Summary
Histone variant H2A.Z has a conserved role in genome stability, although it remains unclear how this is mediated. Here we demonstrate in fission yeast that the Swr1 ATPase inserts H2A.Z (Pht1) into chromatin and Kat5 acetyltransferase (Mst1) acetylates it. Deletion or unacetylatable mutation of Pht1 leads to genome instability, primarily caused by chromosome entanglement/breakage at anaphase. This leads to the loss of telomere-proximal markers, though telomere protection and repeat length are unaffected by the absence of Pht1. Strikingly the chromosome entanglement in pht1Δ anaphase cells can be rescued by forcing chromosome condensation prior to anaphase onset. We show that the condensin complex, required for the maintenance of anaphase chromosome condensation, prematurely dissociates from chromatin in the absence of Pht1. This and other findings suggest an important role for H2A.Z in the architecture of anaphase chromosomes.
Keywords: Chromosome architecture, condensin, H2A.Z, KAT5, RCA, S. pombe
Introduction
The basic repeating unit of chromatin is the nucleosome core particle: 146 bp of DNA wrapped around a core histone octamer composed of a (H3-H4)2 tetramer and two H2A-H2B dimers. One means of nucleosome-specialization is the replacement of a major histone with a specific variant. These single-copy, non-allelic isoforms are generally expressed throughout the cell-cycle, and many enter nucleosomes through the action of specific deposition machineries 1. Both major and variant histones can be further distinguished by the addition of small chemical moieties, including phosphorylation, acetylation, methylation and ubiquitylation 2,3.
H2A.Z is one of the most studied histone variants. In metazoans it is essential 4, regulating chromosome stability 5, gene activation 6,7 and spermatogenesis 8. The histone is subject to multiple N-terminal acetylations in Saccharomyces cerevisiae (Sc) 9–11, Tetrahymena thermophila 12, and metazoans 13,14. Though the role(s) of these modifications remains uncertain, defects in heterochomatin restriction 11 and chromosome stability 9 are observed when unacetylatable H2A.Z alleles are expressed as the sole source of the histone in budding yeast.
A phenotype common to all H2A.Z-deficient species is genomic instability. This manifests in mammalian cells as chromatin bridges in anaphase 5. Deletion of Sc H2A.Z (htz1Δ) results in an increased rate of chromosome loss, sensitivity to microtubule destabilizing agents, and synthetic genetic interactions with components of the kinetochore and spindle checkpoint machineries 15. Many of these phenotypes are shared by an unacetylable htz1 mutant, suggesting a role for acetylation in genomic stability 9. However we still have little mechanistic understanding of what leads to the chromosome segregation defect in these cells, though centromere organization, kinetochore attachment, cohesin recruitment and arm sister chromatid cohesion appear normal 9.
To explain the role of H2A.Z in chromosome stability we turned to the fission yeast Schizosaccharomyces pombe (Sp) as its three large chromosomes (Sc, 16 × 230kb – 1.3 Mb; Sp, 3 × 3.5 – 5.7 Mb) can be more easily monitored during mitosis. We show that Sp H2A.Z (Pht1; Pseudo-histone 1) co-purifies with a complex almost identical in composition to the Sc SWR-C, with this complex required for the insertion of Pht1 into chromatin. Chromatin-associated Pht1 is acetylated on its N-terminus by the KAT5 acetyltransferase (Sp Mst1). This modification is essential for Pht1 function, with unacetylatable mutants phenocopying complete deletion of the histone variant in all analyses, including genome-scale genetic-interaction and gene-expression studies. In addition we show that chromosome loss in pht1 mutants is primarily caused by broad architectural defects and can be suppressed by improving chromosome condensation. Consistent with this we show by Chromatin Immunoprecipitation that the condensin complex is prematurely released from chromatin at anaphase in pht1Δ cells.
Results
Pht1 is acetylated as a component of chromatin
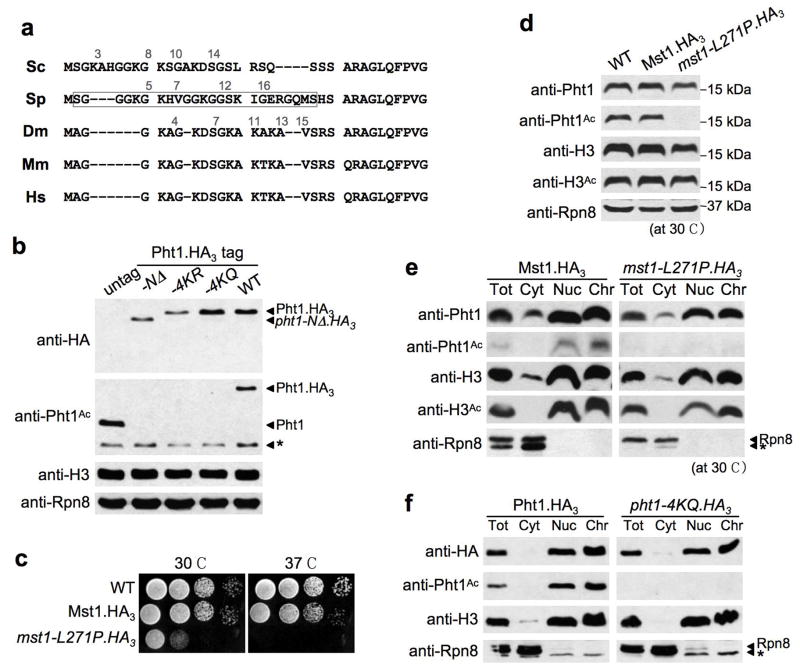
Sp Pht1 has four potential acetylatable lysines (K) on its N-terminus: K5, 7, 12 and 16 (Fig. 1a). We synthesized a peptide containing all four acetyl-lysines and immunized rabbits to create polyclonal anti-Pht1Ac. On immunoblots of Sp whole cell extracts (WCEs) this recognized a protein of the appropriate size for Pht1. This signal disappears in WCEs from pht1Δ, −NΔ (deletion of the N-terminus), or mutants where all four lysines were mutated to unacetylatable arginine (R) or glutamine (Q) (Fig. 1b). Thus the Pht1 N-terminus is acetylated and we have a specific reagent for the modification.
Figure 1. Chromatin-associated Pht1 is acetylated by Mst1.
