Abstract
P68 RNA helicase is a prototypical DEAD box RNA helicase. The protein plays a very important role in early organ development and maturation. In consistence with the function of the protein in transcriptional regulation and pre-mRNA splicing, p68 was found to predominately localize in the cell nucleus. However, recent experiments demonstrate a transient cytoplasmic localization of the protein. We report here that p68 shuttles between the nucleus and the cytoplasm. The nucleocytoplasmic shuttling of p68 is mediated by two nuclear localization signal (NLS) and two nuclear exporting signal (NES) sequence elements. Our experiments reveal that p68 shuttles via a classical RanGTPase dependent pathway.
Keywords: P68 RNA helicase, Nucleocytoplasm Shuttle, NLS, NES, DEAD-box
Introduction
In eukaryotic cells, the nucleus is separated from the cytoplasm. Maintenance of cellular functions requires trafficking of many bio-macromolecules into and out of the nucleus. Proteins that are targeted to the nucleus are marked by one or more sequence elements termed Nuclear Localization Signal (NLS) [1, 2], while the proteins that are transported out of the nucleus carry one or more Nuclear Exporting Signal (NES) sequence tags [3]. Nucleocytoplasmic trafficking occurs via the nuclear pore complex (NPC). Most protein transportation through the NPC is mediated by specific cargo and a nuclear receptor system, importins and exportins [4–6]. Interestingly, many proteins often carry both NLS and NES signals. This characteristic usually leads to shuttling of these proteins between the nucleus and the cytoplasm [7].
The nuclear p68 RNA helicase (hereafter referred to as p68) is a prototypical member of the DEAD box family of RNA helicases [8, 9]. As an early example of a cellular RNA helicase, the ATPase and the RNA unwinding activities of p68 RNA helicase were previously documented [10–12]. Expression of p68 correlates with cell proliferation and early organ maturation [13]. P68 RNA helicase is suggested to function in DNA methylation/demethylation pathways [14], Our laboratory has demonstrated in vitro and in vivo that p68 RNA helicase is an essential splicing factor that plays a role in unwinding the transient U1:5’ splice site duplex [15, 16]. Interestingly, results from several laboratories including our laboratory suggest that p68 RNA helicase may be involved in transcription regulation of a number genes [17–22] [23]. Studies appear to suggest that p68 RNA helicase is involved in transcriptional regulation by different mechanisms of action dependent on each individual regulated gene and biological processes [13, 21, 24–27]. Experiments in our laboratory also demonstrate that p68 is phosphorylated at multiple amino acid residues, including serine/threonine and tyrosine [28, 29]. Tyrosine phosphorylation of p68 correlates with tumor progression [25]. Phosphorylation of p68 at Y593 mediates the effects of growth factors in promoting epithelial-mesenchymal-transition (EMT). The phosphor-p68 promotes EMT by facilitating β-catenin nuclear translocation [30]. In the present study, we demonstrate that p68 shuttles between the nucleus and the cytoplasm. P68 shuttling is mediated by two NLSs and two NESs sequence elements. Our data show that p68 shuttles via the classical RanGTPase dependent pathway.
Results
P68 RNA helicase shuttles between the nucleus and the cytoplasm
We previously reported that Y593 phosphorylated p68 facilitates cytoplasmic β-catenin nuclear translocation by displacing the cytoplasmic β-catenin anchor protein axin [30]. We reasoned that cytoplasmic localization is due to p68 shuttling between the nucleus and the cytoplasm. A number of nuclear localized proteins have been shown to be nucleocytoplasm shuttles [31, 32]. We thus employed a heterokaryon assay [33] using SW620 cells and NIH3T3 cells to test whether p68 shuttles between the nucleus and the cytoplasm. HA-tagged p68s were exogenously expressed in SW620. After fusing the SW620 with NIH3T3 cells, the HA-p68s were detected in the nucleus of NIH3T3 cells (Fig. 1, upper panel). As a negative control, the non-shuttling protein MS2-DEK [34] expressed in SW620 cells could not be detected in the nucleus of NIH3T3 cells (Fig. 1, bottom panel). The experimental results suggest that p68 is a nucleus – cytoplasm shuttling protein with a much longer residence time in the nucleus.
Figure 1. P68 shuttles between the nucleus and the cytoplasm.
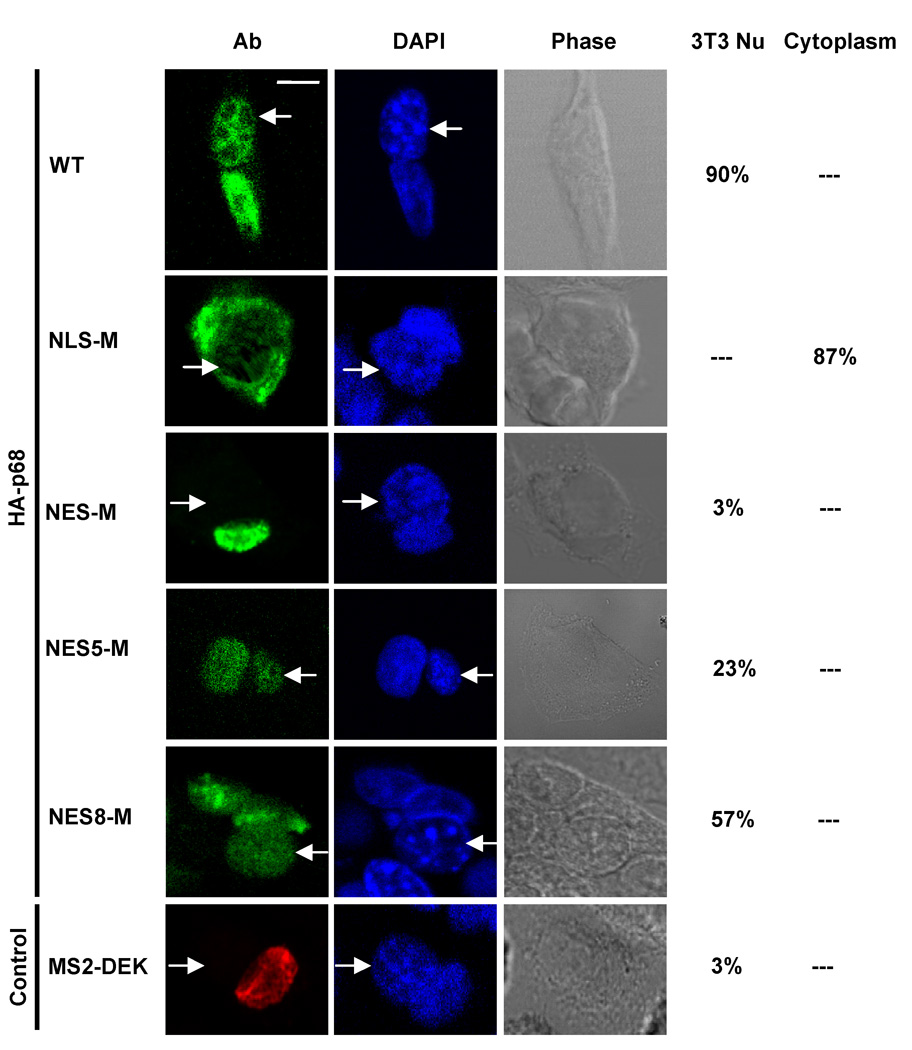
Representative images of SW620 cells expressing HA-p68s (WT, NLS-M, NES-M, NES5-M, and NES8-M). After fusion with NIH3T3 cells, the HA-p68s were immunostained using anti-HA antibody (Ab). The green signal represents staining of HA-p68s. DAPI stains DNA in the cell nucleus of the fused cells (DAPI). The same treated cells were also revealed by phase contrast microscopy (Phase). MS2-DEK (immunostained by antibody against MS2) was a negative control for nucleocytoplasm shuttling assays. Arrows indicate the nucleus of mouse NIH3T3 cells. The numbers on the right side of images are the percentages cells with the HA-p68s detected in NIH3T3 nucleus (HA-3T3/NE) or in the cytoplasm (HA-Cyto) of the fusion cells based on counting a random group of 30 cells.
