Abstract
Purpose
To analyze using hypergeometric probability statistics the impact of performing preimplantation genetic screening (PGS) on a cohort of day 3 cleavage stage embryos.
Methods
Statistical mathematical modeling.
Results
We find the benefit of performing PGS is highly dependent on the number of day 3 embryos available for biopsy. Additional hidden variables that determine the outcome of PGS are the rates of aneuploidy and mosaicism, and the probability of a chromosomally mosaic embryo to test “normal”. If PGS is performed, our analysis shows that many combinations of the number of biopsiable embryos, and the rates of aneuploidy and mosaicism results in a marginal benefit from the intervention. Other combinations are detrimental if PGS is actually undertaken. Finally, increases in PGS error rates lead to a rapid loss in the ability of PGS to provide useful discriminatory information.
Conclusion
We set out the statistical framework to determine the limits of PGS when a specific number of day 3 preimplantation embryos are available for biopsy. In general, PGS cannot be recommended a priori for a specific clinical situation due to the statistical uncertainties associated with the different hidden variable quantitative parameters considered important to the clinical outcome.
Keywords: Preimplantation genetic screening, Mathematical modeling, Hypergeometric statistics
Introduction
Preimplantation genetic screening (PGS) combines two basic techniques to assess the numerical chromosome complement of human cleavage stage embryos: blastomere biopsy of day 3 embryos, typically at the 6–8 cell stage after approximately three cell division cycles, and single cell multicolor fluorescence in situ hybridization (FISH) using complementary DNA sequences tagged with specific fluorochromes. The latter technique allows for the analysis of a limited set of chromosomes, typically 7 to10, of the interphase nucleus from a single blastomere. Early studies of the chromosomal complement of cleavage stage human embryos using FISH technology showed that a significant proportion have numerical chromosomal abnormalities or aneuploidies [1–7]. This observation made for a compelling argument to use PGS in older couples undergoing in vitro fertilization (IVF). It was reasoned if abnormal embryos with limited potential for development and implantation were eliminated (from a cohort of embryos) prior to embryo transfer there would be a corresponding increase in IVF success rates. This intuitively attractive notion was readily embraced and early observational studies reinforced this belief. However, more recent studies have raised doubts on the utility of PGS as presently practiced [8].
Based on the results of observational studies it was proposed that patients of advanced maternal age, typically those greater than 35–38 years of age, might benefit from PGS [9]. Several retrospective studies reported improved implantation rates and ongoing pregnancy in women of advanced maternal age who underwent PGS [10, 11]. The same group also reported improved implantation rates and pregnancy rates in a randomized, control study of women of advanced maternal age who underwent PGS when compared to an age-matched control group [12]. By contrast, a large prospective randomized controlled clinical trial (RCT), with or without PGS, in women greater than 37 years of age did not demonstrate a statistically significant improvement in clinical outcome when there was no restriction on the number of embryos used for transfer [13]. A recent PGS RCT in women of advanced maternal age also failed to show an improved outcome between the treatment and control groups [14]. The results of these two studies were, however, criticized by others on technical and methodological grounds [15–22]. Subsequently, Hadarson et al. [23] published the results of a PGS RCT in women of advanced maternal age which showed that women in the PGS group had lower pregnancy rates. Overall, the results of these studies did not support a role for PGS in women of advanced maternal age. Two recent studies of younger women undergoing PGS also failed to show a benefit [24, 25]. The latter study used trophectoderm biopsy of day 5–6 blastocysts. By contrast, a small RCT conducted by Mersereau et al. [26] in an unselected group of younger patients showed a trend towards a higher pregnancy rate in the PGS group.
Others have also questioned a clinical role for PGS [27–29]. Some clinicians have argued of the need for larger RCTs [25, 26, 30–32]. And yet others have reasoned there is no need to undertake additional RCTs as well-designed studies (cited above) have already demonstrated no benefit of PGS and to undertake additional studies is unethical in light of the findings from some of the studies that performing PGS may, in fact, be harmful [33, 34].
There are two potential errors of chromosome segregation resulting in genetic aneuploidy and mosaicism in cleavage-stage embryos: meiotic and post-zygotic errors. The simple aneuploidies, homogeneous monosomies and trisomies, are typically considered the result of errors of chromosome segregation that occur in meiosis I and II during the process of oocyte maturation and fertilization. By contrast, chromosomal mosaicism is considered a post-zygotic event occurring mainly during the first 3 mitotic cell divisions of preimplantation embryo development (see: “Discussion”).
The aim of the present study is to develop a mathematical model of PGS and describe the statistical framework for determining whether the procedure should or should not be undertaken on a defined number of biopsiable embryos. The current model assumes the biopsy procedure is performed on day 3 of embryo development. We have initially directed our analysis to day 3 biopsied embryos as the majority of appropriately-designed clinical studies described thus far have used day 3 PGS/FISH protocols. We show the full dependence of the probability of success on a number of clinical situations and embryo parameters. The model further assumes that the simple or full aneuploidies and chromosomal mosaicism arise by independent events. Input data on the rates of aneuploidy and mosaicism have been chosen to fall within clinically determined values. In the present analysis we compare the outcome of either performing (treatment) or not performing (control) PGS on a defined number of day 3 embryos, and determine a PGS advantage defined as g; when g is greater than 0.05 there is a PGS advantage. We show the outcome of PGS is dependent on the number of biopsiable day 3 embryos, the rates of aneuploidy and mosaicism, and the probability of a mosaic embryo to test as chromosomally “normal”. For many situations the overall benefits of PGS are shown to be marginal. In other situations performing PGS can actually be harmful. Our mathematical modeling studies were motivated in part by the daunting barriers to full RCTs with acceptably small statistical uncertainties. It should be noted that many of the important quantitative parameters expected to influence the success rate of PGS have been measured with some degree of certainty, or have much lower barriers to measurement. In such a case with well-defined inputs, a mathematical framework may allow us to predict outcomes with low uncertainties than can be obtained by RCTs of practical size. Finally, we discuss some of the limitations of mathematical modeling, particularly with regard to some of the clinical uncertainties that currently exist in relation to the developmental potential and viability of embryos with limited mosaicism.
Methods
We present a model for evaluating the impact of PGS upon the probability that at least two normal diploid embryos are present in utero on day 5 of embryo development. The model is a relatively straightforward application of hypergeometric statistics to a selection problem with hidden variables [35]. Hypergeometric statistics are appropriate for describing the probability to obtain a given number of successes, given a fixed number of draws from a population with a known number of successful and unsuccessful samples. Importantly, the analysis is distinguished from binomial statistics because the hypergeometric samples are done without replacement and therefore the probabilities on successive draws are not equal and independent of one another as in a binomial distribution, ie., the probability distributions are different [35]. In the following, “aneuploid” and “mosaic” have the obvious meanings. By definition we label normal embryos as meaning all cells in the embryo are fully diploid, and thus are neither aneuploid nor mosaic. “Abnormal” embryos are those that are either aneuploid or mosaic. In this section we provide a brief overview of the parameters used to assess the overall benefits of PGS. A more detailed mathematical analysis is summarized in the Addendum.
On day 3 of embryo culture we evaluate the decision to perform PGS based upon the variable input parameters listed below:
∎ α3 : aneuploidy rate for the embryos on day 3.
∎ μ3: mosaicism rate for the embryos on day 3.
∎ μe: probability that a cell taken from a mosaic embryo tests normal.
∎ ηN, ηA: the misdiagnosis rate, for truly normal and truly abnormal embryos, respectively.
∎ T: the desired number of transferred chromosomally (truly) normal embryos that survive to day 5; this parameter defines the success of the transfer.
∎ t3 : the maximum number of embryos that might be transferred on day 3, if PGS is not performed.
∎ t5f, t5p: the maximum number of (apparently) normal embryos to be transferred on day 5, either without (t5f) or with (t5p) PGS.
∎ g: the minimum required gain (in probability to transfer at least T normal embryos) required to make PGS justified relative to its costs and difficulties.
∎ N : the number of biopsiable embryos on day 3.
∎ ρuM, ρvM, ρuAn, ρvAn: the loss rate (mortality) from day 3 to day 5, for mosaic or abnormal (M or An) embryos in utero or in vitro (u or v).
∎ ρuN, ρvN: the loss rate (mortality) from day 3 to day 5, for normal embryos in utero or in vitro (u or v).
For notational ease, we make use of the following parameters, calculated from those above:
∎ μN, μA: mosaicism-induced misdiagnosis rate, for truly normal and truly abnormal embryos, respectively
∎ α: total rate for abnormal (mosaic or aneuploid) embryos on day 3.
∎ ρuA, ρvA: the loss (mortality) rate from day 3 to day 5, for abnormal embryos in utero or in vitro (u or v).
By “truly normal” we refer to those embryos that, if they were to implant produce a normal diploid fetus. To date, there is still some uncertainty of exactly how this is best determined in practice (see “Discussion”). Similarly, by “truly aneuploid” we refer to those embryos that will fail, for chromosomal reasons, to produce a diploid fetus. The mosaicism and misdiagnosis rates should be evaluated by clinical criteria: that is, by the rate at which embryos get assigned to the wrong category of suitability for transfer. For clarity and generality we have separated mosaicism and misdiagnosis, although mathematically in the current model their effects are the same. It should be noted that the primary end-point of the analysis is the probability of having two normal diploid embryos in utero on day 5 of embryo development following PGS. We have not addressed in the present analysis the implantation potential of a biopsied embryo regardless of genetic constitution.
