Abstract
Purpose
A better understanding of photoreceptor fate specification may lead to efficient production of photoreceptors for cell replacement studies. This study investigates the role of proneural bHLH gene neurogenin1 (ngn1) in photoreceptor genesis using the chick retina.
Methods
In situ hybridization was used to delineate the spatial and temporal pattern of ngn1 expression. RCAS retrovirus was used to drive overexpression of ngn1 in retinal cells, and siRNA was used to reduce ngn1 expression in loss-of-function experiments.
Results
Chick ngn1 was transiently expressed during early phases of retinal neurogenesis, from embryonic day 3 (E3) to E6, with cells expressing ngn1 confined to the apical side of the retinal neuroepithelium. The time window and the anatomical location of ngn1 expression coincided with photoreceptor genesis and differed from that of other transiently expressed proneural bHLH genes, such as ash1, ath3, ath5, and ngn2. Most ngn1-expressing cells lacked BrdU incorporation and lacked phosphorylated histone H3. In low density cell culture, ngn1 overexpression increased neuroD expression, expanded the photoreceptor population, but reduced the ganglion population. Treatment of dissociated retinal cells with siRNA against ngn1 mRNA specifically reduced the photoreceptor population. Overexpression of ngn1 in the retina reduced the expression of ash1, ath5, chx10, and ngn2.
Conclusions
The data suggest that ngn1 participates in a complex transcriptional network and may play a role in guiding a progenitor cell to the photoreceptor pathway.
Keywords: gene expression, photoreceptor, retinal development, transcription factors, proneural gene
Introduction
Photoreceptors are specialized sensory neurons in the retina. Because they are terminally differentiated and do not re-enter the cell cycle for regeneration, photoreceptors lost due to various causes cannot be replenished, leading to irreversible blindness. 1,2 One of the potential therapies is cell-replacement with developing photoreceptors. 3 This promising approach faces a major roadblock – the need for a supply of photoreceptors. 4 As a result, attention has been directed at inducing photoreceptor genesis through programming or reprogramming the differentiation of cells that can be propagated in large amounts. Key to this approach is knowledge of the decision-making factor(s) that can guide a progenitor cell to the path of differentiating into a photoreceptor.
The photoreceptors and their “siblings” (i.e., the other five major types of retinal cells) arise from a pool of multipotent progenitors during vertebrate retinal development. 5 What dictates photoreceptor fate over other options during retinal neurogenesis has been a central question in the developmental biology of the retina. Photoreceptor development and maturation requires a number of regulatory genes, such as homeodomain genes crx 6–8 and raxL; 9 the neural retina leucine-zipper gene NRL; 10,11 thyroid hormone receptor TRβ2; 12 and basic helix loop-helix (bHLH) gene neuroD. 13–15 Whether there is a single key factor dictating photoreceptor fate remains elusive.
Proneural bHLH genes have been shown to play important roles in specifying neural fates/diversities in both the central and the peripheral nervous systems (CNS and PNS). The developing vertebrate retina expresses several such genes, such as achaete-scute homologue 1 (ash1), atonal homologue 3 (ath3), ath5, neuroD, neurogenin1 (ngn1), ngn2, ngn3, NSCL1, and NSCL2. Proneural bHLH genes known to be expressed in retinal progenitor cells include ash1, ath3, ngn1, ngn2, and ngn3. 16–21 Analysis of retinal explants derived from ash1-null mice indicated that ash1 participates in the production of late-born neurons, including rod photoreceptors and bipolar cells. 22 In the chick retina, ash1 was proposed to promote amacrine cells, 23 and this was later confirmed experimentally. 24 Studies have indicated ath3 in the production of bipolar and amacrine cells. 25–27 Ngn2 is expressed in the proliferating zone, 25,28 including cells still in the cell cycle. 29,30 In the mouse retina, regions lacking ngn2 expression contain no photoreceptor cells, indicating that ngn2 has a role in photoreceptor genesis. 28 Analyses of retinas from double and triple knockoutsindicate that ngn2 may also play an important role in horizontal cell genesis. 31 A fate mapping study showed that cells expressing ngn2 develop into all major cell types in the mouse retina. 30 Chick ngn3 is transiently expressed during early retinal neurogenesis, and its overexpression increases the population of ganglion cells such that they expand into the territory normally occupied by amacrine cells. Overexpression of ngn3 induces ngn1 expression, while suppressing the expression of ngn2, ash1, and ath3. 21 The role of ngn1 in retinal cell fate specification is not well established, even though in the brain ngn1 is known to play a determinative role in generating neural diversity. 32,33 Perron et al. 16 reported a specific, albeit moderate, increase in the photoreceptor population upon overexpression of a related gene, ngnr1, in Xenopus retina. Thummel et al. 20 found that zebrafish ngn1 is expressed in developing retina and during photoreceptor regeneration after light-induced photoreceptor degeneration. These studies suggest that ngn1 may be involved in photoreceptor generation.
We have investigated the expression of ngn1 in the developing chick retina and the role of ngn1 in retinal cell generation. We report that the expression of chick ngn1 was transient and restricted to early neurogenesis, with a spatial and temporal window of expression coinciding with the generation of photoreceptor precursor cells. In retinal cell culture, ngn1 overexpression increased the photoreceptor population at the expense of ganglion cells, while siRNA against ngn1 reduced the photoreceptor population. Overexpression of ngn1 in the developing retina reduced the expression of other regulatory genes. These results suggest that ngn1 participates in regulatory networks governing retinal neurogenesis and has a role in leading progenitor cells to take the photoreceptor pathway.
Materials and Methods
Chick embryos
Fertilized, pathogen-free White Leghorn chicken eggs were purchased from Spafas and incubated in a Petersime incubator. All use of animals adhered to the ARVO Statement for the Use ofAnimals in Ophthalmic and Vision Research and the procedures and policies set by the Institutional Animal Use and Care Committee at the University of Alabama at Birmingham.
Generation of RCAS-ngn1 retrovirus
Based on published information, 34 we amplified the coding region of chick ngn1 with RT-PCR. After cloning and its sequence verification, the DNA was subcloned into shuttle vector Cla12Nco and then inserted into proviral vector RCAS. 35 Virus particles were produced by transfecting chick embryonic fibroblast cells with the recombinant proviral DNA. Concentrated viral stocks (~1×108 pfu/ml) were prepared as described. 36
Microinjection of retrovirus into chick embryos
Concentrated RCAS-ngn1 virus, or control RCAS-GFP virus, 36 was microinjected into the neural tube and the subretinal space (between the two layers of the optic cup) of day 2.5 chick embryos (E2.5, stage 15–17), as previouslydescribed. 36 Infected eyes were enucleated at various developmental stages and fixed with ice-cold 4% paraformaldehyde, cryoprotected with OCT:sucrose (2:1), frozen with liquid nitrogen, and kept at −80°C. Infection by RCAS viruses (RCAS-ngn1 and RCAS-GFP) was visualized by immunostaining with an antibody against viral protein p27.
