Abstract
Aggregation of proteins containing polyglutamine (polyQ) expansions characterizes many neurodegenerative disorders, including Huntington’s disease. Molecular chaperones modulate Huntingtin (Htt) aggregation and toxicity by an ill-defined mechanism. Here we determine how the chaperonin TRiC suppresses Htt aggregation. Surprisingly, TRiC does not physically block the polyQ tract itself, but rather sequesters a short Htt sequence element N-terminal to the polyQ tract, that promotes the amyloidogenic conformation. The residues of this amyloid-promoting element essential for rapid Htt aggregation are directly bound by TRiC. Our findings illustrate how molecular chaperones, which recognize hydrophobic determinants, can prevent aggregation of polar polyQ tracts associated with neurodegenerative diseases. The observation that the switch of polyQ tracts to an amyloidogenic conformation is accelerated by short endogenous sequence elements provides a novel target for therapeutic strategies to inhibit aggregation.
Expansion of polyglutamine (polyQ) repeats beyond a critical threshold has been linked to protein toxicity in a number of neurodegenerative disorders, including Huntington’s Disease1,2. These expanded polyQ repeat proteins aggregate through both intra- and inter-molecular interactions between polyQ stretches, ultimately leading to the formation of fibrillar beta-sheet rich amyloid inclusions associated with neuronal dysfunction and death3–7. While there is a clear correlation between polyQ length and disease onset and severity, a number of observations suggest that aggregation is profoundly influenced by factors outside the polyQ tract. Thus, the clinical course of disease can vary widely in patients with identical polyQ repeat lengths8. Furthermore, many cellular proteins contain long polyQ tracts, but few trigger neurological disorders. For instance, ataxin-3 remains soluble and non-pathogenic at polyQ lengths well above 50 glutamines1, while Huntingtin is pathogenic above a Q35 threshold. These observations, together with findings that molecular chaperones can suppress aggregation and toxicity raise the question of how context influences behavior of the expanded polyQ tract 9,10.
The intrinsic and extrinsic factors that modulate polyQ aggregation in the cell, other than polyQ-tract length, are not well understood. Studies using Huntingtin-exon1 (Htt-exon1), a naturally-occurring proteolytic fragment of Htt that is responsible for Huntington’s disease (HD) pathogenesis, indicate that the proline-rich domain (PRD) C-terminal to the polyQ tract attenuates its aggregation and subsequent toxicity9,11–13.
Aggregation is also influenced by the cellular environment14–16. Molecular chaperones are important modulators of aggregation and toxicity of polyQ-expanded proteins in the cell. In particular, the hetero-oligomeric chaperonin TRiC, also called CCT, was recently identified as a potent suppressor of Htt aggregation and toxicity17–19. An intriguing question raised by these studies is how chaperones, which generally recognize exposed hydrophobic regions in non-native proteins, can prevent aggregation of the highly polar polyQ stretch.
TRiC is a ring-shaped, hetero-oligomeric chaperonin that uses cycles of ATP-binding and hydrolysis to bind unfolded polypeptides and facilitate their folding20,21. Recent work indicates that individual subunits differ in their recognition specificity22. Indeed, TRiC suppression of Htt-exon1 aggregation is mediated by interactions between specific TRiC subunits, primarily subunit 1 (also called CCT1 or TCP1) and to a lesser extent subunit 4, and early amyloid aggregate precursors18,19. However, the mechanism by which TRiC and other molecular chaperones suppress polyQ aggregation remains to be defined. In the simplest model, TRiC and other chaperones would block aggregation by shielding the polyQ tract, similar to their role preventing aggregation of other unfolded polypeptides10,21,23. However, this model presents several problems. Firstly, TRiC, like other chaperones, appears to preferentially recognize hydrophobic sequences22 while the polyQ tract is primarily polar24. Additionally, since ATPase cycling leads to substrate release, and the polyQ tract is inherently aggregation prone 3,25, the simple shielding model also raises the question of why aggregation does not occur upon ATP-induced Htt release during chaperonin cycling, since in these conditions the natively unfolded polyQ tract is released again to the cellular millieu.
Here we interrogate the molecular basis of the Htt-TRiC interaction and elucidate how it can prevent aggregation in human cells. We demonstrate that TRiC suppresses pathogenic-length Htt-exon1 aggregation by sequestering a cis-acting intrinsic amphipathic-activator of polyQ aggregation contained within the first 17 amino acids of Htt (N17Htt). Our findings describe an unexpected mechanism for how molecular chaperones alleviate Huntingtin aggregation. Instead of simply binding the core structural element of the aggregates, TRiC inhibits a specific element that initiates aggregation. A deeper understanding of this mechanism may provide the foundation for the design of a new class of neurodegenerative disease therapies.