We determine if PGS increases the probability to transfer at least T truly normal embryos surviving to day 5. If this probability gain is greater than g, then PGS is warranted; if less, it is not. Given the relative costs of PGS to a full cycle, and current national average per-cycle success rates, we set g equal to 5%. Thus, the PGS gain needs to exceed 0.05 to show an advantage.
The biological origin of the values of the various parameters α, ρvN, etc, is an interesting question but not directly germane to the calculation at hand. Similarly, we leave largely unaddressed how the values of these various parameters can best be determined. Suffice to mention these are quantifiable parameters and are likely to be clinic-dependent.
Several of the quantities described above are derivatives of other parameters. The quantity α can be straightforwardly calculated as 1-(1-α3)(1-μ3). Defining fM = (1-α3)μ0, fAn = (1-μ3)(α3), and fb = μ3α3/α, the loss (mortality) rates can be formally calculated as:
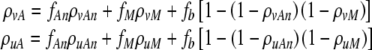 |
1 |
Finally, we can set μA equal to μe(1-α3). For practical purposes, μN may be always set to 0, as mosaic by definition in the current model is not “normal”. However, the value of μN may be set to non-zero should the reader wish to allow for the possibility of truly mosaic embryos that nonetheless end up developing normally (see “Discussion”).
The strategy of the calculation is to calculate the joint probability distribution, given the parameters, to find:
A number
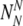 of chromosomally (truly) normal embryos labeled by PGS as normal
of chromosomally (truly) normal embryos labeled by PGS as normal 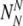
A number
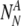 of chromosomally (truly) aneuploid embryos labeled by PGS as normal
of chromosomally (truly) aneuploid embryos labeled by PGS as normal 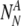
then, given the maximum number of transferred embryos t5p, to calculate the probability to transfer at least T embryos out of 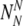 which survive to day five. In the following, we make use of the following notations
which survive to day five. In the following, we make use of the following notations 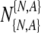 , where the superscript refers to the true normality (N) or aneuploidy (A) of the embryo, and the subscript refers to the label it receives following PGS. Again, these labels are the clinically relevant labels. We also notate the quantity
, where the superscript refers to the true normality (N) or aneuploidy (A) of the embryo, and the subscript refers to the label it receives following PGS. Again, these labels are the clinically relevant labels. We also notate the quantity 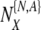 as the number of transferred embryos. In order to avoid the proliferation of subscripts, we sometimes use unsubscripted μ, η, and ρ in argument lists to refer to the set of relevant subscripted variables, as understood both from context and from the equations that follow the argument list.
as the number of transferred embryos. In order to avoid the proliferation of subscripts, we sometimes use unsubscripted μ, η, and ρ in argument lists to refer to the set of relevant subscripted variables, as understood both from context and from the equations that follow the argument list.
The average number of labeled normal and aneuploid embryos (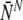 and
and 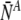 , respectively) can be straightforwardly encapsulated in matrix form as:
, respectively) can be straightforwardly encapsulated in matrix form as:
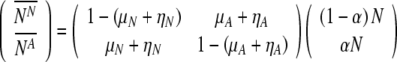 |
2 |
However, the probability to transfer at least T truly normal embryos depends not only on these averages, but also their full distribution; indeed, on their full joint distribution. To enumerate and subsequently maximize this probability, we must consider hypergeometric statistics [35].
We consider first a simple case of a day 3 transfer without PGS.
This first case is relatively simple: calculating the probability of at least T truly normal embryos surviving to day 5 in utero, given a transfer of t3 total embryos. With no information about the embryos, they are considered equally likely to be chosen whether normal or aneuploid. This corresponds to the case in which embryos are chosen on day 3 for transfer, without any benefit of PGS The underlying probability PN that NN of the N embryos are truly normal is given by:
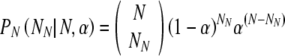 |
3 |
where 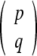 means the number of ways to choose q items from a collection of p without regard to order, calculated as
means the number of ways to choose q items from a collection of p without regard to order, calculated as 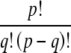 .
.
Given NN truly normal embryos (and NA = N-NN aneuploid), the likelihood of selecting T or more normal embryos for transfer is given by a hypergeometric distribution. Importantly, it is not binomial because the sampling is done without replacement. We notate PH(i|S,N,t) as the hypergeometric distribution describing the probability to select i successes from a sample of size N containing S successful samples, when drawing t samples:
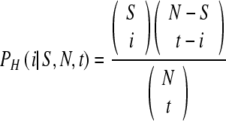 |
4 |
We sum over all possible values of NN, weighting each value with the probability of Eq. 3. For each value of NN, we sum the probability that exactly Tt are selected (out of the sample of size NN and NA); for each value of Tt, we sum the probabilities that exactly Ts survive to day 5, for all Ts ≥ T, and with the probability of Ts given based on ρNu.
First: if we transfer exactly Tt embryos, what is the probability Pts(Tt, Ts,ρ) that exactly Ts of them survive? It is
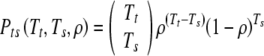 |
5 |
as is common in such equations, 00 is understood to equal 1 (as it is the quantity ρ which can continuously approach zero).
This gives us, for the probability to have T surviving embryos on day 5, given no PGS and day 3 transfer:
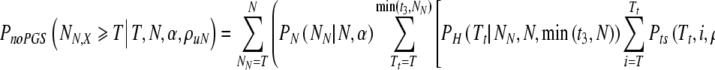 |
6 |
The corresponding equation for a day 5 transfer, without PGS is given as:
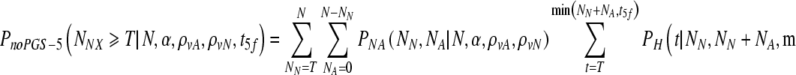 |
9 |
And for transfer on day 5 with PGS as:
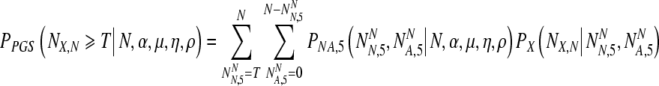 |
19 |
Derivations for Eqs. 9 and 19 are given in the “Addendum”. As shown, culture may be extended to day 5 without performing PGS as a means to “filter out” abnormal embryos, ie., the probability of having T surviving embryos on day 5 after PGS on day 3 and on day 5 without PGS.
Results
Schematically, we define the quantity ΔP ≡ PPGS(NX,N ≥ T | N, α)—Pno PGS(NX,N ≥ T | N, α), where the functions are chosen appropriately to the non-PGS transfer (either day 3 or day 5 day). PGS is warranted if ΔP ≥ g. When ΔP > 0, PGS is superior to not doing PGS. We also define the cost function C to account for the advantages of avoiding a loss (ie., miscarriage or failed cycle):
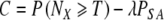 |
21 |
Here the value of λ is chosen subjectively to reflect the relative weighting of desire to transfer sufficient embryos and desire to avoid miscarriages. Again, the function P is chosen appropriate to the options being compared. C can be calculated with and without PGS being performed (with the value of P(NX ≥ T) and PSA, the loss rate, calculated appropriately for the two cases.) We define ΔC as the difference between the two cases. A value of λ near 1/3 seems reasonable since many couples are likely to stop IVF after either a third failed cycle or loss. We use this value throughout the remainder of the paper, along with the value σ = 0.04.
It would be straightforward to extend the definition of C to incur a penalty for the possibility of multiple implantations and births. Under most circumstances, the values of t{3,5f,5p} are chosen to reduce this possibility. Moreover, we have chosen values that conform to reasonable clinical practice. However, these are quantities under clinical control and the effect of varying their values can be easily examined using the expressions given here.
In what follows, we examine a baseline case of clinical relevance that might typically be considered for undertaking PGS for advanced maternal age. We take T = 2, t3 = 3, t5f = 2, t5p = 2, and ηA = ηN = 0.05. We take μN = 0 and μe = 0.75. We take α3 = 0.3 and μ3 = 0.3, implying a value α of 0.51. We set ρvAn = ρuAn = 0.6; ρuM = ρvM = 0.9, and ρuN = 0.4, ρvN = 0.5. Finally, we set N = 6. We examine the dependence of the PGS efficacy on each of the major clinical variables in turn, while holding the remaining quantities in the baseline case fixed.
Theoretically, PGS confers two separate advantages, one of which can be had without performing PGS. The first advantage is the knowledge of the apparent genetic “quality” of embryos when choosing those to be transferred. The second is that when PGS is performed, the embryos are transferred on day 5. One may opt for a day 5 transfer without doing PGS. Thus, it is necessary to measure the effect of PGS per se, and examine the difference between day 5 transfers with and without PGS.
Table 1 shows the relative advantage (both in ΔP and ΔC) for a PGS transfer on day 5, compared to a day 5 transfer without PGS based on the number of biopsiable embryos on day 3. As shown, PGS is unlikely to provide substantial benefit unless there are many biopsiable embryos. It should be noted that the calculation assumes an abnormal embryo rate of 51% (α = 0.51). Increasing or decreasing the aneuploidy rate will shift the number of biopsiable embryos required to show a PGS advantage (see below).
Table 1.