Identification of NLSs and NESs of p68
Most nucleocytoplasm shuttling proteins carry sequence elements of both NLS and NES. We analyzed the amino acid sequence of p68 and found a number of sequence segments that resemble NLSs and NESs (Fig. 2A and Fig. 3A). The NLS sequences were selected based on similarity to the classical SV40 and bipartite NLS sequences [35, 36], while the NES sequences were selected based on similarity to the consensus hydrophobic residue rich NES sequence, ϕX2–3ϕX2–3ϕXϕ where ϕ is a hydrophobic residue and X is any amino acid residue [37]. To test the functionality of these putative NLSs and NESs in p68, we first fused each individual putative NLS or NES with a fluorescent protein DsRed. The fusion proteins were expressed in SW620 cells. It was clear that only NLS3 and NLS4 led to a substantial nuclear accumulation of the fluorescent protein (Fig. 2B). To verify the functionality of NLS3 and NLS4, we made mutations in NLS3 (R352A, R353A, K360A, and R362A) or NLS4 (R484A, R494A and K501A) in the context of full length p68. The HA-tagged mutants were expressed in SW620 cells. Immunostain of the exogenously expressed HA-p68, wt and the mutants, indicated that nuclear localization of the HA-mutants was dramatically reduced (Fig. 2C). Quantification of fluorescence intensity in the nucleus and the cytoplasm of a random group of cells confirmed the reduction of nuclear HA-p68 (the mean average of Cyto/Nu fluorescence intensity ratio were 0.038 +/− 0.026 for wt, 1.477 +/− 0.029 for NLS3-M, and 1.489 +/− 0.097 for NLS4-M). The results suggested that NLS3 and NLS4 indeed functioned as nuclear localization signals of p68. Fusion of NES2 and NES5 with the fluorescent protein resulted in high levels of cytoplasmic fluorescent protein (Fig. 3B). Interestingly, fusion of NES8 with DsRed led to slightly higher levels of fluorescent protein localization in the cytoplasm (Fig. 3B), indicating that NES8 is a weak nuclear export signal. Treatment of cells with leptomycin B (LMB) abolished the cytoplasmic localization patterns of DsRed fusion proteins (NES2, NES5, and NES8) (Fig. 3C). The results suggested that NES2, NES5 and NES8 potentially function as nuclear export signals of p68. To further test the functions of nuclear localization signals and nuclear export signals of p68, we constructed several p68 deletion mutants (Fig. 4A). These deletion mutants were expressed in SW620 cells either as HA-tag proteins or eGFP-fused proteins (due to small sizes of truncates). Locations of each putative NLSs and NESs of p68 in the p68 truncation proteins are indicated. The localizations of these p68 truncates were analyzed by immunostaining with anti-HA antibody or imaging of eGFP. DII and DIII localized to the nucleus, indicating that NLS4 is a functional NLS, while the NES1 and NES 7–8 are not functional NES. Strong cytoplasmic DIV and DV were observed, suggesting that one or more of NES 2 – 6 are functional NES. The stronger fluorescence in DV than that of DIV and certain levels of cytoplasmic localization of DVI indicate function of NLS3. In summery, these results supported that NLS3 and NLS4 may function as nuclear localization signals of p68 while NES2 and NES5 may function as nuclear exporting signals of p68. Consistent with our experimental results, NLS3 is located in an exposed helical secondary structure while NLS4 is located in an exposed loop that is flanked by two α-helixes in a computer simulated p68 model structure (Fig. 4B). Both NES5 and NES8 are also located on the exposed surface in the model structure, while NES2 is buried under an α-helix in the model structure. This structure model provided additional support for the identification of the NLSs and NESs of p68.
Figure 2. NLSs of p68.
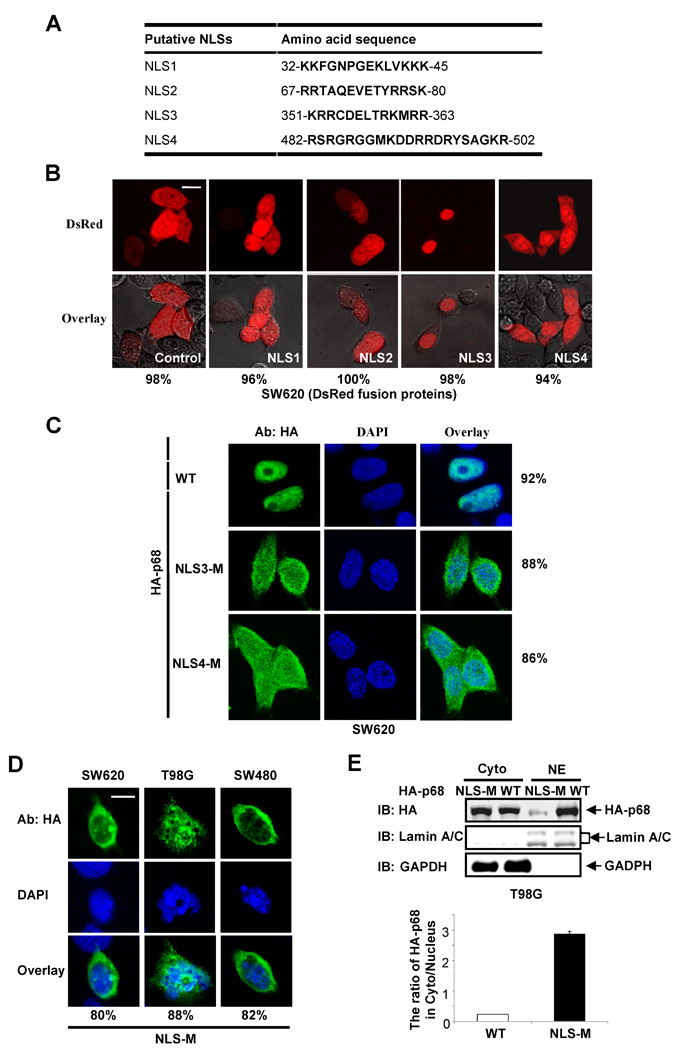
(A) Putative sequence segments of p68 that resemble NLS. (B) Representative of confocal fluorescent microscopy images show the localizations of p68 putative NLSs (N-terminal NLS1, NLS2, NLS3, NLS4) fused with DsRed in SW620 cells. The fused proteins were expressed in the cells. The red signal represents the expressed fusion proteins. The control is the expressed DsRed protein without the NLS fusion. (C) Examples of confocal images show sub-cellular localizations of the exogenously expressed HA-p68s in SW620 cells by immunostaining using anti-HA antibody (HA-p68). The green signal represents staining of HA-tag. The blue signal represents staining of DNA. HA-p68s, wild-type (WT) NLS mutant (NLS3-M, NLS4-M), were expressed in the cells. The right panels are overlay of anti-HA and DAPI stains. (D) HA-p68 NLS mutant (NLS-M) was expressed in different cell lines (indicated). Examples of confocal images of the cells show sub-cellular localizations of the NLS-M by immunostaining using anti-HA antibody. The top panel is the immunostaining using anti-HA antibody. The middle panel is DAPI stain of cell nucleus. The bottom panel is overlay of anti-HA and DAPI stains. In (B), (C), and (D), the numbers are percentage of cells showing similar image pattern (nuclear vs cytoplasm staining) in a random group of 50 cells. (E) The levels of exogenously expressed HA-p68s, wild type (WT) or NLS-Mutant (NLS-M) in the extracts made from the cytoplasm or the nucleus of T98G cells were examined by immunoblot using anti-HA antibody (IB:HA) (upper panel). The IBs with antibodies against lamin A/C and GAPDH are the loading controls. The bottom panel is the quantization of an average four separate experiments of the IB signals and presented as ratio of cytoplasmic over nuclear HA-p68s (WT or NLS-M).