RESULTS
The N-terminal domain is the primary TRiC recognition site within Htt
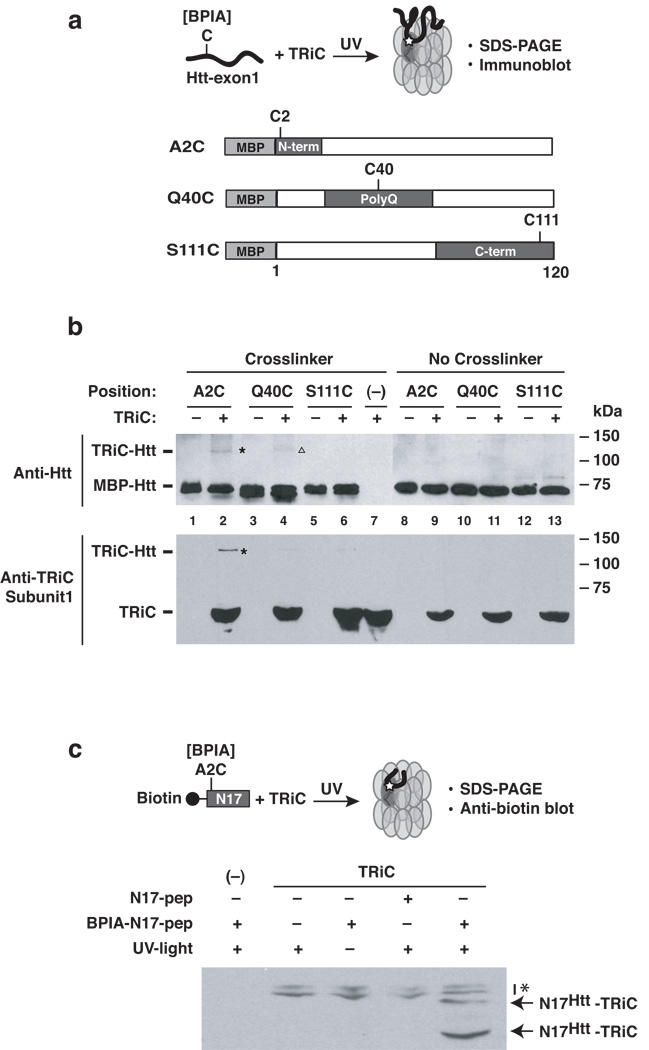
Reconstitution experiments show that TRiC can directly bind and suppress the aggregation of a pathogenic form of Huntingtin-exon1 (Htt-exon1), the naturally-occurring proteolytic fragment found in Huntington’s disease (HD) aggregates26,27. To gain insight into how TRiC modulates Htt aggregation, we used photo-crosslinking to map the contacts between the chaperonin and different regions in Htt (Fig. 1 and Supplementary Fig. 1). Incorporation of photoactivatable probes into a protein or peptide is a well-established and powerful approach to identify chaperonin-substrate binding sites22,28–30. Illumination with UV-light (photolysis) will activate the probe for a very short interval (ns), allowing formation of a covalent bond with proteins located in very close proximity to the probe at the time of excitation. We used the photoactivatable bifunctional cross-linker BPIA (Fig. 1, Supplementary Fig. 1c), since the highly reactive benzophenone moiety of BPIA can form covalent crosslinks to proteins largely independent of the chemical nature of the binding environment. In addition, the short length of BPIA [10 Å]31, equivalent to an extended polypeptide chain of 3 amino acids, increases the likelihood that the crosslinks only occur at sites that directly contact the chaperonin. We introduced unique cysteines into Htt-exon1 either at the N-terminus, the polyQ tract or the C-terminus of Htt-exon1 and coupled them to BPIA (Fig. 1a). Photoactivation generated specific TRiC-Htt photoadducts that were analyzed by immunoblotting with huntingtin-specific antibodies (Fig. 1b, top panel; Supplementary Fig. 1a, b). Surprisingly, the major TRiC-dependent Htt photoadduct was obtained when the crosslinker was placed within the N17 domain (Fig. 1a; Htt-A2C; Fig. 1b, lane 2, see lane 9 for control). A weaker photoadduct was observed with the polyQ tract (Fig. 1a, Htt-Q40C; Fig. 1b, lane 4; we note that in some experiments only the N-terminus was crosslinked to TRiC, e. g. Supplementary Fig. 1b). No TRiC dependent photoadducts were formed when the probe was placed at the C-terminus (Fig. 1a, Htt-S111C; Fig. 1b, lane 6). The molecular mass of the crosslinks was consistent with monomeric Htt adducts with a single subunit, although higher molecular mass species could also be observed (shown in Supplementary Fig. 1a). These results suggest that the N17 domain is the major TRiC binding determinant within Htt.
Figure 1. Mapping the contact sites between Htt-exon1 and the chaperonin TRiC.
(a) TRiC - Htt-Exon1 contacts identified by UV-induced crosslinking. The photoactivatable crosslinker BPIA (benzophenone-4-iodacetamine) was placed at one of three uniquely designed cysteines (A2C, Q40C or S111C) within MBP (maltose binding protein)-Htt-Exon1. (b) Photo-crosslinked adducts were detected by anti-Htt (upper panel) and anti-TRiC subunit 1 immunoblot analyses (lower panel). A major crosslink product is observed when BPIA is positioned at the N-terminus (A2C; asterisk). The minor crosslink with the polyglutamine region (Q40C) is indicated by an open triangle. (c) Direct interaction of TRiC with the N-terminal domain of Htt. Crosslinking to TRiC was tested for an isolated peptide comprising the N-terminal 17 amino acids of Htt (N17). Photo-adducts were detected probing for the biotin moiety of the peptide. Non-specific background signals are labeled with an asterisk. (−): no TRiC control. Results representative of at least three independent experiments are shown.
Individual TRiC subunits differ in their substrate recognition specificity19,22. Analysis using TRiC subunit-specific antibodies indicated that the Htt N-terminal domain crosslinks to subunit 1, i. e. TCP1/CCT1 (Fig. 1b, bottom panel). No crosslinks were detected between CCT1 and either the Q-tract or the C-terminal domain. Of note, the interaction of TCP1/CCT1 with the N-terminus of Htt may be physiologically relevant given that previous findings showed that TRiC subunit 1 suppresses by itself aggregation and toxicity of pathogenic-length Htt-exon1 in vivo and in vitro 19.
Our analysis suggests that a major point of contact for TRiC within Htt resides within the N-terminal domain. To confirm this finding, as well as to assess whether the TRiC-N17 interaction is independent of the context of the remainder of Htt-exon1, we next examined whether purified TRiC can interact directly with the isolated N-terminal 17 amino acids of Huntingtin (herein N17). To this end, TRiC was incubated with a synthetic N17 peptide containing BPIA and an N-terminal biotin tag to facilitate detection and analysis of the photo-adducts. Following incubation of the N17 peptide with TRiC and photolysis, efficient TRiC-N17 crosslinks were detected in a crosslinker and UV-dependent manner (Fig. 1c), confirming that indeed, TRiC interacts directly with the N-terminal domain of Htt.
The N-terminus of huntingtin promotes rapid polyQ aggregation
The unexpected observation that TRiC primarily contacts the N-terminal sequence element flanking the polyQ tract of Htt led us to examine whether this region plays a role in mediating Htt aggregation.