(a) Alignment of the H2A.Z N-termini from Sc (Htz1, YOL012C), Sp (Pht1, SPBC11B10.10c), Drosophila melanogaster (Dm; H2Av, P028985), Mus musculus (Mm; NP_058030) and Homo sapiens (Hs; NP_002097). Pht1 is mis-annotated in the literature by the addition of 32 N-terminal residues (MILRHAPRVHESAFSLTHKTFAFCNCNNRFKM-). This would give a protein of 18kDa rather than the 14kDa observed. The acetylated residues on Sc Htz1 (K3, 8, 10 and 14) correspond to Sp Pht1 K5, 7, 12 and 16. A synthetic tetra-Ac peptide covering this region (boxed) was immunized into rabbits to create anti-Pht1Ac. (b) Pht1 is acetylated on the N-terminus. Immunoblotting was performed on strains expressing C-terminally HA3-tagged Pht1 (or indicated mutants) at the endogenous locus. Total Pht1 was detected by anti-HA (12CA5; Supplementary Table 3). * in anti-Pht1Ac panel refers to cross-reaction with H4Ac. H3 and Rpn8 serve as loading controls. (c) The mst1-L271P allele is slow and ts. Spot tests are 10-fold dilutions onto YES plates. (d) WCEs from mst1-L271P show a profound reduction in Pht1Ac without affecting total Pht1 or H3Ac levels. H3 and Rpn8 serve as loading controls. (e – f) Pht1 acetylation by Mst1 is not required for assembly of the histone into chromatin. In f strains express Pht1.HA3 or unacetylatable pht1-4KQ.HA3 at the endogenous locus. Cells were spheroblasted (Total) and fractionated into Cytoplasm, Nucleus and Chromatin. H3 and Rpn8 controls (* cross-reacting species) indicate efficient fractionation: the former is primarily localized in insoluble chromatin, the latter in soluble cytoplasm.
The primary acetyltransferase for Sc Htz1 is Esa1, the catalytic subunit of the NuA4 complex 9–11. Esa1 is a member of the KAT5 family, with the most likely Sp homolog the essential protein Mst1 16. We created a temperature sensitive (ts) allele of Mst1 (mst1-L271P), that was slow at the permissive temperature (25°C) and lethal >34°C (Fig. 1c). WCEs from mst1-L271P cells showed no appreciable change in total Pht1 levels but a profound reduction in Pht1Ac (Fig. 1d), indicating that Pht1 acetylation is Mst1-dependent. Cell fractionation showed that Pht1Ac is chromatin-associated, though acetylation is not required for entry to this cellular compartment (Figs. 1e–f).
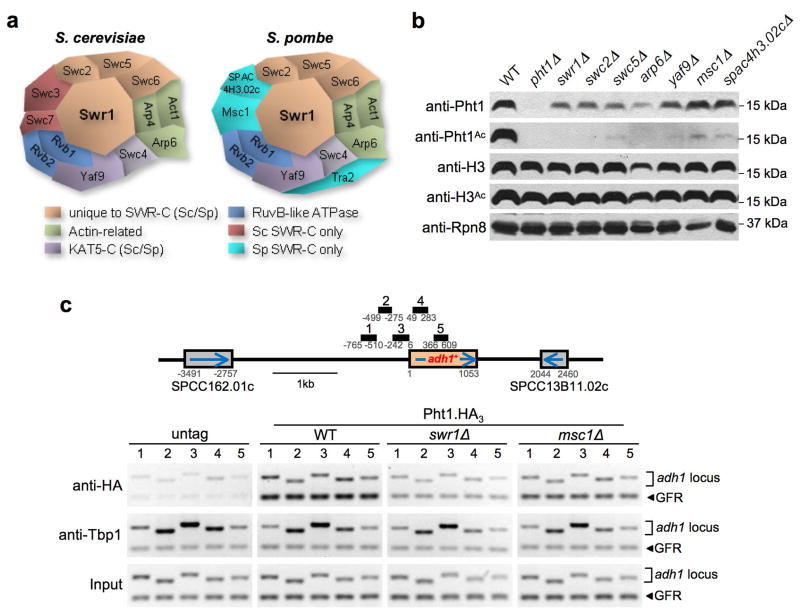
Sc Htz1 associates with the SWR-Complex, containing the Swr1-ATPase, and incorporation of the histone into chromatin is markedly reduced in swr1Δ cells 17–19. We subjected WCEs from Pht1.TAP to sequential affinity purification and identified the associated proteins by Mass Spectrometry (Supplementary Table 1). Reciprocal tagging and purification of these factors delineates the Sp SWR-C. This is almost identical in composition to the Sc SWR-C, even to the level where subunits shared between the Sp SWR and Mst1-acetyltransferase complexes are those shared by their Sc counterparts, the SWR-C and NuA4 (Fig. 2a). Western analysis with anti-Pht1Ac distinguished those subunits of the Sp SWR-C required for the efficient acetylation of the histone (Fig. 2b), most likely because of inefficient assembly of the variant into chromatin in each background (Fig. 2c). Thus a pathway first identified in Sc also operates in Sp: SWR-C inserts Pht1 into chromatin, where it is acetylated by Mst1.
Figure 2. Pht1 is inserted into chromatin by the SWR-C.
(a) The SWR-C is highly conserved from Sp to Sc. Multiple subunits of each complex were C-terminally TAP-tagged and associated factors identified by MS (see Supplementary Table 1 and 17). In this schematic location does not indicate direct interaction. (b) SWR-C components are individually required for the efficient acetylation of Pht1, suggesting chromatin-insertion defects. Total Pht1 levels are also reduced in many of these mutants. Immunoblotting of H3, H3Ac and Rpn8 serve as loading controls. (c) Pht1 is inefficiently assembled to chromatin in swr1Δ, and to a lesser extent, msc1Δ. Schematic depicts the neighborhood of constitutively expressed adh1+. Black boxes depict the location of ChIP primers (Supplementary Table 5) relative to the start codon (+1). Pht1.HA3 was detected by anti-HA. Appropriate localization of Tbp1 at the adh1+ promoter confirms sample integrity. In these duplex reactions upper band (adh1 locus) are the specific primers numbered in the schematic, lower band (GFR) is a non-transcribed Gene-Free Region included as a loading control. Lower panel (Input) is used to normalize the PCR amplification efficiency of each primer pair.
Acetylation is integral to Pht1 function
To determine the relevance of N-terminal acetylation to Pht1 function, we compared the gene expression and genetic interaction profiles of various unacetylatable mutant alleles (pht1- NΔ, −4KR, and −4KQ) to pht1Δ.