The day 5 PGS advantage ΔP and ΔC as a function of the number of biopsiable embryos N when comparing day 5 transfer without PGS to day 5 transfer with PGS. The baseline case is bolded
| N | ΔP | ΔC |
|---|---|---|
| 2 | -0.01 | -0.00 |
| 3 | -0.01 | -0.00 |
| 4 | -0.00 | +0.00 |
| 5 | +0.01 | +0.01 |
| 6 | +0.02 | +0.03 |
| 7 | +0.04 | +0.04 |
| 8 | +0.06 | +0.06 |
| 9 | +0.07 | +0.07 |
| 10 | +0.08 | +0.09 |
| 11 | +0.10 | +0.10 |
| 12 | +0.10 | +0.11 |
Table 2 shows the advantage of day 5 transfer without PGS relative to a day 3 transfer. As clearly shown transfer on day 5 is generally a nearly equal contributor to success as PGS. This can be understood by recognizing that by waiting to transfer on day 5, embryos that are lost (arrest) between day 3 and day 5 can effectively be "replaced" (when there are sufficient embryos, N); this is not possible with a day 3 transfer. We note that for low numbers of day 3 embryos waiting to day 5 is harmful (see “Discussion”).
Table 2.
The advantage ΔP and ΔC as a function of the number of biopsiable embryos N, when comparing day 5 transfer without PGS to day 3 transfer. This table separates out the passive filtering that occurs with day 5 transfer without PGS. The baseline case is bolded
| N | ΔP | ΔC |
|---|---|---|
| 2 | -0.03 | -0.03 |
| 3 | -0.07 | -0.07 |
| 4 | 0.00 | 0.00 |
| 5 | +0.07 | +0.07 |
| 6 | +0.12 | +0.12 |
| 7 | +0.16 | +0.16 |
| 8 | +0.19 | +0.19 |
| 9 | +0.22 | +0.21 |
| 10 | +0.23 | +0.23 |
| 11 | +0.24 | +0.24 |
| 12 | +0.25 | +0.25 |
The total advantage of day 5 transfer following PGS relative to a day 3 transfer is shown in Table 3.
Table 3.
The advantage ΔP and ΔC as a function of the number of biopsiable embryos N, when comparing day 3 transfer to day 5 transfer with PGS. This is essentially a summation of the data of Tables 1 and 2. The baseline case is bolded
| N | ΔP | ΔC |
|---|---|---|
| 2 | -0.03 | -0.03 |
| 3 | -0.08 | -0.08 |
| 4 | +0.00 | +0.00 |
| 5 | +0.08 | +0.08 |
| 6 | +0.15 | +0.15 |
| 7 | +0.20 | +0.20 |
| 8 | +0.25 | +0.25 |
| 9 | +0.29 | +0.29 |
| 10 | +0.32 | +0.31 |
| 11 | +0.34 | +0.34 |
| 12 | +0.36 | +0.36 |
For the remaining cases, we examine only the PGS advantage (ΔP and ΔC) between day 5 transfer with or without PGS. Very roughly, the additional effect of waiting from day 3 to day 5 can be approximated by simply adding the appropriate quantity from Table 2.
We examine the dependence on the number of embryos N for high and low values of aneuploidy as summarized in Table 4. We modify the baseline case, and set for low aneuploidy (α3 = 0.15, μ3 = 0.15); for high aneuploidy we set (α3 = 0.4, μ3 = 0.5); these correspond to α = 0.28 and 0.7, respectively. The PGS advantage is greater at larger N and higher aneuploidy, as might be expected. However, as shown, performing PGS with low numbers of biopisable embryos incurs a penalty.
Table 4.
The advantage ΔP and ΔC as a function of the number of biopsiable embryos N, for cases of low and high abnormality α = 0.28 and 0.7, when comparing day 5 transfer with or without PGS. The baseline case is bolded
| N | ΔP (low α) | ΔC (low α) | ΔP (high α) | ΔC (high α) |
|---|---|---|---|---|
| 2 | -0.01 | -0.01 | -0.00 | +0.00 |
| 3 | -0.02 | -0.02 | -0.00 | +0.00 |
| 4 | -0.01 | -0.01 | -0.00 | +0.00 |
| 5 | -0.01 | -0.01 | +0.01 | +0.01 |
| 6 | +0.01 | +0.01 | +0.01 | +0.02 |
| 7 | +0.02 | +0.02 | +0.03 | +0.03 |
| 8 | +0.03 | +0.03 | +0.04 | +0.04 |
| 9 | +0.04 | +0.04 | +0.05 | +0.06 |
| 10 | +0.05 | +0.05 | +0.07 | +0.07 |
| 11 | +0.05 | +0.05 | +0.08 | +0.08 |
| 12 | +0.05 | +0.06 | +0.09 | +0.10 |
For a particular case of clinical interest, with N = 6, we also calculate ΔP and ΔC as a function of the misdiagnosis rate η for high and low aneuploidy rates. We use the same settings for high and low aneuploidy rates as in the previous section. Again, to isolate the effect of PGS, we compare day 5 success rates with and without PGS. We set both ηA and ηN to a single value. Table 5 shows ΔP and ΔC as functions of this value. For practical purposes the misdiagnosis rate will be dependent on technical factors associated with the PGS analysis, and will be different for each center performing PGS. Low error rates show a PGS advantage, whereas high rates are detrimental.
Table 5.
The day 5 PGS advantage ΔP and ΔC as function of the misdiagnosis rate η. The baseline case is bolded
| η | ΔP | ΔC |
|---|---|---|
| 0 | +0.05 | +0.06 |
| 0.0125 | +0.05 | +0.05 |
| 0.0250 | +0.04 | +0.04 |
| 0.0375 | +0.03 | +0.03 |
| 0.0500 | +0.02 | +0.03 |
| 0.0625 | +0.02 | +0.02 |
| 0.0750 | +0.01 | +0.01 |
| 0.0875 | +0.00 | +0.00 |
| 0.1000 | -0.01 | -0.00 |
For the cases with biopsiable day 3 embryos, N = 6 and N = 9, we examine ΔP and ΔC as functions of α3 (the aneuploidy rate) in Table 6. The concavity of the advantage is easily understood. When aneuploidy rates are low, PGS is unnecessary for selecting good embryos and provides little or no benefit. By contrast, when abnormalities are high PGS can do nothing other than inform the clinician that indeed, the abnormalities are high. Thus, high aneuploidy rates may provide diagnostic information, but offers little therapeutic advantage. In between, PGS provides discriminatory benefit. That the advantage must decline to zero is made immediately clear by considering the case as α approaches 100%-where the probability of success is 0% whether PGS is performed or not.
Table 6.
The day 5 PGS advantage ΔP and ΔC as functions of the aneuploidy rate α3, for cases of low and high numbers N of biopsiable embryos. The baseline case is bolded
| α3 | ΔP (N = 6) | ΔC (N = 6) | ΔP (N = 9) | ΔC (N = 9) |
|---|---|---|---|---|
| 0.0 | -0.02 | -0.02 | -0.00 | -0.00 |
| 0.1 | -0.01 | -0.00 | +0.02 | +0.02 |
| 0.2 | +0.01 | +0.01 | +0.05 | +0.05 |
| 0.3 | +0.02 | +0.03 | +0.07 | +0.07 |
| 0.4 | +0.03 | +0.04 | +0.09 | +0.09 |
| 0.5 | +0.04 | +0.04 | +0.10 | +0.10 |
| 0.6 | +0.04 | +0.04 | +0.10 | +0.10 |
| 0.7 | +0.03 | +0.04 | +0.08 | +0.09 |
| 0.8 | +0.02 | +0.03 | +0.05 | +0.06 |
| 0.9 | +0.01 | +0.02 | +0.02 | +0.04 |
For the cases N = 6 and N = 9, we examine ΔP and ΔC as functions of μ3 (the mosaicism rate) in Table 7. Firstly, we do not observe the same concavity effect as α3 for the mere reason that in this case we have held α3 at 30%, so that even for low values of μ3 there are substantial abnormalities for PGS to usefully discriminate against; this has important clinical consequences. As mosaicism rates increase the PGS gain is lost even with high numbers of biopsiable embryos.
Table 7.
The day 5 PGS advantage ΔP and ΔC as functions of the mosaicism rate μ3, for cases of low and high numbers N of biopsiable embryos. The baseline case is bolded
| μ3 | ΔP (N = 6) | ΔC (N = 6) | ΔP (N = 9) | ΔC (N = 9) |
|---|---|---|---|---|
| 0.000 | +0.05 | +0.05 | +0.10 | +0.10 |
| 0.075 | +0.04 | +0.05 | +0.09 | +0.10 |
| 0.150 | +0.04 | +0.04 | +0.09 | +0.09 |
| 0.225 | +0.03 | +0.03 | +0.08 | +0.08 |
| 0.300 | +0.02 | +0.03 | +0.07 | +0.07 |
| 0.375 | +0.02 | +0.02 | +0.06 | +0.06 |
| 0.450 | +0.01 | +0.02 | +0.05 | +0.05 |
| 0.525 | +0.01 | +0.01 | +0.04 | +0.04 |
| 0.600 | +0.01 | +0.01 | +0.03 | +0.03 |
| 0.675 | +0.00 | +0.01 | +0.02 | +0.02 |
| 0.750 | +0.00 | +0.01 | +0.01 | +0.02 |
| 0.825 | +0.00 | +0.00 | +0.01 | +0.01 |
| 0.900 | +0.00 | +0.00 | +0.00 | +0.01 |
For the cases N = 6 and N = 9, we examine ΔP and ΔC as functions of μe (the probability that a mosaic embryo tests “normal”) in Table 8. Over the clinically relevant range the variation is rather weak.
