Abstract
Purpose
Methylation of sperm DNA is impaired in many infertile men potentially adversely effecting reproductive outcomes. In somatic cells oxidative damage to DNA and hyperhomocysteinaemia are linked with DNA hypomethylation. The objective of this study was to investigate if these pathologies also impair sperm DNA methylation.
Methods
The relationship between sperm DNA quality, oxidative stress and serum homocysteine was analysed at study entry and after 3 months of antioxidant treatment.
Results
Overall a significant negative correlation was observed between sperm DNA methylation and sperm DNA fragmentation, as well as seminal reactive oxygen species (ROS) production. Sperm DNA methylation was not significantly related to serum homocysteine concentrations. Administration of an antioxidant supplement produced a significant fall in seminal ROS levels and sperm DNA fragmentation, while increasing sperm DNA methylation.
Conclusions
These results suggest that oxidative stress related damage to sperm DNA impedes the process of methylation, while antioxidant supplementation appears to have the potential to reduce DNA damage and normalize sperm DNA methylation.
Keywords: Antioxidant, DNA methylation, Homocysteine, Sperm
Introduction
Since reports emerged linking IVF conceived pregnancies with the epigenetic disorders Angelman and Beckwith-Wiedemann Syndromes [1–4], there has been intense scientific interest in epigenetic processes involved in reproduction. Disorders of genomic imprinting occur when there is a failure of the correct pattern of DNA parental-origin-dependant monoallelic gene expression [4]. The methylation of cytosine residues in DNA by DNA methyltransferase is considered to be one of the major epigenetic mechanisms controlling gene expression and imprinting [5]. Hypomethylation of DNA is associated with gene transcriptional activity while hypermethylation is associated with gene silencing.
While several studies have suggested that adverse embryo culture environments are responsible for the majority of epigenetic defects observed in IVF conceived pregnancies [3, 4, 6], other observations have suggested that sperm abnormalities may also play a role. Firstly, investigators have reported that aberrant sperm DNA methylation, principally in the form of hypomethylation, is more commonly seen in infertile men compared to their normozoospermic counterparts [7–11]. Secondly, experimental inhibition of sperm epigenetic programming by exposure of male rodents to endocrine disruptors results in reduced sperm fertilization capacity, altered embryonic gene expression, an increase in pre-implantation embryonic loss [12–16] and cancer in future generations [17, 18]. Finally, studies have also linked sperm DNA hypomethylation in men with a reduction in IVF pregnancy rates [9, 19, 20]. Therefore, infertility related anomalies in sperm epigenetic programming may have serious clinical consequences and certainly warrant further investigation.
Epigenetic programming of sperm DNA occurs at several key stages in the spermatogenesis cycle. In rodents it is reported that DNA methyltransferase 1 (DMT1) mRNA and protein are expressed at high levels in mitotic and early meiotic male germ cells, with the enzyme then being translationally down regulated in pachytene spermatocytes [21–23]. Human studies have demonstrated that the paternally imprinted gene H19 has its methylation pattern erased in early fetal life; with remethylation being initiated as spermatogonia enter meiosis and is effectively complete by the primary spermatocyte stage of differentiation [24]. Testicular tissue samples taken from fertile men show DMT1 gene expression is restricted to spermatogonia, pachytene spermatocytes and round spermatids; with DMT1 protein only being expressed in the nucleus of spermatogonia and in the cytoplasm of round spermatids [25]. Interestingly, Ariel et al. reported that spermatogenesis-specific genes can also undergo quite late epigenetic re-programming while maturing within the epididymis [26]. Therefore, pathology within both the testicular and epididymal environment has the potential to disrupt the establishment of normal sperm DNA methylation patterns.
To date only one study has reported on any underlying mechanism for abnormal sperm DNA methylation seen in infertile men [27]. This study identified DNA sequence variations in the gene encoding the DNA methyltransferase enzyme DNMT3L in several infertile men, which inturn was associated with abnormal paternal DNA methylation. However, studies investigating the link between somatic cell DNA hypomethylation and cancer have suggested other possible mechanisms for sperm DNA hypomethylation may exist such as defects in the folate / homocysteine pathway and oxidative stress.
Oxidative stress occurs when the body’s production of reactive oxygen species (ROS) exceeds its own production of protective anti-oxidants, leading to oxidative attack on cellular structures such as DNA. Oxidative attack leads to the generation of DNA strand breaks and the formation of DNA base adducts such as 8-hydroxyl-2′-deoxyguanosine (8-OH-dG) and O6-methylguanine, both reported to interfere with the DNA’s ability to act as a substrate for DNA methyltransferases [28]. The presence of 8-OHdG in CpG dinucleotide sequences [29–31] or O6-methylguanine [32], strongly inhibits methylation of adjacent cytosine residues and ultimately leads to global DNA hypomethylation.