Figure 3. NESs of p68.
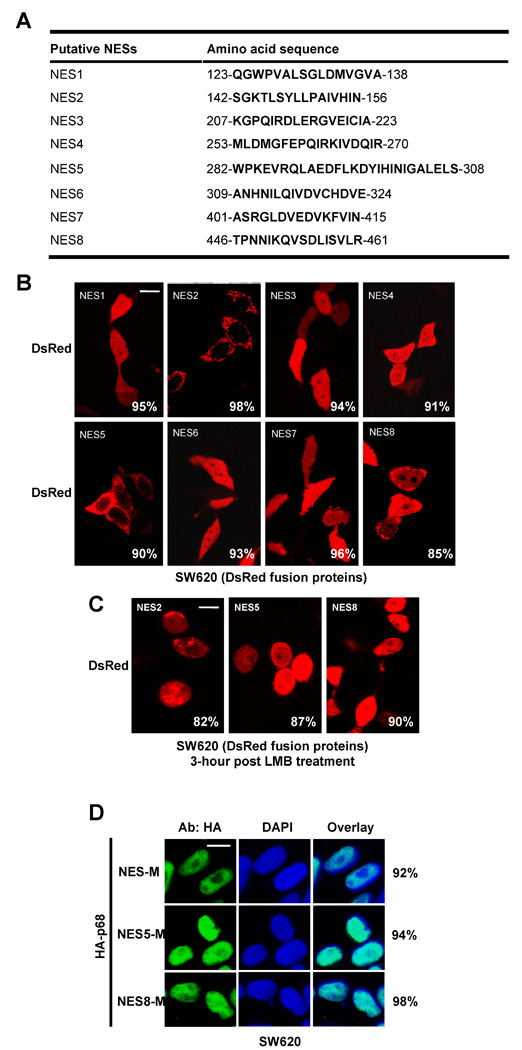
(A) Putative sequence segments of p68 that resemble NES. (B) Examples of confocal fluorescent microscopy images of SW620 cells show localizations of p68 putative NESs (N-terminal fusion) fused DsRed. The fused proteins were expressed in the cells. The red signals are the fluorescence of the expressed fusion proteins. (C) Inhibition of the exporting of selected putative p68 NESs (NES2, NES5, and NES8) fused with DsRed protein by LMB. The fusion proteins were expressed in SW620 cells. The cells were treated with LMB 24 hrs post fusion protein expression. The confocal florescence images were taken 3 hours post drug treatments. (D) Examples of confocal fluorescent microscopy images of SW620 cells show the sub-cellular localizations of the p68 NES mutants (NES-M, NES5-M, and NES8-M) in by immunostaining using anti-HA antibody. The green signal represents staining of HA-tag. The blue signal represents staining of DNA. In (B), (C) and (D), the numbers are percentage of cells showing similar localization pattern in a random group of 50 cells.
Figure 4. NESs and NLSs of p68.
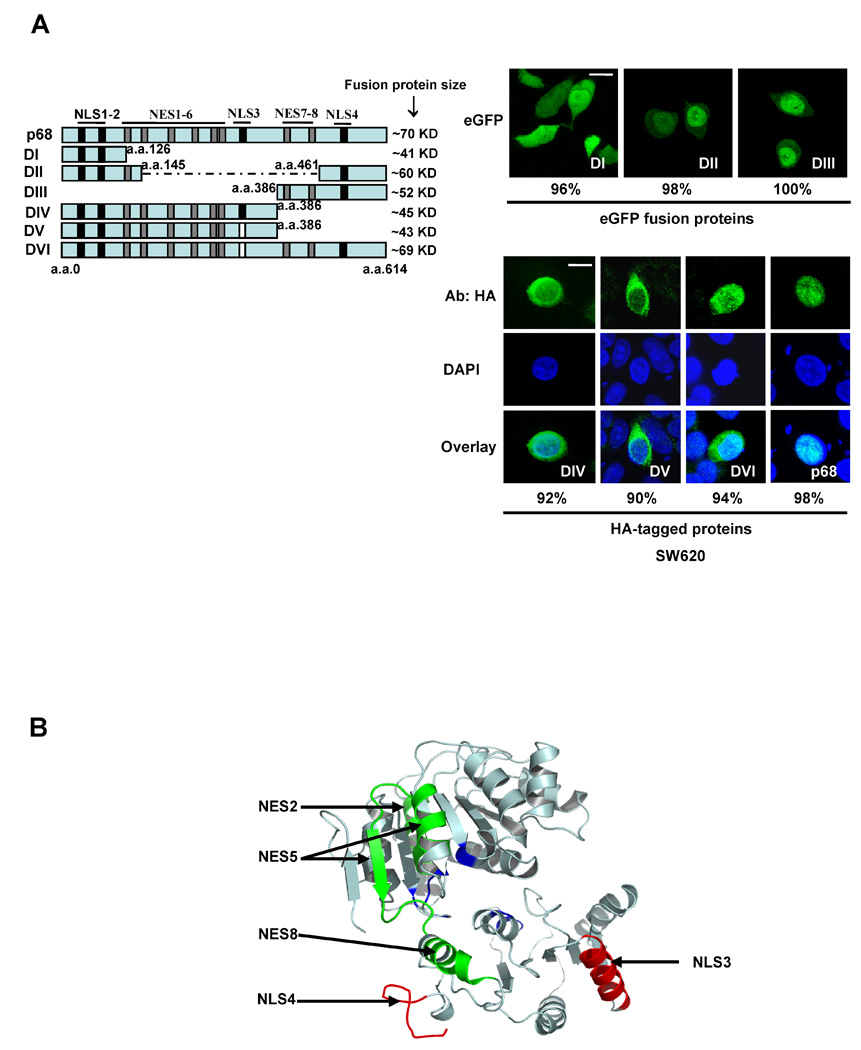
(A) P68 truncates (illustrated in the left panel) containing different putative NLSs and NESs either fusion C-terminal flank eGFP or N-terminal HA-tag were expressed in SW620 cells. The numbers indicate the molecular weight of each fusion protein. The black bars indicate the location of NLSs. The gray bars indicate NESs. The white bars indicate that the corresponding NLSs were deleted. (right panel) Examples of confocal fluorescent microscopy images show the localizations of eGFP fused (up) or HA-tagged p68 truncates (down) in SW620 cells. The HA-tagged p68 truncates in SW620 cells were immunostained with anti-HA antibody (Green). The blue is DAPI stain of the cell nucleus and overlay of anti-HA and DAPI stains. The numbers are percentage of cells showing similar localization pattern in a random group of 50 cells. (B) Model structure of p68 RNA helicase core domain. The model structure was constructed based on the X-ray crystal structure of another DEAD box RNA helicase drosophila vasa. The positions of NLS3 and NLS4, NES2, NES5, and NES8 are indicated by arrows.