Low density retinal cell culture
Retinas (n=3–16) were dissected from E4.5 – E8.5 chick embryos infected with RCAS-ngn1 or RCAS-GFP as control. Retinal cells were dissociated with trypsin-EDTA and seeded into the wells of 24-well plates treated with polyornithine at a density that covered <1/5 of the surface area. After 4 days in culture with Medium 199 supplemented with 10% fetal calf serum, cells were fixed with ice-cold 4% paraformaldehyde, the subjected to immunostaining or in situ hybridization. For experiments with E4.5 and E8.5 retinas, double-labeling for viral (p27) and retinal markers was carried out. The number of the positive cells and the number of total cells were scored from 9 view areas from each culture well, each with 40–300 cells, under a 20x objective. The percentage of positive cells was calculated for one well, and the means and SDs from 3 wells were calculated using the computer program Origin 7.0 (OriginLab Corp.).
Low density retinal cell culture with siRNA
Retinal cells from E4.5 chick embryos were seeded at low density, as described in the previous subsection. Poly-ornithine-treated glass coverslips were coated with siRNA immediately before cell seeding. Two Silencer® Select pre-designed siRNAs against ngn1, uucgauuuuggugaguuugGT and uaagggugugcagcaaagcCT, were selected and synthesized by Ambion (Applied Biosystems). Silencer® Select Negative Control #1 siRNA from Ambion was used as a control. SiRNA and the transfection agent (siPORT™ NeoFX™ transfection agent, Ambion) were mixed following the manufacturer’s instructions, and added to the cell culture at a final concentration of 300 nM. The culture was maintained for 2 or 4 days before fixation for immunocytochemistry, in situ hybridization, or TUNEL analysis. Scoring cell numbers and statistical analysis were as described in the previous subsection.
In situ hybridization
Digoxigenin (Dig) labeled antisense RNA probe against ngn1 was prepared from linearized plasmids harboring the coding sequence using the Genius kit (Roche Molecular Biochemicals) following the manufacturer’sinstructions. Dig-labeled antisense RNA probes against chick ash1, 21 ath3, 37 ath5, 38 chx10, 37 neuroD, 36 ngn2, 29 and ngn3 21 were prepared as described. Retinal cryosections of 10 μm thick were used for in situ hybridization following procedures described previously. 37
Immunocytochemistry
The following monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank (University ofIowa): anti-bromodeoxyuridine (BrdU, clone G3G4,1:100; developed by Dr. Stephen J. Kaufman), anti-islet-1 (clone39.4D5, 1:100; developed by Dr. Thomas Jessell), anti-AP2α(3B5, 1:50; developed by Dr. Trevor Williams), and anti-visinin(clone 7G4, 1:500; developed by Dr. Constance Cepko). Antibodies obtained from a commercial source included: monoclonal antibody against Brn3a (1:200; Chemicon); polyclonal antibody against red opsin (1:200; Chemicon); polyclonal antibody against calretinin (1:500; Chemicon), anti-phosphorylated histone H3 (1:200; Upstate Biotechnology), and polyclonal antibody against an RCAS viral protein, p27 (1:500; Spafas). Monoclonal antibody RA4 (1:1000dilution) was a gift from Dr. Steven McLoon (University of Minnesota). A monoclonal antibody against chick NeuroD was produced in our laboratory. Standard immunocytochemistry was performed with secondary antibodies conjugated with peroxidase, alkaline phosphatase (Vector Laboratories), or fluorophore (Molecular Probes).
TUNEL assay
TUNEL assay was used to detect the presence of apoptotic cells in E4.5 retinal cell culture, using the In Situ Cell Death Detection Kit (RocheMolecular Biochemicals) following the manufacturer’s instruction.
Pulse-labeling chick embryos with BrdU
BrdU (50 μg in 50 μlof HBSS) was dropped through an opening in the shell onto the vitelline membrane of E5 and E7.5 chick embryos. The embryos were incubated for 3 hours before the eyes were harvested and fixedwith 4% paraformaldehyde. For double labeling, cryosections were first subjected to in situ hybridization with Dig-labeled anti-ngn1 RNA probesand then to BrdU detection using a specific antibody as previouslydescribed. 37
Results
Transient ngn1 expression in early retinal development
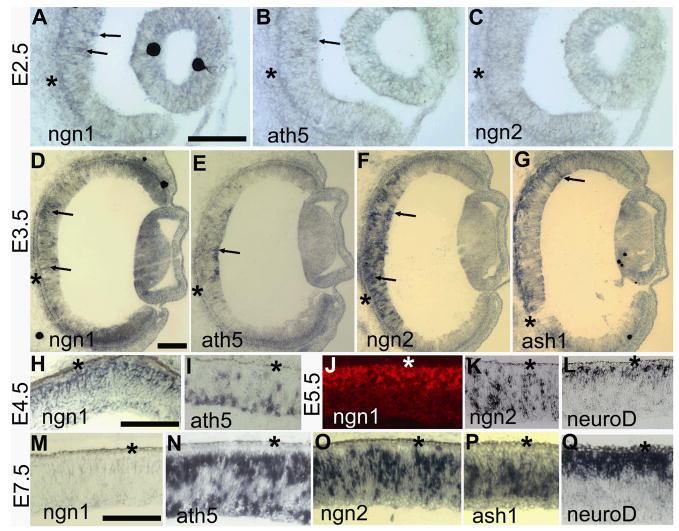
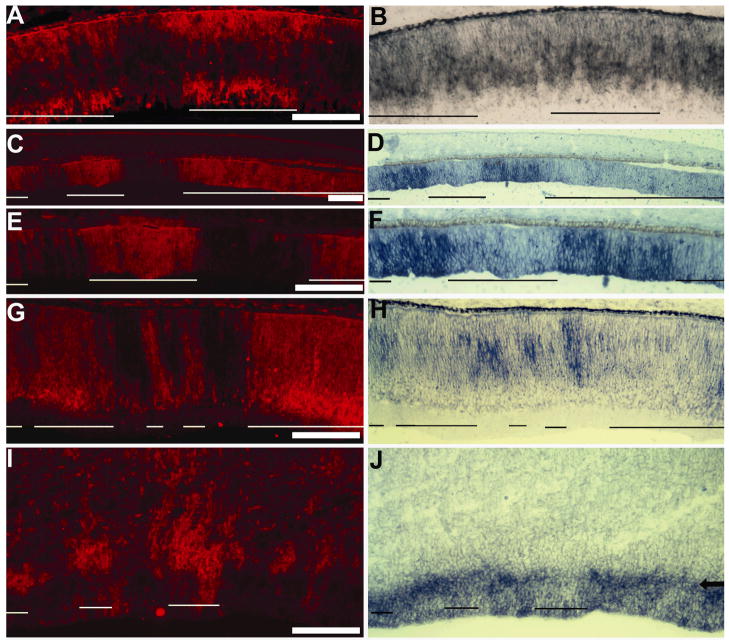
To gain a perspective on how ngn1 might function in the retina, its expression was examined and compared with the expression of some better-studied proneural bHLH genes. The earliest retinal expression of ngn1 detected with Dig-labeled antisense RNA probes was embryonic day 2.5 (E2.5), when some positive cells were observed at the central region (Fig. 1A). At this time, ath5 expression was visible in only a few cells (Fig. 1B), while no ngn2 expression was evident (Fig. 1C). Expression of ngn1 (Fig. 1D), ath5 (Fig. 1E), ngn2 (Fig. 1F), and ash1 (Fig. 1G) was visible in the E3.5 retina. Starting from E4.5, differences emerged in the spatial expression patterns of the genes. Cells expressing ngn1 localized to the outermost zone in the pseudostratified retinal neuroepithelium at E4.5 (Fig. 1H) and E5.5 (Fig. 1J), an anatomical location of newly born photoreceptor precursors. This spatial pattern is similar to neuroD expression (Fig. 1L). Chick neuroD is expressed in young photoreceptor cells or their precursors. 14,36 However, neuroD expression is not evident in retinas younger than E5. 36 Thus, ngn1 expression preceded that of neuroD. Cells expressing ath5 mostly localized to the vitreal side (Fig. 1I), and cells expressing ngn2 were distributed across the neuroepithelium (Fig. 1K). Expression of ngn1 began to decrease in the E6 central retina (data not shown) and by E7.5 the expression of ngn1 was no longer detectable in the central region (Fig. 1M). This temporal pattern coincided with photoreceptor genesis, which in the chick retina mostly takes place between E5 and E7. 39 The E7.5 retina continued the expression of ath5 (Fig. 1N), ngn2 (Fig. 1O), ash1 (Fig. 1P), and neuroD (Fig. 1Q), each with its distinctive spatial pattern maintained, except ath5, which had an additional zone of expression in the outer portion of the retinal neuroepithelium. Overall, while partially overlapping with ngn3 expression (Ma et al., 2009), ngn1 expression differed spatially and temporally from that of ath5, ngn2, neuroD, and ash1.