The folate/homocysteine pathway is responsible for the generation of methyl donors and therefore is central to the process of DNA methylation for all cells [33]. Defects in the folate cycle are responsible for depleting S-adenosylmethionine (SAM) levels and have been linked with DNA hypomethylation in somatic cells [33, 34]. Methylenetetrahydrofolate reductase (MTHFR) is a key folate-metabolizing enzyme which catalyses the conversion of 5,10-methylene tetrahydrofolate to 5-methyl tetrahydrofolate, the later which provides methyl groups for the methionine synthase-mediated remethylation of homocysteine to methionine. Studies in mice have linked polymorphisms in the MTHFR gene with reduced availability of methionine / SAM resulting in hypomethylation of testicular DNA [35]. Interestingly, while polymorphisms in the MTHFR gene limiting its enzymatic activity are more common in infertile men [36–39], no study has yet examined the link between defects in the folate pathway and sperm DNA methylation.
Since oxidative stress and hyper-homocysteinaemia have been linked with somatic cell DNA hypomethylation, we propose that similar mechanisms may operate in sperm. The principal aim of this study was to investigate the possible link between seminal oxidative stress related DNA fragmentation, serum homocysteine and methylation of sperm DNA.
Materials and methods
Subjects and study design
Participants in the study were recruited from men with known male factor infertility (“infertile subjects”) defined as the presence of abnormal WHO semen quality criteria (WHO 1999) and the inability to conceive despite more than 12 months of unprotected intercourse. “Fertile controls” were men who were acting as sperm donors at an academic affiliated ART unit (Repromed, South Australia) and who had proven fertility within the last 12 months and normal semen parameters according to WHO criteria. All infertile participants were asked to take one capsule of Menevit® (Bayer Australia Ltd, Sydney, Australia) per day for a period of 3 months and to provide a semen and serum sample both at study entry and exit. Three month duration of antioxidant therapy was considered appropriate as this would cover one full spermatogenesis cycle (∼ 70 days). Each capsule of Menevit contained 500 μg of folate and various anti-oxidants (Vitamin C 100 mg, Vitamin E 400 IU, Lycopene 6 mg, zinc 25 mg, selenium 26 μg, and garlic oil 333 μg). Fifty men entered the study, with five withdrawing over the next 3 months due to a lack of continuing interest (4) or perceived side effects (1). The twelve “fertile controls” produced only a single semen sample for the purposes of this study and were not administered any antioxidant therapy. The study was prospectively approved by the Human Research and Ethics Committee, Women’s and Children’s Hospital (approval REC 1942/4/10), with all participants giving written informed consent for their involvement.
Sample collection and preparation
Semen samples were produced by masturbation after a period of 3–5 days abstinence and then analysed for sperm count, motility and morphology as per WHO guidelines. After liquefaction, a portion of semen was centrifuged at 300 g for 10 min to enable removal of seminal plasma, followed by washing of the sperm twice in Dulbecco’s Phosphate Buffered Saline (PBS) (JRH Biosciences, Kansas, USA). Sperm were then suspended in 1 ml of PBS and used in either the Nitro Blue Tetrazolium (NBT) assay or smeared on poly-L-lysine coated slides for fixation and later TUNEL and global DNA methylation analysis. All serum samples were obtained by venipuncture between the hours of 9 and 11 am, and frozen at −70°C until homocysteine was measured.
Assessment of sperm global DNA methylation
Measurement of sperm global DNA methylation was made using the immunohistochemical 5-methylcytosine staining technique [7]. After fixation of cells with ethanol (96%) the slides were rinsed twice in PBS containing 0.25% Triton X (Sigma-Aldrich, St. Louis, USA). Decondensation of sperm DNA was carried out by decondensing buffer (1 M HCL, 10 mM Tris Buffer, pH 9.5 containing dithiotretiol) (Sigma-Aldrich, St. Louis, USA) at room temperature for 20 min. Sperm were then washed twice in PBS / Tween 0.5% buffer and then the sperm DNA was denatured by 6 N HCl followed by neutralisation with 100 mM TRIS HCl (pH 8.5) for 30 min at room temperature. Cells were then incubated with a dilution of 1:50 monoclonal primary specific antibody against 5-methylcytidine (5mC) (Eurogentec S.A., BI-MECY-1000, Seraig, Belgium) for 1 h at 37C. After rinsing twice in PBS, sperm were incubated with Fluorescein Affinipure goat anti-mouse IgG (Jackson Immunoresearch Laboratories Inc, West Grove, PA, USA) at a dilution of 1:100 in 0.05% Tween 20 in PBS for 20 min. The slides were then rinsed in PBS buffer and mounted in ProLong® Gold antifade reagent (Invitrogen Molecular Probes, Oregon, USA). The degree of global DNA methylation was determined by measuring mean value of intensity of the fluorescence and expressed as Arbitrary Unit (a.u.) on at least 300 cells. The primary antibody step was omitted on the negative control slides.
In order to minimize qualitative error related to fading of flourencent staining intensity over time, all slides were captured for later image analysis within 1 h of completion of the immunochemistry process. Furthermore, all measurements were conducted on a single microscope at a set fluorescent light exposure intensity. Finally, individual infertile patient’s entry and exit methylation slides were always analysed in the same assay run, so as to minimize skewing of results by inter-assay variation. Using these precautions the intra and inter-assay CV was 5 and 7% respectively.
