applying this information to affected patients will therefore require an understanding of the basic pathophysiology: in patients with “gain of function” mutation, potassium channel blockers would be logical therapy, whereas drugs to increase potassium current would be more appropriate in patients with loss of function mutations.
Variation in genes controlling the magnitude of the cardiac sodium current has also been associated with AF. AF is a common feature in the Brugada syndrome [55], a relatively rare monogenic disorder characterized by structurally normal heart, a distinctive electrocardiographic phenotype (J-point elevation in the right precordial leads that is either constant or can be provoked by sodium channel blocker therapy), and a high incidence of SCD due to VF [56,57]. Loss of function mutations in the cardiac sodium channel gene SCN5A are identified in 25% of patients with Brugada syndrome, and mutations in other genes (whose function is to modify cardiac sodium current) have been reported albeit much less commonly [56,58,59].
Mutations in genes encoding two sodium channels ancillary subunits, SCN1B and SCN2B, have been reported in patients with AF [60]. Interestingly, the affected patients showed ECG abnormalities reminiscent of the Brugada syndrome, although SCD did not occur. Indeed, a Finnish study reported a remarkably high prevalence of this ECG pattern (approximately 10%) in patients with lone AF [61], suggesting that abnormal sodium channel signaling could be responsible for a substantial proportion of cases of AF. Further support for this idea has come from a study in which resequencing the SCN5A coding region in 375 patients with AF yielded previously reported mutations (long QT syndrome, Brugada syndrome) in 3.2% and novel mutations in 2.7% [62]. Further exploration of the role of variation in the sodium channel gene and its modulators in AF seems warranted since drugs with sodium channel blocking properties (such as flecainide) are widely used in the therapy of AF. The use of such drugs in patients in whom the fundamental electrophysiologic lesion arises from loss of sodium current is not only predicted to be ineffective, but may in fact, increase SCD risk [20,63].
Case series of patients with AF have reported that common ion channel polymorphisms (e.g. S38G in KCNE1 [64], H558R in SCN5A [65]) increase AF risk, but these have been small and not reproduced. A small study reported an association between lower fibrillatory rate and the 38GG KCNE1 genotype (n=13; 392±36 vs 443±49 fpm, p=.006) but no association with the SCN5A H558R polymorphism. Thus, these data suggest that the KCNE1 S38G genotype exerts functional effects on atrial electrophysiology [27].
A genome-wide approach, initially in Iceland and extended to European and Asian populations, identified polymorphisms on chromosome 4 (at 4q25), that confer an increased risk of AF with an odds ratio of ~1.4/allele [66], and this finding has now been independently replicated [32]. The variants are located in an intergenic region, and the closest gene is PITX2, which encodes a cardiac transcription factor important for left-right differentiation and for the development of atrial myocardium that invaginates the pulmonary veins [67]. 4q25 risk alleles seem prevalent regardless of the subtype of atrial fibrillation; the risk has been identified in subsets with AF after cardiac surgery [68], and appears to apply regardless of age or presence or absence of left ventricular hypertrophy [32,66]. These findings suggest the possibility is that variation in PITX2 function lays down an “AF-prone” substrate as early as embryonic development, and that AF occurs later in life in susceptible individuals when as-yet-poorly understood triggers occur. Variant 4q25 alleles also associate with cases of stroke in which usual etiologies are absent (termed “cryptogenic stroke”), suggesting undiagnosed AF may play a prominent role in these patients [69].
Studies examining variability in drug response during AF have been hampered by variable endpoints used to gauge drug efficacy, the empiric and non-randomized nature in which drugs are selected for individual patients, and lack of very large cohorts of patients with well-characterized drug responses. Because renin-angiotensin activation has been implicated in AF, one study examined the role of the common angiotensin converting enzyme insertion/deletion (I/D) polymorphism in drug response. The D allele is associated with higher angiotensin II levels, and in 229 patients the D allele blunted response to antiarrhythmic drug therapy: subjects with DD/ID genotypes (71%) were more likely to have recurrent AF during therapy (P<0.005). Other studies using a candidate gene approach have implicated common variants in the interleukin-6 gene [70,71] in AF after surgery.