Fig. 1.
The expression pattern of ngn1 in the developing chick retina in comparison with other proneural bHLH genes. A–C: In situ hybridization of serial sections to detect the expression of ngn1 (A), ath5 (B), and ngn2 (C) in E2.5 eye. D–G: In situ hybridization of serial sections to detect the expression of ngn1 (D), ath5 (E), ngn2 (F), and ash1 (G) in E3.5 eye. H–I: Spatial expression patterns of ngn1 (H) and ath5 (I) in E4.5 retinas. J–L: Spatial expression patterns of ngn1 (J), ngn2 (K), and neuroD (L) in E5.5 retinas. M–Q: Spatial expression patterns of ngn1 (M), ath5 (N), ngn2 (O), ash1 (P), and neuroD (Q) in E7.5 retinas. In situ hybridization signals were developed with nitroblue tetrazolium, except in J, where rhodamine-tyramide was used. Arrows in A–G point to retinal cells expressing the respective gene. The RPE layer is indicated by an asterisk. Scale bars: 100 μm.
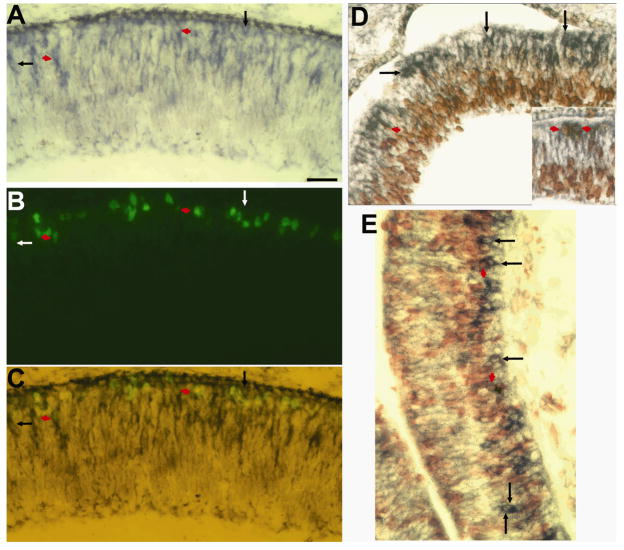
In addition to coinciding with the prospective location of photoreceptor cells, the localization of ngn1-expressing cells to the outermost region of the retinal neuroepithelium also coincided with the location of M-phase cells. To determine whether ngn1 expression defined M-phase cells, double-labeling for phosphorylated histone H3 (phospoH3) and for ngn3 mRNA was carried out. Double-labeled cells were detected (Fig. 2A–C), but accounted for ~10% of the phospoH3+ cells and ~5% of the ngn1 mRNA+ cells. A typical view area under a 40 x objective contained 3 double-labeled cells from 26 phospoH3+ cells and 60 ngn1 mRNA+ cells. To determine whether ngn1-expressing cells were cycling or postmitotic, double-labeling for ngn1 expression and BrdU incorporation was carried out after pulsing the embryo with BrdU. Only a small number of double-labeled cells were detected in the retina (Fig. 2D) and the brain (Fig. 2E). In a typical view area with a 40 x objective, 4 were double-labeled, representing 7% of the 61 ngn1 mRNA+ cells and 3% of the 156 BrdU+ cells. Thus, the majority of ngn1-expressing cells lacked phospoH3 or BrdU incorporation.
Fig. 2.
Double-labeling for ngn1 expression and cell proliferation. A–C: Double-labeling for ngn1 mRNA (A, blue) and phosphorylated histone H3 (B, green) in E6 retina. Panel C shows a simultaneous view of both. D,E: Double-labeling for ngn1 mRNA (blue) and BrdU incorporation (red) in E5 retina (D) and brain (E). Inset in D provides a clearer image of double-labeled cells. Arrows point to cells positive for ngn1 mRNA but negative for BrdU or phospoH3. Short arrows (red) point to double-labeled cells. Scale bar: 50 μm.
Effect of ngn1 overexpression on retinal neurogenesis
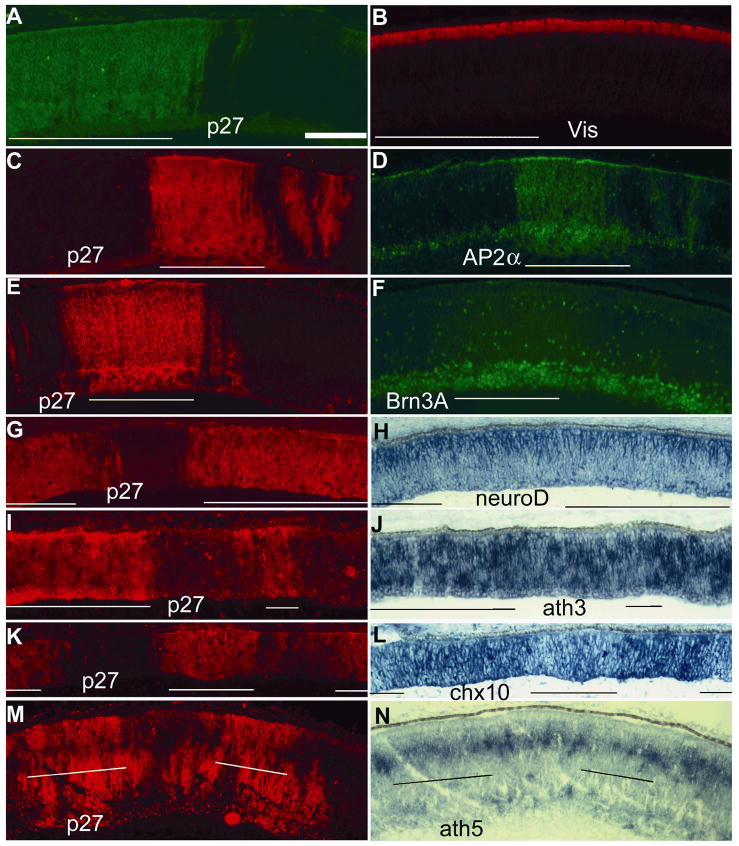
The replication-competent avian leukosis virus RCAS 35 was used to drive the overexpression of ngn1 in chick embryos. This virus infects an increasing number of cells over time in a population of proliferating cells, because viruses released from infected cells can infect other cells. 40,41 RCAS infection and transduction do not affect retinal development; we have observed no abnormalities either at the gross level or at the microscopic level from hundreds of embryos infected with RCAS or RCAS-GFP. 21,24,42,43 Analyses of cell populations in regions infected with RCAS-GFP against the adjacent, uninfected regions revealed no differences in the numbers of AP2+ cells, Pax6+ cells, chx10+ cells, visinin+ cells, BrdU+ cells, and ngn2+ cells. 24 RCAS-ngn1, or RCAS-GFP, was microinjected into the lumen of the neural tube and the subretinal space at E2.5 (~64 hours of incubation) through ~E3 (70 hours of incubation), 24 because during this time the neural tube and the subretinal space remain connected. Expression of the transgene was expected to begin at ~E3.5 and to persist thereafter until the retina was harvested for analysis, based on the fact that transgene expression becomes apparent 21–24 hours after infection. 40,41 Previous studies showed that RCAS-driven overexpression of bHLH genes may alter retinal neurogenesis in a gene-specific manner. 21,24,44–46 Retinal sections from embryos infected with RCAS-ngn1 were examined histologically, followed by immunohistochemistry with antibodies that recognize specific cell types. Retinas from embryos (n>50) microinjected with RCAS-ngn1 displayed no obvious alterations in histology or in retinal neurogenesis. For unequivocal evidence, we compared immunostaining of regions infected by RCAS-ngn1 with adjacent, uninfected regions on the same retinal section. This is to minimize ambiguities arising from comparing retinas that may vary due to differences in developmental stages and/or the angles at which the retinas were sectioned. Molecular analyses with specific antibodies showed no changes in the populations of photoreceptor (visinin+, Fig. 3A, B), amacrine (AP2α+, Fig. 3C, D), or ganglion cells (Brn3A+, Fig. 3E, F).