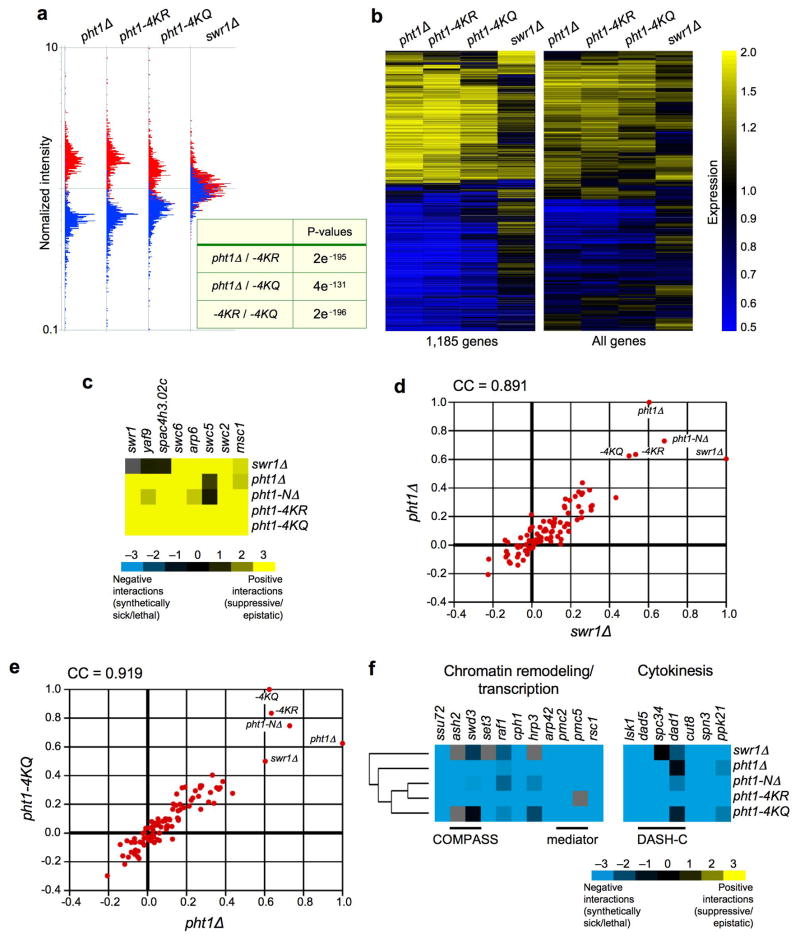
The transcription profiles of the unacetylatable mutants were almost identical to pht1Δ (e.g. relative P-value: Δ to −4KR = 2e−195; Fig. 3a). A striking observation was that almost all genes were modulated to some degree in pht1 mutants (Fig. 3b), suggesting that the histone acts as a general transcriptional regulator. By comparison swr1Δ showed weaker changes in gene expression (Fig. 3b). Since loss of the ATPase strongly reduces, but does not completely ablate chromatin-associated Pht1 (Fig. 2c), it is likely that the remainder is sufficient to mask most H2A.Z-dependent phenotypes.
Figure 3. Unacetylatable pht1 mutants phenocopy pht1Δ.
(a) Panel compares induced (red) and repressed (blue) genes (>1.5-fold in at least one condition) for whole genome array data from eachstrain. Relative P-values for expression overlaps among induced genes between strain pairs are as indicated. (b) Heat map of induced (yellow) and repressed (blue) genes in each mutant. Left: 1,185 genes are >1.5-fold changed in at least one mutant background. Right: all genes on the array. (c) Unacetylatable pht1 mutants display positive genetic interactions with deletions of SWR-C subunits. (d) Correlation coefficient (CC) plot comparing the genetic interactions of 101 query mutants (including pht1Δ, −NΔ, −4KR, −KQ and swr1Δ) mated against a library of 2,161 non-essential deletions. Red dots indicate the CC of each genetic screen to the mutants on the X- or Y-axes. (e) CC plot of pht1Δ Vs pht1-4KQ. (f) pht1Δ, −NΔ, −4KR, −KQ and swr1Δ share a large number of synthetic genetic interactions, including with members of the COMPASS, RSC, SET3-C, Mediator and DASH complexes.
We then used the Pombe Epistatic Mapper 2 (PEM-2) system to quantitatively analyze genetic interaction patterns 20,21. To this end we individually crossed pht1Δ, −NΔ, −4KR, −4KQ and swr1Δ strains to a library of 2161 non-essential Sp deletions (the Bioneer collection), and derived scores covering each negative (e.g. synthetic sick/lethal) and positive (e.g. suppression) genetic interaction using colony size as a quantitative read-out 20,22–24. Positive interactions enrich for factors that are co-complexed or function in the same pathway 20,22,23. Consistent with this, pht1Δ, unacetylatable pht1, or swr1Δ each gave rise to positive genetic interactions in combination with deletions of all non-essential subunits of the SWR-C (20 and Fig. 3c). This implies that preventing Pht1 acetylation disables the primary function of the SWR-C/Pht1 pathway in fission yeast.
The genetic interaction profile of a particular mutant can be used as a high-resolution phenotype. Comparison of this profile to those generated from other mutants can identify functionally related factors 20,22,23. When compared to data from >100 genetic screens (not shown), pht1Δ and swr1Δ were highly correlated (Fig. 3d), confirming that they function in the same pathway. Indeed, of all mutants analyzed, the most highly correlated to pht1Δ were −NΔ, −4KR and −4KQ, suggesting that N-terminal acetylation is critical for Pht1 function (Fig. 3e). Inspection of individual interactions showed that all unacetylatable pht1 alleles were synthetic with deletions of factors involved in chromatin modification/remodeling (e.g. COMPASS, RSC, SET3-C), transcription (e.g. Mediator) and chromosome segregation/cytokinesis (e.g. cut8, DASH complex) (e.g. Fig. 3f). These are reminiscent of the synthetic interactions displayed by Sc htz1Δ 15,17, further suggesting strong conservation of H2A.Z function in each organism.
Chromosome loss in pht1Δ is caused by entanglement at anaphase
Knockout or depletion of H2A.Z in Sc 15 or mammalian cells 5 leads to increased rates of chromosome loss. This phenotype was also observed if any component of the Sp Pht1Ac pathway is disrupted, including mutants in swr1 (and msc1), pht1 (pht1Δ, −4KR or −4KQ), or mst1 (Supplementary Table 2 and 16,25,26). One possible explanation for chromosome instability is disruption of the centromere 27. However as in htz1Δ cells 9, centromere structure and function appeared normal in pht1 cells (Supplementary Fig. 1).