Current status of SCD genetics and pharmacogenetics
The major problem with identifying genetic contributors to SCD risk is patient ascertainment, since large scale accrual of samples from patients suffering SCD is a cumbersome undertaking. Thus, only a handful of studies have been conducted to examine the role of genetic risk markers for SCD, and these have examined candidate genes, e.g. those in which mutations cause the congenital monogenic syndromes or that encode pathways strongly modulating cardiovascular electrophysiology, such as the adrenergic system. In general, these studies have involved small numbers of cases of SCD, and have not been replicated [72]. A number of reports indicate that that monogenic arrhythmia syndromes can present as the Sudden Infant Death Syndrome (SIDS) [73–75].
The non-synonymous SCN5A variant resulting in S1103Y – identified almost exclusively in African-Americans – generates a dysfunctional channel and has been associated with increased susceptibility to a range of arrhythmia phenotypes, including SCD, diLQTS, and SIDS [76–79]. Other studies have also examined variation in SCN5A in SCD, but the results have been conflicting [80,81]. Single studies have reported associations between SCD and common variants in genes for the beta-2 adrenergic receptor [82], alpha-2 [83] adrenergic receptor, and hepatic lipase [84].
As described above, single nucleotide polymorphisms in NOS1AP have been identified as modulators of the normal QT interval. Two studies have examined the association between NOS1AP SNPs and SCD. In the Rotterdam Heart Study, involving 5374 subjects age >55, there was a strong association between NOS1AP SNPs and QT duration, but no relationship to SCD (which occurred in 233 subjects) [33]. By contrast, in 14,737 European-American subjects in ARIC and CHS (with SCD in 334), NOS1AP alleles were identified that conferred a relative risk of 1.3 (95% CI: 1.10–1.56, P=0.002), using an additive model [34]; interestingly, not all the NOS1AP SNPs associated with SCD predicted QT prolongation. No associations were identified in 4394 African-American subjects (with 164 cases of SCD). Prolongation of the QT interval has been implicated as a risk factor for SCD in general [85,86], so this association has biologic plausibility; it also possible that NOS1AP modulates SCD risk through other pathways. GWAS approaches will likely be reported soon in these and similar large cohorts.
diLQTS
Studies of genetic contributors to diLQTS risk have focused on logical candidate genes and are limited, in general, by small study size. As with many other rare adverse drug effects, accrual of very large numbers of affected patients and controls presents immense logistical challenges and generally requires participation from multiple centers. As discussed above, case ascertainment can be a particular problem in diLQTS.
Summary
The study of arrhythmia pharmacogenetics presents an interesting set of challenges and opportunities. Arrhythmias are common, extract a large toll in public health terms, and response to drug therapy is variable among individuals. Studies to identify mechanisms underlying variability in arrhythmia phenotypes, and thus point to subset-specific drug or other therapeutic strategies, are underway using both candidate gene and genome-wide association approaches.