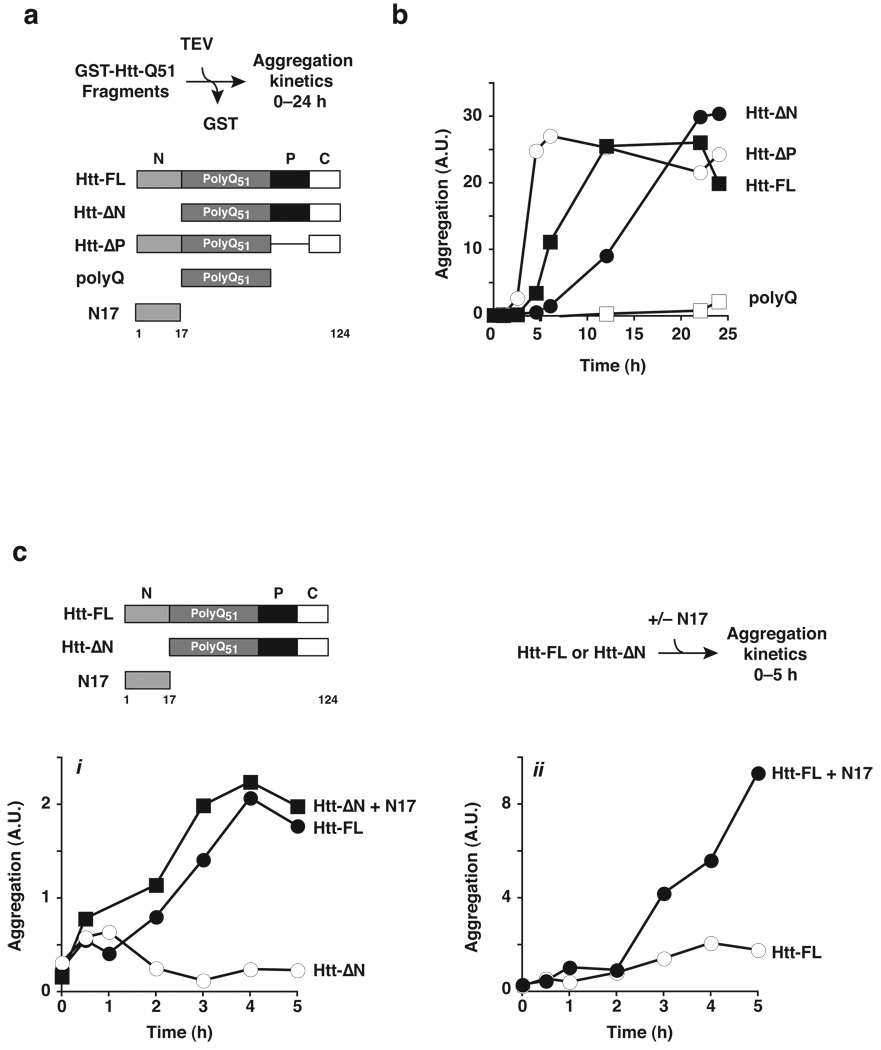
While the relationship between polyQ tract length and aggregation propensity has been extensively documented3,25,32, the role of the flanking sequences in the Htt aggregation process is less well understood. To directly examine whether Htt determinants outside the polyQ tract intrinsically modulate Htt aggregation, we compared the effect of various Htt domains on the in vitro aggregation rates and yields of purified Htt-exon1 bearing a pathogenic length Q51 tract16,19,33,34 (Fig. 2 and Supplementary Fig. 2–5). Amyloid fibril formation was assessed using a previously described filter-trap assay, modified to enable quantification over four orders of magnitude by infrared imaging (Fig. 2a; Supplementary Figs. 2 and 3). The contribution of the N-terminal flanking region was assessed comparing aggregation of full length Htt-exon1 (herein Htt-FL, Fig. 2a) with that of an Htt-exon1 variant lacking the N17 domain (Htt-ΔN; Fig. 2a; Supplementary Figs. 3 and 4). Since previous studies indicated that a proline-rich domain (PRD) C-terminal to the polyQ tract attenuates Htt aggregation and toxicity in vivo9,11,12, we also assessed whether deleting the proline-rich flanking region C-terminal to the polyQ tract had a direct effect on Htt aggregation in vitro (Htt-ΔP, Fig. 2a). To correlate our findings on Htt with previous biophysical studies focused on the aggregation propensities of polyQ-only tracts, we also compared the aggregation rates of the various Htt variants lacking any Htt flanking sequences and only bearing a polyQ tract of identical length (polyQ, Fig. 2a and Supplementary Fig. 4). Maximal aggregation of full-length Htt-exon1 (Htt-FL, Fig. 2b) was observed after 12 hours. Deleting the proline-rich domain (Htt-ΔP) greatly enhanced the Htt aggregation kinetics (Fig. 2b). Notably, previous in vivo analyses showed that the Pro-rich region suppresses Htt aggregation and cytotoxicity in cells and suggested suppression may involve recruitment of cellular factors, such as SH3 domain proteins or prolyl isomerases, to this region9,11,12,35,36. However, our biochemical analysis indicates that the suppression observed in vivo can be explained by a direct effect of the Pro-rich region on the conformational equilibrium of Htt itself. Strikingly, deletion of the N-terminal domain of Htt strongly delayed the kinetics of aggregation (Htt-ΔN; Fig. 2b). This result suggests that the N17 domain, which contains the primary TRiC recognition site in Htt, is an endogenous intrinsic enhancer of Htt-exon1 aggregation. Of note, the N-terminal region of Htt accelerated aggregation but did not affect the final yield, since Htt-ΔN reached Htt-FL aggregation levels after 24 hrs of incubation (Fig. 2b).
Figure 2. The N-terminus of huntingtin promotes rapid polyQ aggregation.
(a) Htt-Exon1 fragments used to analyze the contribution of individual domains towards aggregation. Formation of SDS-insoluble, heat-stable aggregates was determined by filter-trapping and quantified by infrared Li-Cor imaging as described in Online Methods19,24 (Supplementary Figs. 2 and 3). (b) Aggregation kinetics of Htt-exon1 fragments (Supplementary Fig. 4). Aggregation was initiated by TEV protease addition (time = 0 h) to GST- Htt-exon1 forms. All Htt variants were efficiently cleaved by TEV protease with similar kinetics (Supplementary Fig. 5). (c) Trans addition of N17 peptide enhances Htt-exon1 aggregation kinetics. A synthetic peptide containing 17 amino acids of the N-terminal region (N17Htt) was added to Htt-ΔN (i) or to full-length Htt-exon1 (ii) and assayed as in (b). Similar results were obtained when N17 was generated by TEV cleavage from a GST fusion (data not shown). Of note, CD and NMR indicate that N17 by itself was fully soluble at the concentrations used here (data not shown).
We next compared the aggregation of pathogenic Q51 Htt-exon1 with that of the Q51 tract without any flanking sequences (polyQ; Fig. 2a). Strikingly, the aggregation rate of the Q51 tract was dramatically slower than those of Htt-exon1 with the same polyQ tract; indeed marked Q51 aggregation was only observed after several days of incubation (Fig. 2b, Fig. 3c; and data not shown). Importantly, the aggregation rates observed for Q51 with this assay were similar to those previously determined for synthetic polyQ peptides13. It thus appears that isolated, pathogenic length polyQ tracts aggregate with exceedingly slow kinetics indicating that the natural flanking sequences within the Htt protein, and perhaps other polyQ-expanded proteins, play a critical role modulating the aggregation rate.
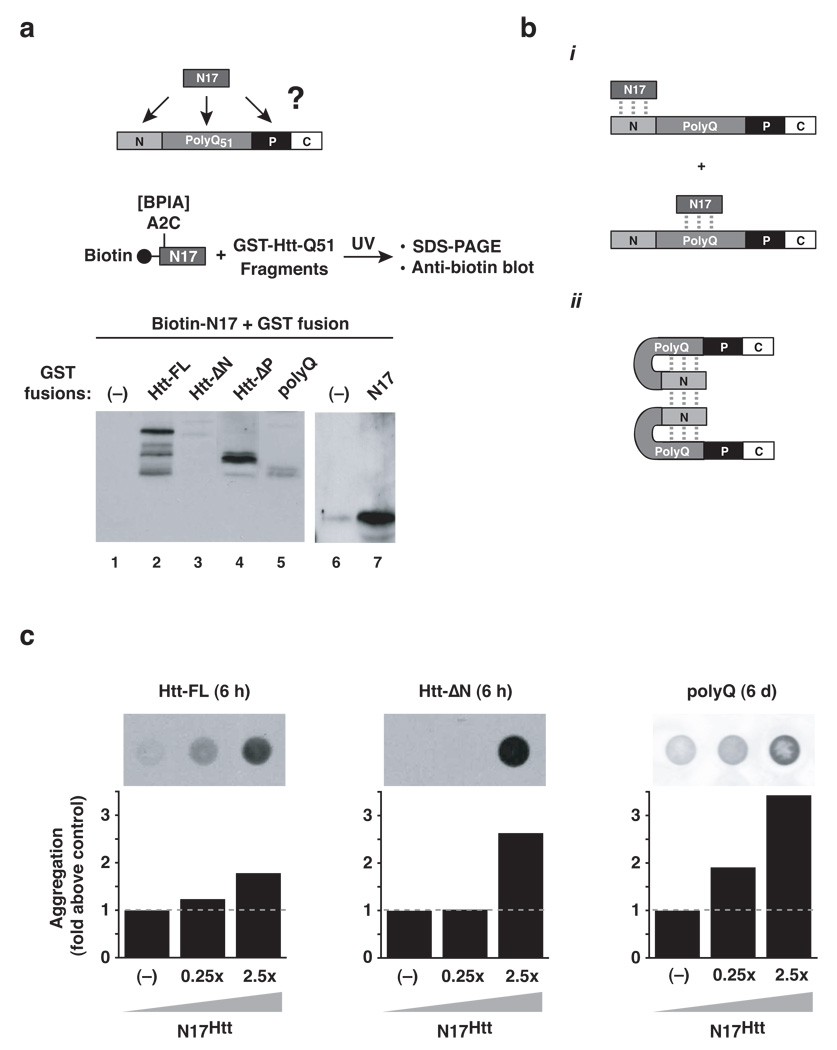
Figure 3. The N-terminus of huntingtin interacts with N17 and the polyQ region in Htt-exon1.