We next mutated both NLS3 and NLS4 by the same mutations described above (hereafter referred to as NLS-M). The mutant was expressed in SW620, T98G, and SW480 cells. Immunofluorescence staining demonstrated that no significant levels of p68 mutant localized in the cell nucleus (Fig. 2D). The results were further verified by immunoblot analyses of exogenously expressed HA-p68s (wt and the mutant) in the cytoplasmic and nuclear extracts made from HA-p68s expressing T98G cells (Fig. 2E). It was also clear that the mutant no longer shuttled between the nucleus and the cytoplasm as demonstrated by the heterokaryon assay (Fig. 1, 2nd panel from top). The distribution pattern did not change significantly when the heterokaron cells were treated with LMB (data not shown). In our structure model, NES5 and NES8 are well exposed, while NES2 is buried. Thus, we constructed the p68 NES mutant by mutations at both NES5 and NES8, F293A, L294A, L298A, L305A, L456A, and I457A (hereafter referred to as NES-M). Immuno fluorescence staining of SW620 cells that expressed the mutant showed an exclusive nuclear localization of the mutant (Fig. 3D). Heterokaryon assay demonstrated that the nucleocytoplasmic shuttling of p68 was almost completely abolished by the mutations (Fig. 1, 3rd panel from top). In contrast, mutations separately on either NES5 or NES8 (same mutations) did not completely abolish but certainly reduced the shuttle of HA-p68 to the NIH3T3 cell nucleus (Fig. 1, 4th and 5th panels from top). Our mutational analyses confirmed that the NLS3/NLS4 and NES5/NES8 were functional NLSs and NESs, and these NLSs and NESs were required for p68 nuceocytoplasmic shuttling.
P68 shuttles via a RanGTPase dependent pathway
CRM1 is an export receptor mediating nuclear export of proteins that carry leucine-rich nuclear exporting signals [38, 39]. We demonstrated that p68 RNA helicase carries NLSs and NESs sequences that mediate nucleocytoplasmic shuttling. We reasoned whether overexpression of CRM1 would affect the localization of p68. We exogenously expressed CRM1 in SW620 cells (Fig. 5A). Immunoblot of p68 indicated that there were significantly higher levels of cytoplasmic p68 after CRM1 was exogenously expressed in the cells, and the increases in cytoplasmic p68 were inhibited by LMB treatment (Fig. 5B). Increases in cytoplasmic p68 by expression of CRM1 were not observed with p68 mutant that carries mutations at both NES5 and NES8 (Fig. 5C). To further confirm the effects of CRM1 on export of p68, we probed the interaction between p68 and endogenous CRM1 via co-immunoprecipitation. It was clear that p68 co-immunoprecipitated with endogenous CRM1 (Fig. 5D). The results suggest that p68 RNA helicase was exported from the nucleus mediated by exportin pathway. The effects of exogenous expression of CRM1 on p68 export were further confirmed by immunostaining analyses. It was evident that staining of p68 in the cytoplasm was significantly increased upon the expression of CRM1, and the increases were inhibited by LMB (Fig. 5E). To further verify that the p68 nucleocytoplasmic shuttling is mediated by RanGTPase pathway, we tested whether p68 interacts with importin in vitro. We used a commercially available his-tag importin α2. When his-importin α2 was incubated with recombinant GST-p68, GST-p68 could not be pulled-down by his-importin α2 (Fig. 6A). It is known that importin α2 functions as a hetero-dimer with importin β [40, 41]. Therefore, we added another commercially available recombinant importin β1. It was clear that GST-p68 co-precipitated with his-importin α2 when importin β1 was also present, but the p68 NLS mutant (NLS-M) did not co-precipitate with his-importin α2 under any condition (Fig. 6A). We also carried out co-precipitation experiments with immunopurified HA-p68 from HEK293 cells. The his-importin α2/importin β1 co-immunoprecipitated with the purified HA-p68 using anti-HA antibody. However, the importins did not immunoprecipitate with HA-p68 NLS mutant (Fig. 6B). The interaction of p68 with importins was verified by co-immunoprecipitation experiment with endogenous importin α2/β1 (Fig. 6C). The observations suggested that p68 interacts with the importin α2/β1 dimer, providing additional support for the nucleocytoplasmic shuttling of p68 via the RanGTPase pathway.
Figure 5. P68 nucleocytoplasm shuttle is RanGTPase pathway dependent.