References
- 1.Benjamin EJ, Wolf PA, D'Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham Heart Study. Circulation. 1998;98:946–952. doi: 10.1161/01.cir.98.10.946. [DOI] [PubMed] [Google Scholar]
- 2.Spooner PM, Albert C, Benjamin EJ, Boineau R, Elston RC, George AL, Jr, et al. Sudden cardiac death, genes, and arrhythmogenesis : consideration of new population and mechanistic approaches from a national heart, lung, and blood institute workshop, part I. Circulation. 2001;103:2361–2364. doi: 10.1161/01.cir.103.19.2361. [DOI] [PubMed] [Google Scholar]
- 3.Fox CS, Parise H, D'Agostino RB, Sr, Lloyd-Jones DM, Vasan RS, Wang TJ, et al. Parental Atrial Fibrillation as a Risk Factor for Atrial Fibrillation in Offspring. The Journal of the American Medical Association. 2004;291:2851–2855. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 4.Darbar D, Herron KJ, Ballew JD, Jahangir A, Gersh BJ, Shen WK, et al. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol. 2003;41:2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 5.Chung MK, Martin DO, Sprecher D, Wazni O, Kanderian A, Carnes CA, et al. C-Reactive Protein Elevation in Patients With Atrial Arrhythmias: Inflammatory Mechanisms and Persistence of Atrial Fibrillation. Circulation. 2001;104:2886–2891. doi: 10.1161/hc4901.101760. [DOI] [PubMed] [Google Scholar]
- 6.Murray KT, Rottman JN, Arbogast PG, Shemanski L, Primm RK, Campbell WB, et al. Inhibition of angiotensin II signaling and recurrence of atrial fibrillation in AFFIRM. Heart Rhythm. 2004;1:669–675. doi: 10.1016/j.hrthm.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Pedersen OD, Bagger H, Kober L, Torp-Pedersen C. Trandolapril reduces the incidence of atrial fibrillation after acute myocardial infarction in patients with left ventricular dysfunction. Circulation. 1999;100:376–380. doi: 10.1161/01.cir.100.4.376. [DOI] [PubMed] [Google Scholar]
- 8.Madrid AH, Bueno MG, Rebollo JM, Marin I, Pena G, Bernal E, et al. Use of irbesartan to maintain sinus rhythm in patients with long-lasting persistent atrial fibrillation: a prospective and randomized study. Circulation. 2002;106:331–336. doi: 10.1161/01.cir.0000022665.18619.83. [DOI] [PubMed] [Google Scholar]
- 9.Carnes CA, Chung MK, Nakayama T, Nakayama H, Baliga RS, Piao S, et al. Ascorbate attenuates atrial pacing-induced peroxynitrite formation and electrical remodeling and decreases the incidence of postoperative atrial fibrillation. Circ Res. 2001;89:E32–E38. doi: 10.1161/hh1801.097644. [DOI] [PubMed] [Google Scholar]
- 10.Steinberg JS. Postoperative atrial fibrillation: a billion-dollar problem. J Am Coll Cardiol. 2004;43:1001–1003. doi: 10.1016/j.jacc.2003.12.033. [DOI] [PubMed] [Google Scholar]
- 11.Wyse DG, Waldo AL, DiMarco JP, Domanski MJ, Rosenberg Y, Schron EB, et al. A comparison of rate control and rhythm control in patients with atrial fibrillation. N Engl J Med. 2002;347:1825–1833. doi: 10.1056/NEJMoa021328. [DOI] [PubMed] [Google Scholar]
- 12.Oral H, Chugh A, Good E, Sankaran S, Reich SS, Igic P, et al. A tailored approach to catheter ablation of paroxysmal atrial fibrillation. Circulation. 2006;113:1824–1831. doi: 10.1161/CIRCULATIONAHA.105.601898. [DOI] [PubMed] [Google Scholar]
- 13.Hsu LF, Jais P, Sanders P, Garrigue S, Hocini M, Sacher F, et al. Catheter ablation for atrial fibrillation in congestive heart failure. N Engl J Med. 2004;351:2373–2383. doi: 10.1056/NEJMoa041018. [DOI] [PubMed] [Google Scholar]
- 14.Singer DE. Overview of the randomized trials to prevent stroke in atrial fibrillation. Annals of Epidemiology. 1993;3:563–567. doi: 10.1016/1047-2797(93)90117-m. [DOI] [PubMed] [Google Scholar]