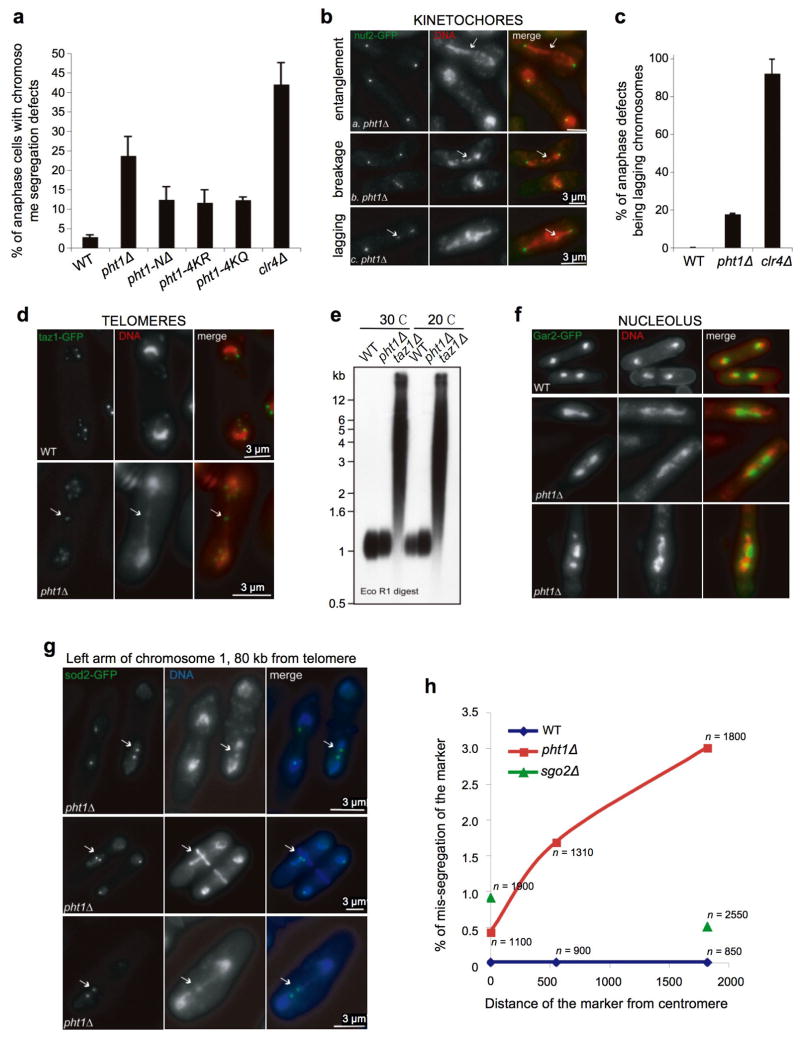
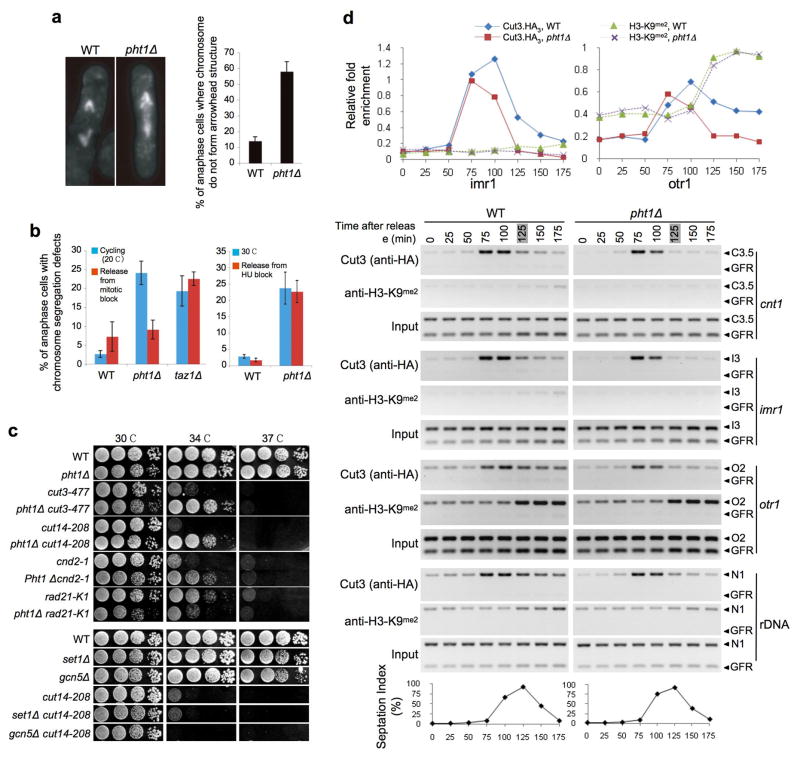
On cytological analysis of individual Sp cells we observed a >8-fold increase (relative to WT) in the number of pht1Δ cells with anaphase chromosome segregation defects (Fig. 4a). Three specific categories of anaphase defects were distinguished using the kinetochore marker Nuf2-GFP: (i) lagging chromosomes, where the mis-segregating chromatin contains at least one kinetochore, (ii) chromosome entanglement, where the mis-segregating chromatin is stretched between the spindle poles but contains no kinetochore, and (iii) entanglement leading to breakage, where broken pieces of chromatin with no kinetochore lag on the spindle (Fig. 4b). The primary segregation defect in pht1Δ cells was entanglement: >80% of defective anaphases. This was clearly distinct from the primary anaphase segregation defect in heterochromatin-deficient clr4Δ cells: >90% lagging chromosomes (Fig. 4c). Lagging chromosomes result from merotely, a defect where a single kinetochore attaches to microtubules emanating from opposite spindle poles 28. Thus, to a large extent, kinetochore - microtubule attachments appear normal in pht1Δ cells, supporting our ChIP observation of a WT-like centromere in pht1 mutants (Supplementary Fig. 1c). Consistent with this result, pht1Δ cells did not activate the spindle checkpoint, nor did they rely on spindle checkpoint genes for survival (not shown).
Figure 4. Lack of Pht1Ac leads to chromosome segregation defects in anaphase.
(a) The percentage (mean ± s.d.) of late anaphase cells with chromosome segregation defects (see succeeding panels) was visually scored. All strains were grown in YES at 30°C. (b) Kinetochore segregation was monitored by distribution of Nuf2-GFP 49. Three chromosome mis-segregation defects (arrowed) are distinguished in pht1Δ anaphase cells: lagging, entanglement, and entanglement leading to breakage. (c) The primary chromosome segregation defect in pht1Δ (entanglement) is distinct from that in heterochromatin-deficient clr4Δ (lagging). Bars are mean ± s.d. (d) Telomere segregation was monitored by distribution of Taz1-GFP 29. In the example two individual telomeres (two Taz1 spots) are entangled in the middle of a pht1Δ anaphase cell, leading to stretched chromosome arms. (e) Pht1 (in contrast to Taz1) does not regulate telomere repeat length. (f) Nucleolar segregation was monitored by distribution of Gar2-GFP 50. In many pht1Δ cells the nucleolus appears stretched or fragmented. (g – h) Chromosome marker loss in pht1Δ cells increases with distance from the centromere. GFP-marked loci locations (cen2-lacO, 5 kb from CEN2; ade3-lacO, 1350 kb from TEL1-L; sod2-lacO, 80 kb from TEL1-L 30) were used to monitor and quantify chromosome segregation defects. n represents the number of binucleate cells counted for each marker in each background.