Table 8.
The day 5 PGS advantage ΔP and ΔC as functions of the probability μe for a mosaic to test as normal, for cases of low and high numbers N of biopsiable embryos. The baseline case is bolded
| μe | ΔP (N = 6) | ΔC (N = 6) | ΔP (N = 9) | ΔC (N = 9) |
|---|---|---|---|---|
| 0.000 | +0.09 | +0.09 | +0.22 | +0.22 |
| 0.125 | +0.08 | +0.08 | +0.19 | +0.19 |
| 0.250 | +0.07 | +0.07 | +0.16 | +0.17 |
| 0.375 | +0.05 | +0.06 | +0.14 | +0.14 |
| 0.500 | +0.04 | +0.05 | +0.13 | +0.14 |
| 0.625 | +0.03 | +0.04 | +0.11 | +0.12 |
| 0.750 | +0.02 | +0.03 | +0.07 | +0.07 |
| 0.875 | +0.01 | +0.02 | +0.05 | +0.05 |
| 1.000 | +0.00 | +0.01 | +0.03 | +0.03 |
Finally, we examine the effect of modifying the loss (mortality rates) for mosaic embryos. For high and low numbers of embryos N, we vary ρuM = ρvM, with the results shown in Table 9. This examines the filtering effect of selecting against mosaic embryos by transferring on day 5.
Table 9.
The day 5 PGS advantage ΔP and ΔC as functions of the loss (mortality) rate ρ(u/v)M for mosaic embryos, for cases of low and high number N of biopsiable embryos. The baseline case is bolded
| ρ(u/v)M | ΔP (N = 6) | ΔC (N = 6) | ΔP (N = 9) | ΔC (N = 9) |
|---|---|---|---|---|
| 0.0 | +0.07 | +0.08 | +0.11 | +0.11 |
| 0.1 | +0.07 | +0.07 | +0.11 | +0.11 |
| 0.2 | +0.07 | +0.07 | +0.11 | +0.11 |
| 0.3 | +0.06 | +0.07 | +0.11 | +0.11 |
| 0.4 | +0.06 | +0.06 | +0.11 | +0.11 |
| 0.5 | +0.05 | +0.06 | +0.10 | +0.11 |
| 0.6 | +0.05 | +0.05 | +0.10 | +0.10 |
| 0.7 | +0.04 | +0.04 | +0.09 | +0.09 |
| 0.8 | +0.03 | +0.04 | +0.08 | +0.09 |
| 0.9 | +0.02 | +0.03 | +0.07 | +0.07 |
| 1.0 | +0.01 | +0.02 | +0.06 | +0.06 |
We also note that the formulae can be used to decide how many chromosomes should be tested for in an individual case. For purposes of analysis we simply define an error rate that is independent of the number of chromosome probes. However, in the case of FISH, as more test probes are used the value of μA decreases, but the value of μN increases. Decreasing the number of chromosomes probes may, depending on the value of the other parameters, increase the chance of success by decreasing the probability of a false positive. Whether this outweighs the loss of detection ability can be evaluated directly, if one so wishes (see “Discussion”).
Though attention often focuses on “false abnormals” (which reduces the pool of available normal embryos for transfer), we note that often effects that "false normals" can also be damaging. Because one typically wishes to transfer only a limited number of embryos that have tested normal (in order to avoid high multiples), in practice such “false normals” can "take slots away" from truly normal embryos. For this reason both kinds of errors can have a clinical impact (see “Discussion”).
Finally, we note that most standard clinical practice accords relatively well with an implied value for g of roughly 5%.
Discussion
We herein present a mathematical model to assess the impact of PGS on the probability of transferring two “normal” fully diploid embryos on day 5 of preimplantation embryo development. We have analyzed the PGS advantage, the gain (g), by assessing the influence of the number of biopsiable embryos (N), the aneuploidy rate (α), the mosaicism rate (μ) and the misdiagnosis rate (η) on the probability of transferring two, day 5 embryos to the uterine cavity. We have further analyzed the cost function of transferring abnormal embryos on day 5. We define a PGS advantage when the value of g is greater than 0.05. The transfer of two, day 5 embryos is probably appropriate for women of advanced maternal age who undergo PGS. A recent report from the European Society of Human Reproduction and Embryology (ESHRE) PGD Consortium [31] shows 1.8 embryos on average are transferred in PGS cycles for the clinical indication of advanced maternal age (2342 embryos in 1296 embryo transfer cycles). The current model assumes embryo biopsy is performed on a cohort of day 3 embryos. To date, most well-designed PGS RCT have used day 3 biopsy protocols. We are, of course, aware there will be a distribution of cell counts in a cohort of day 3 biopsied embryos, typically 6–10 cells. Mathematically, the actual embryo cell count on day 3 is not important to the analysis since the model compares outcomes following the transfer of day 5 embryos that are either biopsied or not biopsied on day 3 of embryo development. The end-point of the analysis is to determine the probability of transferring “normal” and “abnormal” embryos with or without the intervention of PGS. Clinically, however, the distribution of embryos cell counts will determine the probability of obtaining viable day 5 blastocysts with or without a day 3 biopsy being performed. The optimal range of cell counts for embryo viability on day 3 in the absence of cell biopsy is probably 7–9 cells [36–38]. For example, Racowsky et al. [38] showed a 3% viability rate for embryos with 6 cells or less on day 3 of culture. Consequently, there is probably little to be gained in undertaking the biopsy of 6-cell embryos regardless of embryo quality unless being performed either for diagnostic purposes or research studies. Recently published PGS studies reported approximately 80–85% of the biopsied day 3 embryos were comprised of 6–8 cells [13, 24, 39]. Interestingly, the viability of biopsied 6-cell embryos was noted to be poor producing a low percentage (approximately 5% in total) of high and good quality day 5 blastocysts, prompting the authors to question the utility of performing blastomere biopsy on 6-cell embryos [39]. This is consistent with the earlier observations of embryology morphology of Racowsky et al. [38]. Additionally, the overall distribution of chromosomally abnormal embryos was not significantly different in a cohort of 6–8 cell embryos, indicating no clear cut correlation between chromosomal abnormalities and dysmorphism in good quality, biopsy-grade embryos [24]. For our analysis, we have further assumed day 3 biopsy does not impact subsequent embryo development. This assumption seems reasonable for those centers performing large numbers of embryo biopsy procedures [39–41]. For example, Schoolcraft et al. [41] concluded from their studies that day 3 embryo biopsy did not affect blastocyst development rates, or the proportion of perfect blastocysts. Practically, however, this may not be true when more than a single blastomere is removed for analysis, or as noted above, a day 3 embryo has a low cell count. Importantly, these are quantifiable parameters under clinical control. Patient selection, ovarian stimulation protocol, embryo biopsy and extended embryo culture can all impact the number and quality of day 3 embryos developing to high quality blastocysts available for transfer. For the purposes of mathematical modeling any change in the rates of blastocyst development after a day 3 biopsy simply requires a change in the value of ρ; this would be the case, for example, if one wished to analyze the outcome of performing PGS on a cohort of embryos comprised of either 6-cell or high quality 8-cell embryos, and so on. Similarly, if some clinics performing PGS were to observe lower rates of blastocyst development with or without a day 3 biopsy this can be easily analyzed by making changes to the appropriate parameters. In the present model, we have assumed rates of blastocyst development of normal diploid embryos are approximately 10–20% greater than aneuploid embryos. This value can be determined clinically and will be dependent on the center performing both embryo biopsy and culture. Schoolcraft et al. [41] estimated an overall selective improvement of approximately 15% in biopsied diploid embryos developing to blastocysts, although the rate was slightly higher for perfect blastocysts. Similar ranges were reported by others using microarray data [42]. Importantly, our purpose, from the standpoint of mathematical modeling, is not to define an outcome for every possible clinical situation involving PGS, but rather to demonstrate specific trends using input parameters that fall within quantitatively acceptable ranges derived from published data in the clinical literature. This, however, does not preclude analyzing a unique clinical situation, if so desired, as long as the appropriate input parameters, N, α, μ, η are known.
In general, we find, depending on the relative weighting of embryo loss and the expected aneuploidy rate, the benefit of undertaking PGS is strongly dependent on the actual number, N of biopsiable embryos on day 3 (Table 1). When day 3 or day 5 transfers are compared with PGS (day 3 biopsy, day 5 transfer) the aneuploidy rate determines the minimum required number of biopsiable embryos for PGS to be of benefit (g = 0.05) and can vary from 5 to 11. Even for PGS to show no gain, g = 0, the minimum number of biopsiable embryos can vary from 4 to 6. Thus, when N is known, the outcome depends strongly on the aneuploidy rate, a hidden variable that is not known on a case by case basis in the clinical setting of PGS. Any benefit of PGS is only realized when there are many day 3 embryos available for biopsy (Table 3). In addition, we find few situations where the gain is greater than 10%. A small gain perhaps explains why the different PGS studies described in the recent clinical literature have produced such contradictory results [8]. The different reported outcomes are probably explained by such variable factors as the number of biopsied embryos, the distribution of the embryo cell counts of the biopsied embryos on day 3, the number of cells biopsied (1 versus 2), the impact of the biopsy procedure on embryo viability, and the number of FISH chromosome probes used for assessing the numerical composition of the biopsied embryos. In the recent PGS RCT reported by Staessen et al. [13], Mastenbroek et al. [14] and Hadarson et al. [23] 7–8 chromosome probes were used, 4–6 embryos biopsied and approximately 2 embryos transferred per cycle. Our analysis suggests women of advanced maternal age with a limited ovarian response may never reach the required number of good quality biopsiable day 3 embryos for PGS to show an advantage. Parenthatically, Munné et al. [11] reported that women of advanced maternal age undergoing PGS with eight or more 2PN zygotes showed the highest implantation and pregnancy rates. However, the actual numbers and quality of the biopsied embryos in the different treatment groups in this non-randomized study were not provided.