- 15.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 Haplotypes on Transcriptional Regulation and Warfarin Dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 16.Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shurin SB, Nabel EG. Pharmacogenomics -- Ready for Prime Time? N Engl J Med. 2008;358:1061–1063. doi: 10.1056/NEJMe0800801. [DOI] [PubMed] [Google Scholar]
- 18.Davies MJ, Thomas A. Thrombosis and acute coronary-artery lesions in sudden cardiac ischemic death. N Engl J Med. 1984;310:1137–1140. doi: 10.1056/NEJM198405033101801. [DOI] [PubMed] [Google Scholar]
- 19.Vreede-Swagemakers JJM, Gorgels AP, Duboisarbouw WI, Vanree JW, Daemen MJAP, Houben LGE, et al. Out-Of-Hospital Cardiac Arrest In The 1990s - A Population-Based Study In The Maastricht Area On Incidence, Characteristics And Survival. J Am Coll Cardiol. 1997;30:1500–1505. doi: 10.1016/s0735-1097(97)00355-0. [DOI] [PubMed] [Google Scholar]
- 20.The CAST Investigators. Preliminary Report: Effect of encainide and flecainide on mortality in a randomized trial of arrhythmia suppression after myocardial infarction. N Engl J Med. 1989;321:406–412. doi: 10.1056/NEJM198908103210629. [DOI] [PubMed] [Google Scholar]
- 21.Zipes DP, Camm AJ, Borggrefe M, Buxton AE, Chaitman B, Fromer M, et al. ACC/AHA/ESC 2006 Guidelines for Management of Patients With Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death: a report of the American College of Cardiology/American Heart Association Task Force and the European Society of Cardiology Committee for Practice Guidelines (writing committee to develop Guidelines for Management of Patients With Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death): developed in collaboration with the European Heart Rhythm Association and the Heart Rhythm Society. Circulation. 2006;114:e385–e484. doi: 10.1161/CIRCULATIONAHA.106.178233. [DOI] [PubMed] [Google Scholar]
- 22.Priori SG, Napolitano C, Schwartz PJ. Low penetrance in the long-QT syndrome: clinical impact. Circulation. 1999;99:529–533. doi: 10.1161/01.cir.99.4.529. [DOI] [PubMed] [Google Scholar]
- 23.Roden DM. Clinical practice. Long-QT syndrome. N Engl J Med. 2008;358:169–176. doi: 10.1056/NEJMcp0706513. [DOI] [PubMed] [Google Scholar]
- 24.DaTorre SD, Creer MH, Pogwizd SM, Corr PB. Amphipathic lipid metabolites and their relation to arrhythmogenesis in the ischemic heart. J Mol Cell Cardiol. 1991;23 Suppl 1:11–22. doi: 10.1016/0022-2828(91)90019-i. [DOI] [PubMed] [Google Scholar]
- 25.Roden DM, Viswanathan PC. Genetics of acquired long QT syndrome. J Clin Invest. 2005;115:2025–2032. doi: 10.1172/JCI25539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makkar RR, Fromm BS, Steinman RT, Meissner MD, Lehmann MH. Female gender as a risk factor for torsades de pointes associated with cardiovascular drugs. JAMA. 1993;270:2590–2597. doi: 10.1001/jama.270.21.2590. [DOI] [PubMed] [Google Scholar]
- 27.Husser D, Stridh M, Sornmo L, Roden DM, Darbar D, Bollmann A. A Genotype Dependent Intermediate ECG Phenotype in Patients with Persistent Lone Atrial Fibrillation. Circ Arrhythmia Electrophysiol. 2009 doi: 10.1161/CIRCEP.108.799098. CIRCEP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 29.Chang KC, Barth AS, Sasano T, Kizana E, Kashiwakura Y, Zhang Y, et al. CAPON modulates cardiac repolarization via neuronal nitric oxide synthase signaling in the heart. Proc Natl Acad Sci U S A. 2008;105:4477–4482. doi: 10.1073/pnas.0709118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milan DJ, Peterson TA, Ruskin JN, Peterson RT, Macrae CA. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation. 2003;107:1355–1358. doi: 10.1161/01.cir.0000061912.88753.87. [DOI] [PubMed] [Google Scholar]