Chromosome entanglement in pht1Δ cells was particularly obvious when we followed telomere segregation at anaphase. To do so, we used the telomere-binding protein Taz1-GFP as a marker. In a normal mitosis, six Taz1-GFP spots (identifying the left and right telomeres on the three Sp chromosomes) migrate to opposite poles with the bulk of DNA. However in pht1Δ cells we often observed two or more Taz1-GFP foci entangled in the centre, leading to stretched chromosome arms (Fig. 4d). One possible explanation for this entanglement is a defect in telomere function comparable to that observed in cells lacking the protection factor Taz1 29. However many of the phenotypes of taz1Δ were not shared by pht1Δ: e.g. cold-sensitivity, size of the chromatin bridge, presence of Rad22 foci, rqh1-SM suppression (Supplementary Fig. 2). In addition, and most striking, while telomere-repeat length was dramatically increased in taz1Δ, it was comparable to WT in pht1Δ (Fig. 4e).
To quantify how frequently the entanglement in pht1Δ cells leads to chromosome loss, we employed a series of strains containing: (i) lacOperator (lacO) repeats integrated at different chromosomal locations, and (ii) LacI-GFP, the fluorophore marked lacO binding protein 30. Surprisingly, marker loss in mitotic pht1Δ cells increased with distance from the centromere, suggesting that they generally lost broken pieces of chromosomes rather than whole chromatids (Fig. 4g–h). This is in sharp contrast to sgo2Δ cells, which are unable to correct chromosome bi-orientation defects 31,32. In sgo2Δ cells marker loss was similar irrespective of chromosome position, suggesting the loss of whole chromatids (Fig. 4h). Together these observations suggest that the primary role of Pht1 in chromosome transmission is to maintain overall chromosome architecture rather than to regulate the function of a specific region (such as the centromere or telomere).
Pht1 plays a role in chromosome architecture/compaction
Chromosomes condense to a folded rod-shaped structure upon mitotic entry. As the chromatids are pulled to the spindle poles in early anaphase, the chromosome arms trail behind the centromeres, forming a distinctive “arrowhead” structure. This was lost in >50% of cells lacking pht1 (Fig. 5a). Such a generalized defect further supports the idea that Pht1 regulates overall chromosome architecture rather than that of specific loci.
Figure 5. Pht1 plays a role in chromosome architecture/compaction.
(a) pht1Δ (and −4KR or −4KQ; not shown) cells frequently (mean ± s.d.) lose the “arrowhead” structure of segregating chromosomes in anaphase, indicating a disruption of chromatin architecture. In the example each anaphase (WT and pht1Δ) is at roughly the same stage: 6.5 μm between the poles. (b) The chromosome segregation defects of pht1Δ, but not taz1Δ, can be rescued by chromosome hyper-condensation induced by prolonged mitotic arrest. WT, pht1Δ or taz1Δ cells in the background of a cold-sensitive tubulin mutant nda3-KM311 (red) or not (blue) were placed at the restrictive temperature (20°C, six hours), during which nda3-KM311 fails to assemble microtubules and undergoes spindle-dependent checkpoint arrest. Cells were released into anaphase at the permissive temperature (32°C) and scored for chromosome segregation defects. Arresting with HU has no effect on the phenotype. (c) pht1Δ partially rescues mutants in three subunits of condensin (each SMC and the kleisin: cut3-477, cut14-208 and cnd2-1), but is synthetic with a mutant in cohesin (kleisin: rad21-K1). The ability to rescue condensin is not shared by deletions of the Set1 methyltransferase or Gcn5 acetyltransferase. Spot tests are 10-fold dilutions onto YES plates. (d) WT or pht1Δ (additionally containing Cut3.HA3 and cdc25-22) were arrested in G2 and released into the cell cycle (with samples taken every 25 minutes as in Supplementary Methods). Septation index confirms that each population completes mitosis by 125 minutes (peak septation; shaded in grey). ChIP was used to monitor condensin and H3-K9Me2 at various locations, including the Chr I centromere (for primer schematic see Supplementary Fig. 1c) and rDNA. While H3-K9Me2 at imr1 and otr1 follows similar creation kinetics in WT and pht1Δ, Cut3.HA3 drops prematurely at all locations in the absence of the histone variant (see also Supplementary Fig. 3d). The “Relative fold enrichment” in the ChIP quantitation graphs is the ratio between the specific signal at each location and the respective Input.
The primary chromosome segregation defect in pht1Δ cells (i.e. chromosome entanglement despite accurate centromere segregation; Fig. 4) is reminiscent of that observed in mutants of condensin, a five-subunit complex instrumental to the architecture and segregation of chromosomes in mitosis 33–36 (Supplementary Fig. 3a). We thus speculated that pht1 mutants may mis-regulate chromosome condensation during anaphase. If so, it may be possible to suppress their defect by forcing chromosome hyper-condensation. To this end we used the cold-sensitive tubulin mutant, nda3-KM311, which fails to assemble microtubules at the restrictive temperature (20°C), leading to a spindle-dependent checkpoint arrest 37. This prolonged early mitotic arrest induces chromosome hyper-condensation 37, and when shifted to the permissive temperature (32°C) these cells enter a synchronous anaphase 31,37. Strikingly, this prolonged mitotic arrest specifically rescued the chromosome segregation defect of pht1Δ but not taz1 cells (Fig. 5b). Arresting pht1Δ cells in S-phase with hydroxyurea (HU, depletes nucleotide pools) had no effect on the chromosome segregation defect, showing that merely prolonging the cell-cycle was not sufficient to rescue pht1Δ defects (Fig. 5b).
A functional relationship would be indicated by genetic interaction, so we tested that between pht1 and condensin. Unacetylatable pht1 or pht1Δ each raised the restrictive temperature of ts alleles of three complex subunits: cut3-477 (smc4-S1147P), cut14-208 (smc2-S861P) and cnd2-1 (cnd2-A114T) (Fig. 5c and Supplementary Fig. 3b). This partial rescue was specific, as pht1Δ was synthetic with rad21-K1, a mutant in the condensin-related complex cohesin, which holds sister-chromatids together prior to anaphase onset. This suppression is unlikely to be mediated through indirect transcriptional effects (e.g. increased expression of condensin), since the mRNA levels (by gene expression microarray) of all tested complex subunits and known regulators (e.g. Cut17, Ark1, Top2, Fin1, Pim1, Pic1, Acr1, Nuc1) were comparable to WT in pht1Δ and unacetylatable pht1 cells (Supplementary Fig. 3c and not shown). In addition the partial rescue was not observed on deletion of two other transcriptional regulators: the Set1 methyltransferase or Gcn5 acetyltransferase (Fig. 5c).