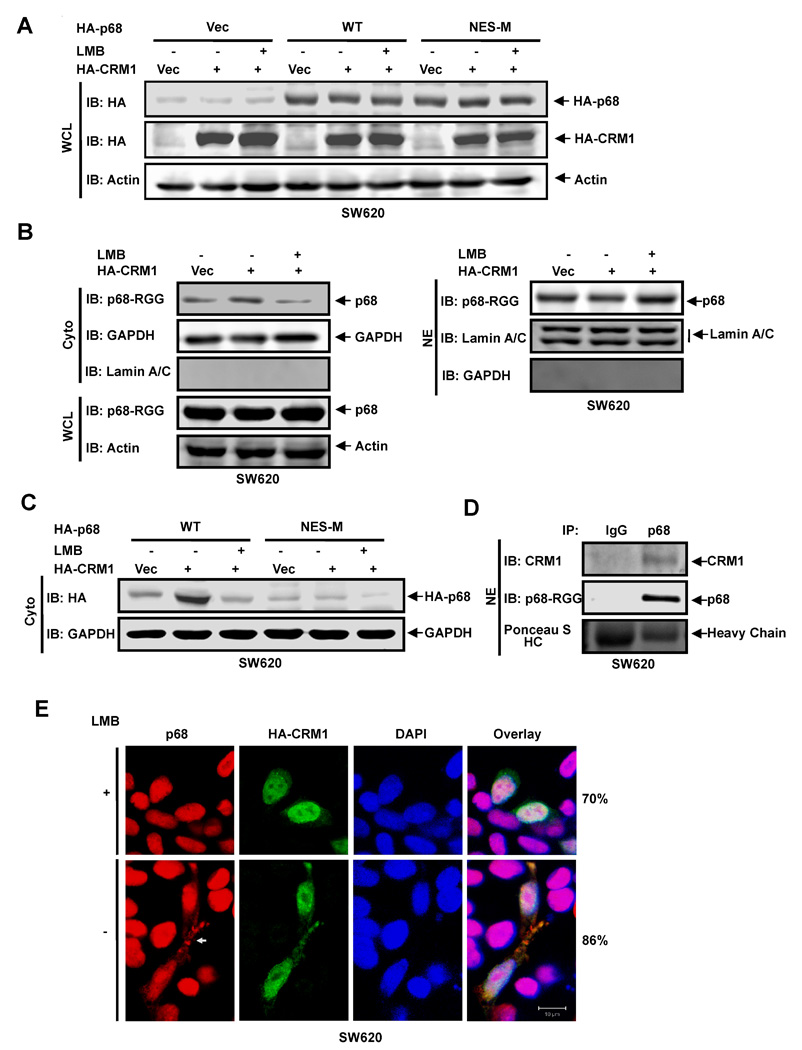
(A) Exogenous expression of HA-p68s, wild-type (WT) and mutant (NES-M), and HA-CRM1 in SW620 cells were analyzed by immunoblot of whole cell lysate (WCL) using anti-HA antibody (IB:HA). (B) (Left panel) Cytoplasmic (Cyto) p68 levels of SW620 cells were analyzed by immunoblot using the antibody p68-rgg (IB:p68-RGG). Immunoblots of p68 in whole cell lysate (IB:p68-RGG) indicate cellular levels of p68. (Right panel) Nuclear (NE) levels of p68 in SW620 cells were analyzed by immunoblot using the antibody p68-rgg (IB:p68-RGG). (C) The cytoplasmic (Cyto) levels of exogenously expressed HA-p68s (WT and NES-M) were analyzed by immunoblot of cytoplasmic extracts using anti-HA antibody (IB:HA). In (A), (B), and (C), IBs of β-actin (IB:actin), GAPDH (IB:GAPDH), Lamin A/C (IB:Lamin A/C) are controls. The SW620 cells were transfected with vector alone (Vec), the vector for CRM1 (HA-CRM1), and the vector for CRM1 plus treatment with LMB (HA-CRM1 + LMB). (D) Co-immunoprecipitation of p68 with endogenous CRM1 in nuclear extracts (NE) of SW620 cells was analyzed by immunoprecipitation using the antibody Pabp68 (IP, p68). The immunoprecipitates were examined by immunoblot using antibody against CRM1 (IB:CRM1) and the antibody p68-RGG (IB:p68-RGG). Ponceau S staining the IgG haeve chain is the loading control (Ponceau S HC). Immunoprecipitation using rabbit IgG (IP, IgG) is negative control for co-immunoprecipitation. (E) Examples of confocal fluorescent microscopy images of SW620 cells show the cellular localizations of endogenous p68 (P68) and exogenously expressed HA-CRM1 (HA-CRM1). The cells were treated (+) or untreated (−) with LMB. Red signals are staining p68. The green signal represents staining of HA-tag. The blue signal represents staining of DNA. Overlay images are overlay of staining of p68, HA-CRM1, and DNA. The numbers are percentage of cells showing similar image pattern in a random group of 50 cells.
Figure 6. P68 interacts with importins in vitro.
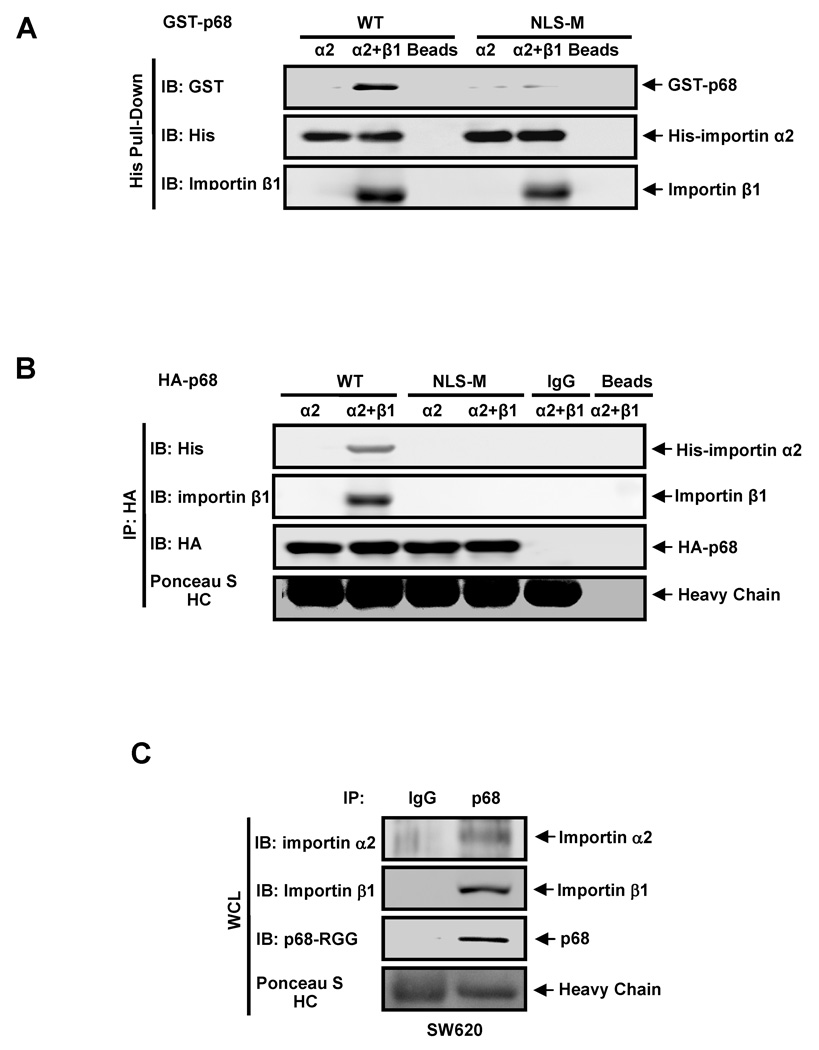
(A) Immunoblot analyses of GST-p68 (IB:GST) and his-importin α2 (IB:His) in His-pull-down proteins (His Pull-down) from a protein mixture of GST-p68 wild-type (WT) or GST-p68 NLS-M with; his-importin α2 (α2), his-importin α2 and importin β1 (α2+β1), and Ni-TED beads alone (Beads). Importin β1 is visualized by immunoblot using antibody against importin β (IB: Importin β) (B) Immunoblot analyses of his-importin α2 (IB:His) and HA-p68 (IB:HA) in co-immunoprecipitates (IP:HA) from a protein mixture of HA-p68 wild-type (WT), HA-p68 mutant NLS-M (NLS-M), rabbit IgG (IgG), or protein G beads (Beads) with; his-importin α2 (α2), his-importin α2 and importin β1 (α2+β1). (C) Co-immunoprecipitation of p68 with endogenous importin α2 and β1 in whole cell lysate (WCL) of SW620 cells was analyzed by immunoprecipitation using the antibody Pabp68 (IP, p68). The immunoprecipitates were examined by immunoblot using antibodies against importin α2 (IB: Importin α2), importin β1 (IB: Importin β1), and the antibody p68-RGG (IB:p68-RGG). Immunoprecipitation using rabbit IgG (IP, IgG) is negative control for co-immunoprecipitation. In (B) and (C), Ponceau S staining IgG heavy chain (Ponceasu S:HC) are the loading controls.