Assessment of sperm DNA fragmentation (TUNEL)
Sperm DNA fragmentation was detected by the In Situ Cell Death Detection Kit (Roche Diagnostics GmbH, Mannheim, Germany) which is based on the labelling of DNA strand breaks. This assay was performed on the washed sperm sample using a modification of the microscopic (Tdt-mediated Terminal dUTP Nick-end Labelling) TUNEL technique as previously described [40, 41]. Briefly, sperm were smeared on poly-L-lysine coated slides, air-dried and fixed with 3:1 Methanol/Glacial Acetic acid fixative. The sperm were then permeabilised with 0.1% Triton X-100/0.1% sodium citrate and washed with PBS before being incubated with FITC-labelled terminal deoxyribonucleotidyl transferase (TdT), followed by fluorescein labeling with Propidium Iodide. The smear was rinsed in PBS buffer and mounted in a 1:1 mixture of ProLong® Gold antifade reagent and glycerol. Stained cells were quantified on Olympus BX51 fluorescence microscope, with a minimum of 300 sperm per slide being assessed using image analysis software (MacProbe V 4.3, Perceptive Scientific Instruments, League, Texas). The percentage of sperm DNA fragmentation was calculated as the number of TUNEL positive nuclei (FITC-labelled, green) per total number of sperm nuclei (Propidium Iodide, red). For a positive control sperm cells were incubated with 3 U/ μL DNAse prior to incubation with the TUNEL reagents and for a negative control the terminal transferase was omitted from the reaction.
Measurement of reactive oxygen species production
A modified colorimetric Nitro Blue Tetrazolium (NBT) test was used to evaluate reactive oxygen species (ROS) production of both leukocytes and sperm cells within semen [42]. After washing in PBS, sperm were resuspended in 200 μL of PBS and incubated with NBT Reagent (0.01% NBT in PBS, Sigma-Aldrich, St. Louis, USA) at 37°C for 45 min. Following incubation the samples were washed and centrifuged at 500 g for 10 min in PBS to remove all residual NBT solution, leaving only a cell pellet. Formazan, a blue water insoluble crystal produced from the yellow water-soluble tetrazolium salt by the action of cellular superoxide anions, is then deposited inside the sperm and leukocytes. The amount of formazan crystal present within a cell is closely related to its production of free radicals [43]. In order to quantify the formazan product, the intra-cellular formazan was solubilized in 60 μL of 2 M KOH and Dimethyl Sulphoxide (DMSO) (Sigma-Aldrich) and the colour reaction was measured spectrophotometrically on a microplate reader (Bio-Tek, USA, Model EL×800) at 630 nm. ROS production was expressed as μg formazan per 107 sperm, derived from a standard curve of absorbance values for known amounts of formazan substrate.
Homocysteine analysis
Serum samples for hormone assessment were separated within 1 h of collection and frozen at − 70°C until assayed. Homocysteine level in serum was measured by using Bio-Rad Microplate Enzyme Immunoassay Homocysteine Kit (Bio-Rad Laboratories, Inc., Hercules, CA,USA) which is based on competition between S-adenosyl-L-homocysteine (SAH) in the sample and immobilized SAH bound to the walls of the microtitre plate for binding sites on a monoclonal anti-SAH antibody.
Identification of leukocytes in semen by using immunocytochemical CD45 measurement
Leukocyte density was determined by immunocytochemistry using the Leukocyte Common Antigen marker CD45. A monoclonal antibody against the CD45 (Rat Anti-human CD45- Serotec Ltd, UK, Code: MCA 345) was applied in a humidified chamber for a period of 1 h, then the slides were washed three times in PBS buffer before application of a secondary antibody for 30 min (Goat anti-rat IgG) (Jackson Immuno Research Laboratories, Inc, USA). After applying Texas Red Avidin D conjugate (Vector Laboratories Inc., Burlingame, CA ) slides were rinsed in PBS buffer and mounted in a 1:1 mixture of ProLong® Gold antifade reagent (Invitrogen Molecular Probes, Eugene, Oregon, USA) and glycerol. Subsequently, the leukocyte cells were visualized by using on Olympus BX51 fluorescence microscope with the 595 nm filter for Texas Red Dye and leukocyte concentration determined by analysing the relative ratio of CD45 positive cells to sperm in at least 300 high power fields.
Statistical analysis
Data were analysed using GraphPad Prism (GraphPad Software Inc.,La Jolla, CA,USA). Correlations between variables were analysed using the Spearman Rank Order correlation test. Sperm quality parameters before and after antioxidant therapy were expressed as mean ± SD or median (inter-quartile ranges) and analysed using the pair t-test or Wilcoxon Signed Rank test, depending on whether the data followed a normal distribution. A P value <0.05 was considered statistically significant.
Results
Table 1 depicts sperm quality parameters for both the fertile and infertile groups. As only men with male factor infertility were recruited into the study, it was expected that the infertile group would have lower sperm quality than the fertile controls. There was a statistically significantly increased level of sperm DNA fragmentation (TUNEL) and semen reactive oxygen species production (NBT) in the infertile group compared to the fertile men. The relationship between sperm global DNA methylation and semen parameters such as sperm concentration (r = −0.330, p = 0.0289) and morphology (r = −0.394, p = 0.008) was significant, but no significant relationship was observed between levels of DNA methylation and sperm motility (r = 0.075, p = 0.624) or leukocyte concentration (r = −.21, p = 0.12).