- 31.Milan DJ, Jones IL, Amsterdam AH, Rosenbaum DS, Roden D, MacRae CA. Abstract 637: A Pharmacogenetic Screen for Modifiers of Drug Induced QT Prolongation Reveals 15 Novel Genes. Circulation. 2007;116:II. [Google Scholar]
- 32.Kaab S, Darbar D, van NC, Dupuis J, Pfeufer A, Newton-Cheh C, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009 doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aarnoudse AJ, Newton-Cheh C, de Bakker PIW, Straus SMJM, Kors JA, Hofman A, et al. Common NOS1AP Variants Are Associated With a Prolonged QTc Interval in the Rotterdam Study. Circulation. 2007;116:10–16. doi: 10.1161/CIRCULATIONAHA.106.676783. [DOI] [PubMed] [Google Scholar]
- 34.Kao WH, Arking DE, Post W, Rea TD, Sotoodehnia N, Prineas RJ, et al. Genetic Variations in Nitric Oxide Synthase 1 Adaptor Protein Are Associated With Sudden Cardiac Death in US White Community-Based Populations. Circulation. 2009 doi: 10.1161/CIRCULATIONAHA.108.791723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jouven X, Desnos M, Guerot C, Ducimetiere P. Predicting sudden death in the population: the Paris Prospective Study I. Circulation. 1999;99:1978–1983. doi: 10.1161/01.cir.99.15.1978. [DOI] [PubMed] [Google Scholar]
- 36.Friedlander Y, Siscovick DS, Weinmann S, Austin MA, Psaty BM, Lemaitre RN, et al. Family history as a risk factor for primary cardiac arrest. Circulation. 1998;97:155–160. doi: 10.1161/01.cir.97.2.155. [DOI] [PubMed] [Google Scholar]
- 37.Dekker LRC, Bezzina CR, Henriques JPS, Tanck MW, Koch KT, Alings MW, et al. Familial Sudden Death Is an Important Risk Factor for Primary Ventricular Fibrillation: A Case-Control Study in Acute Myocardial Infarction Patients. Circulation. 2006;114:1140–1145. doi: 10.1161/CIRCULATIONAHA.105.606145. [DOI] [PubMed] [Google Scholar]
- 38.Kaab S, Pfeufer A, Hinterseer M, Nabauer M, Yalilzadeh S, George AL, Norris KJ, Wilde AA, Bezzina CR, Schulze-Bahr E, et al. Common Gene Variants Associated with drug induced Long-QT Syndrome. Circulation. 2005 Oct. 2005: (Abstr.) [Google Scholar]
- 39.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crawford DC, Ritchie MD, Denny JC, Havens A, Weiner J, Pulley J, Basford M, Masys DR, Roden DM, Haines JL. Electronic Medical Records Linked to DNA: A Valuable Resource for Large-Scale Genetic Association Studies. American Society for Human Genetics, accepted for presentation. 2008 (Abstr.) [Google Scholar]
- 41.Brugada R, Tapscott T, Czernuszewicz GZ, Marian AJ, Iglesias A, Mont, et al. Identification of a genetic locus for familial atrial fibrillation. N Engl J Med. 1997;336:905–911. doi: 10.1056/NEJM199703273361302. [DOI] [PubMed] [Google Scholar]
- 42.Ellinor PT, Shin JT, Moore RK, Yoerger DM, Macrae CA. Locus for Atrial Fibrillation Maps to Chromosome 6q14–16. Circulation. 2003;107:2880–2883. doi: 10.1161/01.CIR.0000077910.80718.49. [DOI] [PubMed] [Google Scholar]