Our results are consistent with a model where an acetylated form of Pht1 regulates condensin loading and/or localization in mitosis. To test this, we arrested WT or pht1Δ cells expressing C-terminally tagged Cut3 (Cut3.HA3) at the G2/M boundary, and then monitored condensin occupancy kinetics through a synchronous mitosis (Fig. 5d). To first ensure that mitotic progression in WT and pht1Δ cells followed similar kinetics, we examined the peak septation index and H3-K9Me2 in each population. A newly formed septum indicates the completion of mitosis and can be visualized by specific staining with Calcofluor (Supplementary Methods). H3-K9Me2 is markedly reduced at centromeric repeats (otr) during mitosis, increasing as cells enter G1/S 38. Each pattern was indistinguishable between WT and pht1Δ cells (Fig. 5d). When we examined condensin recruitment, Cut3.HA3 initially (≤75 mins) followed similar kinetics at the centromere (cnt, inr and otr) and rDNA of WT and pht1Δ cells (Fig. 5d and Supplementary Fig. 3d). However significant differences were then observed: Cut3.HA3 levels continued to increase in WT cells, peaking at 100 mins before dropping at 125 mins (the latter corresponding to the peak septation index time point; Fig. 5d). However in pht1Δ condesin delocalization began by 100 mins, indicating the premature dissociation of the complex from chromatin. This likely explains many of our observations in these cells: loss of the anaphase arrowhead structure leading to chromosome entanglement and the loss of telomere-proximal regions (Fig 5).
Discussion
In this work we identify and characterize a previously unknown role for the histone variant H2A.Z (Sp Pht1) in chromosome architecture at anaphase. In the absence of Pht1, chromosomes frequently entangle in anaphase, which can lead to breakage and loss, particularly of telomere-proximal regions. We provide evidence that chromosome entanglement in pht1Δ is most likely due to premature dissociation of the condensin complex in anaphase. We also demonstrate that the factors involved in the chromatin-loading and acetylation of H2A.Z are highly conserved in Sc and Sp. The last common predecessor of these yeasts was ~ 380 million years ago (mya): by comparison that of the entire mammalian class existed ~ 165 mya 39, while the primate line split to humans and gorillas ~ 8 mya 40. We propose that since the pathways regulating H2A.z localization, modification, and function are so well conserved, the role of the histone variant in chromosome architecture will be equally penetrant.
Extensive conservation of the Swr1-C
Comprehensive proteomic analyses identify two subunits in the Sp SWR-C not seen in its Sc counterpart: Msc1 and SPAC4H3.02c (Fig. 2a, Supplementary Table 1, and 41). Both deletions are epistatic with pht1 (Fig. 3c and 20), further suggesting a functional relationship. Msc1 was originally identified as a multi-copy suppressor of cells defective for checkpoint kinase Chk1 (Rad27) function 42. The Walworth lab have demonstrated an epistatic chromosome loss phenotype in pht1Δ and msc1Δ, and suggested that Msc1 was upstream of Pht1 in this pathway 26, a relationship our data supports (e.g. Fig. 2c). Msc1 was recently shown to interact with the Mst1 acetyltransferase by yeast-2-hybrid analysis 16. Despite this, any physical interaction between Mst1 and Msc1 is likely transient, as Msc1.TAP purification did not identify Mst1 (or any non-shared subunit of the Swr1 and Mst1 Complexes: Supplementary Table 1), and unique members of the Mst1-C do not co-purify Msc1 (not shown). However this is an interesting link between the Swr1 and Mst1 complexes, and may suggest that Msc1 directly recruits Mst1 to sites of Pht1 integration.
N-terminal acetylation is integral to Pht1 function
The Swr-C is required for the assembly of Pht1 into chromatin, where it is acetylated by Mst1 (Figs. 1 and 2). This relationship between the ATPase (Swr1), histone variant (H2A.Z) and KAT5-family acetyltransferase (Mst1) appears to be widely conserved 9,15,43–46. Unacetylatable pht1 phenocopies pht1Δ throughout this work, but is perhaps most striking in large-scale gene expression and genetic analyses (Fig. 3). This strongly suggests that acetylation of the histone is integral to its function. Based on Sc Htz1, it is likely that all four lysine residues in the Pht1 N-terminus are modified 10,11. However our pht1Ac antibody was raised to a tetra-acetylated peptide (Fig. 1a) and the unacetylatable pht1 used throughout this work has mutations at all four N-terminal lysines, so this is as yet unconfirmed. mst1-L271P reduces Pht1Ac below the threshold of detection (Fig. 1d), suggesting we have identified the major enzyme for this modification. In this regard, we note that mst1 mutants also show increased rates of chromosome loss 16, likely due in part to reduced Pht1Ac.
Pht1 regulates chromosome architecture at anaphase
Knockout or depletion of H2A.Z in Sc 15, Sp (this work and 25,26), or mammalian cells 5 leads to increased rates of chromosome loss. An acetylated form of H2A.Z mediates the chromosome stability role in both Sc 9 and Sp (this work), strongly suggesting that this will prove to be the case in other organisms. Directly monitoring chromosome segregation in individual Sp cells allowed us to show that lack of Pht1 induces chromosome arm entanglements in anaphase that can lead to chromosome breaks (Fig. 4). Furthermore, Pht1 is required for the stable association of condensin with chromatin through anaphase (Fig. 5d), and the chromosome entanglement in pht1Δ cells can be rescued by a pre-anaphase arrest where chromosomes hyper-condense (Fig. 5b). Thus the lack of Pht1 interferes with chromosome architecture in anaphase. However it also improves the viability of mutants that regulate this process, such as those in condensin (cut3-477, cut14-208, cnd2-1) or topoisomerase II (top2-191) (Fig. 5c and Supplementary Fig. 4). To resolve this apparent contradiction, we propose that Pht1 actually plays a dual role in mitotic chromosome architecture (see below).