Table 1.
Comparison of sperm quality between the infertile and fertile study groups. All values are expressed as mean ± standard deviation and analysed using the unpaired t-test
| Fertile n = 12 | Infertile n = 45 | ||
|---|---|---|---|
| Sperm quality parameters | Mean ± SD | Mean ± SD | P |
| Sperm Concentration (×106/mL) | 122.1 ± 79.8 | 36.17 ± 46.2 | <0.0001 |
| Motility (%) | 52.6 ± 7.0 | 36.5 ± 12.3 | <0.0001 |
| Normal Morphology (%) | 21.0 ± 6.6 | 6.7 ± 5.0 | <0.0001 |
| CD 45 (×106 cell/ml) | 0.89 ± 0.51 | 1.53 ± 1.30 | 0.136 |
| ROS Production (μg Formazan/107 sperm) | 15.4 ± 12.1 | 77.8 ± 58.9 | 0.0006 |
| DNA Fragmentation (TUNEL Positive %) | 10.3 ± 3.8 | 22.9 ± 9.1 | <0.0001 |
| Sperm Global DNA Methylation (au) | 104.7 ± 11.3 | 93.96 ± 30.99 | 0.095 |
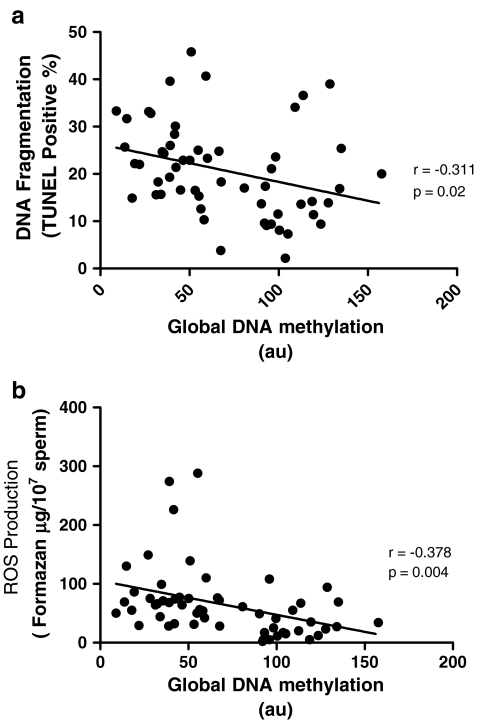
The results in Fig. 1a illustrate the correlation between levels of sperm DNA methylation and DNA fragmentation (TUNEL). It can be seen that a significant negative correlation exists between these two parameters (r = −0.311, p = 0.02), supporting a possible link between DNA damage and impaired capacity for methylation. The observation of a similar statistically significant negative correlation between sperm DNA methylation and seminal ROS production (r = −0.378, p = 0.004; Fig. 1b), together with a positive correlation between TUNEL and NBT results (r = 0.336, p = 0.022) suggests that oxidative damage to sperm DNA is at least in part responsible for modifying sperm global DNA hypomethylation.
Fig. 1.
Relationship between sperm global DNA methylation with sperm DNA fragmentation (a) and seminal ROS production (b). Statistical analysis was performed using the Spearman Rank Order correlation test
No significant correlation was observed between serum homocysteine and global sperm DNA methylation in the infertile cohort (r = 0.26, p = 0.09). Furthermore, sperm global DNA methylation levels did not differ significantly between those infertile men with “normal” serum homocysteine range (<11.4 mmol/L—95th percentile in folate replete men aged 20–45 years)[44] and those with hyper-homocysteinaemia (mean sperm DNA methylation 91.9 ± 29.7 v 94.8 ± 35.9 a.u. respectively, p = 0.378).
Three months supplementation with antioxidant produced a significant fall in seminal ROS levels and DNA damage (TUNEL), combined with an increase in sperm global DNA hypomethylation (Table 2). Sperm count, motility and morphology did not change significantly over the 3 month supplementation period, while serum homocysteine levels decreased a small but statistically significant amount.
Table 2.