(a) N17 peptide interactions with Htt-Exon1 were assayed by crosslinking. The indicated purified GST-Htt-Exon1 domain deletion fragments (5 µM) or GST alone were incubated with N17 peptide (12.5 µM) carrying the crosslinker BPIA and crosslinked by UV irradiation. Adducts were resolved by SDS-PAGE and probed for the biotin moiety of the peptide. (−): GST only control. (b) (i) Summary of N17 interaction sites within Htt-Exon1; (ii) Predicted model for how N17 inter- and intramolecular interactions within Htt-Exon1 promote aggregation. (c) Dose-dependent effect of N17 addition on polyQ (−): buffer only control. Results representative of at least three independent experiments are shown.
To gain insight into the mode of action of N17 on Htt we examined its ability to promote aggregation in trans. Strikingly, an isolated N17 peptide promoted aggregation of Htt-ΔN in trans (Fig. 2c and Supplementary Fig. 4a). This effect was not observed upon addition of peptides bearing the scrambled N17 sequence nor with another CCT1 binding peptide derived from another TRiC substrate (Supplementary Fig. 4b). Stoichiometric addition of N17 to Htt-ΔN yielded aggregation kinetics comparable to Htt-FL (Fig. 2c, i; Supplmentary Fig. 4a). Even N17 addition to Htt-FL itself further enhanced aggregation in a concentration dependent manner (Fig. 2c, ii; Supplementary Fig. 4a and Fig. 3c). These results indicate that the N-terminal domain is indeed a key intrinsic modulator of Htt aggregation that appears to act on a rate-limiting step leading to polyQ aggregation.
N17 interacts with two distinct Htt domains
Our finding that the isolated polyQ tract aggregates very slowly suggests that the conformational change leading to amyloid-like fibrillization is a rare event, which is promoted by the N-terminal N17 element of Htt. We hypothesized that N17 may stabilize the amyloidogenic conformer by direct interaction with specific regions within Htt (Fig. 3a). To test this hypothesis, the interaction sites of N17 within Htt were mapped using an N17 peptide bearing the photoactivatable crosslinker BPIA and a set of GST-fused Htt variants lacking various domains. Interactions were stabilized by photolysis and analyzed by SDS-PAGE and immunoblot analysis against N17 (Fig. 3a). N17 did not crosslink to GST alone, which served as a control (Fig. 3a, lanes 1 and 6). The distinct pattern of interactions observed for the various Htt fragments indicated that N17 has two distinct interaction sites within Htt-exon1. First, prominent photoadducts were obtained for all Htt fragments containing the N17 domain (Fig. 3a, lanes 2 and 4) suggesting N17 engages in homotypic interactions with itself. Confirming this idea, N17 crosslinked directly to GST fused to the N17 domain (Fig. 3a, lane 7), but not to GST alone (Fig. 3a, lane 6). A second site of N17 interaction appears to be in the polyQ tract. Indeed, we found marked N17 photoadducts with the polyQ tract alone as well as minor crosslink products with the Htt fragment lacking the N17 domain (polyQ and ΔN, Fig. 3a, lane 5 and 3; respectively).
The observation that the N-terminal element contacts two regions in Htt, itself and the polyQ tract (Fig. 3bi), suggest a possible mechanism by which it could stabilize an amyloidogenic conformation (Fig. 3bii). In this model, the interaction of N17 with the polyQ tract could serve to promote an aggregation prone conformation in the polyQ polymer, whereas the homotypic N17 interactions may serve to promote self-association of Htt molecules into oligomers or fibers, facilitating elongation. The idea that N17 directly promotes an amyloidogenic conformation in the polyQ tract itself predicts that N17 addition to an isolated Q51 tract should stimulate aggregation. Indeed, as observed for FL-Htt, trans addition of N17 peptide stimulated aggregation of the isolated polyQ tract as well as all constructs containing the polyQ region (Fig. 3c, and data not shown).