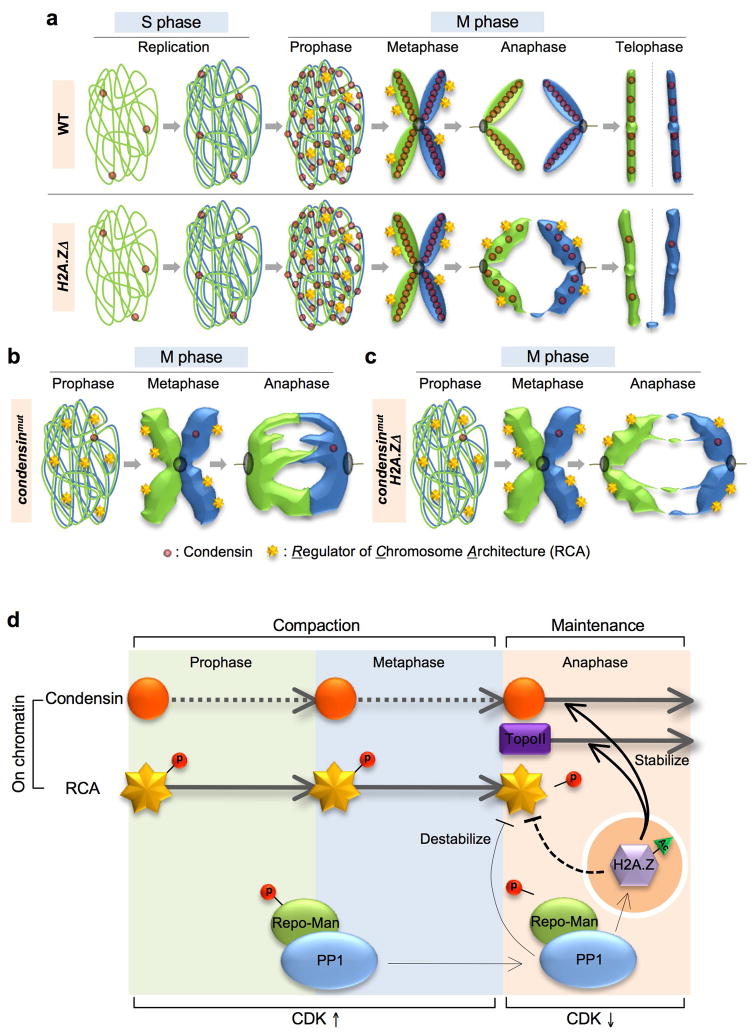
Mitotic defects in a chicken condensin mutant primarily occur in anaphase, such that chromosomes prematurely lose their compact organization as they move to the poles 47. Thereafter individual chromatids can no longer be distinguished and prominent chromatin bridges are visible in >90% of cells (e.g. Fig. 6b and Supplementary Fig. 3a). These phenotypes can be overcome if Repo-Man, a targeting subunit for protein phosphatase 1 (PP1), is unable to direct the phosphatase to chromosomes in anaphase 47. In WT cells Repo-Man is subject to CDK-dependent phosphorylation(s) to abrogate chromosome binding at early mitosis when CDK levels are high. Chromosome compaction at this stage is only slightly affected in condensin mutants, prompting the Earnshaw lab to propose that an as-yet-uncharacterized activity, Regulator of Chromosome Architecture (RCA), drives initial condensation. RCA is then inhibited in a Repo-Man/PP1-dependent manner after CDK levels fall at anaphase onset and condensin steps in to stabilize chromosome architecture until mitosis completes 47 (all modeled in Fig. 6d).
Figure 6. H2A.Z plays a role in higher order chromosome architecture.
(a). Upper row: The localization/association of condensin with chromosomes is tightly regulated. The complex is primarily (but not exclusively) found in the cytoplasm through interphase, and imported into the nucleus at prophase for chromosome loading, with levels peaking at anaphase 51,52. Lower row: the pathway in pht1Δ cells. Condensin loads normally but prematurely dissociates (Fig. 5d). This explains the poorly resolved structure observed in anaphase (Fig. 5a), which likely leads to chromosome entanglement and loss (Fig. 4). (b) Sister chromatids in condensin mutants are “fuzzy” and resolve poorly at prophase. The sisters remain connected by chromatin bridges as they pull apart in anaphase (see Supplementary Fig. 3a). (c) The rescue by pht1Δ (and unacetylatable mutants) of various chromosome architecture mutants (condensin and Topo II; Fig. 5c and Supplementary Fig. 3b and 4) indicates that a partially compensatory condensed topology exists in pht1Δ cells. (d) Model separates mitotic chromosome condensation/architecture into two stages: initial compaction and maintenance. The first stage is dependent on the poorly characterized activity (or factor) RCA, the second on the condensin complex and topoisomerase II. H2A.Z may have a dual regulatory role: somehow opposing RCA, yet stabilizing the association of condensin with chromatin.
The above model suggests that there are at least two steps in chromosome condensation. The first is RCA-dependent compaction to the characteristic X-shaped mitotic chromosome. The second is the condensin-dependent maintenance of a robust architecture that can withstand the pulling forces of microtubules in anaphase. Our data suggests that Pht1 regulates both steps: promoting the inhibition of RCA, and stabilizing the association of condensin. As above, artificially maintaining RCA activity in anaphase by preventing the loading of PP1 to chromatin partly rescues the chromosome segregation defects of chicken smc2 mutants 47. This is reminiscent of the genetic interactions between pht1 and condensin (e.g. Fig. 5c). It is highly likely that RCA is conserved in Sp as deletion of PP1Dis2, the fission yeast PP1 ortholog that localizes on chromatin, also partly rescues cut3-477 (Supplementary Fig. 4b). Finally we have recently shown that Sc Bud14, a regulator of Sc PP1 (Glc7), is required for the efficient loading of Htz1 onto chromatin (and its subsequent acetylation) 48. This suggests that H2A.Z could act downstream of PP1 to inhibit RCA in anaphase (Fig. 6d). Further studies are underway to resolve this model.
Supplementary Material
Acknowledgments
We thank Robin Allshire (University of Edinburgh), Marc Bühler (Friedrich Miescher Institute for Biomedical Research), Julie Cooper (Cancer Research UK), Dan Finley (Harvard Medical School), Susan Forsburg (University of Southern California, Los Angeles), Keith Gull (University of Oxford), Charles Hoffmann (Boston College), Junko Kanoh (Kyoto University), Rich Maraia (NIH), Danesh Moazed (Harvard Medical School), Toru Nakamura (University of Illinois at Chicago), Frank Neumann (Rockefeller University), Paul Nurse (Rockefeller University), Matthew O’Connell (Mount Sinai School of Medicine), Janet Partridge (St. Judes Childrens’s Research Hospital), Mitsuhiro Yanagida (Kyoto University) and the Yeast Genome Resource Center (Osaka City University) for the generous supply of antibodies and yeast strains (detailed in Supplementary Tables 3 – 4). We also thank Guoquing Zhong, Shamanta Chandran, Thanuja Punna (all University of Toronto), and Mike Shales (UCSF) for technical support. Finally we are grateful to Gwyneth Ingram (University of Edinburgh) for expertise with the Telomere Repeat length assay. Work in the J.B. lab is funded by Cancer Research UK, T.K. lab by a start-up grant from the OCU, K.G.H. lab by a program grant from the Wellcome Trust, and M-C.K. lab by the Speaker’s Fund for Biomedical Research: Toward the science of Patient Care, awarded by the City of New York.