Sperm quality before and after 3 months of anti-oxidant supplementation. Sperm parameters are expressed as mean ± standard deviation or median (inter-quartile range) depending of their normal distribution. Parametric data was analyzed by paired t-test and non-parametric data with the Wilcoxon signed rank test
| Sperm quality parameters n = 45 | Pre-antioxidant supplementation | On anti-oxidant supplementation | P |
|---|---|---|---|
| Sperm Concentration (×106/mL) | 21.7(12.08–38.33) | 21.4(12.75–44.1) | 0.304 |
| Motility (%) | 36.5 ± 12.3 | 37.1 ± 11.5 | 0.778 |
| Normal Morphology (%) | 6.7 ± 5.0 | 6.71 ± 4.5 | 0.938 |
| Homocysteine (μmol/L) | 10.00 (9.13–11.68) | 9.38 (8.50–10.90) | 0.001 |
| ROS Production (μg Formazan/107 sperm) | 66.4 (43.5–88.0) | 44.4 (33.3–81.4) | 0.027 |
| DNA Fragmentation (TUNEL Positive%) | 22.2 (16.5–26.6) | 18.2 (13.4–23.1) | 0.002 |
| Sperm Global DNA Methylation (a.u.) | 93.96 ± 30.99 | 108.61 ± 35.54 | 0.0017 |
Discussion
While several papers have now reported that infertile men’s sperm are more likely to express aberrant DNA methylation patterns [7–11, 40], this study is one of the first to report on the underlying mechanisms behind such observations. The results of our study suggest that oxidative damage to sperm DNA integrity inhibits methylation, while abnormalities in the methyl donor pathway do not appear to play a significant role in sperm DNA methylation. These results are consistent with the small body of evidence already existing. Firstly, a recent report [45] also found a statistically significant negative correlation between sperm DNA fragmentation, as assessed by the Sperm Chromatin dispersion Test, and sperm DNA methylation. A similar negative trend (r = −0.45, P > 0.05) had earlier been reported between sperm DNA fragmentation (TUNEL) and methylation [20]. Unfortunately neither group of researchers measured seminal ROS levels in their study, making it impossible to confirm that oxidative stress was the underlying cause of the observed DNA damage/hypomethylation. Interestingly, Tavalaee et al. (2009) explained their results by suggesting that hypomethylated sperm may be more prone to DNA damage [45]. While it is possible that normally methylated DNA may be less susceptible to DNA damage, we are unaware of any evidence supporting the concept that methylation of DNA protects it from apoptotic or oxidative damage, the two principal causes of sperm DNA damage [46]. Furthermore, Tavalaee et al. [45] observed no link between protamination of the sperm (CMA3 staining) and global DNA methylation, making a non specific defect in chromatin remodelling unlikely. As oxidative stress effects a significant proportion of infertile men [47] and is widely believed to be the primary cause of sperm DNA fragmentation [46, 48], it is likely that oxidative DNA damage played some role in sperm DNA hypomethylation reported in these earlier studies.
The link between oxidative DNA damage and hypomethylation is already established for somatic cells, with several investigators reporting a link between the presence of oxidative DNA adducts in somatic cells and impaired DNA methyltrasferase activity [29, 30]. Furthermore, incorporation of 8-OHdG in the methyl-CpG binding protein (MBP) recognition sequence has been reported to result in significant inhibition of MBP binding, further impeding the process of DNA methylation [31]. The observation of a statistically significant negative correlation between sperm DNA methylation and seminal ROS production, together with a strong positive correlation between TUNEL and NBT results strongly suggests that oxidative damage to sperm DNA is responsible for sperm global DNA hypomethylation. Furthermore, the observed improvement in sperm DNA methylation with 3 months antioxidant therapy also suggests that oxidative damage to sperm impairs DNA methylation. However, the link between sperm oxidative DNA damage and hypomethylation may best be confirmed by studies correlating the generation of the oxidative specific DNA base adduct 8-hydroxyl-2′-deoxyguanosine (8-OH-dG) with sperm DNA methylation.
While we did observe a very significant correlation between total semen ROS production and sperm DNA methylation, no significant correlation was observed between semen leukocyte concentration and sperm DNA methylation. This would imply that sperm themselves, not seminal leukocytes, are the primary source of ROS production interfering with the DNA methylation process. As none of the men in this study had any history suggestive of active genital tract infection, it is likely that the seminal leukocytes were relatively inactive and therefore not a dominant source of ROS production. Furthermore, it makes biological sense that intrinsic ROS production within the sperm cytoplasm is more likely to interfere with the process of sperm DNA methylation in the adjacent nucleus than extrinsic ROS released into the extra-cellular environment by leukocytes.
A weakness of our study was that it did not include a concurrent placebo control in the infertile subgroup. Some sperm parameters such as concentration and motility are prone to large fluctuations on different sampling occasions, even within the same individual, and therefore have a tendency for spontaneous “improvement” if subjects are recruited into a trial based on an initial low result. This non-treatment related improvement in sperm quality over time is termed “regression to the mean”. Conversely, failure to see significant fluctuations in sperm quality in a placebo control group over time suggests that regression to the mean is not an important determinant for that particular sperm parameter. As we did not have a placebo control in the infertile group, we can not state for certain that the observed improvements in sperm DNA methylation over three months of antioxidant therapy are not the result of spontaneous “regression to the mean”. However, since infertile participants were not selected for enrolment in the trial based on low initial sperm DNA methylation results, regression to the mean is unlikely to be a major cause for the observed improvement in sperm DNA methylation. Furthermore, the observed significant correlation between ROS production and sperm DNA methylation suggests a true biological cause-effect association between oxidative stress and impaired sperm DNA methylation.