Discussion
P68 RNA helicase was shown to predominately localize in the cell nucleus [42]. However, recent experiments carried out in Janknecht’s and our laboratories showed a transient cytoplasmic localization of the protein [19, Gao and Liu, unpublished observations]. In this report, we presented data demonstrating the nucleocytoplasm shuttling of p68 RNA helicase. Our experiments showed that p68 shuttling is mediated by two NLSs and two NESs sequence elements. The p68 nuclear export and import follow a RanGTPase dependent pathway. Interestingly, a number of DEAD/DExH box RNA helicases have also been shown to shuttle between the nucleus and cytoplasm, including eIF-4AIII, An3, GRTH/Ddx25, and RNA helicase A [34, 43–45]. However, the functional role of the shuttling of these RNA helicase is not well known. P68 RNA helicase was previously shown to interact with mRNA/mRNP [16]. Thus, one possibility is that the p68 export and shuttle is associated with mRNP export. It was shown that shuttling of GRTH/Ddx25 is dependent upon mRNP exporting [43]. However, the interaction of p68 with CRM1 is RNA independent, as p68 still co-immunoprecipitated with CRM1 in the nuclear extracts treated by RNase (data not shown), indicating that p68 nuclear export may not dependent upon mRNA exporting.
The cellular function(s) of the p68 RNA helicase nucleocytoplasmic shuttling is an open question. In general, proteins that shuttle between the nucleus and the cytoplasm usually have distinct cellular functions in these two separated sub-cellular compartments. They often re-localize to the nucleus or the cytoplasm to fulfill specific functional role(s) in response to a particular cellular signal stimuli [46, 47]. Alternatively, nucleocytoplasmic shuttling of a protein can function as a ‘chaperons’ to help nuclear import or export of other proteins or RNAs. [48, 49]. Cellular functions of p68 helicase in the cell nucleus are well documented. The protein is functionally involved in gene transcription [21–23, 50], pre-mRNA [15, 16], pre-rRNA, and pre-miRNA processing [51, 52]. Whether p68 RNA helicase has any potential function(s) in the cytoplasm is currently not very clear. Goh et al. detected the interaction of p68 with HCV-NS5B in the cytoplasm in the viral infected or NS5B expressing 293 cells and the interaction is essential for viral replication [53]. In another independent study, D. Harris and co-workers found that the 3’-nontranslated region of HCV interact with p68 in an RNA affinity capture experiments with the cytoplasmic extracts of 293 cells that express viral replicon [54]. These observations are consistent with our experimental results indicating that p68 localizes both in the nucleus and the cytoplasm. However, despite the demonstration of the function of p68 in HCV replication in the cytoplasm of virus infected cells, the evidence that suggest the involvement of p68 in any cellular process in the cytoplasm is currently lack. We have observed a significant increases in cytoplasmic p68 levels upon treatment of cells with several growth factors and chemokines [30, Wang, unpublished observations], indicating that p68 may functions in the cytoplasm under specific cellular conditions, such as abnormal growth or cell migration in response to growth factor or chemokine stimulations.
Localization of p68 is intriguing. The protein is shuttling between the nucleus and the cytoplasm, while the protein predominantly resides in the cell nucleus, and is nearly undetectable outside of the nucleus by immunostaining or by immunoblot analyses of cytoplasmic extracts. The phenomenon suggests that the function of the NESs of p68 is tightly regulated. It is possible that an interacting partner(s) in the nucleus masks the p68 NESs sequences. Nuclear export of p68 depends on the dissociation of the interaction partner(s). Our data demonstrated that p68 has three sequence elements that potentially function as NESs. On the other hand, structural modeling of p68 helicase core showed that one of the potential NES (NES2) is buried under an α-helix. Therefore, it is possible that a specific post-translational modification drives conformational changes which subsequently expose this buried NES sequences for p68 export [55]. Very similar regulatory mechanisms were observed in nuclear exporting and importing of Dok1 and NF-AT1 [56, 57]. In consistent with this notion, we observed a substantially increased cytoplasmic p68 levels in cells when the cells were treated with several growth factors and chemokines [30, and data not shown], suggesting a possibility that the growth factors and chemokines treatments trigger the exposure of the NES sequences for nuclear export of p68. There are two NLS sequences. Mutation of either NLS reduced nuclear localization (increased cytoplasmic p68, Fig. 2C), indicating that both NLSs contribute partly to nuclear localization of p68.
Materials and Methods
Reagents, antibodies, and cells
Leptomycin B (LMB), PEG3350, cycloheximide, HA peptide, and protease inhibitor cocktail were purchased from Calbiochem, Sigma, and Roche respectively. Antibodies against HA and His tags, GAPDH, Lamin A/C, β-actin, and Histone 2A were purchased from Roche, Cell signaling, Upstate, and Chemicon respectively. The monoclonal antibody P68-RGG and polyclonal antibody Pabp68 against p68 were produced in our laboratory. Cell lines SW480, SW620, HT29, NIH3T3, and T98G were purchased from ATCC and cultured by following the vendor’s instructions.
Plasmids construction
The HA-tagged p68s expression plasmids (wild type, and Y593F, Y595F, LGLD, and NLS-M mutants) were constructed in pHM6 vectors as previously reported [30]. The vectors for expression of eGFP and DsRed fusion proteins were constructed using eGFP-pcDNA-3.1(+) or pDsRed1–N1 vectors. Full length p68 or deletion mutants of p68 variants were sub-cloned into the vector by EcoR321 and NotI restriction sites. The p68 or deletion mutants were fused at the C-terminal of the fluorescent protein tag. Different putative NLSs or NESs tagged DsRed were constructed by cloning into pDsRed-N1 vectors at 5’ (BamHI and EcoRI sites) with the addition of a starting methionine at the N-terminal of each NLS or NES respectively. Site directed mutagenesis was performed using QuikChange® Multi Site-Directed Mutagenesis Kit (Stratagene). All the DNA clones and mutations were verified by auto-DNA sequencing at GSU. The procedures for transfection of expression vectors of HA-p68, fluorescent proteins, and Crm1 and duplex RNAi were similar to our previous reports [30, 50].
Expression and purification of recombinant GST-p68
Recombinant GST-p68 was expressed and purified as described in our previous report [58].