An amphipathic region within N17 is essential for rapid Htt aggregation
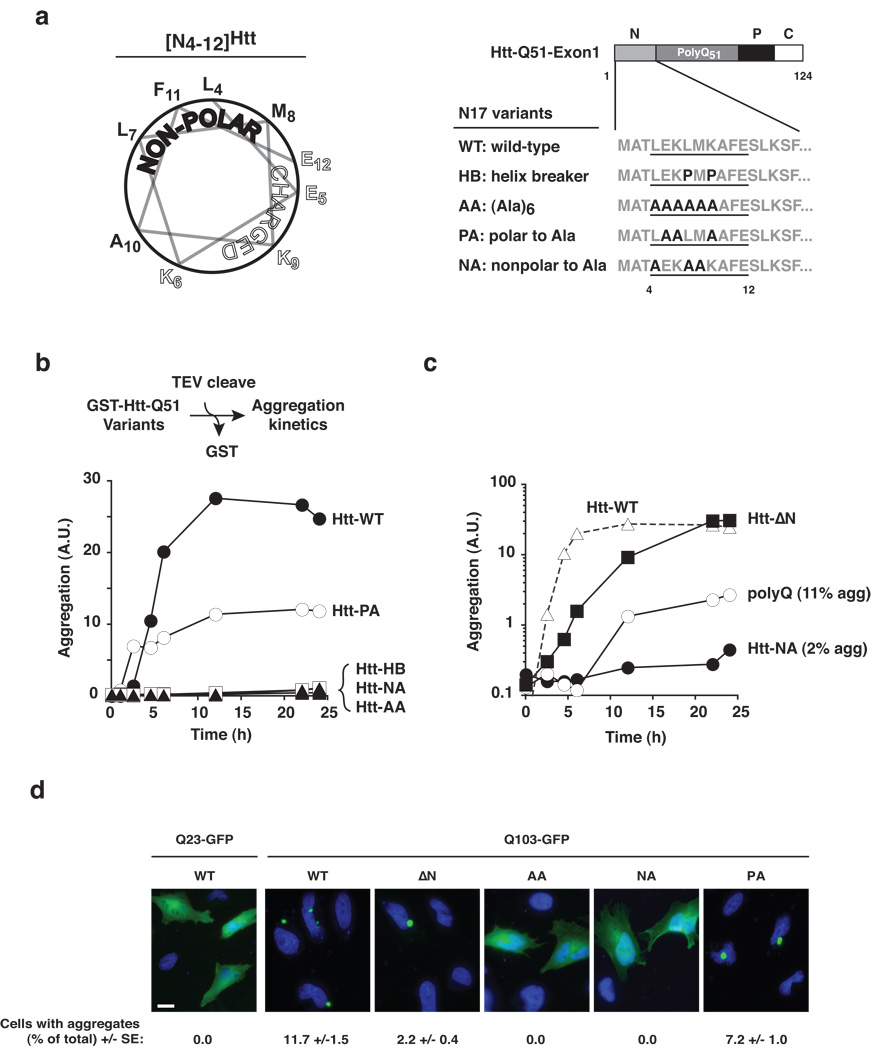
Next, we examined in more detail the structural properties of the N17 region. The AGADIR algorithm37 indicated that amino acids 4–12 within N17 exhibit alpha-helix propensity, consistent with previous circular dichroism measurements38. Projection of this sequence onto a helical wheel revealed the amphipathic nature of the helix (Fig. 4a, left panel). We found that conservation of the helical character in this N-terminal element is important for promoting polyQ aggregation, as introducing two helix-breaking prolines at positions 7 and 9 within the intact Htt-exon1 (HB: helix-breaker; Fig. 4a, right panel) dramatically blocked aggregation even after prolonged incubation (Fig. 4b). Furthermore, replacing the natural Htt sequence by a poly-alanine helix (AA: all alanine) also blocked aggregation (Fig. 4b), indicating that specific determinants within the N17 helix stabilize the amyloidogenic Htt conformation.
Figure 4. The amphipathic N-terminal helix of Htt is necessary for rapid aggregation.
(a) Secondary structure prediction (see Online Methods) identifies amino acids 4 to 12 of Htt-Exon1 [N4–12Htt] as alpha-helical, which are projected as a helical wheel to illustrate its amphipathic nature (left panel). Schematic representation of helical variants of Htt-exon1 analyzed for effects on aggregation (right panel). (b) Aggregation kinetics of Htt-exon1 bearing the indicated N-terminal helix variants assessed as in Fig. 2b. The mutants did not affect the efficiency of TEV protease mediated cleavage (Supplementary Fig. 5). (c) Modification of the hydrophobic face of the N-terminal Htt helix reduces aggregation below the levels observed for isolated polyQ only. Results representative of at least three independent experiments are shown. (d) Fluorescence microscopy of helical variants of Qn-GFP (n = 25, 103) expressing HeLa cells. Cells were scored for foci by visual inspection of GFP aggregates. Statistical analysis was performed using the one-sided, paired Student’s t-test: Means + SE of three independent experiments counting at least 200 cells each are shown. Scale bar represents 20 µm.
We next examined the contribution of the amphipathic nature of N17 to aggregation (Fig. 4b). Alanine replacement of the polar residues (Htt-PA: polar to alanine) reduced the maximal Htt aggregation yield to ~50% of Htt-FL. Importantly, replacing the hydrophobic face with alanines (Htt-NA: non-polar to alanine) completely abrogated Htt aggregation (Fig. 4b). A remarkable outcome from this analysis is that mutations blocking the N17 hydrophobic surface (i. e. HB, AA, and NA; Fig. 4b) dramatically impaired aggregation whereas deletion of the entire N17 only slowed this process (Fig. 2b). This is underscored by directly comparing the aggregation kinetics of Htt bearing the N17 helix mutants with those of Htt-ΔN or a polyQ tract of identical length (Fig. 4c). Indeed, blocking the hydrophobic surface of N17 suppressed aggregation to levels even below those of isolated polyQ (Fig. 4c, note logarithmic scale). Furthermore, disruption of the hydrophobic face of N17 rendered Htt refractory to the aggregation enhancing activity of added N17 peptide (Supplementary Fig. 6). It thus appears that a cis acting block of the hydrophobic surface of N17 is dominant over the effect of trans activators of aggregation.
The dramatic effect of blocking the hydrophobic surface of N17 on Htt aggregation was also observed in vivo upon expression of GFP-tagged Htt-exon1 Q103 variants in HeLa cells (Fig. 4d). As expected, Htt-Q103 aggregated, whereas the non-pathogenic Q23 variants remained soluble and diffuse. Deletion of the N17 region resulted in a reproducible reduction in the number of cells bearing aggregates, as expected from its lower aggregation kinetics in vitro (Fig. 4d). Strikingly, disruption of the N17 hydrophobic determinant by the NA or AA mutation completely abrogated Htt aggregation, despite expression of a Htt form containing a 103-long Q-tract. We conclude that rapid Htt aggregation is critically dependent on the hydrophobic surface of the amphipathic N17 helix.
In sum, blocking the hydrophobic face of the N17 helix has a dominant negative effect on aggregation both in vitro and in vivo. We propose that the mutated NA-N17 helix sterically hinders the oligomerization events leading to aggregation whereas the absence of the N-terminus allows the formation of a new polyQ-polyQ interface between beta-sheet monomers, albeit at a slower rate.
TRiC sequesters the hydrophobic surface of Htt-N17
Our experiments reveal that the N17 element, which contains the major Htt binding site for TRiC, plays a key role directly promoting efficient Htt aggregation. Furthermore, our N17 analysis showed that blocking the hydrophobic face of this amphipathic helix has a dominant negative effect of Htt aggregation. Given the preference of chaperones for hydrophobic recognition determinants39, we considered whether TRiC inhibits Htt aggregation through interaction with the hydrophobic face of the N17 element. We next tested directly the effect of disrupting the hydrophobic surface of N17 on its interaction with TRiC. Whereas an N17 peptide bearing the wild type sequence efficiently crosslinked to TRiC (Fig. 1c and Fig. 5a), introducing the alanine substitutions in the hydrophobic face of N17 (N17-NA) dramatically reduced interaction with TRiC, indicating that the hydrophobic residues are indeed required for TRiC binding (Fig. 5a; Supplementary Fig. 7). This conclusion was supported by whole cell extract TRiC association experiments using either N17-WT or N17-NA Htt-exon1 (Supplementary Fig. 8). While WT-Htt exon1 associates with TRiC upon incubation with yeast or mammalian cell extracts, as previously described19, this interaction was abrogated by the N17-NA mutation in Htt-exon1 (Supplementary Fig. 8 for yeast extracts; not shown for mammalian lysates).