Our finding of no significant correlation between serum homocysteine and sperm DNA methylation does not support a significant role for abnormalities in the folate/ homocysteine pathway as a cause of sperm DNA hypomethylation. Infertile men are more prone to inefficient folate cycle reconversion of homocysteine to methionine as polymorphisms in their MTHFR gene are more common [36–39, 49, 50], resulting in an up to 70% reduction in MTHFR activity. However, the lack of a significant negative correlation between sperm DNA methylation and homocysteine makes mutations in the MTHFR gene and hyperhomocysteinaemia unlikely candidates for causing sperm DNA hypomethylation in infertile men. A weakness of our study was that we did not specifically target men with MTHFR homozygous mutations and poor dietary folate intake who are likely to have extremely high levels of serum homocysteine. Such extreme abnormalities in homocysteine metabolism may still potentially be associated with alterations in sperm DNA methylation, despite the evidence suggesting that minor elevations in homocystine do not impact on sperm DNA methylation status.
The clinical implications of impaired sperm DNA methylation are presently uncertain. Animal studies have shown that chemically blocking sperm DNA methylation results in reduced sperm fertilization capacity, altered embryonic gene expression and an increase in pre-implantation embryonic loss [12–16] . Furthermore, the creation of mouse embryos using sperm with high degrees of DNA damage has been shown to result in epigenetic abnormalities in the resulting progeny with major physical and behavioural abnormalities later in life [51]. Aitken and De Iuliis have speculated that aberrant sperm DNA methylation may also lead to epigenetic defects that adversely affecting the health of the next generation of children [48]. Imprinting disorders such as Beckwith-Wiedermann and Angelman syndromes are relatively rare, making epidemiological linkage between infertility and the development of these imprinting disorders extremely difficult. As such, the link between sperm DNA methylation and the development of childhood imprinting disorders is still far from proven. Russell-Silver Syndrome, a rare disorder characterized by growth restriction, limb and facial anomalies and learning difficulties has been shown to be primarily caused by hypomethylation on the paternal allele of DMR1 at 11p15 [52, 53]. It is interesting to note that hypomethylation of the 11p15 DMR1 has also been reported to be more common in oligospermic infertile men [8], and at least one case report has linked the use of IVF-ICSI for severe male factor infertility with the development of Russell-Silver Syndrome. However, the rare association of IVF-ICSI treatment with any form of imprinting syndrome [54] suggests that aberrant sperm DNA methylation is not a significant cause for any classical imprinting syndrome.
The majority of human studies have suggested a negative link between sperm DNA methylation status and chances of pregnancy [9, 19, 20], with only one study failing to report such a link [45]. A complicating factor in determining the direct effect of sperm DNA methylation on pregnancy outcome is its positive association with sperm DNA integrity [20, 45]. Since sperm DNA fragmentation is clearly linked with pregnancy outcome [55] it is virtually impossible to determine if sperm DNA fragmentation alone or hypomethylation are primarily responsible for pregnancy outcome. Future experiments correlating sperm DNA methylation status with the embryonic epigenetic profile may shed light on the role that sperm DNA methylation plays in early embryo development.
The results of this study suggest that antioxidant supplementation in infertile men can result in significant improvements in sperm DNA integrity and methylation. Improvements in sperm DNA integrity with antioxidant therapy has been reported by many previous investigators, yet the ability of antioxidants to boost pregnancy rates is still under considerable debate [47]. Antioxidant supplements will not be capable of normalizing sperm DNA hypomethylation in all infertile men as in some individuals hypomethylation of individual gene loci is related to mutations within the DNMT3L gene [27], not oxidative stress. Furthermore, while the role that impaired sperm DNA integrity and methylation plays in the health of the next generation has yet to be determined, we can speculate that antioxidant mediated improvements in sperm DNA quality has at least the potential to benefit reproductive outcomes [55]. Large prospective studies correlating sperm DNA quality in the insemination sample with epigenetic profiles and the heath outcomes in the resulting children are urgently required. If these studies do confirm a link between poor sperm DNA quality and adverse child health outcomes, pre-conception anti-oxidant supplements may become standard clinical practice, just as pre-conception folate supplementation is the standard for women. Until these studies are conducted, the absolute value of male pre-conception antioxidant supplementation will be unknown.
Acknowledgements
The authors wish to thank all the participants and the staff of Repromed, especially Margaret Szemis for contribution to data collection and Dr. Megan Mitchell for her support in the conduct of the methylation assays. The authors acknowledge financial support from The Colin Matthews Research Fund (University of Adelaide) and Bayer Consumer Care, Australia. Ms Tunc is a recipient of a Faculty of Health Sciences Postgraduate Scholarship from The University of Adelaide.
Footnotes
Capsule
Oxidative stress impedes sperm DNA methylation, while antioxidant supplementation can reduce DNA damage and normalize sperm DNA methylation.