We further observe by comparing day 3 transfers and day 5 transfers without PGS that extended embryo culture is an equal contributor to success as PGS (Table 2). This can be understood by recognizing that by waiting to transfer on day 5 any embryos that are lost between day 3 and day 5 can effectively be “replaced” with surviving embryos if there are sufficient embryos available in the cohort. The actual benefits of culture to day 5 depend on the ability of extended culture to select against both aneuploidy and mosaic embryos and on the proportion of abnormal embryos. By comparing the data in Tables 1 and 2 the benefit of day 5 transfer is not due to PGS. Importantly, extending the culture (or performing PGS) with low numbers of day 3 embryos results in a penalty. Any gain is only seen when there are high numbers of embryos available on day 3. This has been observed clinically [37, 43]. We note that the results of our analysis summarized in Tables 1 and 2 have outcomes described in the clinical literature and point to the robustness of our mathematical modeling studies and of our underlying assumptions enuciated in the “Methods” section.
Previous estimated rates for simple or constitutional aneuploidies as a result of meiotic errors are highly variable. Several studies analyzing human metaphase II oocytes with either CGH-FISH or CGH showed aneuploidy rates as high as 40–60% [44]. Aneuploidy rates in human embryos have been reported to vary between 15–85% [28]. Studies using CGH showed 51% of cleavage stage embryos were aneuploidy, whereas 24% were mosaics and only 25% displayed diploidy in all the cells [45, 46].The precision afforded by CGH is not achievable with a limited number of FISH chromosome probes, precluding the possibility of an accurate determination of the true incidence of aneuploidy. Moreover, there is an intrinsic failure rate for each FISH probe. For example, a failure rate of 1% per probe will give error rates of approximately 5% and 10% for 5 and 10 probes, respectively. A 2% failure rate per probe would result in error rates of approximately 9% and 18% for 5 and 10 probes, respectively. Most studies report misdiagnosis rates of 5–15% indicating an approximately 1–2% failure rate per probe. Increasing the number of probes merely increases the error rate and the benefit of detecting additional numerical chromosomal abnormalities is lost. A failure rate of 1% for 24 FISH probes results in a prohibitively high error rate of 22%. A lesser or greater number of probes will result in a shift in the rates of detection of truly normal and abnormal embryos and change the proportion of normal and abnormal blastocysts available for transfer. If, for example, PGS were performed using a sufficient number of FISH probes to detect an estimated 70% of chromosomally abnormal embryos, 30% of abnormal embryos go undetected. The actual proportion of undetected chromosomally abnormal embryos in a cohort of embryos will then depend on the individual rates of segregation errors of those chromosomes resulting in aneuploidy not included in the FISH panel; these are tagged as “normal”. In this case the only selection against transfer would be determined by the value of ρvAn. The assigned false-normal embryos are in effect errors of diagnosis, increasing the misdiagnosis rate, ηA. However, as noted in the data in Table 5, an increase in η leads to a rapid decrease in the gain from PGS. Importantly, these are measurable clinical parameters, and will be different for each center performing PGS, since most centers use a different FISH panel. Our current statistical model assumes all chromosomes are detected; this is expected to favor a PGS gain, whereas the use of fewer chromosome probes is expected to decrease the PGS gain. Mathematically, the method of assessing the numerical chromosome complement of the biopsied embryos is not relevant. Of importance are the values we assign to the analysis, such as α, μ, ρ and η. As an aside, our analysis indicates that the application of different technologies, such as CGH, to detect additional chromosomal abnormalities, following biopsy of day 3 embryos may not provide a clinically useful PGS advantage.
The PGS gain increases as the aneuploidy rate increases, although it is low, approximately 3–5 % unless there are large numbers of biopsiable embryos (Table 4). For low numbers of embryos PGS incurs a penalty when compared to a day 5 transfer without PGS. PGS is unnecessary for selecting normal embryos when the aneuploidy rate is low and provides little benefit. Also, the PGS gain is marginal with high aneuploidy rates even when there are a large numbers of embryos available for biopsy (Table 6). Detecting high aneuploidy rates with PGS can at best inform the clinician that indeed the abnormalities are high. How this information is used clinically is open to debate being dependent on the indication for performing PGS. For in-between rates of aneuploidy PGS provides some discriminatory benefit. Some clinicians, depending on the indication, have advocated using the findings of PGS to advise patients on their treatment options. We, however, have reservations concerning the diagnostic utility of PGS except when the proportion of abnormal embryos is either high or low, and with adequate numbers of biopsied embryos. We note that many combinations of rates of aneuploidy and number of biopsied embryos provide little discriminatory benefit and for this group there is considerable degree of diagnostic uncertainty. Moreover, as η increases the PGS gain is rapidly lost and provides no advantage when compared to a day 5 transfer without PGS (Table 5). Munné and colleagues have consistently argued the importance of the need for low error rates on the diagnostic utility of PGS and have expressed concern regarding the consistently high error rates reported in some clinical studies [21]. We agree with their position. From the standpoint of mathematical modeling we can precisely define misdiagnosis and error rates to determine if PGS provides a diagnostic advantage. We cannot, however, take account of the foibles of human activity. In the clinical setting it is the responsibility of each clinic to ensure that the highest standards are met and good practice guidelines are followed for blastomere biopsy, fixation and FISH analysis [47]. The lack of rigorous laboratory practices and quality control measures have been suggested as possible reasons for the reported outcomes from previous PGS RCT [22].
Mosaic aneuoploidy is a confounding factor when a day 3 embryo biopsy is performed for the purposes of undertaking PGS [48]. It has been argued that the first few cell division cycles during early embryonic development may be particularly sensitive to mitotic errors when all the necessary cell cycle check points are not activated and maternal cytoplasmic factors are needed during the early phase of embryonic development [44, 49, 50]. The relationship between the mix of cells that are diploid/aneuploidy and diploid/mosaic and of their ability to develop to morphologically normal appearing blastocysts is not well established, although chaotic mosaics display high rates of developmental arrest [28, 51]. However, blastocyst development, per se, is not an absolute filter for eliminating aneuploidy/mosaic embryos, although blastocyst development rates are lower when compared to fully diploid embryos [41, 52–55]. In general it is assumed that day 3 embryos with greater than 50% mosaicism have limited developmental capacity and would be selected against following day 3 biopsy and culture to day 5 [21]. Los et al. [48] have previously described the probability of biopsying a mosaic cell for the idealized situation of 8-cell embryos undergoing biopsy with the removal of one or two blastomeres. If, for example, an 8-cell embryo is 50% mosaic (ie, 4/4 diploid/aneuploid) and following biopsy is tagged “normal” resulting in a 57% mosaic (7 cells, 3/4 diploid/aneuploid embryo) there is likely a low probability that such an embryo continues normal development. Conversely, the same embryo after biopsy if now tagged “abnormal” is now composed of a 43% mosaic embryo (7 cells. 4/3 diploid/aneuploid embryo); this embryo may or may not develop to a blastocyst, but nevertheless is typically discarded. Consequently, both false-negative and false-positive rates have clinical impact. The issue of mosaic aneuploidy and misdiagnosis becomes more problematic as the proportion of mosaicism drops to less than 50% since it increases the likelihood of diagnosing a mosaic embryo as “normal”. These embryos may or may not develop to blastocysts. Considerable uncertainty exists as to the precise developmental potential and viability of cleavage-stage embryos with limited mosaicism, consisting of perhaps 2–3 chromosomally abnormal blastomeres. Some of these embryos may in fact develop normally and potentially implant resulting in a normal pregnancy. For example, the remaining diploid cells may have a growth advantage and progressively “dilute out” the few abnormal cells, or alternatively some chromosomal mosaic cells may be developmentally lethal and be eliminated with further development of the embryo, whereas in other cases there may be self-correction producing a normal diploid embryo [56]. The actual proportion of mosaic embryos in a cohort of IVF-generated embryos that undergo self-correction to give a normal diploid embryo is largely unknown. Mathematically we can assign the proportion of mosaic cells in an embryo and determine the probability of testing either normal or abnormal. In the current analysis, we have averaged together the effects of aneuploidy and mosaics into a single “abnormal” category. On average this is probably correct and seems a reasonable approximation, although it neglects the detailed correlations between developmental arrest and mosaic/diploid and mosaic/aneuploidy mix in a sample. We believe, however, the effect is likely to be small. Munné et al. have recently reported that approximately 2.5% of mosaic embryos consisted of limited mosaicism, whereas most are extensive mosaics and are detected following a day 3 biopsy [21]. Similarly, Los et al. [48] estimated that the actual number of embryos containing limited mosaicism is low, as is the proportion of such embryos that can give rise to a viable pregnancy. They have also concluded that the majority of mosaic embryos undergo developmental arrest at the morula stage of preimplantation embryo development. A recent report showed there was no difference in the rates of either mosaicism or confined placental mosaicism from late first trimester chorionic villus samplings from natural conceptions and pregnancies following in vitro fertilization [57]. This provides strong evidence to indicate that in vitro-generated embryos have effective selection mechanisms against mosaic embryos with few surviving to result in a viable pregnancy. In the current analysis, we have labeled fully diploid embryos, “normal”, and thus by definition mosaic embryos are “abnormal”. We have therefore set μA equal to μe(1-α3) and μN to zero. However, μN may be greater than zero if, for example, it is determined that certain diploid/limited mosaic embryos have the same viability as fully diploid embryos. In this case we can straightforwardly reassign these embryos as “normal” by changing the value of μN. Recall, the end-point of our current analysis is the probability of transferring two, day 5 fully diploid embryos. Importantly, the detailed numerical results of our modeling studies are subject to refinements as we accrue more information on the developmental fate and viability of embryos with limited mosaicism, although, as noted above, current evidence indicates that the proportion of such embryos capable of producing a viable pregnancy is probably quite small.