- 43.Darbar D, Hardy A, Haines JL, Roden DM. Prolonged signal-averaged P-wave duration as an intermediate phenotype for familial atrial fibrillation. J Am Coll Cardiol. 2008;51:1083–1089. doi: 10.1016/j.jacc.2007.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oberti C, Wang L, Li L, Dong J, Rao S, Du W, et al. Genome-wide linkage scan identifies a novel genetic locus on chromosome 5p13 for neonatal atrial fibrillation associated with sudden death and variable cardiomyopathy. Circulation. 2004;110:3753–3759. doi: 10.1161/01.CIR.0000150333.87176.C7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, et al. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135:1017–1027. doi: 10.1016/j.cell.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 46.Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 47.Rhodes TE, Manderfield LJ, Lundquist AL, George AL, Donahue BS, Roden DM, Darbar D. Gain-of-function KCNQ1 and KCNE3 Mutations Associated with Atrial Fibrillation. Circulation. 2006 Accepted for presentation, AHA 2006 Scientific sessions: (Abstr.) [Google Scholar]
- 48.Otway R, Vandenberg JI, Guo G, Varghese A, Castro ML, Liu J, et al. Stretch-Sensitive KCNQ1 Mutation: A Link Between Genetic and Environmental Factors in the Pathogenesis of Atrial Fibrillation? J Am Coll Cardiol. 2007;49:578–586. doi: 10.1016/j.jacc.2006.09.044. [DOI] [PubMed] [Google Scholar]
- 49.Priori SG, Pandit SV, Rivolta I, Berenfeld O, Ronchetti E, Dhamoon A, et al. A Novel Form of Short QT Syndrome (SQT3) Is Caused by a Mutation in the KCNJ2 Gene. Circ Res. 2005 doi: 10.1161/01.RES.0000162101.76263.8c. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y, Xia M, Jin Q, Bendahhou S, Shi J, Chen Y, et al. Identification of a KCNE2 Gain-of-Function Mutation in Patients with Familial Atrial Fibrillation. Am J Hum Genet. 2004;75:899–905. doi: 10.1086/425342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olson TM, Alekseev AE, Moreau C, Liu XK, Zingman LV, Miki T, et al. KATP channel mutation confers risk for vein of Marshall adrenergic atrial fibrillation. Nat Clin Pract Cardiovasc Med. 2007;4:110–116. doi: 10.1038/ncpcardio0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antzelevitch C, Pollevick GD, Cordeiro JM, Casis O, Sanguinetti MC, Aizawa Y, et al. Loss-of-Function Mutations in the Cardiac Calcium Channel Underlie a New Clinical Entity Characterized by ST-Segment Elevation, Short QT Intervals, and Sudden Cardiac Death. Circulation. 2007;115:442–449. doi: 10.1161/CIRCULATIONAHA.106.668392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hodgson-Zingman DM, Karst ML, Zingman LV, Heublein DM, Darbar D, Herron KJ, et al. Atrial Natriuretic Peptide Frameshift Mutation in Familial Atrial Fibrillation. N Engl J Med. 2008;359:158–165. doi: 10.1056/NEJMoa0706300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–2191. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- 55.Kusano KF, Taniyama M, Nakamura K, Miura D, Banba K, Nagase S, et al. Atrial Fibrillation in Patients With Brugada Syndrome: Relationships of Gene Mutation, Electrophysiology, and Clinical Backgrounds. J Am Coll Cardiol. 2008;51:1169–1175. doi: 10.1016/j.jacc.2007.10.060. [DOI] [PubMed] [Google Scholar]
- 56.Antzelevitch C, Brugada P, Borggrefe M, Brugada J, Brugada R, Corrado D, et al. Brugada Syndrome. Report of the Second Consensus Conference. Endorsed by the Heart Rhythm Society and the European Heart Rhythm Association. Circulation. 2005;111:429–440. doi: 10.1161/01.CIR.0000152479.54298.51. [DOI] [PubMed] [Google Scholar]