Footnotes
Author Contributions
H-S.K. and V.V. contributed equally to this work. H-S.K. was responsible for the data in Fig. 1 (with A.T. contributing Fig. 1c), Fig. 2 (with Mass Spectrometry help from J.F., T.K., A.E. and J.F.G.), Fig. 5c–d and Supplementary Figs 1 and 3. V.V. (with help from K.G.H.) was responsible for the data in Fig. 4, Fig. 5a–c and Supplementary Figs 2, 3a and 4. S.W. and J.B. performed and analyzed the microarrays in Fig. 3a–b; A.R. and N.J.K. performed and analyzed the genetic screens in Fig. 3c–f. L.R.C and C.S.B. created the anti-Pht1 antibodies used in Fig. 1. H-S.K., V.V. and M-C.K. planned experiments, analyzed the data and wrote the manuscript.
References
- 1.Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. TiGs. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Latham JA, Dent SYR. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 3.Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 4.Faast R, et al. Histone variant H2A.Z is required for early mammalian development. Curr Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- 5.Rangasamy D, Greaves I, Tremethick DJ. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat Struct Mol Biol. 2004;11:650–655. doi: 10.1038/nsmb786. [DOI] [PubMed] [Google Scholar]
- 6.Leach TJ, et al. Histone H2A.Z is widely but non-randomly distributed in chromosomes of Drosophila melanogaster. J Biol Chem. 2000;275:23267–23272. doi: 10.1074/jbc.M910206199. [DOI] [PubMed] [Google Scholar]
- 7.Rangasamy D, Berven L, Ridgway P, Tremethick DJ. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 2003;22:1599–1607. doi: 10.1093/emboj/cdg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ. The X and Y chromosomes assemble into H2A.Z, containing facultative heterochromatin, following meiosis. Mol Cell Biol. 2006;26:5394–5405. doi: 10.1128/MCB.00519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keogh MC, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ lysine 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren Q, Gorovsky MA. Histone H2A.Z acetylation modulates an essential charge patch. Mol Cell. 2001;7:1329–1335. doi: 10.1016/s1097-2765(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 13.Bruce K, et al. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucl Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonenfant D, Coulot M, Towbin H, Schindler P, van Oostrum J. Characterization of histone H2A and H2B variants and their post-translational modifications by mass spectrometry. Mol Cell Proteomics. 2006;5:541–552. doi: 10.1074/mcp.M500288-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Krogan NJ, et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex and the Histone AcetylTransferase NuA4. Proc Natl Acad Sci USA. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez EB, Nugent RL, Laria S, Forsburg SL. S. pombe histone acetyltransferase Mst1 (KAT5) is an essential protein required for damage response and chromosome segregation. Genetics. 2008;179:757–71. doi: 10.1534/genetics.107.085779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogan NJ, et al. A Snf2-Family ATPase complex required for recruitment of the Histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 18.Kobor MS, et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLOS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 20.Roguev A, et al. Conservation and rewiring of functional modules revealed by an epistasis map (E-MAP) in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- 22.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 23.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 24.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carr AM, et al. Analysis of a histone H2A variant from fission yeast: evidence for a role in chromosome stability. Mol Gen Genet. 1994;245:628–635. doi: 10.1007/BF00282226. [DOI] [PubMed] [Google Scholar]
- 26.Ahmed S, Dul B, Qiu X, Walworth NC. Msc1 acts through histone H2A.Z to promote chromosome stability in Schizosaccharomyces pombe. Genetics. 2008;177:1487–1497. doi: 10.1534/genetics.107.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ekwall K. Epigenetic control of centromere behaviour. Ann Rev Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- 28.Gregan J, et al. The Kinetochore Proteins Pcs1 and Mde4 and Heterochromatin Are Required to Prevent Merotelic Orientation. Curr Biol. 2007;17:1190–1200. doi: 10.1016/j.cub.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 30.Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell. 2004;6:329–341. doi: 10.1016/s1534-5807(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 31.Vanoosthuyse V, Prykhozhij S, Hardwick KG. Shugoshin 2 regulates localization of the chromosomal passenger proteins in fission yeast mitosis. Mol Biol Cell. 2007;18:1657–1669. doi: 10.1091/mbc.E06-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawashima SA, et al. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 2007;21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakazawa N, et al. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J Cell Biol. 2008;180:1115–1131. doi: 10.1083/jcb.200708170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saka Y, et al. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutani T, et al. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 2002;16:729–742. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 38.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 39.O’Brien SJ, et al. The promise of comparative genomics in mammals. Science. 1999;286:458–481. doi: 10.1126/science.286.5439.458. [DOI] [PubMed] [Google Scholar]
- 40.Takahata N, Satta Y. Evolution of the primate lineage leading to modern humans: Phylogenetic and demographic inferences from DNA sequences. Proc Natl Acad Sci USA. 1997;94:4811–4815. doi: 10.1073/pnas.94.9.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shevchenko A, et al. Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 2008;9:R167. doi: 10.1186/gb-2008-9-11-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed S, Palermo C, Wan S, Walworth NC. A novel protein with similarities to Rb binding protein 2 compensates for loss of chk1 function and affects histone modification in fission yeast. Mol Cell Biol. 2004;24:3660–3669. doi: 10.1128/MCB.24.9.3660-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi K, et al. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development. 2007;134:1931–1941. doi: 10.1242/dev.001891. [DOI] [PubMed] [Google Scholar]
- 44.Ruhl DD, et al. Purification of a human SCRAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- 45.Kusch T, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 46.Updike DL, Mango SE. Temporal regulation of foregut development by HTZ-1/H2A.Z and PHA-4/FoxA. PLoS Genetics. 2006;2:e161. doi: 10.1371/journal.pgen.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vagnarelli P, et al. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat Cell Biol. 2006;8:1133–1142. doi: 10.1038/ncb1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fiedler D, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 2001;110:322–334. doi: 10.1007/s004120100153. [DOI] [PubMed] [Google Scholar]
- 50.Gulli MP, et al. gar2 is a nucleolar protein from Schizosaccharomyces pombe required for 18S rRNA and 40S ribosomal subunit accumulation. Nucl Acids Res. 1995;23:1912–1918. doi: 10.1093/nar/23.11.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagstrom KA, Meyer BJ. Condensin and cohesin: more than chromosome compactor and glue. Nature Rev Genet. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- 52.Hirano T. Condensins: Organizing and segregating the genome. Curr Biol. 2005;15:R265–275. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 53.Dunaway S, Walworth NC. Assaying the DNA damage checkpoint in fission yeast. Methods. 2004;33:260–263. doi: 10.1016/j.ymeth.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 54.Lyne R, et al. Whole-genome microarrays of fission yeast: characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niwa O, Matsumoto T, Yanagida M. Construction of a mini-chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol Genet Genomics. 1986;203:397–405. [Google Scholar]
- 56.Krogan NJ, et al. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.