Mosaicism significantly influences the gain of PGS. For low levels of mosaicism PGS offers advantages, whereas increasing mosaicism decreases the PGS advantage (Table 7). The “tipping point” depends on the rate of mosaicism and the number of biopsiable embryos. We also show that for typical ranges of mosaicism described in clinical studies there are substantial abnormalities for PGS to usefully discriminate against and the gain of PGS is marginal. The PGS advantage is lost with increasing rates of misdiagnosis of mosaic embryos (Table 8). This can be understood clinically because in most situations a limited number of day 5 embryos are transferred to the uterus. As the proportion of misdiagnosed embryos in the cohort of embryos targeted for transfer increases the greater the probability that abnormal embryos will be chosen for transfer. The data summarized in Tables 7 and 8 were determined using a fixed aneuploidy rate of 30% (α3 = 0.3). If the aneuploidy rates were either increased or decreased the proportion of abnormal embryos is shifted as is the PGS gain, ceteris parabus. Increasing the number of embryos for biopsy increases the gain of PGS, but as noted previously the advantage is lost if the aneuploidy rate is high. The selective filtering of mosaic embryos due to extended culture will likely depend on the proportion of abnormal cells in the embryo. Selective loss can in theory vary from 0 to 100%. The filtering effect of extended culture for mosaic aneuploidies and its impact on PGS are dependent on the number of biopsiable day 3 embryos (Table 9). If 100% of mosaic aneuploidies were effectively filtered out by extended culture the effectiveness of PGS is decreased. This is easily understood. For a 0% loss rate gains are seen with PGS. In this case, PGS will detect some of the abnormal mosaic embryos and be discarded, notwithstanding the uncertainties raised in the previous paragraph regarding limited mosaicism. In practice, the actual survival is likely to fall in the mid-ranges where PGS becomes less discriminatory. One can, for example, consider embryos with limited mosaicism and tagged “normal” equivalent to a low loss rate, ρ(u/v)M. In this case a PGS gain is seen. In this setting the benefits of PGS will then depend on the true aneuploidy rate (α3). However, with increases in α3 the numerical advantage of having more embryos to biopsy on day 3 is lost.
In summary, we find whether or not PGS is warranted is a strong function of the aneuploidy rate, mosaicism rate, misdiagnosis rate and the number of biopsiable embryos. Even for large aneuploidy rates, PGS may not be of benefit if there are small numbers of biopsiable embryos, or when the misdiagnosis rate is high. By taking into account the rates of aneuploidy, mosaicism and misdiagnosis, and the number of biopsiable embryos, and the number of embryos to transfer on day 3 or 5, we have shown, subject to the assumptions enumerated in the Methods and Results sections of this article, how to calculate directly whether PGS provides any benefit. Depending on the values of these parameters PGS can vary from actively harmful to beneficial. However, our analysis indicates that PGS shows a gain with specific combinations of numbers of biopsied embryos and rates of aneuploidy and mosaicism, whereas other combinations produce no benefit or are detrimental. This suggests that some patients will and others will not show a gain from the use of PGS. The challenge is to identify those patients who might benefit from the intervention of PGS. Whether or not these patients can be identified through the use of RCTs remains to be seen given the statistical uncertainties associated with the hidden variables considered important to the outcome of PGS. Other challenges remain, however, such as when to perform a biopsy procedure, polar body, cleavage stage embryo or blastocyst, and to identify the most appropriate diagnostic technology with which to undertake PGS. A recent Practice Committee opinion on preimplantation genetic testing did not support the use of PGS to improve the pregnancy outcome in patients with advanced maternal age, recurrent pregnancy loss and repeat implantation failure [58]. As noted in the “Introduction” (and references cited therein) there is a lack of agreement of whether additional PGS RCTs based on day 3 FISH are even justified. Moreover, high rates of mosaicism in day 3 embryos may preclude the use of PGS at this stage of embryonic development independent of the method of analysis. It remains to be determined if other technologies provide any diagnostic advantage. Finally, we have given the statistical expectations in closed form. However, the model is quite amenable to Monte Carlo simulation. Such a simulation would calculate the average success rate over many clinical trials with a given set of inputs (α, µ, N, etc.). Moreover, as more precise data are obtained on the various parameters used in our analysis, particularly on the issue of limited mosaicism, further refinements can be made to the mathematical model.
Addendum
In the “Methods” section we outlined the basic parameters used to calculate the benefits of PGS. We also considered the impact of aneuploidy rates on day 3 embryo transfers without performing PGS. A more mathematical treatment will be presented in a separate publication. Below, we present in more detail the impact of PGS using day 3 biopsied embryos. The discussion that follows considers the intervention of PGS and its impact on the probability of transferring fully diploid embryos.
The calculations in this addendum form the basis for the data summarized in Tables 1, 2, 3, 4, 5, 6, 7, 8 and 9.
The loss rate (spontaneous abortion)
We define the spontaneous abortion rate σ, the rate at which chromosomally aneuploid embryos that survive to day five implant and cause a spontaneous miscarriage. (As most aneuploidies do not go to term, this is essentially equivalent to the implantation rate for aneuploid embryos.) In parallel to PN, we define PA the probability that exactly NA of the N embryos are truly aneuploid. We can then calculate the probability PSA to undergo a spontaneous miscarriage:
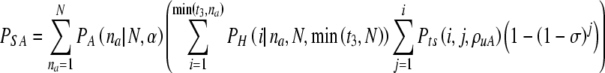 |
7 |
Depending on the purpose of the calculation, one in principle should be careful to properly correlate the spontaneous miscarriage and successful transfer cases.
We have not done so here.
Day 5 transfer, without PGS
Day five transfer without PGS differs from the day three case due to the fact that some actionable filtering (based on the differential between ρvA and ρvN) of normal and abnormal embryos can take place. Formally, when written out fully one will find that the indices on the sums are entwined in a slightly different way. We reiterate that the value α is the aneuploidy rate observed at day 3; due to the ρvA-ρvN differential the aneuploidy rate is different on day five and must be calculated anew.
First we calculate the joint probability PNA(NN, NA, N,α,ρvA,ρvN) that on day five there are exactly NN normal and NA aneuploid embryos. We find this by summing joint survival probabilities products Pts over all possible values of NN3 and NA3 consistent with N, NN, and NA:
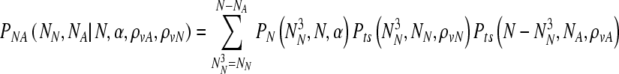 |
8 |
Choosing now to transfer at most t5f embryos, what is the probability PnoPGS5(N,α,ρvA,ρvN, t5f) that at least T of them are normal? It is the probability to select at least T normal embryos from a set of NN normal and NA abnormal embryos, summed over all possible values of NN and NA weighted by their joint probabilities:
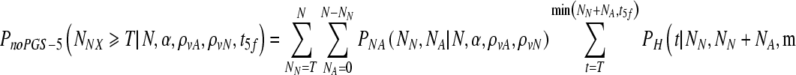 |
9 |
Loss rate (spontaneous abortion) day 5 without PGS
Again, the spontaneous abortion rate σ is defined as the rate at which chromosomally aneuploid embryos that survive to day five implant and cause a spontaneous abortion. The probability of a spontaneous miscarriage is given by summing over all possible values of NA and all possible values of the number of transferred aneuploid embryos:
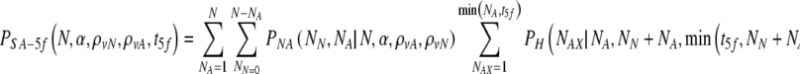 |
10 |
Day 3 PGS
The probability PnoPGS5(NN,X ≥ T| T, N, α, ρvN = 0) just calculated must be compared to the probability PPGS that at least T normal embryos are transferred on day five following a day three PGS biopsy. In this case, the PGS analysis labels embryos as normal or aneuploid, but with some error rate due to the combined effects of laboratory uncertainties and embryonic mosaicism.