References
- 1.Cox GF, et al. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet. 2002;71(1):162–4. [DOI] [PMC free article] [PubMed]
- 2.Gosden R, et al. Rare congenital disorders, imprinted genes, and assisted reproductive technology. Lancet. 2003;361(9373):1975–7. [DOI] [PubMed]
- 3.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72(1):156–60. [DOI] [PMC free article] [PubMed]
- 4.Lawrence LT, Moley KH. Epigenetics and assisted reproductive technologies: human imprinting syndromes. Semin Reprod Med. 2008;26(2):143–52. [DOI] [PubMed]
- 5.Schaefer CB, et al. Epigenetic decisions in mammalian germ cells. Science. 2007;316(5823):398–9. [DOI] [PubMed]
- 6.Thompson JG, et al. Epigenetic risks related to assisted reproductive technologies: short- and long-term consequences for the health of children conceived through assisted reproduction technology: more reason for caution? Hum Reprod. 2002;17(11):2783–6. [DOI] [PubMed]
- 7.Benchaib M, et al. Quantitation by image analysis of global DNA methylation in human spermatozoa and its prognostic value in in vitro fertilization: a preliminary study. Fertil Steril. 2003;80(4):947–53. [DOI] [PubMed]
- 8.Marques CJ, et al. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363(9422):1700–2. [DOI] [PubMed]
- 9.Kobayashi H, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16(21):2542–51. [DOI] [PubMed]
- 10.Houshdaran S, et al. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS ONE. 2007;2(12):e1289. [DOI] [PMC free article] [PubMed]
- 11.Marques CJ, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14(2):67–74. [DOI] [PubMed]
- 12.Doerksen T, Trasler JM. Developmental exposure of male germ cells to 5-azacytidine results in abnormal preimplantation development in rats. Biol Reprod. 1996;55(5):1155–62. [DOI] [PubMed]
- 13.Doerksen T, Benoit G, Trasler JM. Deoxyribonucleic acid hypomethylation of male germ cells by mitotic and meiotic exposure to 5-azacytidine is associated with altered testicular histology. Endocrinology. 2000;141(9):3235–44. [DOI] [PubMed]
- 14.Kelly TL, Li E, Trasler JM. 5-aza-2′-deoxycytidine induces alterations in murine spermatogenesis and pregnancy outcome. J Androl. 2003;24(6):822–30. [DOI] [PubMed]
- 15.Oakes CC, et al. Adverse effects of 5-aza-2′-deoxycytidine on spermatogenesis include reduced sperm function and selective inhibition of de novo DNA methylation. J Pharmacol Exp Ther. 2007;322(3):1171–80. [DOI] [PubMed]
- 16.Pathak S, et al. Effect of tamoxifen treatment on global and insulin-like growth factor 2-H19 locus-specific DNA methylation in rat spermatozoa and its association with embryo loss. Fertil Steril. 2008;91(Suppl 5):2253–63. [DOI] [PubMed]
- 17.Anway MD, Skinner MK. Epigenetic programming of the germ line: effects of endocrine disruptors on the development of transgenerational disease. Reprod Biomed Online. 2008;16(1):23–5. [DOI] [PMC free article] [PubMed]
- 18.Anway MD, et al. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308(5727):1466–9. [DOI] [PMC free article] [PubMed]
- 19.Cisneros FJ. DNA methylation and male infertility. Front Biosci. 2004;9:1189–200. [DOI] [PubMed]
- 20.Benchaib M, et al. Influence of global sperm DNA methylation on IVF results. Hum Reprod. 2005;20(3):768–73. [DOI] [PubMed]
- 21.Trasler JM, et al. DNA methyltransferase is developmentally expressed in replicating and non-replicating male germ cells. Nucleic Acids Res. 1992;20(10):2541–5. [DOI] [PMC free article] [PubMed]
- 22.Numata M, Ono T, Iseki S. Expression and localization of the mRNA for DNA (cytosine-5)- methyltransferase in mouse seminiferous tubules. J Histochem Cytochem. 1994;42(9):1271–6. [DOI] [PubMed]
- 23.Jue K, et al. Developmental and hormonal regulation of DNA methyltransferase in the rat testis. Biol Reprod. 1995;52(6):1364–71. [DOI] [PubMed]
- 24.Kerjean A, et al. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum Mol Genet. 2000;9(14):2183–7. [DOI] [PubMed]
- 25.Omisanjo OA, et al. DNMT1 and HDAC1 gene expression in impaired spermatogenesis and testicular cancer. Histochem Cell Biol. 2007;127(2):175–81. [DOI] [PubMed]
- 26.Ariel M, Cedar H, McCarrey J. Developmental changes in methylation of spermatogenesis-specific genes include reprogramming in the epididymis. Nat Genet. 1994;7(1):59–63. [DOI] [PubMed]
- 27.Kobayashi H, et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet. 2009; in press [DOI] [PMC free article] [PubMed]
- 28.Franco R, et al. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266(1):6–11. [DOI] [PubMed]
- 29.Weitzman SA, et al. Free radical adducts induce alterations in DNA cytosine methylation. Proc Natl Acad Sci USA. 1994;91(4):1261–4. [DOI] [PMC free article] [PubMed]