Fig. 3.
Double-staining of E7.5 retinas partially infected by RCAS-ngn1 for alterations in retinal neurogenesis. A, B: Double-labeling for RCAS viral protein p27 (A) and photoreceptor protein visinin (Vis, B). C, D: Double-labeling for viral protein p27 (C) and amacrine protein AP2α (D). E, F: Double-labeling for viral protein p27 (E) and ganglion protein Brn3A (F). G, H: Double-labeling for viral protein p27 (G) and neuroD mRNA (H). I, J: Double-labeling for viral protein p27 (I) and ath3 mRNA (J). K, L: Double-labeling for viral protein p27 (K) and chx10 mRNA (L). M, N: Double-labeling for viral protein p27 (M) and ath5 mRNA (N). Infected regions are underlined. Scale bar: 100 μm. Of note, in some of the double-fluorescent images (C–F), the anti-p27 signals were so strong that they were also visible under the filter used to view the anti-AP2α (D) or anti-Brn3A (F) signals, resulting in optical spillover. The genuine anti-AP2α (D) or anti-Brn3A (F) signals can be distinguished by their nucleus localization from the optical spillover of anti-P27 signals in the cytoplasm.
Effects on regulatory gene expression by ngn1 overexpression
To determine whether ngn1 overexpression might have altered gene expression without concomitant alterations in cell production, we examined the retinas infected with RCAS-ngn1 for the expression of a number of regulatory genes involved in different retinal cell populations. For the immunohistochemical analysis, uninfected regions in the same retinal sections were used as internal controls to avoid ambiguities from comparing different retinal sections that potentially differed in developmental stage or angle of sectioning. In retinas infected with RCAS-ngn1, expression of neuroD (Fig. 3G, H) remained unchanged, despite the spatial similarity of their expression. Expression of ath3, which plays an important role in the development of progenitor cells and bipolar cells, 27 also remained unaltered with ngn1 overexpression (Fig. 3I, J; Fig. 5G). On the other hand, expression of chx10, a homeodomain gene with important roles in the development of progenitor cells and bipolar cells, 47 was essentially abolished in the region infected with RCAS-ngn1, whereas in the adjacent, uninfected region a high level of chx10 expression was observed (Fig. 3K, L; Fig. 5G). Ngn1 overexpression also reduced the expression of ath5 (Fig. 3M, N; Fig. 5G), which is required for ganglion cell development (for review see Mu and Klein 48) and may also participate in the production of other retinal cells. 49
Fig. 5.
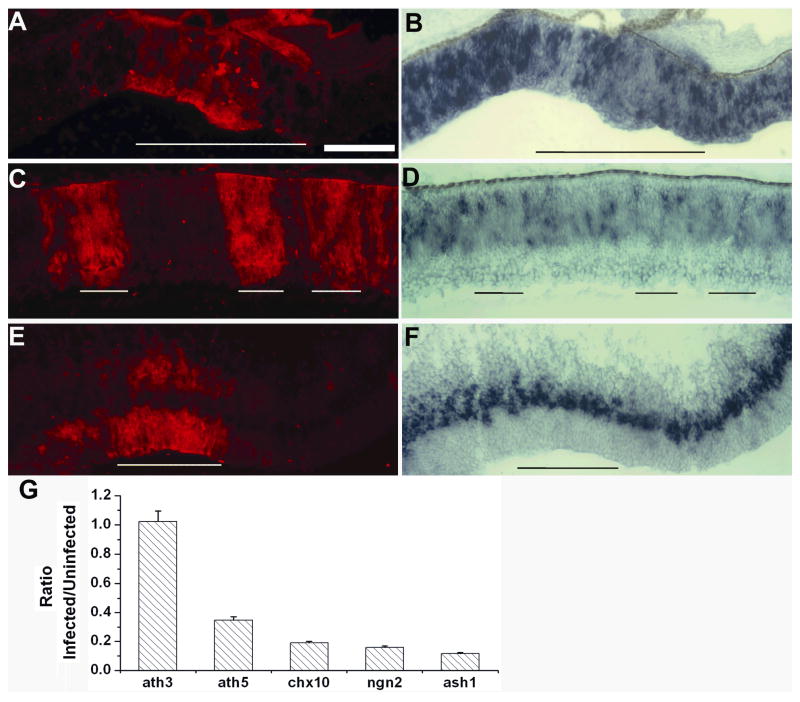
Down-regulation of ngn2 expression in retinas infected with RCAS-ngn1. A–D: Double-labeling for RCAS viral protein p27 (A, C) and for ngn2 mRNA (B, D) in E7.5 peripheral retina (A, B) and central retina (C, D). E, F: Double-labeling for viral protein p27 (E) and ngn2 expression (F) in E7.5 brain partially infected with RCAS-ngn1. Infected regions are approximately underlined. Scale bar: 100 μm. G: Means and SDs of the ratio of the number of cells expressing each of the markers in infected regions over the number in uninfected regions.
Ash1 plays a determinative role in generating neural diversity in other regions of the CNS. 32,33 In the retina, ash1 expression is detected during both early and late phases of neurogenesis in a subpopulation of retinal progenitor cells, and it may participate in the generation of amacrine cells and/or rod photoreceptors and bipolar cells, which are the last born neurons in the retina. In E7.5 retina, ash1 expression is detected in cells within the proliferating zone, and the expression was unaffected by RCAS-GFP infection (Fig. 4A, B). In E7.5 retinas infected with RCAS-ngn1, expression of ash1 was abolished at the periphery (Fig. 4C–F), where neurogenesis was active, as reflected by the presence of a large number of ash1-expressing cells. Expression of ash1 was also decreased in the central retina (Fig. 4G, H), where neurogenesis was subsiding. The lack of ash1 expression was specific to the infected regions, while the adjacent, uninfected region maintained ash1 expression. Suppression of ash1 expression was also observed in the brain (Fig. 4I, J).