- 57.Chen PS, Priori SG. The Brugada Syndrome. J Am Coll Cardiol. 2008;51:1176–1180. doi: 10.1016/j.jacc.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe H, Koopmann TT, Le SS, Yang T, Ingram CR, Schott JJ, et al. Sodium channel beta1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest. 2008;118:2260–2268. doi: 10.1172/JCI33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.London B, Michalec M, Mehdi H, Zhu X, Kerchner L, Sanyal S, et al. Mutation in Glycerol-3-Phosphate Dehydrogenase 1 Like Gene (GPD1-L) Decreases Cardiac Na+ Current and Causes Inherited Arrhythmias. Circulation. 2007;116:2260–2268. doi: 10.1161/CIRCULATIONAHA.107.703330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watanabe H, Darbar D, Ingram CR, Jiramongkolchai K, Chopra SS, Kucera G, Stubblefield T, Wang J, Roden DM. Loss of Function Mutations in Sodium Channel Beta Subunits Associated with Atrial Fibrillation and ST-segment elevation. Accepted for presentation, American Heart Association annual scientific sessions, 2007. Circulation. 2007;116:II–54. (Abstr.) [Google Scholar]
- 61.Junttila MJ, Raatikainen MJ, Perkiomaki JS, Hong K, Brugada R, Huikuri HV. Familial clustering of lone atrial fibrillation in patients with saddleback-type ST-segment elevation in right precordial leads. Eur Heart J. 2007;28:463–468. doi: 10.1093/eurheartj/ehl474. [DOI] [PubMed] [Google Scholar]
- 62.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, et al. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–1935. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Priori SG, Napolitano C, Schwartz PJ, Bloise R, Crotti L, Ronchetti E. The elusive link between LQT3 and brugada syndrome : the role of flecainide challenge. Circulation. 2000;102:945–947. doi: 10.1161/01.cir.102.9.945. [DOI] [PubMed] [Google Scholar]
- 64.Lai LP, Su MJ, Yeh HM, Lin JL, Chiang FT, Hwang JJ, et al. Association of the human minK gene 38G allele with atrial fibrillation: evidence of possible genetic control on the pathogenesis of atrial fibrillation. Am Heart J. 2002;144:485–490. doi: 10.1067/mhj.2002.123573. [DOI] [PubMed] [Google Scholar]
- 65.Chen LY, Ballew JD, Herron KJ, Rodeheffer RJ, Olson TM. A common polymorphism in SCN5A is associated with lone atrial fibrillation. Clin Pharmacol Ther. 2007;81:35–41. doi: 10.1038/sj.clpt.6100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 67.Mommersteeg MTM, Brown NA, Prall OWJ, de Gier-de Vries C, Harvey RP, Moorman AFM, et al. Pitx2c and Nkx2–5 Are Required for the Formation and Identity of the Pulmonary Myocardium. Circ Res. 2007;101:902–909. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 68.Body SC, Collard CD, Sherman SK, Fox AA, Liu KY, Ritchie MD, Perry TE, Muehlschlegel JD, Arnaki S, Seidman CE, et al. Variation in the 4q25 Chromosomal Locus Predicts New-onset Atrial Fibrillation after Cardiac Surgery. American Heart Association 2008 Scientific Sessions. Circulation. 2009;118:S-882. (Abstr.) [Google Scholar]
- 69.Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, Gschwendtner A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–409. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 70.Gaudino M, Andreotti F, Zamparelli R, Di Castelnuovo A, Nasso G, Burzotta F, et al. The -174G/C Interleukin-6 Polymorphism Influences Postoperative Interleukin-6 Levels and Postoperative Atrial Fibrillation. Is Atrial Fibrillation an Inflammatory Complication? Circulation. 2003;108:195II–199II. doi: 10.1161/01.cir.0000087441.48566.0d. [DOI] [PubMed] [Google Scholar]
- 71.Motsinger AA, Donahue BS, Brown NJ, Roden DM, Ritchie MD. Risk factor interactions and genetic effects associated with post-operative atrial fibrillation. Pac Symp Biocomput. 2006:584–595. 584-595. [PubMed] [Google Scholar]
- 72.Prutkin JM, Sotoodehnia N. Genetics of sudden cardiac arrest. Prog Cardiovasc Dis. 2008;50:390–403. doi: 10.1016/j.pcad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 73.Arnestad M, Crotti L, Rognum TO, Insolia R, Pedrazzini M, Ferrandi C, et al. Prevalence of Long-QT Syndrome Gene Variants in Sudden Infant Death Syndrome. Circulation. 2007;115:361–367. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]