Interactions between p68 and importins
Recombinant GST-p68 (8 µg) was incubated with commercial his-importin α2 (8 µg) and/or importin β1 (Calbiochem) overnight at 4°C in 500 µl in PBS buffer. After incubation, the protein complex was pulled-down by Ni-TED silica beads. The pull-down proteins were separated by 10% SDS-PAGE, and were analyzed by immunoblot using anti-GST or anti-his-tag antibodies. The presence of importin β1 was visualized by ponceau S staining. For co-immunoprecipitation of HA-p68 with importin α2, HA-p68s, wt or mutant, were expressed in HEK cells and were immunoprecipated using 30 µl anti-HA antibody. Protein G agarose (50 µl) was added to the mixture. After extensive washing, HA-p68s were eluted from the beads by competition using HA peptide (100 µg/ml). The eluted HA-p68s were dialyzed against PBS. The recombinant importin α2 and/or β1 was incubated with the purified HA-p68 in PBS for 4 hours. The protein mixtures were immunoprecipated using anti-HA antibody. The precipitated complex was separated in 10% SDS-PAGE followed by immunoblot using anti-his-tag antibody. The presence of HA-p68 was detected by immunoblot using the antibody p68-rgg.
Heterokaryon analyses and immunofluoresence imaging
The experimental procedures for immunofluoresence staining and imaging were similar to our previous reports [30, 59]. For heterokaryon analyses, SW620 cells were first transfected with HA-p68s (wt or mutants) or MS2-DEK. The cells were mixed with an equal number of mouse NIH3T3 cells 24 hour post-transfection and reseeded in 4-well chambers. Subsequently, 50 µg/ml of cycloheximide was added to the culture medium to inhibit protein synthesis. After 3h, the co-cultured cells were fused using 50% PEG3350 for 2 minutes, washed, and incubated with the medium containing 75 µg/ml cycloheximide for 3 hours. The treated cells were then fixed and immunostained as described above. Rabbit polyclonal anti-MS2 antibody was employed to stain MS2-DEK, followed by Alex Fluor 555 goat anti-rabbit IgG antibody. In a random group of 30 cells, the numbers of cells with both NIH3T3 and SW620 nucleus and with HA-p68 in NIH3T3 nucleus or in the cytoplasm of fused cells were counted. The percentage of HA-p68 in the nucleus or cytoplasm of fused cells equals fused cells with HA-p68 in NIH3T3 nucleus or cytoplasm / total fused cells with expression of HA-p68.
Computational homology structure modeling the helicase core of p68
The sequence alignment of p68 RNA helicase core with homologous RNA helicase cores of several DEAD box family proteins (e.g. vasa, eIF4A, Dhhlp, and UAP56) was performed by the program ClustalW. The secondary structure elements were predicted based on the consensus analysis using computational programs JPRED, PHD, and PSIPRED [60, 61]. The homology modeling of the p68 RNA helicase structure was constructed using the homology-modeling server SWISS-MODEL based on X-ray crystal structure of Drosophila vasa [62], which has the highest alignment score with p68 RNA helicase core. The putative NLSs and NESs were indicated in the modeling structure.
Acknowledgments
We thank Drs. Joan A. Steitz, Melisa J. Moore, and Hung-Ying Kao for providing the vectors for expression of MS2-DEK and human CRM1. We are grateful to Professor Peter Stockley for providing antibody against MS2-DEK. We also thank Birgit Neuhaus for assistance in confocal imaging. This manuscript is greatly improved by comments from Christie Carter, Michael Kirberger, and Heena Dey. This work is supported in part by research grants from National Institute of Health (GM063874), (CA118113), and Georgia Cancer Coalition to ZR Liu. X. Gao is supported by a MBD fellowship, GSU.
References
- 1.Goldfarb DS, Gariepy J, et al. Synthetic peptides as nuclear localization signals. Nature. 1986;322(6080):641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- 2.Schneider J, Schindewolf C, et al. A mutant SV40 large T antigen interferes with nuclear localization of a heterologous protein. Cell. 1988;54(1):117–125. doi: 10.1016/0092-8674(88)90185-7. [DOI] [PubMed] [Google Scholar]
- 3.Wen W, Meinkoth JL, et al. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82(3):463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 4.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99(7):677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 5.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 6.Hill CS. Nucleocytoplasmic shuttling of Smad proteins. Cell Res. 2009;19(1):36–46. doi: 10.1038/cr.2008.325. [DOI] [PubMed] [Google Scholar]
- 7.Lusk CP, Blobel G, et al. Highway to the inner nuclear membrane: rules for the road. Nat Rev Mol Cell Biol. 2007;8(5):414–420. doi: 10.1038/nrm2165. [DOI] [PubMed] [Google Scholar]
- 8.Crawford L, Leppard K, et al. Cellular proteins reactive with monoclonal antibodies directed against simian virus 40 T-antigen. J Virol. 1982;42(2):612–620. doi: 10.1128/jvi.42.2.612-620.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lane DP, Hoeffler WK. SV40 large T shares an antigenic determinant with a cellular protein of molecular weight 68,000. Nature. 1980;288(5787):167–170. doi: 10.1038/288167a0. [DOI] [PubMed] [Google Scholar]
- 10.Iggo RD, Lane DP. Nuclear protein p68 is an RNA-dependent ATPase. Embo J. 1989;8(6):1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford MJ, Anton IA, et al. Nuclear protein with sequence homology to translation initiation factor eIF-4A. Nature. 1988;332(6166):736–738. doi: 10.1038/332736a0. [DOI] [PubMed] [Google Scholar]
- 12.Hirling H, Scheffner M, et al. RNA helicase activity associated with the human p68 protein. Nature. 1989;339(6225):562–564. doi: 10.1038/339562a0. [DOI] [PubMed] [Google Scholar]
- 13.Stevenson RJ, Hamilton SJ, et al. Expression of the 'dead box' RNA helicase p68 is developmentally and growth regulated and correlates with organ differentiation/maturation in the fetus. J Pathol. 1998;184(4):351–359. doi: 10.1002/(SICI)1096-9896(199804)184:4<351::AID-PATH1235>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 14.Jost JP, Schwarz S, et al. A chicken embryo protein related to the mammalian DEAD box protein p68 is tightly associated with the highly purified protein-RNA complex of 5- MeC-DNA glycosylase. Nucleic Acids Res. 1999;27(16):3245–3252. doi: 10.1093/nar/27.16.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu ZR. p68 RNA Helicase Is an Essential Human Splicing Factor That Acts at the U1 snRNA-5' Splice Site Duplex. Mol Cell Biol. 2002;22(15):5443–5450. doi: 10.1128/MCB.22.15.5443-5450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C, Yang L, et al. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol Cell Biol. 2005;25(17):7484–7493. doi: 10.1128/MCB.25.17.7484-7493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita T, Kobayashi Y, et al. Full activation of estrogen receptor alpha activation function-1 induces proliferation of breast cancer cells. J Biol Chem. 2003;278(29):26704–26714. doi: 10.1074/jbc.M301031200. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe M, Yanagisawa J, et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF- 1) with an RNA coactivator, SRA. Embo J. 2001;20(6):1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Rossow KL, Janknecht R. Synergism between p68 RNA helicase and the transcriptional coactivators CBP and p300. Oncogene. 2003;22(1):151–156. doi: 10.1038/sj.onc.1206067. [DOI] [PubMed] [Google Scholar]