When undergoing PGS and transferring embryos on day 5 theoretically two separate effects favor successful transfer of normal embryos. The first, naturally, is the diagnostic information from PGS itself, separating normal from aneuploid embryos. The second is the filtering effect of culturing embryos from day 3 to day 5, as demonstrated in the previous section above. To preserve clarity in the strategy of calculation, we first go through the calculation of successful transfer in the case that all values of ρ are zero. We then introduce the complications of in vitro filtering in the following section, which are essentially the same as in the case discussed above in day 5 transfer without PGS.
Success probability, neglecting in vitro loss (mortality)
The strategy we take is to break the calculation into the following pieces:
The probability to find a particular number of
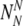 and
and 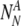
For each set of
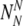 and
and 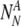 , the probability to select at least T of the truly normal (out of
, the probability to select at least T of the truly normal (out of 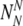 ) to transfer
) to transfer
The probability Ptruth that the N embryos contain exactly NN truly normal embryos is:
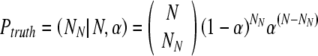 |
11 |
If there are NA(=N-NN) truly aneuploid embryos, the probability PNA to observe 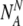 of them labeled as normal is:
of them labeled as normal is:
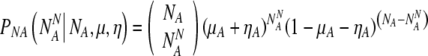 |
12 |
Similarly, if there are NN truly normal embryos, the probability PNN to observe 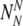 of them labeled as normal is:
of them labeled as normal is:
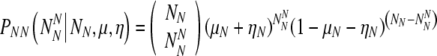 |
13 |
So we can write that the probability Plabel that the final normally-labeled sample consists of 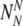 truly normal embryos and
truly normal embryos and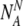 aneuploid embryos is:
aneuploid embryos is:
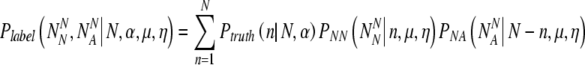 |
14 |
Once again, we must use the hypergeometric distribution to assign the probability
PX to select at least T truly normal embryos out of the collection of normally-labeled embryos:
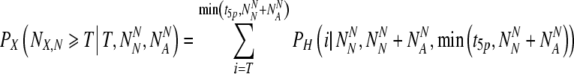 |
15 |
We now have all the expressions needed to evaluate the desired quantity PPGS:
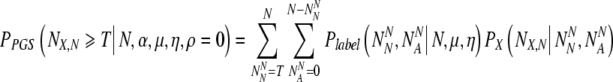 |
16 |
Spontaneous miscarriage rate, neglecting in vitro loss (mortality), day 5 transfer with PGS
We may straightforwardly define the probability to select for transfer a number of aneuploid embryos PXA in parallel to PX. We then calculate the probability for a spontaneous abortion PSAPGS (ρ = 0) while undergoing PGS:
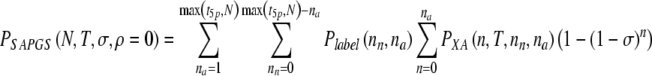 |
17 |
Successful transfer probability accounting for in vitro loss (mortality), day 5 transfer with PGS
As in the earlier section, we must now consider all possible values of 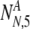 and
and 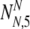 , the number of PGS-labeled "normal" embryos that are truly normal and aneuploid, respectively, surviving to day five. We find the probability PNA,5(
, the number of PGS-labeled "normal" embryos that are truly normal and aneuploid, respectively, surviving to day five. We find the probability PNA,5(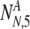 ,
, 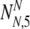 | N, NN, NA, α, ρ, μ,η) (where μ, ρ, and η are a notational shorthand for all relevant members of the parameter families with the various subscripts):
| N, NN, NA, α, ρ, μ,η) (where μ, ρ, and η are a notational shorthand for all relevant members of the parameter families with the various subscripts):
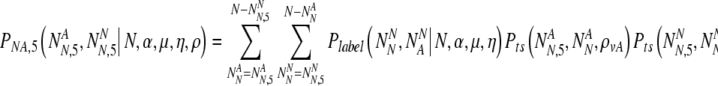 |
18 |
We now sum over values of the transferred number of truly normal embryos for the probability to transfer that number, to find:
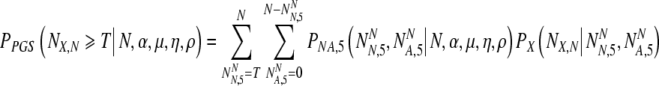 |
19 |
Miscarriage rate accounting for in vitro loss (mortality)
Now accounting properly for the loss rates (mortality) we find:
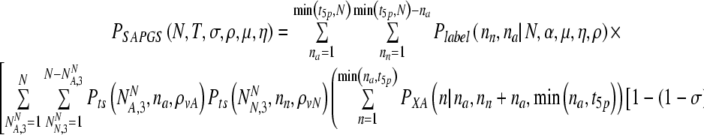 |
20 |
where for clarity the internal summands NN and NA stand for 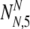 and
and 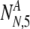 , respectively.
, respectively.
ΔP and ΔC
Displaying now the full functional dependence, we have described the proper framework for calculating the increase in the probability of success:
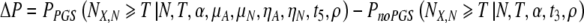 |
22 |
We can also calculate the increase in the cost function accounting for embryo loss:
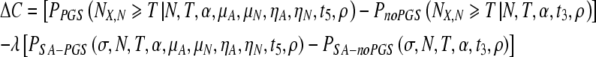 |
23 |
Footnotes
Capsule Hypergeometric statistical modeling is used to define the limits of preimplantation genetic screening.
References
- 1.Munné S, Grifo J, Cohen J, Weier H. Chromosome abnormalities in human arrested preimplantation embryos: a multiple probe FISH study. Am J Hum Genet. 1994;55:150–9. [PMC free article] [PubMed]
- 2.Munné S, Daily T, Sultan KM, Cohen J. The use of fist polar bodies for preimplantation diagnosis of aneuploidy. Mol Hum Reprod. 1995;10:1014–20. [DOI] [PubMed]
- 3.Munné S, Marquez C, Reing A, Garrisi J, Alikani M. Chromosome abnormalities in embryos obtained after conventional in vitro fertilization and intracytoplasmic sperm injection. Fertil Steril. 1998;69:904–8. [DOI] [PubMed]
- 4.Munné S, Magli C, Bahce M, Fung J, Legator M, Morrison L, et al. Preimplantation diagnosis of the aneuploidies most commonly found in spontaneous abortions and live births: XY, 13, 14, 15, 16, 18, 21, 22. Prenat Diagn. 1998;18:1459–66. [DOI] [PubMed]
- 5.Harper JC, Dawson K, Delhanty JDA, Winston RML. The use of fluorescent in-situ hybridization (FISH) for analysis of in-vitro fertilization embryos: a diagnostic tool for the infertile couple. Hum Reprod. 1995;10:3255–8. [DOI] [PubMed]
- 6.Magli MC, Gianaroli L, Ferraretti AP. Chromosomal abnormalities in embryos. Mol Cell Endocrinol. 2001;183:S29–34. [DOI] [PubMed]
- 7.Abdelhadi I, Colls P, Sandalinas M, Escudero T, Munné S. Preimplantation genetic diagnosis of numerical abnormalities for 13 chromosomes. Repro Biomed Online. 2003;6:226–31. [DOI] [PubMed]
- 8.Fritz MA. Perspectives on the efficacy and indications for preimplantation screening: where are we now? Hum Reprod. 2008;23:2617–21. [DOI] [PubMed]
- 9.Munné S, Lee A, Rozenwaks Z, Grifo J, Cohen J. Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum Reprod. 1993;8:2185–91. [DOI] [PubMed]
- 10.Munné M, Magli C, Cohen J, Morton P, Sadowy S, Gianaroli L, et al. Positive outcome after preimplantation diagnosis of aneuploidy in human embryos. Hum Reprod. 1999;14:2191–9. [DOI] [PubMed]
- 11.Munné S, Mireia S, Escudero T, Velilla E, Walmsley R, Sadowy S, et al. Improved implantation after preimplantation genetic diagnosis of anueploidy. Repro Biomed Online. 2003;7:91–7. [DOI] [PubMed]
- 12.Gianaroli L, Magli C, Ferraretti AP, Munné S. Preimplantation diagnosis for aneuploidies in patients undergoing in vitro fertilization with a poor prognosis: identification of the categories for which it should be proposed. Fertil Steril. 1999;72:837–44. [DOI] [PubMed]
- 13.Staessen C, Platteau P, Van Assche E, Michiels A, Tournaye H, Camus M, et al. Comparison of blastocyst transfer with or without preimplantation genetic diagnosis for aneuploidy screening in couples with advanced maternal age: a prospective randomized controlled trial. Hum Reprod. 2004;19:2849–58. [DOI] [PubMed]
- 14.Mastenbroek S, Twisk M, van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, et al. In vitro fertilization with preimplantation screening. N Engl J Med. 2007;357:9–17. [DOI] [PubMed]
- 15.Cohen J, Munné S. Two-cell biopsy and PGD outcome. Hum. Reprod. 2005;20:2363–4. [DOI] [PubMed]
- 16.Wilton LJ. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:1770. [PubMed]
- 17.Handyside A, Thornhill AR. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:1770. [PubMed]