Fig. 4.
Diminished ash1 expression in E7.5 retinas infected by RCAS-ngn1. A, B: Control RCAS-GFP retina with double-labeling for RCAS viral protein p27 (A) and for ash1 mRNA (B). C–H: RCAS-ngn1 partially-infected retinas showing double-labeling for RCAS viral protein p27 (C, E, G) and for ash1 mRNA (D, F, H) in peripheral retina (C–F) and central retina (G, H). C and D are lower magnifications of E and F. I, J: Double-labeling for viral protein p27 (I) and ash1 expression (J) of E7.5 brain partially infected with RCAS-ngn1. Arrow in J points to the zone with diminished ash1 expression. In all panels, infected regions are approximately underlined. Scale bars: 100 μm.
Ngn2 is another regulatory gene involved in cell fate specification during neurogenesis in other regions of the CNS. 32,33 In the retina, ngn2 expression spans early and late phases of cell neurogenesis, and it is believed to participate in the development of progenitor cells that eventually differentiate into all major types of retinal neurons. 28–31 Infection with RCAS-GFP does not alter ngn2 expression. 21 In RCAS-ngn1 infected retinas, ngn2 was down-regulated (Fig. 5A–D, G). The down-regulation was observed at places where neurogenesis was active, as reflected by the presence of a large number of ngn2-expressing cells (Fig. 5A, B). The down-regulation was also observed at places where neurogenesis was tapering off, as reflected by the presence of a relatively smaller number of ngn2-expressing cells (Fig. 5C, D). Notably, no suppression of ngn2 expression was apparent in the brain (Fig. 5E, F). This resistance to ngn1’s suppression could be due to either ngn1 alone being insufficient to suppress ngn2 expression or other genes/factors promoting ngn2 in these particular cells, or both.
Ngn3, the third member of the ngn subfamily, promotes early retinal neurogenesis in the chick. 21 Infection by RCAS-ngn3 results in ectopic expression of ngn1, suggesting an inductive relation of ngn3→ngn1. 21 To examine whether ngn1 could reciprocally induce ngn3, we examined E7.5 retinas infected with RCAS-ngn1 for ngn3 expression, considering that at this developmental stage the level of endogenous ngn3 expression is undetectable in normal retina. 21 No induction of ngn3 was found (data not shown).
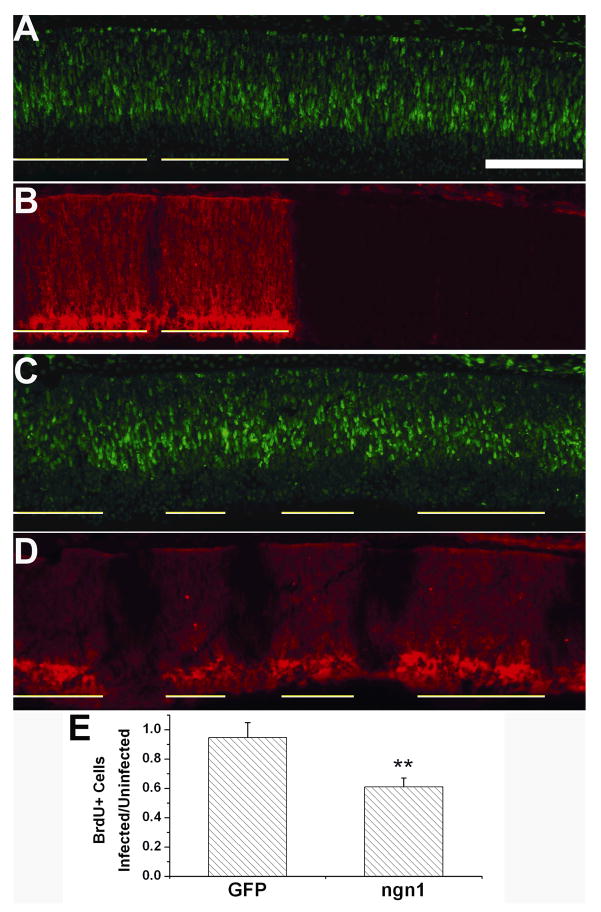
To examine the effect of ngn1 overexpression on cell proliferation, BrdU incorporation analysis was carried out. To minimize ambiguities arising from comparing retinas having different cell proliferation activities due to difference in developmental stages, the number of BrdU+ cells in regions infected by RCAS-ngn1 was compared to that in adjacent, uninfected regions of the same retinal section. Infection with RCAS-GFP did not change the number of BrdU+ cells (Fig. 6A, B, E). However, in regions infected with RACS-ngn1, fewer BrdU+ cells were present compared with the internal control (adjacent, uninfected region; Fig. 6C, D). Scoring the number of BrdU+ cells in regions of the similar sizes showed a 40% reduction in the infected region (Fig. 6E).
Fig. 6.
Analyses of cell proliferation with BrdU incorporation of retinas infected with RCAS-ngn1. A, B: Double-labeling for RCAS viral protein p27 (A) and for BrdU incorporation (B) of a control E7.5 retina with partial infection by RCAS-GFP. C, D: Double-labeling for the viral protein p27 (C) and for BrdU incorporation (D) of an E7.5 retina with partial infection by RCAS-ngn1. Infected regions are underlined. Scale bar (100 μm) applies to all panels.
Effects on retinal populations from manipulating ngn1 expression in vitro
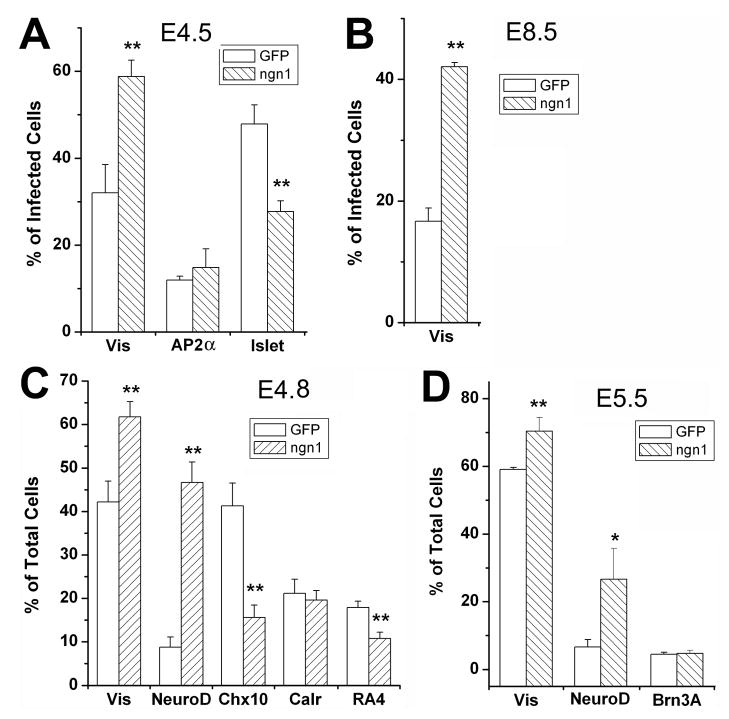
One reason for the lack of changes in retinal cell populations from ngn1 overexpression could be cell contact-mediated feedback mechanisms circumventing the effect of the experimental manipulation on cell production. Ngn1 has been shown to induce and then be a target of Notch/Delta-mediated lateral inhibition, 50–52 which plays an important role in regulating retinogenesis. 53–56 To minimize cell contact-mediated feedback modulation, low-density retinal cell culture 38,53,57,58 was used to examine whether ngn1 could steer retinal progenitors to a particular path. Retinal cells were isolated at E4.5, when expression of ngn1 was high (Fig. 1H), and cultured at low density for 4 days (4 DIV). We found that ngn1 induced an 84% increase in photoreceptor population (visinin+/p27+), from 32% of the total P27+ (infected) cells in the control (infected with RCAS-GFP) to 59% in the experimental retina (Fig. 7A; p<0.01). This strong photoreceptor-promoting effect was still observed at E8.5 (Fig. 7B), when the retina normally no longer expressed ngn1. When the number of total cells, instead of infected cells, was used in the analysis, a milder effect was observed (Fig. 7C, D), likely due to a dilution effect by uninfected cells. Consistent with the expansion of the photoreceptor population, the number of cells expressing neuroD was also increased (Fig. 7C, D). No significant change in AP2α+ or calretinin+ amacrine cells was observed (Fig. 7A, C). The number of cells expressing chx10 was reduced (Fig. 7C). The ganglion cell population (Islet-1+, RA4+, or Brn3A+) was also reduced. With retinal cells of E4.5, when a major portion of the cells born will take on a ganglion fate, the ganglion population was decreased by 42%, from 48% of the infected cells in the control to 28% in the experimental retinas (p<0.01; Fig. 7A). A milder, yet significant (p<0.01), reduction in ganglion population was observed with E4.8 cells (Fig. 7C). With E5.5 retinal cells, no significant reduction in the ganglion population was observed (Fig. 7D).