- 20.Wilson BJ, Bates GJ, et al. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol. 2004;5:11. doi: 10.1186/1471-2199-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endoh H, Maruyama K, et al. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol. 1999;19(8):5363–5372. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Bates GJ, Nicol SM, et al. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. Embo J. 2005;24(3):543–553. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buszczak M, Spradling AC. The Drosophila P68 RNA helicase regulates transcriptional deactivation by promoting RNA release from chromatin. Genes Dev. 2006;20(8):977–989. doi: 10.1101/gad.1396306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahlina K, Goren I, et al. p68 dead box RNA helicase expression in keratinocytes: Regulation, nucleolar localization, and functional connection to proliferation and VEGF gene expression. J Biol Chem. 2004 doi: 10.1074/jbc.M402467200. [DOI] [PubMed] [Google Scholar]
- 25.Yang L, Lin C, et al. Phosphorylations of DEAD box p68 RNA helicase are associated with cancer development and cell proliferation. Mol Cancer Res. 2005;3(6):355–363. doi: 10.1158/1541-7786.MCR-05-0022. [DOI] [PubMed] [Google Scholar]
- 26.Wei Y, Hu MH. [The study of P68 RNA helicase on cell transformation] Yi Chuan Xue Bao. 2001;28(11):991–996. [PubMed] [Google Scholar]
- 27.Warner DR, Bhattacherjee V, et al. Functional interaction between Smad, CREB binding protein, and p68 RNA helicase. Biochem Biophys Res Commun. 2004;324(1):70–76. doi: 10.1016/j.bbrc.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, Liu ZR. Bacterially expressed recombinant p68 RNA helicase is phosphorylated on serine, threonine and tyrosine residues. Protein Expr Purif. 2004;35(2):327–333. doi: 10.1016/j.pep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Yang L, Lin C, et al. Signaling to the DEAD box-Regulation of DEAD-box p68 RNA helicase by protein phosphorylations. Cell Signal. 2005 doi: 10.1016/j.cellsig.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Yang L, Lin C, et al. P68 RNA Helicase Mediates PDGF-Induced Epithelial Mesenchymal Transition by Displacing Axin from beta-Catenin. Cell. 2006;127(1):139–155. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 31.Cartwright P, Helin K. Nucleocytoplasmic shuttling of transcription factors. Cell Mol Life Sci. 2000;57(8–9):1193–1206. doi: 10.1007/PL00000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J, McKeon F. Nucleocytoplasmic shuttling and the control of NF-AT signaling. Cell Mol Life Sci. 2000;57(3):411–420. doi: 10.1007/PL00000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. Embo J. 1998;17(12):3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibuya T, Tange TO, et al. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11(4):346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 35.Kalderon D, Roberts BL, et al. A short amino acid sequence able to specify nuclear location. Cell. 1984;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 36.Robbins J, Dilworth SM, et al. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64(3):615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 37.Neumann G, Hughes MT, et al. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. Embo J. 2000;19(24):6751–6758. doi: 10.1093/emboj/19.24.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fornerod M, Ohno M, et al. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90(6):1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe M, Fukuda M, et al. Involvement of CRM1, a nuclear export receptor, in mRNA export in mammalian cells and fission yeast. Genes Cells. 1999;4(5):291–297. doi: 10.1046/j.1365-2443.1999.00259.x. [DOI] [PubMed] [Google Scholar]
- 40.Goldfarb DS, Corbett AH, et al. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14(9):505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 41.Lindsay ME, Plafker K, et al. Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import. Cell. 2002;110(3):349–360. doi: 10.1016/s0092-8674(02)00836-x. [DOI] [PubMed] [Google Scholar]
- 42.Nicol SM, Causevic M, et al. The Nuclear DEAD Box RNA Helicase p68 Interacts with the Nucleolar Protein Fibrillarin and Colocalizes Specifically in Nascent Nucleoli during Telophase. Exp Cell Res. 2000;257(2):272–280. doi: 10.1006/excr.2000.4886. [DOI] [PubMed] [Google Scholar]
- 43.Sheng Y, Tsai-Morris CH, et al. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is a transport protein involved in gene-specific mRNA export and protein translation during spermatogenesis. J Biol Chem. 2006;281(46):35048–35056. doi: 10.1074/jbc.M605086200. [DOI] [PubMed] [Google Scholar]
- 44.Aratani S, Oishi T, et al. The nuclear import of RNA helicase A is mediated by importin-alpha3. Biochem Biophys Res Commun. 2006;340(1):125–133. doi: 10.1016/j.bbrc.2005.11.161. [DOI] [PubMed] [Google Scholar]
- 45.Askjaer P, Bachi A, et al. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol. 1999;19(9):6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu L, Massague J. Nucleocytoplasmic shuttling of signal transducers. Nat Rev Mol Cell Biol. 2004;5(3):209–219. doi: 10.1038/nrm1331. [DOI] [PubMed] [Google Scholar]
- 47.Kau TR, Way JC, et al. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4(2):106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 48.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677(1–3):3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 49.Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60(8):1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L, Lin C, et al. Phosphorylation of p68 RNA helicase plays a role in platelet-derived growth factor-induced cell proliferation by up-regulating cyclin D1 and c-Myc expression. J Biol Chem. 2007;282(23):16811–16819. doi: 10.1074/jbc.M610488200. [DOI] [PubMed] [Google Scholar]
- 51.Davis BN, Hilyard AC, et al. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukuda T, Yamagata K, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9(5):604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 53.Goh PY, Tan YJ, et al. Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J Virol. 2004;78(10):5288–5298. doi: 10.1128/JVI.78.10.5288-5298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harris D, Zhang Z, et al. Identification of cellular factors associated with the 3'-nontranslated region of the hepatitis C virus genome. Mol Cell Proteomics. 2006;5(6):1006–1018. doi: 10.1074/mcp.M500429-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Wrighton KH, Lin X, et al. Phospho-control of TGF-beta superfamily signaling. Cell Res. 2009;19(1):8–20. doi: 10.1038/cr.2008.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niu Y, Roy F, et al. A nuclear export signal and phosphorylation regulate Dok1 subcellular localization and functions. Mol Cell Biol. 2006;26(11):4288–4301. doi: 10.1128/MCB.01817-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okamura H, Aramburu J, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6(3):539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 58.Huang Y, Liu ZR. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J Biol Chem. 2002;277(15):12810–12815. doi: 10.1074/jbc.M200182200. [DOI] [PubMed] [Google Scholar]
- 59.Wang H, Liu Y, et al. The recombinant beta subunit of C-phycocyanin inhibits cell proliferation and induces apoptosis. Cancer Lett. 2007;247(1):150–158. doi: 10.1016/j.canlet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 60.Cuff JA, Clamp ME, et al. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14(10):892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 61.McGuffin LJ, Bryson K, et al. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 62.Sengoku T, Nureki O, et al. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125(2):287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]