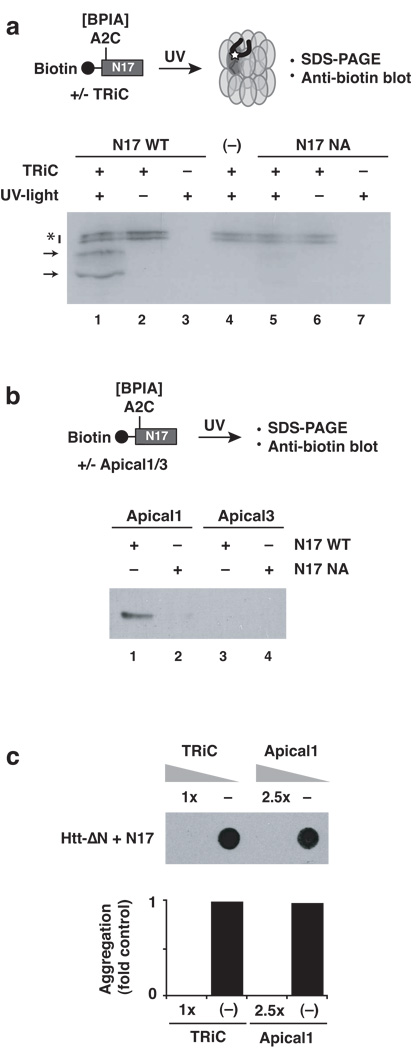
Figure 5. The hydrophobic surface of the N17 helix is the major Htt binding site for the chaperonin TRiC.
(a) Direct interaction of TRiC with N17 requires the hydrophobic face of the helix. Crosslinking to TRiC was tested for the wild type N17 peptide (N17-WT) or with a peptide carrying alanine substitutions in the hydrophobic side of the N17-helix (N17-NA). TRiC- and UV-dependent adducts were detected probing for the biotin moiety of the peptide. Non-specific background signals are labeled with an asterisk. (−): no peptide control. Results representative of at least three independent experiments are shown. (b) The substrate binding apical domain of TRiC subunit 1 mediates binding to the hydrophobic surface of N17. In vitro crosslinking using N17-WT indicate an interaction with the isolated apical domain of TRiC subunit 1. The interaction is based on similar contacts with intact TRiC, as crosslinking was perturbed by the NA mutation within N17. No interaction is observed to the apical domain of TRiC subunit 3. As with other TRiC substrates, specific subsets of TRiC subunits interact with different substrates. (c) Purified TRiC and Apical 1 domain neutralize the aggregation promoting effect of trans- addition of N17 to Htt-ΔN. (−): Ovalbumin non-specific negative control. The lower panel shows the quantification of signal observed on the membrane (upper panel).
Previous in vivo experiments indicated that overexpression of TRiC subunit 1, also called TCP1 or CCT1, can suppress aggregation and toxicity of Htt-exon119. Furthermore, the purified substrate binding domain of subunit 1 (herein Apical 1) alone can suppress Htt-exon1 aggregation in vitro19. In contrast, other TRiC subunits, such as subunit 3, also called CCT3, did not affect Htt aggregation either in vivo or in vitro19, indicating that TRiC subunit 1 contains a major chaperonin binding site for Htt. Intriguingly, our analysis of Htt crosslinks with intact TRiC (Fig. 1) also implicated subunit 1 as a major contact site within TRiC for the Htt N-terminus (Fig. 1b, bottom panel). We next tested whether TRiC suppresses Htt aggregation through the interaction of subunit 1 with the hydrophobic surface of helix N17. Indeed, isolated Apical 1 domain efficiently crosslinked to the isolated N17 domain with the wild type sequence (Fig. 5b, N17-WT). In contrast Apical 1 did not interact with an N17 domain containing a disrupted hydrophobic face (N17-NA, Fig. 5b, compare lanes 1 and 2). Importantly, interaction with N17 was TRiC-subunit specific, since the apical domain of subunit 3, herein Apical 3, showed not detectable crosslinks with either N17 peptide (Fig. 5b, lanes 3 and 4). Thus, the substrate-binding domain of TRiC subunit 1 interacts directly with the hydrophobic face of the N17 helix. We next examined whether both TRiC or Apical 1 can neutralize the aggregation promoting action of N17. To this end, either chaperonin was added to the trans-activation assay whereby N17 stimulates Htt-ΔN aggregation (Fig. 5c). Indeed, both TRiC and Apical 1 dramatically suppressed the aggregation promoting effect of N17 addition (Fig. 5c).
DISCUSSION
Here we define the mechanism by which the chaperonin TRiC suppresses Htt aggregation. Our results do not support the simple model whereby the chaperonin directly sequesters the aggregation prone polyQ tract, but instead indicate that TRiC acts primarily by blocking a short amphipathic Htt sequence element that promotes aggregation. Our analysis resolves the conundrum of how chaperonins, which recognize hydrophobic determinants, can suppress aggregation of the polar polyQ tract. Furthermore, our finding that aggregation of isolated polyQ tracts occurs very slowly, and is strongly accelerated by an endogenous flanking region in Htt-exon1 has important implications for understanding and treatment of polyQ diseases.
Htt conformational equilibrium is modulated by the N17 domain
The correlation between polyQ tract length and disease severity has lead to the concept that positive regulation of polyQ aggregation kinetics is solely a function of polyQ repeat length. Our understanding of Huntington’s Disease must include the critical contribution of the N17 amphipathic helix, which we unequivocally demonstrate promotes rapid Htt aggregation by direct modulation of Htt conformation (Fig. 6a). Notably, the N17 domain is highly conserved across species. The N-terminus of Htt was previously implicated in Htt trafficking in vivo40 and promoting membrane association of native Htt41, suggesting that the cellular function of Htt maintains this aggregation promoting element despite its potential to disrupt protein homeostasis.
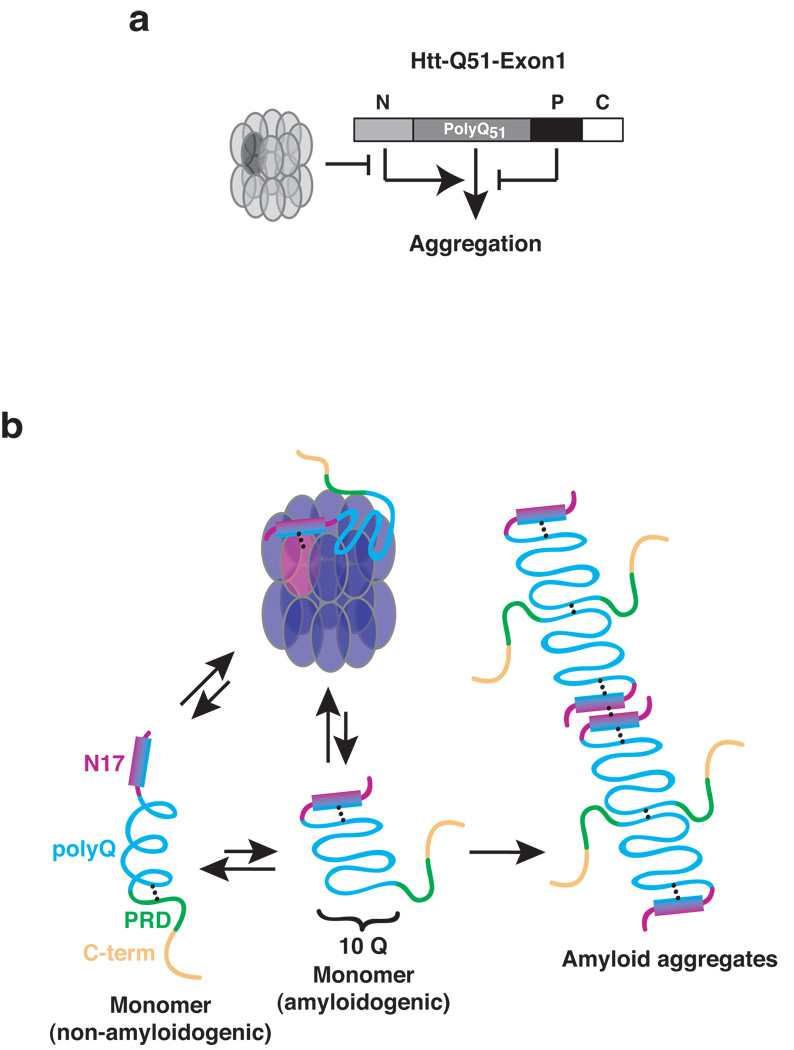
Figure 6. Huntingtin polyQ aggregation is controlled by the interplay of positive and negative regulatory sequence elements with the chaperone machinery.