Fig. 7.
Quantitative analysis of alterations in retinal neurogenesis from ngn1 overexpression in dissociated retinal cells. Shown are the means ± SDs of the calculated percentage of cells identified with each marker. Statistically significant differences from the control are shown at the 0.05 (*) and at 0.01 (**) levels.
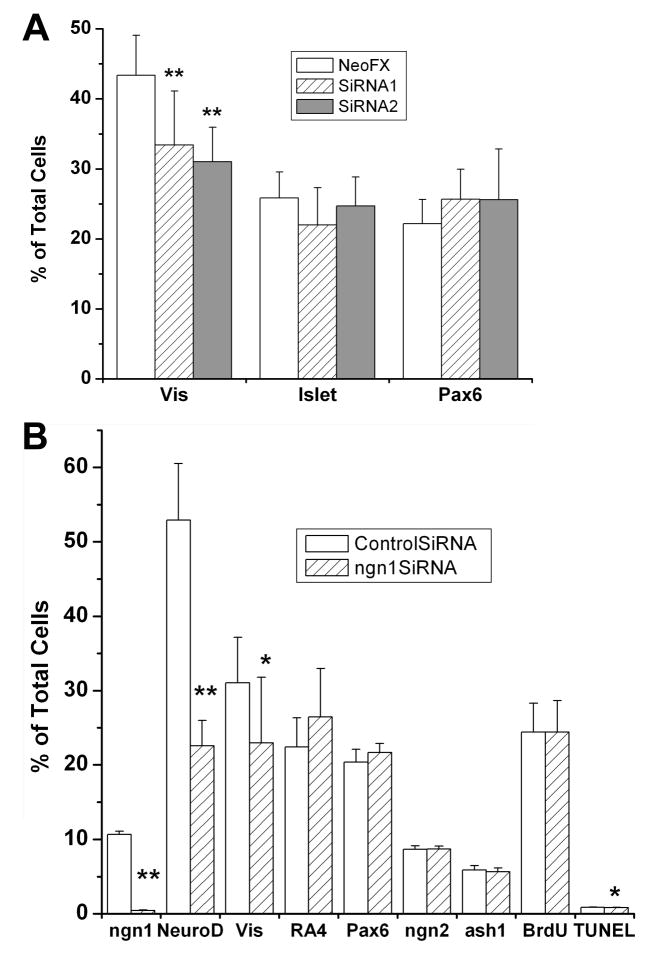
Low density retinal cell culture was also used in siRNA knockdown experiments. Dissociated cells from E4.5 retinas were cultured in the presence, or absence, of siRNA against ngn1. In initial experiments, two siRNAs against ngn1 were used separately. We found that either of the two reduced the number of visinin+ cells significantly (P<0.01), albeit mildly (Fig. 8A), while neither siRNA affected the number of Islet-1+ cells or Pax6+ cells (ganglion and amacrine). In subsequent experiments, the siRNAs were used in combination. To verify that the siRNA reduced ngn1 mRNA, we used in situ hybridization to identify cells with detectable levels of ngn1 mRNA in E4.5 retinal cell cultures 48 hours after siRNA treatment. In cultures receiving a control siRNA, ~10% of the cells were ngn1 mRNA+. The number was reduced by 25 fold, to 0.4% of total cells in cultures receiving siRNA against ngn1 (p<0.01; Fig. 8B). At the same time, the number of NeuroD+ cells was reduced by >50%, from 52.9±7.6% of the total cells in the culture treated with control siRNA to 22.6±3.4% in the experimental culture (p<0.01; Fig. 8B). The number of visinin+ cells was significantly (p<0.05), yet mildly, reduced. There were no significant changes in the numbers of cells that were positive for RA4 (ganglion) or Pax6, BrdU incorporation, or expression of ngn2 or ash1 (Fig. 8B). There was a small, yet statistically significant (p<0.5), reduction in the number of TUNEL+ cells, from 0.9% of the total cells in the control culture to 0.8% in the experimental culture. The results suggest that the reductions in the number of NeuroD+ cells and the number of visinin+ cells from treatment with siRNA against ngn1 were unlikely due to reduced cell proliferation or increased cell death.
Fig. 8.
Effect of siRNA against ngn1 on ngn1 mRNA and on retinal cell populations. Shown are the means ± SDs of the percentage of cells identified with each marker among dissociated E4.5 retinal cells cultured for 2 (for data on ngn1, NeuroD, and TUNEL) or 4 days (for data on the other markers) under the treatments specified. Vis, visinin. Statistically significant differences from the control are shown at the 0.05 (*) and at 0.01 (**) levels.
Discussion
In the chick retina, ngn1 was expressed during the early phases of retinal neurogenesis, and the expression became undetectable during the later phases, when neurogenesis is still active. Expression of ngn1 likely occurred in both proliferating and postmitotic cells, because only some of the ngn1-expressing cells incorporated BrdU or expressed phospoH3. Thus, it is plausible that ngn1 is expressed as a cell is undergoing the transition from proliferation to differentiation.