- 30.Turk PW, et al. DNA adduct 8-hydroxyl-2′-deoxyguanosine (8-hydroxyguanine) affects function of human DNA methyltransferase. Carcinogenesis. 1995;16(5):1253–5. [DOI] [PubMed]
- 31.Valinluck V, et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 2004;32(14):4100–8. [DOI] [PMC free article] [PubMed]
- 32.Hepburn PA, Margison GP, Tisdale MJ. Enzymatic methylation of cytosine in DNA is prevented by adjacent O6-methylguanine residues. J Biol Chem. 1991;266(13):7985–7. [PubMed]
- 33.Yi P, et al. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem. 2000;275(38):29318–23. [DOI] [PubMed]
- 34.Jamaluddin MD, et al. Homocysteine inhibits endothelial cell growth via DNA hypomethylation of the cyclin A gene. Blood. 2007;110(10):3648–55. [DOI] [PMC free article] [PubMed]
- 35.Chen Z, et al. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum Mol Genet. 2001;10(5):433–43. [DOI] [PubMed]
- 36.Bezold G, Lange M, Peter RU. Homozygous methylenetetrahydrofolate reductase C677T mutation and male infertility. N Engl J Med. 2001;344(15):1172–3. [DOI] [PubMed]
- 37.Stuppia L, et al. The methylenetethrahydrofolate reductase (MTHFR) C677T polymorphism and male infertility in Italy. J Endocrinol Investig. 2003;26(7):620–2. [DOI] [PubMed]
- 38.Singh K, et al. Mutation C677T in the methylenetetrahydrofolate reductase gene is associated with male infertility in an Indian population. Int J Androl. 2005;28(2):115–9. [DOI] [PubMed]
- 39.Paracchini V, Garte S, Taioli E. MTHFR C677T polymorphism, GSTM1 deletion and male infertility: a possible suggestion of a gene-gene interaction? Biomarkers. 2006;11(1):53–60. [DOI] [PubMed]
- 40.Benchaib M, et al. Sperm DNA fragmentation decreases the pregnancy rate in an assisted reproductive technique. Hum Reprod. 2003;18(5):1023–8. [DOI] [PubMed]
- 41.Gandini L, et al. Study of apoptotic DNA fragmentation in human spermatozoa. Hum Reprod. 2000;15(4):830–9. [DOI] [PubMed]
- 42.Tunc O, Thompson J, Tremellen K. Development of the NBT assay as a marker of sperm oxidative stress. Int J Androl. 2008. doi:10.1111/j.1365-2605.2008.00941.x. [DOI] [PubMed]
- 43.Esfandiari N, et al. Utility of the nitroblue tetrazolium reduction test for assessment of reactive oxygen species production by seminal leukocytes and spermatozoa. J Androl. 2003;24(6):862–70. [DOI] [PubMed]
- 44.Selhub J, et al. Serum total homocysteine concentrations in the third National Health and Nutrition Examination Survey (1991–1994): population reference ranges and contribution of vitamin status to high serum concentrations. Ann Intern Med. 1999;131(5):331–9. [DOI] [PubMed]
- 45.Tavalaee M, Razavi S, Nasr-Esfahani MH. Influence of sperm chromatin anomalies on assisted reproductive technology outcome. Fertil Steril. 2009;91(4):1119–26. [DOI] [PubMed]
- 46.Aitken RJ. Founders’ lecture. Human spermatozoa: fruits of creation, seeds of doubt. Reprod Fertil Dev. 2004;16(7):655–64. [DOI] [PubMed]
- 47.Tremellen K. Oxidative stress and male infertility—a clinical perspective. Hum Reprod Updat. 2008;14(3):243–58. [DOI] [PubMed]
- 48.Aitken RJ, De Iuliis GN. Origins and consequences of DNA damage in male germ cells. Reprod Biomed Online. 2007;14(6):727–33. [DOI] [PubMed]
- 49.Lee CR, et al. Genetic variation in soluble epoxide hydrolase (EPHX2) and risk of coronary heart disease: the Atherosclerosis Risk in Communities (ARIC) study. Hum Mol Genet. 2006;15(10):1640–9. [DOI] [PMC free article] [PubMed]
- 50.Dhillon VS, Shahid M, Husain SA. Associations of MTHFR DNMT3b 4977 bp deletion in mtDNA and GSTM1 deletion, and aberrant CpG island hypermethylation of GSTM1 in non-obstructive infertility in Indian men. Mol Hum Reprod. 2007;13(4):213–22. [DOI] [PubMed]
- 51.Fernandez-Gonzalez R, et al. Long-term effects of mouse intracytoplasmic sperm injection with DNA-fragmented sperm on health and behavior of adult offspring. Biol Reprod. 2008;78(4):761–72. [DOI] [PubMed]
- 52.Gicquel C, et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat Genet. 2005;37(9):1003–7. [DOI] [PubMed]
- 53.Netchine I, et al. 11p15 imprinting center region 1 loss of methylation is a common and specific cause of typical Russell-Silver syndrome: clinical scoring system and epigenetic-phenotypic correlations. J Clin Endocrinol Metab. 2007;92(8):3148–54. [DOI] [PubMed]
- 54.Amor DJ, Halliday J. A review of known imprinting syndromes and their association with assisted reproduction technologies. Hum Reprod. 2008;23(12):2826–34. [DOI] [PubMed]
- 55.Zini A, et al. Sperm DNA damage is associated with an increased risk of pregnancy loss after IVF and ICSI: systematic review and meta-analysis. Hum Reprod. 2008;23(12):2663–8. [DOI] [PubMed]