(a) Control of polyQ aggregation in huntingtin. The proline-rich domain (P) attenuates polyQ-aggregation propensity, while the N-terminal region (N) promotes the amyloid conformation. In turn, the chaperonin TRiC counteracts the positive effect of the N region, thereby suppressing aggregate formation. (b) Proposed model for the molecular events regulating Htt conformation. The polyQ domain in the Htt monomer populates predominantly a helical, non-amyloidogenic conformation likely stabilized by the proline-rich region42. The amphipathic N-terminal helix of Htt-Exon1 interacts with both the polyQ region and itself. N17-PolyQ interactions likely stabilize the amyloidogenic beta-sheet conformation. Homotypic N-terminal interactions may facilitate intermolecular contacts linking amyloidogenic species to higher ordered structures. TRiC sequesters N17 by binding the hydrophobic side of the N17-helix thus blocking amyloid formation and growth.
PolyQ aggregation and cytotoxicity is thought to arise from a conformational transition whereby a polyQ protein monomer switches from an non-toxic, extended alpha-helical conformation to a more compact toxic beta-sheet conformation that assembles into amyloid-like fibrils (Fig. 6b)26,34,42. Our finding that a pathogenic length polyQ tract aggregates exceedingly slowly (Fig. 2b and 3c) indicates that the polyQ tract only rarely switches to the amyloidogenic conformation. For huntingtin, this conformational equilibrium is directly regulated by sequence elements outside the polyQ tract (Fig. 6a). The proline-rich region, which slows aggregation, likely stabilizes the non-amyloidogenic conformation (Fig. 6b)13. Our data comparing the aggregation kinetics of various Htt variants also suggest that the C-terminus of Htt exon1 also plays a yet to be defined role in the aggregation process (Fig. 2 and data not shown). The N17 aggregation-promoting element likely accelerates the rate-limiting conformational switch to the amyloidogenic conformation (Fig. 6b).
How does N17 promote the amyloidogenic conformation? We find that N17 interacts with itself and the polyQ tract (Fig. 3). We propose these interactions serve to trigger the initial conformational switch to the beta-sheet conformation, as well as facilitate self-assembly and incorporation into fibrils (Fig. 6b). Our model is consistent with the proposed molecular architecture of amyloid fibrils, whereby amyloidogenic monomers are thought to polymerize in a head-to-head and tail-to-tail fashion6,43. Our model is also supported by the observed in vivo effects of N17 on Htt aggregation (Fig. 4d)11,40. Furthermore, our results provide a mechanistic explanation for the previously observed suppression of Htt aggregation using N17 region-specific antibodies44. Recently, Thakur et al utilized “Htt-exon1”-like peptides to demonstrate that expanded polyQ tract induce N17 unfolding and subsequent homo-oligomerization with neighboring Htt-exon1 N17 regions 45. Although carried out with synthetic peptides rather than the entire Htt-exon1 sequence, these findings are consistent with our model, whereby homotypic N17-N17 and polyQ-polyQ interactions contribute to rapid polyQ aggregation through stabilization of an amyloidogenic conformation. A scenario whereby the N-terminal helix mediates head-to-head interactions could also explain why blocking the hydrophobic face of the amphipathic N17 helix, e. g. by the N17-NA-Htt mutation, inhibits aggregation in a dominant negative manner, beyond what is observed for Htt-ΔN or even for the polyQ tract itself; thus our model would predict the blocked N17 helix inhibits new polyQ-polyQ interactions. Our results explain how TRiC binding to this region could effectively block aggregation of even very long polyQ tracts.
TRiC suppresses Htt aggregation by blocking an amyloid promoting switch
Our identification of the hydrophobic face of N17 as both critical for Htt aggregation as well as the major TRiC-binding determinant within Htt suggests an unexpected mechanism for how the chaperonin can prevent Htt aggregation. Because blocking the hydrophobic face of N17 strongly inhibits Htt aggregation to levels below those observed for polyQ only, it is likely that TRiC binding to this element acts similar to the N17-NA mutations. This model can explain previous observations that TRiC potently inhibits Htt aggregation at an early step and at substoichiometric ratios17–19. TRiC binding to N17 would both stabilize the monomeric non-toxic conformation and impose a large steric “cap” on the polyQ oligomers that prevent fibril formation (Fig. 6b). Since TRiC also crosslinks weakly to the polyQ tract, additional TRiC-polyQ contacts could further modulate aggregation, perhaps through interaction with TRiC subunits other than CCT1. Of note, the photocrosslinking assay detects soluble and largely non-amyloidogenic Htt molecules, and thus failure to detect strong TRiC-polyQ interactions might be due to the possibility that TRiC binds preferentially to the amyloidogenic conformation of polyQ. Nonetheless, we find that TRiC prevents full length Htt-exon1 aggregation much more effectively that aggregation of polyQ-only (Supplementary Fig. 9), supporting the notion that the presence of N17 is key for efficient TRiC mediated suppression of aggregation. Taken together, our study indicates that molecular chaperones can modulate formation of toxic aggregates by interacting with short sequence elements that regulate the amyloidogenic conformational transition rather than just by sequestration of the polyQ tract alone.