Spatially, cells expressing ngn1 were confined to the outermost portion of the retinal neuroepithelium, the prospective location of photoreceptor cells. Transient expression has been considered a signature of genes specifying neural types. 33 Thus, the temporal and spatial patterns of ngn1 expression are consistent with a role in photoreceptor fate specification. In support of this, overexpression of ngn1 in low-density cell culture resulted in an expansion of the photoreceptor population and an increase in number of cells expressing neuroD, which participates in photoreceptor production likely through promoting differentiation and survival. 13–15,36 Conversely, the photoreceptor population was reduced when retinal cells were cultured in the presence of siRNA against ngn1. Not totally unexpected, in vivo overexpression of ngn1 did not produce noticeable alterations in retinal cell populations. This could result from confounding factors in the retina, including bHLH factors cross-regulating one another through networks and/or cascades. 21,31 Indeed, we have observed that overexpression of ngn1 diminished the expression of ash1, ath5, and ngn2, all of which participate in the genesis of different types of retinal cells, including photoreceptors. 22,28,30,38,49 Another potential reason for the lack of alteration in retinal cell populations from ngn1 overexpression is the presence of a cell contact-mediated feedback mechanism theorized to participate in the global regulation over retinogenesis that results in a balanced production of all cell types. This could explain why with low density cell culture, we detected ngn1’s photoreceptor-promoting activity. Ngn1 promoting photoreceptor production is consistent with the limited information that is available in the literature. In Xenopus retina, the ngn1-related gene, ngnr1, specifically increases the photoreceptor population. 16 In zebrafish retina, ngn1 is expressed both in development and during photoreceptor regeneration after light-induced photoreceptor degeneration. 20 Little is known about the function of ngn1 in mouse retinal development. In the brain, ngn1 plays a determinative role in generating neural diversity. 32,33
During retinal neurogenesis, a number of proneural bHLH genes are expressed. Among them, ngn1 showed a spatial and temporal pattern of expression similar only to that of ngn3. Both were switched off early, when retinal neurogenesis in the chick is still active and the expression other bHLH regulatory genes (ath5, ngn2, and ash1) remains high. Temporally, expression of ngn3 precedes the expression of ngn1. 21 Spatially, expression of ngn3, but not ngn1, was also detected on the vitreal side, the prospective location of ganglion cells. Overexpression of ngn3 results in an expansion of the ganglion population into the territory otherwise occupied by amacrine cells. 21 Overexpression of ngn1, however, reduced the ganglion cell population. Overexpression of ngn3 leads to ectopic induction of ngn1. Yet, no induction of ngn3 was observed with ngn1 overexpression. These findings suggest ngn1 as a downstream genetic target of ngn3 and a linear inductive relationship of ngn3→ngn1.
How and why ngn1 suppressed the expression of ash1, ath5, and ngn2 is unclear. One simple speculation is rivalry. It is plausible that ngn1 is expressed during the phase of photoreceptor genesis not only to promote photoreceptor production but also to suppress the expression of ash1, ath5, and ngn2 and thus attenuate the production of other cell types. Previous studies with similar experimental approaches showed that in the chick retina, overexpression of ath5 promotes ganglion cell production, 46 overexpression of ash1 increases the amacrine cell population, 24 and overexpression of ngn2 has no effects on retinal cell populations. 24 Thus, ngn1 differed from these bHLH genes not only in its expression pattern but also in its effect on retinal neurogenesis upon overexpression.
The chick ngn1 displayed a distinctive pattern of expression coinciding with photoreceptor genesis, exhibited a photoreceptor-promoting activity, and repressed other regulatory genes whose expression persist through late phases of retinal neurogenesis. These properties support ngn1 to play a role in guiding a progenitor cell to the path of developing into a photoreceptor.
Acknowledgments
Grant support: NIH/NEI grant EY11640 and unrestricted grant to UAB Ophthalmology from Research to Prevent Blindness.
The authors thank Dr. Stephen Hughes for retroviral vector RCAS (B/P) and shuttle vector Cla12Nco. This work was supported in part by NIH/NEI grant EY11640 and an unrestricted grant to UAB Department of Ophthalmology from Research to Prevent Blindness.
Footnotes
Section code: BI or RC.
Commercial relationships: None.
References
- 1.Curcio CA, Medeiros NE, Millican CL. Photoreceptor loss in age-related macular degeneration. Invest Ophthalmol Vis Sci. 1996;37:1236–1249. [PubMed] [Google Scholar]
- 2.Milam AH, Li ZY, Fariss RN. Histopathology of the human retina in retinitis pigmentosa. Prog Retin Eye Res. 1998;17:175–205. doi: 10.1016/s1350-9462(97)00012-8. [DOI] [PubMed] [Google Scholar]
- 3.MacLaren RE, Pearson RA, MacNeil A, Douglas RH, Salt TE, Akimoto M, Swaroop A, Sowden JC, Ali RR. Retinal repair by transplantation of photoreceptor precursors. Nature. 2006;444:203–207. doi: 10.1038/nature05161. [DOI] [PubMed] [Google Scholar]
- 4.Bennett J. Retinal progenitor cells--timing is everything. N Engl J Med. 2007;356:1577–1579. doi: 10.1056/NEJMcibr070209. [DOI] [PubMed] [Google Scholar]
- 5.Adler R. A model of retinal cell differentiation in the chick embryo. Prog Retina Eye Res. 2000;19:529–557. doi: 10.1016/s1350-9462(00)00008-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Wang Q-L, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Kenkins NA, Zack D. Crx, a novel otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 7.Furukawa Y, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 8.Peng GH, Chen S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum Mol Genet. 2007;15(16):2433–2452. doi: 10.1093/hmg/ddm200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CM, Cepko CL. The chicken RaxL gene plays a role in the initiation of photoreceptor differentiation. Development. 2002;129:5363–5375. doi: 10.1242/dev.00114. [DOI] [PubMed] [Google Scholar]
- 10.Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 11.Daniele LL, Lillo C, Lyubarsky AL, Nikonov SS, Philp N, Mears AJ, Swaroop A, Williams DS, Pugh EN., Jr Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest Ophthalmol Vis Sci. 2005;46:2156–2167. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 13.Pennesi ME, Cho JH, Yang Z, Wu SH, Zhang J, Wu SM, Tsai MJ. BETA2/NeuroD1 null mice: a new model for transcription factor-dependent photoreceptor degeneration. J Neurosci. 2003;23:453–461. doi: 10.1523/JNEUROSCI.23-02-00453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan R-T, Wang S-Z. Requirement of neuroD for photoreceptor formation in the chick retina. Invest Ophthalmol Vis Sci. 2004;45:48–58. doi: 10.1167/iovs.03-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Etter P, Hayes S, Jones I, Nelson B, Hartman B, Forrest D, Reh T. NeuroD1 regulates expression of thyroid hormone receptor β2 and cone opsins in the developing mouse retina. J Neurosci. 2008;28:749–756. doi: 10.1523/JNEUROSCI.4832-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perron M, Opdecamp K, Butler K, Harris WA, Bellefroid EJ. X-ngnr-1 and Xath3 promote ectopic expression of sensory neuron markers in the neurula ectoderm and have distinct inducing properties in the retina. Proc Natl Acad Sci USA. 1999;96:14996–15001. doi: 10.1073/pnas.96.26.14996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vetter ML, Brown NL. The role of basic helix-loop-helix genes in vertebrate retinogenesis. Semin Cell Dev Biol. 2001;12:491–498. doi: 10.1006/scdb.2001.0273. [DOI] [PubMed] [Google Scholar]