- 18.Cohen J, Wells D, Munné S. Removal of 2 cells from cleavage stage embryos is likely to reduce the efficacy of chromosomal tests that are used to enhance implantation rates. Fertil Steril. 2007;87:496–503. [DOI] [PubMed]
- 19.Munné S, Cohen J, Simpson JL. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:1769–70. [DOI] [PubMed]
- 20.Munné S, Gianaroli L, Tur-Kaspa I, Magli C, Sandalinas M, Grifo J, et al. Substandard application of preimplantation genetic screening may interfere with its clinical success. Fertil Steril. 2007;88:781–4. [DOI] [PubMed]
- 21.Munné S, Wells D, Cohen J. Technology requirements for preimplantation genetic diagnosis to improve assisted reproduction outcomes. Fertil Steril. 2009; Article in press. [DOI] [PubMed]
- 22.Simpson JL. What next for preimplantation genetic screening? Randomized clinical trial in assessing PGS: necessary but not sufficient. Hum Reprod. 2008;23:2179–81. [DOI] [PubMed]
- 23.Hadarson T, Hanson C, Lundin K, Hillensjo T, Nilsson L, Stevic J, et al. Preimplantation genetic screening in women of advanced maternal age caused a decrease in clinical pregnancy rate. Hum Reprod. 2008;23:2806–12. [DOI] [PubMed]
- 24.Staessen C, Verpoest W, Donoso P, Haentjens P, Van der Elst J, Liebaers I, et al. Pre-implantation genetic screening does not improve delivery rate in women under the age of 36 following single-embryo transfer. Hum Reprod. 2008;23:2818–25. [DOI] [PubMed]
- 25.Jansen RPS, Bowman MC, de Boer KA, Leigh DA, Lieberman DB, McArthur SJ. What next for preimplantation genetic screening (PGS)? Experience with blastocyst biopsy and testing for aneuploidy. Hum Reprod. 2008;23:1476–8. [DOI] [PubMed]
- 26.Mersereau JE, Pergament E, Xhang X, Milad MP. Preimplantation genetic screening to improve in vitro fertilization pregnancy rates: a prospective randomized controlled trial. Fertil Steril. 2008;90:1287–9. [DOI] [PubMed]
- 27.Shahine LK, Cedars MI. Preimplantation genetic diagnosis does not increase pregnancy rates in patients at risk for aneuploidy. Fertil Steril. 2006;85:51–6. [DOI] [PubMed]
- 28.Donoso P, Staessen C, Fauser BCLM, Devroey P. Current value of preimplantation genetic aneuploidy screening in IVF. Hum Reprod Update. 2007;13:15–25. [DOI] [PubMed]
- 29.Gleicher N, Weghofer A, Barad D. Preimplantation genetic screening: “established” and ready for prime time? Fertil Steril. 2008;89:780–8. [DOI] [PubMed]
- 30.Yakin K, Urman B. What next for preimplantation genetic screening? A clinician’s perspective. Hum Reprod. 2008;23:1686–90. [DOI] [PubMed]
- 31.Goossens V, Harton G, Moutou PN, Traeger-Synodinos J, Sermon K, Harper JC. ESHRE PGD Consortium data collection VIII: cycles from January to December 2005 with pregnancy follow-up to October 2006. Hum Reprod. 2008;23:2629–45. [DOI] [PubMed]
- 32.Harper JC, Sermon K, Geraedts J, Vesela K, Harton G, Thornhill A, et al. What next for preimplantation genetic screening? Hum Reprod. 2008;23:478–80. [DOI] [PubMed]
- 33.Fauser BCJM. Preimplantation genetic screening: the end of an affair? Hum Reprod. 2008;23:2622–5. [DOI] [PubMed]
- 34.Mastenbroek S, Scriven P, Twisk M, Vivelle S, Van der Veen F, Repping S. What next for preimplantation screening? More randomized controlled trials needed? Hum Reprod. 2008;23:2626–8. [DOI] [PubMed]
- 35.Hodges JL, Lehman EL. Basic concepts of probability and statistics. Philadelphia: Society for Industrial and Applied Mathematics; 2004. p. 173–7.
- 36.Alikani M, Calderon G, Tomkin G. Cleavage anomalies in early human embryos and survival after prolonged culture in vitro. Hum Reprod. 2000;15:2634–43. [DOI] [PubMed]
- 37.Racowsky C, Jackson KV, Cekleniak NA, Fox JH, Hornstein MD, Ginsburg AS. The number of eight-cell embryos is a key determinant for selecting day 3 or day 5 transfer. Fertil Steril. 2000;73:558–64. [DOI] [PubMed]
- 38.Racowsky C, Combelles CMH, Nureddin A, Pan Y, Finn A, Miles L, et al. Day 3 and day 5 morphologic predictors of embryo viability. Repro Biomed Online. 2003;6:323–31. [DOI] [PubMed]
- 39.Goossens V, De Ryke D, De Vos A, Staessen C, Michiels A, Verpoest W, et al. Diagnostic efficiency, embryonic development and clinical outcome after the biopsy of one or two blastomeres for preimplantation genetic diagnosis. Hum Reprod. 2008;23:481–92. [DOI] [PubMed]
- 40.Debrock S, Melotte C, Spiessens C, Peeraer K, Vanneste E, Meeuwis L, et al. Preimplantation genetic screening for aneuploidy of embryos after in vitro fertilization in women aged at last 35 years: a prospective randomized trial. Fertil Steril. 2009; Article in press. [DOI] [PubMed]
- 41.Schoolcraft WB, Katz-Jaffe MG, Stevens J, Rawlins M, Munné S. Preimplantation aneuploidy testing for infertile patients of advanced maternal age: a randomized prospective trial. Fertil Steril. 2009;92:157–62. [DOI] [PubMed]
- 42.Miller KA, Li X, Lambrese K, Su J, Treff N, Scott RT. Blastocyst formation rates in chromosomally normal versus abnormal embryos as analyzed by 24 chromosome microarray-based aneuploidy screening (MPGD). Fertil Steril. 2008;90(suppl 1):S346. [DOI]
- 43.Papanikolaou EG, D’haeseleer E, Verheyen G, Van de Velde H, Camus M, Van Steirteghem A, et al. Live birth rate is significantly higher after blastocyst transfer than after cleavage-stage embryo transfer when at least four embryos are available on day of embryo culture. A randomized prospective study. Hum Reprod. 2005;20:3198–203. [DOI] [PubMed]
- 44.Delhanty JDA. Mechanisms of aneuploidy induction in human oogenesis in early development. Cytogenet Genome Res. 2005;111:237–44. [DOI] [PubMed]
- 45.Wells D, Delhanty JDA. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod. 2000;6:1055–62. [DOI] [PubMed]
- 46.Voullaire L, Slater H, Williamson R, Wilton L. Chromosome analysis of blastomeres from human embryos by using comparative genomic hybridization. Hum Genet. 2000;106:210–7. [DOI] [PubMed]
- 47.The Preimplantation Genetic Diagnosis International Society (PGDIS). Guidelines for good practice PGD: programme requirements and laboratory quality assurance. Repro Biomed Online. 2008;16:134–47. [DOI] [PubMed]
- 48.Los JF, Van Opstal D, van der Berg C. The development of cytogenetically normal, abnormal and mosaic embryos: a theoretical model. Hum Reprod Update. 2004;10:79–94. [DOI] [PubMed]
- 49.Delhanty JDA, Handyside A. The origin of genetic defects in man and their detection in the preimplantation embryo. Hum Reprod Update. 1995;1:201–15. [DOI] [PubMed]
- 50.Handyside A, Delhanty JDA. Preimplantation genetic diagnosis: strategies and surprises. Trends Genet. 1997;13:270–5. [DOI] [PubMed]
- 51.Delhanty JDA, Harper JC, Ao A, Handyside A, Winston RML. Multi-colour FISH detects chromosomal mosaicism and chaotic division in normal pre-implantation embryos from fertile patients. Hum Genet. 1997;99:755–60. [DOI] [PubMed]
- 52.Magli MC, Jones GM, Gras L, Gianaroli L, Korman I, Trounson AO. Chromosome mosaicism in day 3 embryos that develop to morphologically normal blastocysts in vitro. Hum Reprod. 2000;15:1781–6. [DOI] [PubMed]
- 53.Bielanska M, Tan SL, Ao A. Chromosomal mosaicism throughout human preimplantation development in vitro: incidence, type and relevance to embryo outcome. Hum Reprod. 2002;17:413–9. [DOI] [PubMed]
- 54.Sandalinas M, Sadowy S, Alikani M, Calderon G, Cohen J, Munné S. Developmental ability of chromosomally abnormal human embryos to develop to the blastocyst stage. Hum Reprod. 2001;16:1954–8. [DOI] [PubMed]
- 55.Fragouli E, Lenzi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular analysis of the human blastocyst stage. Hum Reprod. 2008;23:2596–608. [DOI] [PubMed]
- 56.Hernandez ER. What next for preimplantation genetic screening? Beyond aneuploidy. Hum Reprod. 2009;24:1538–41. [DOI] [PubMed]
- 57.Huang A, Adusumalli J, Patel S, Liem J, Williams J, Pisarska MD. Prevalence of chromosomal mosaicism in pregnancies with infertility. Fertil Steril. 2009;91:2355–1260. [DOI] [PubMed]
- 58.The Practice Committee of the Society for Assisted Reproductive Technology and the Practice Committee of the American Society for Reproductive Medicine. Preimplantation genetic testing: a practice committee opinion. Fertil Steril. 2007;88:1497–504. [DOI] [PubMed]