- 74.Priori SG, Napolitano C, Giordano U, Collisani G, Memmi M. Brugada syndrome and sudden cardiac death in children. Lancet. 2000;355:808–809. doi: 10.1016/S0140-6736(99)05277-0. [DOI] [PubMed] [Google Scholar]
- 75.Donofrio MT, Gullquist SD, O'Connell NG, Redwine FO. Fetal Presentation of Congenital Long QT Syndrome. Pediatr Cardiol. 1999;20:441–444. doi: 10.1007/s002469900510. [DOI] [PubMed] [Google Scholar]
- 76.Splawski I, Timothy KW, Tateyama M, Clancy CE, Malhotra A, Beggs AH, et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297:1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- 77.Van Norstrand DW, Tester DJ, Ackerman MJ. Overrepresentation of the proarrhythmic, sudden death predisposing sodium channel polymorphism S1103Y in a population-based cohort of African-American sudden infant death syndrome. Heart Rhythm. 2008;5:712–715. doi: 10.1016/j.hrthm.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Plant LD, Bowers PN, Liu Q, Morgan T, Zhang T, State MW, et al. A common cardiac sodium channel variant associated with sudden infant death in African Americans, SCN5A S1103Y. J Clin Invest. 2006;116:430–435. doi: 10.1172/JCI25618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen S, Chung MK, Martin D, Rozich R, Tchou PJ, Wang Q. SNP S1103Y in the cardiac sodium channel gene SCN5A is associated with cardiac arrhythmias and sudden death in a white family. J Med Genet. 2002;39:913–915. doi: 10.1136/jmg.39.12.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen J, Xie X, Zhu J, Tao Q, Wang X. Single-nucleotide polymorphisms in SCN5A gene in Chinese Han population and their correlation with cardiac arrhythmias. Genet Med. 2004;6:159. doi: 10.1097/00125817-200405000-00009. [DOI] [PubMed] [Google Scholar]
- 81.Stecker EC, Sono M, Wallace E, Gunson K, Jui J, Chugh SS. Allelic variants of SCN5A and risk of sudden cardiac arrest in patients with coronary artery disease. Heart Rhythm. 2006;3:697–700. doi: 10.1016/j.hrthm.2006.01.029. [DOI] [PubMed] [Google Scholar]
- 82.Sotoodehnia N, Siscovick DS, Vatta M, Psaty BM, Tracy RP, Towbin JA, et al. {beta}2-Adrenergic Receptor Genetic Variants and Risk of Sudden Cardiac Death. Circulation. 2006;113:1842–1848. doi: 10.1161/CIRCULATIONAHA.105.582833. [DOI] [PubMed] [Google Scholar]
- 83.Snapir A, Mikkelsson J, Perola M, Penttila A, Scheinin M, Karhunen PJ. Variation in the alpha2B-adrenoceptor gene as a risk factor for prehospital fatal myocardial infarction and sudden cardiac death. J Am Coll Cardiol. 2003;41:190–194. doi: 10.1016/s0735-1097(02)02702-x. [DOI] [PubMed] [Google Scholar]
- 84.Fan YM, Lehtimaki T, Rontu R, Ilveskoski E, Goebeler S, Kajander O, et al. Age-dependent association between hepatic lipase gene C-480T polymorphism and the risk of pre-hospital sudden cardiac death: the Helsinki Sudden Death Study. Atherosclerosis. 2007;192:421–427. doi: 10.1016/j.atherosclerosis.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 85.Schwartz PJ, Wolf S. QT interval prolongation as a predictor of sudden death in patients with myocardial infarction. Circulation. 1978;56:1074–1077. doi: 10.1161/01.cir.57.6.1074. [DOI] [PubMed] [Google Scholar]
- 86.Straus SM, Kors JA, De Bruin ML, van der Hooft CS, Hofman A, Heeringa J, et al. Prolonged QTc interval and risk of sudden cardiac death in a population of older adults. J Am Coll Cardiol. 2006;47:362–367. doi: 10.1016/j.jacc.2005.08.067. [DOI] [PubMed] [Google Scholar]