- 18.Yan R-T, Ma W, Liang L, Wang S-Z. bHLH genes in retinal cell fate specification. Mol Neurobiol. 2005;32:157–171. doi: 10.1385/MN:32:2:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matter-Sadzinski L, Puzianowska-Kuznicka M, Hernandez J, Ballivet M, Matter JM. A bHLH transcriptional network regulating the specification of retinal ganglion cells. Development. 2005;132:3907–3921. doi: 10.1242/dev.01960. [DOI] [PubMed] [Google Scholar]
- 20.Thummel R, Kassen SC, Enright JM, Nelson CM, Montgomery JE, Hyde DR. Characterization of Muller glia and neuronal progenitors during adult zebrafish retinal regeneration. Exp Eye Res. 2008;87:433–444. doi: 10.1016/j.exer.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma W, Yan R-T, Mao W, Wang S-Z. Neurogenin3 promotes early retinal neurogenesis. Mol Cell Neurosci. 2009;40:187–198. doi: 10.1016/j.mcn.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomita K, Nakanishi S, Guillemot F, Kageyama R. Mash1 promotes neuronal differentiation in the retina. Genes Cells. 1996;1:765–774. doi: 10.1111/j.1365-2443.1996.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 23.Jasoni CL, Walker MB, Morris MD, Reh TA. A chicken achaete-scute homolog (CASH-1) is expressed in a temporally and spatially discrete manner in the developing nervous system. Development. 1994;120:769–783. doi: 10.1242/dev.120.4.769. [DOI] [PubMed] [Google Scholar]
- 24.Mao W, Yan R-T, Wang S-Z. Proneural gene ash1 promotes amacrine cell production in the chick retina. Dev Neurobiol. 2009;69:88–104. doi: 10.1002/dneu.20693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- 26.Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 2000;19:5460–5472. doi: 10.1093/emboj/19.20.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatakeyama J, Tomita K, Inoue T, Kageyama R. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 2001;128:1313–1322. doi: 10.1242/dev.128.8.1313. [DOI] [PubMed] [Google Scholar]
- 28.Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 29.Yan R-T, Ma W, Wang S-Z. neurogenin2 elicits the genesis of retinal neurons from cultures of non-neural cells. Proc Natl Acad Sci USA. 2001;98:15014–15019. doi: 10.1073/pnas.261455698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma W, Wang S-Z. The final fates of neurogenin2-expressing cells include all major neuron types in the mouse retina. Mol Cell Neurosci. 2006;31:463–469. doi: 10.1016/j.mcn.2005.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akagi T, Inoue T, Miyoshi G, Bessh Y, Takahashi M, Lee JE, Guillemot F, Kageyama R. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem. 2004;279:28492–28498. doi: 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- 32.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 33.Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- 34.Perez SE, Rebelo S, Anderson DJ. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development. 1999;126:1715–1728. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- 35.Hughes SH, Greenhouse JJ, Petropoulos CJ, Sutrave P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan R-T, Wang S-Z. neuroD induces photoreceptor cell overproduction in vivo and de novo generation in vitro. J Neurobiol. 1998;36:485–496. [PMC free article] [PubMed] [Google Scholar]
- 37.Li C-M, Yan R-T, S-Z Misexpression of cNSCL1 disrupts retinal development. Mol Cell Neurosci. 1999;14:17–27. doi: 10.1006/mcne.1999.0765. [DOI] [PubMed] [Google Scholar]
- 38.Ma W, Yan R-T, Xie Wang S-Z. A role of ath5 in inducing neuroD and the photoreceptor pathway. J Neurosci. 2004;24:7150–7158. doi: 10.1523/JNEUROSCI.2266-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belecky-Adams T, Cook B, Adler R. Correlations between terminal mitosis and differentiated fate of retinal precursor cells in vivo and in vitro: analysis with the “window-labeling” technique. Dev Biol. 1996;178:304–315. doi: 10.1006/dbio.1996.0220. [DOI] [PubMed] [Google Scholar]
- 40.Reddy ST, Stoker AW, Bissell MJ. Expression of Rous sarcoma virus-derived retroviral vectors in the avian blastoderm: potential as stable genetic markers. Proc Natl Acad Sci USA. 1991;88:10505–10509. doi: 10.1073/pnas.88.23.10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fekete DM, Cepko CL. Replication-competent retroviral vectors encoding alkaline phosphatase reveal spatial restriction of viral gene expression/transduction in the chick embryo. Mol Cell Biol. 1993;13:2604–2613. doi: 10.1128/mcb.13.4.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan R-T, Wang S-Z. Embryonic abnormalities from misexpression of cNSCL1. Biochem Biophys Res Cummun. 2001;287:949–955. doi: 10.1006/bbrc.2001.5690. [DOI] [PubMed] [Google Scholar]
- 43.Li C-M, Yan R-T, Wang S-Z. A novel homeobox gene and its role in the development of retinal bipolar cells. Mech Dev. 2002;116:85–94. doi: 10.1016/s0925-4773(02)00148-x. [DOI] [PubMed] [Google Scholar]
- 44.Liu W, Mo Z, Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc Natl Acad Sci USA. 2001;98:1649–1654. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C-M, Yan R-T, Wang S-Z. Misexpression of chick NSCL2 causes atrophy of Muller glia and photoreceptor cells. Invest Ophthalmol Vis Sci. 2001;42:3103–3109. [PMC free article] [PubMed] [Google Scholar]
- 46.Xie W, Yan R-T, Ma W, Wang S-Z. Enhanced retinal ganglion cell differentiation by ath5 and NSCL1 coexpression. Invest Ophthalmol Vis Sci. 2004;45:2922–2928. doi: 10.1167/iovs.04-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burmeister M, Novak J, Liang MY, Basu S, Ploder L, Hawes NL, Vidgen D, Hoover F, Goldman D, Kalnins VI, Roderick TH, Taylor BA, Hankin MH, McInnes RR. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 48.Mu X, Klein WH. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Semin Cell Dev Biol. 2004;15:115–1231. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cornell RA, Eisen JS. Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development. 2002 Jun;129(11):2639–48. doi: 10.1242/dev.129.11.2639. Erratum in: Development 2002;129:3279. [DOI] [PubMed] [Google Scholar]
- 52.Cau E, Wilson SW. Ash1a and Neurogenin1 function downstream of Floating head to regulate epiphysial neurogenesis. Development. 2003;130:2455–2466. doi: 10.1242/dev.00452. [DOI] [PubMed] [Google Scholar]
- 54.Ahmad I, Dooley CM, Polk DL. Delta-1 is a regulator of neurogenesis in the vertebrate retina. Dev Biol. 1997;185:92–103. doi: 10.1006/dbio.1997.8546. [DOI] [PubMed] [Google Scholar]
- 53.Austin CP, Feldman DE, Ida JA, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- 55.Dorsky RI, Chang WS, Rapaport DH, Harris WA. Regulation of neuronal diversity in the Xenopus retina by Delta signaling. Nature. 1997;385:67–70. doi: 10.1038/385067a0. [DOI] [PubMed] [Google Scholar]
- 56.Silva AO, Ercole CE, McLoon SC. Regulation of ganglion cell production by Notch signaling during retinal development. J Neurobiol. 2003;54:511–524. doi: 10.1002/neu.10156. [DOI] [PubMed] [Google Scholar]
- 57.Adler R, Hatlee M. Plasticity and differentiation of embryonic retinal cells after terminal mitosis. Science. 1989;243:391–393. doi: 10.1126/science.2911751. [DOI] [PubMed] [Google Scholar]
- 58.Kelley MW, Turner JK, Reh TA. Ligands of steroid/thyroid receptors induce cone photoreceptors in vertebrate retina. Development. 1995;121:3777–3785. doi: 10.1242/dev.121.11.3777. [DOI] [PubMed] [Google Scholar]