Implications for other polyQ-linked diseases
Given that polyQ itself aggregates much more slowly than Htt with an equivalent polyQ length (Fig. 2b and Fig. 3c), it is tempting to speculate that N17-like aggregation-promoting elements also contribute to aggregation of other disease-causing pathogenic polyQ proteins. Importantly, N17 can promote Htt aggregation even when added in trans, suggesting that aggregation-promoting elements need not be physically linked to the N-terminus of the polyQ tract. This finding raises the possibility that elements acting similarly to N17 also exist for other polyQ proteins, but might be located in different regions of the polyQ protein or even in an interaction partner. Of note, such aggregation-promoting elements for different pathogenic polyQ proteins could be regulated by different sets of chaperones. As a result, the balance between individual aggregation-promoting elements and specific chaperone interactions would vary for different polyQ containing proteins, perhaps accounting in part for the age-of-onset, tissue specificity and clinical course variations observed for polyQ diseases16. Our findings illustrate the emerging concept that cellular protein homeostasis is controlled by the interplay of cellular chaperones and defined elements within proteins46,47. The idea that such interactions play an important regulatory role beyond polypeptide folding has important implications for a large number of human pathologies, including aging and neurodegeneration.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Frydman lab and Brigit Riley for advice and stimulating discussions, and Dr. Raul Andino, Jackie Benjamin, Sheila Jaswal and Erik Miller for useful discussions and comments on the manuscript. This work was supported by NIH grant GM74074 and the NIH Nanomedicine Roadmap.
Footnotes
AUTHOR CONTRIBUTIONS
S. T., C. S., and J. F. designed the research. S. T. performed the aggregation and cell culture experiments. C. S. performed the crosslinking experiments. W. A. cloned mutational Htt constructs, and purified Htt variants. M. A. P. provided the original Htt-exon1 cysteine mutant construct. B. C. and L. J. helped with various experimental aspects. S. T. , C. S. , and J. F. wrote the paper.
References
- 1.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci. 2003;4:49–60. doi: 10.1038/nrn1007. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Berthelier V, Yang W, Wetzel R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J Mol Biol. 2001;311:173–182. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- 4.Yang W, Dunlap JR, Andrews RB, Wetzel R. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum Mol Genet. 2002;11:2905–2917. doi: 10.1093/hmg/11.23.2905. [DOI] [PubMed] [Google Scholar]
- 5.Dobson CM. Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Nelson R. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glabe CG, Kayed R. Common structure and toxic function of amyloid oligomers implies a common mechanism of pathogenesis. Neurology. 2006;66:S74–S78. doi: 10.1212/01.wnl.0000192103.24796.42. [DOI] [PubMed] [Google Scholar]
- 8.Brinkman RR, Mezei MM, Theilmann J, Almqvist E, Hayden MR. The likelihood of being affected with Huntington disease by a particular age, for a specific CAG size. Am J Hum Genet. 1997;60:1202–1210. [PMC free article] [PubMed] [Google Scholar]
- 9.Duennwald ML, Jagadish S, Giorgini F, Muchowski PJ, Lindquist S. A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci U S A. 2006;103:11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 11.Dehay B, Bertolotti A. Critical role of the proline-rich region in Huntingtin for aggregation and cytotoxicity in yeast. J Biol Chem. 2006;281:35608–35615. doi: 10.1074/jbc.M605558200. [DOI] [PubMed] [Google Scholar]
- 12.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci U S A. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharyya A, et al. Oligoproline effects on polyglutamine conformation and aggregation. J Mol Biol. 2006;355:524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- 14.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellis RJ, Hartl FU. Principles of protein folding in the cellular environment. Curr Opin Struct Biol. 1999;9:102–110. doi: 10.1016/s0959-440x(99)80013-x. [DOI] [PubMed] [Google Scholar]
- 16.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 17.Behrends C, et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Kitamura A, et al. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 19.Tam S, Geller R, Spiess C, Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leroux MR, Hartl FU. Protein folding: versatility of the cytosolic chaperonin TRiC/CCT. Curr Biol. 2000;10:R260–R264. doi: 10.1016/s0960-9822(00)00432-2. [DOI] [PubMed] [Google Scholar]
- 21.Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spiess C, Miller EJ, McClellan AJ, Frydman J. Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol Cell. 2006;24:25–37. doi: 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartl FUaMH-H. Molecular Chaperones in the Cytosol: from Nascent Chain to Folded Protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 24.Muchowski PJ, et al. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci U S A. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Berthelier V, Hamilton JB, O'Nuallain B, Wetzel R. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry. 2002;41:7391–7399. doi: 10.1021/bi011772q. [DOI] [PubMed] [Google Scholar]
- 26.Poirier MA, Jiang H, Ross CA. A structure-based analysis of huntingtin mutant polyglutamine aggregation and toxicity: evidence for a compact beta-sheet structure. Hum Mol Genet. 2005;14:765–774. doi: 10.1093/hmg/ddi071. [DOI] [PubMed] [Google Scholar]
- 27.Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 28.Etchells SA, et al. The cotranslational contacts between ribosome-bound nascent polypeptides and the subunits of the hetero-oligomeric chaperonin TRiC probed by photocross-linking. J Biol Chem. 2005;280:28118–28126. doi: 10.1074/jbc.M504110200. [DOI] [PubMed] [Google Scholar]
- 29.Kramer G, et al. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- 30.Mayhew M, et al. Protein folding in the central cavity of the GroEL-GroES chaperonin complex. Nature. 1996;379:420–426. doi: 10.1038/379420a0. [DOI] [PubMed] [Google Scholar]
- 31.Buskiewicz I, et al. Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc Natl Acad Sci U S A. 2004;101:7902–7906. doi: 10.1073/pnas.0402231101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S, Wetzel R. Solubilization and disaggregation of polyglutamine peptides. Protein Sci. 2001;10:887–891. doi: 10.1110/ps.42301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherzinger E, et al. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology. Proc Natl Acad Sci U S A. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaffar G, et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 35.Darnell G, Orgel JP, Pahl R, Meredith SC. Flanking Polyproline Sequences Inhibit beta-Sheet Structure in Polyglutamine Segments by Inducing PPII-like Helix Structure. J Mol Biol. 2007;374:688–704. doi: 10.1016/j.jmb.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 36.Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- 37.Lacroix E, Viguera AR, Serrano L. Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J Mol Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 38.Atwal RS, et al. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16 doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 39.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 40.Rockabrand E, et al. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 41.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 42.Nagai Y, et al. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 43.Liebman SW. Structural clues to prion mysteries. Nat Struct Mol Biol. 2005;12:567–568. doi: 10.1038/nsmb0705-567. [DOI] [PubMed] [Google Scholar]
- 44.Colby DW, et al. Potent inhibition of huntingtin aggregation and cytotoxicity by a disulfide bond-free single-domain intracellular antibody. Proc Natl Acad Sci U S A. 2004;101:17616–17621. doi: 10.1073/pnas.0408134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thakur AK, et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monsellier ECF. Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 2007;8:737–742. doi: 10.1038/sj.embor.7401034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 48.Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17:351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 49.Poirier MA, et al. Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization. J Biol Chem. 2002;277:41032–41037. doi: 10.1074/jbc.M205809200. [DOI] [PubMed] [Google Scholar]
- 50.Ferreyra RG, Frydman J. Purification of the cytosolic chaperonin TRiC from bovine testis. Methods Mol Biol. 2000;140:153–160. doi: 10.1385/1-59259-061-6:153. [DOI] [PubMed] [Google Scholar]
- 51.Frydman J, et al. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. Embo J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.