Abstract
Purpose
Association of ESR1 gene PvuII, XbaI and (TA)n microsatellite polymorphisms and woman infertility was evaluated.
Methods
Infertile(n = 104) and fertile(n = 107) women were included in this study. We performed polymerase chain reaction-restriction fragment-length polymorphism analysis for detecting ESR1 polymorphisms.
Result(s)
PvuII and XbaI polymorphisms confered risk for infertility in a simple dominant manner in which a significant relationship was observed between infertile and control women. Infertile women had fewer(<18) short repeat alleles in promotor region. ESR1 genotypes were compared concerning maturation, fertilization, pregnancy rates and embryo quality. Although no difference was found in terms of pregnancy rates, maturation and fertilization rates were significantly smaller in pp and xx genotypes. Also, pp genotypes had significantly lower number of good quality embryos. Long TA repeat in promotor was found to be associated with low fertilization rate.
Conclusion(s)
Polymorphisms at the ESR1 gene are associated with infertility in this Turkish infertile women population.
Keywords: Estrogen receptor gene, Infertility, IVF, Microsatellite polymorphism, Pregnancy rate
Introduction
Estrogen is synthesized by granulosa cells under the control of follicle stimulating hormone (FSH) and luteinizing hormone (LH). Estrogen and FSH act in synergism in the ovary to increase the number of FSH receptors in the granulosa cells, resulting in follicular growth and maturation [1, 2]. It is believed that estrogen plays crucial roles in oocyte maturation and fertilization [3].
Estrogen signaling is mediated via binding to estrogen receptors (ERs), which are ligand-dependent transcription factors. Two subtypes of estrogen receptors exist in humans; ERα [4] and ERβ [5], coded by ESR1 and ESR2 genes, respectively. ERα and ERβ are both members of nuclear hormone receptor family. The ESR1 gene (140 kilo base) is located on chromosome 6q25.1 and consists of 8 exons; and intron 1 contains two single-nucleotide polymorphisms (SNPs) named the PvuII (T/C) and XbaI (A/G) [6].
Recent studies have tried to evaluate the distribution of various ESR1 gene polymorphisms, associated with female and male infertility. ESR1 PvuII polymorphism in women was found to affect pregnancy rate following in vitro fertilization (IVF) [7, 8], whereas in males ESR1 XbaI polymorphism was suggested to have an effect on azoospermia or idiopathic severe oligospermia [9].
ER genes harbour several polymorphisms that may influence the risk for certain infertility-associated gynecological disorders and IVF outcome [10]. PvuII polymorphism is reported to be associated with the susceptibility to endometriosis [11] and controlled ovarian hyperstimulation (COH)/pregnancy outcome of IVF [7, 8].
An additional ESR1 promoter (TA)n dinucleotide repeat polymorphism is suggested to increase the risk of premature ovarian failure (POF) in a simple dominant manner in which women carrying a long (TA)n repeat allele were suggested to have approximately 10 times the risk of POF compared to women homozygous for short ESR1 (TA)n repeats [8]. These previous findings indicate that improving our understanding of ER gene polymorphisms may be important associations for infertility diagnoses and treatments [7, 8, 10]. Therefore, the purpose of the present study was to determine the importance of ESR1 PvuII, XbaI and (TA)n polymorphisms in the etiology of unexplained infertility and to find an association of these polymorphisms with oocyte maturation, fertilization, pregnancy rates and embryo quality.
Materials and methods
Subjects
One hundred and four women who underwent an IVF-ET procedure were retrospectively recruited for this study. Serum FSH levels (≤8.0 IU/ml) were measured for all participants between day 3 and 5 of the spontaneous menstrual cycle using chemiluminescence immunoassay (Immulite 2000w station, Diagnostic Products Corporation, Los Angeles, CA, USA). Females with infertility because of unexplained factor between 28 and 35 ages were enrolled as study group. The age-matched control group consisted of 107 proven fertile healthy females with a history of regular menstrual cycle. Informed consent was obtained from all participants and Gazi University Medical Faculty Local Ethics Committee, the Institutional Review Board (IRB) approved the study.
Stimulation protocol and oocyte retrieval
All the IVF women were administrated the same ovulation stimulation protocol [12] in Gen-Art Woman Health and Reproductive Biotechnology Center. When the leading follicle reached 18 mm in mean diameter with a serum estradiol (E2) level of 200 pg/ml per mature follicle, 10,000 U of hCG (Profasi, Serono, Switzerland) was administered. Oocyte retrieval was performed 36 h after the human chorionic gonadotropin (hCG) administration injection. The ICSI intracytoplasmic sperm injection) was performed according to conventional protocols and the number of mature oocytes was calculated. The oocytes were considered mature if they reached MII stage by 2–3 h after oocyte retrieval. The total number of embryos was calculated by counting the embryos with two pronuclei (2PN-embryos). Routine examination of embryo quality included the number of blastomeres, the degree of fragmentation, and the uniformity of the blastomeres. Embryos were classified according to a simplified system based on Veeck’s morphological criteria: Grade I embryos have equal-sized blastomeres and no cytoplasmic fragmentation, grade II embryos have blastomeres of equal size and minor cytoplasmic fragmentation covering 10% of the preembryo surface, grade III embryos have blastomeres of distinctly unequal size and variable fragmentation, grade IV embryos have blastomeres of equal or unequal size and moderate-to-significant cytoplasmic fragmentation covering >10% of the preembryo surface, and grade V embryos have few blastomeres of any size and severe fragmentation covering >50% of the preembryo surface. None of the embryos were classified as grade V in this study. Depending on the woman’s age and the embryo quality up to three embryos were transferred on the third day after retrieval. Biochemical pregnancy was established when serum ß-HCG was found >20 IU/L on the 12th day of embryo transfer, and clinical pregnancy was defined as the presence of a gestational sac on ultrasound at six gestational weeks.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using Invisorb DNA extraction kit (Invitek, Berlin, Germany). Patients and controls were genotyped for PvuII (T/C, rs2234693, c.454-397) and XbaI (A/G, rs9340799, c.454-351 A>G) SNPs in ESR1 intron 1, using restriction fragment length polymorphism (RFLP) analysis. For the ESR1 PvuII and XbaI SNPs, the forward and reverse primers were: 5′-CTG CCA CCC TAT CTG TAT CTT TTC CTA TTC TCC-3′ and 5′-TCT TTC TCT GCC ACC CTG GCG TCG ATT ATC TGA-3′, respectively. The total volume of the polymerase chain reaction (PCR) reaction mixture was 50 μL and contained 0.2 mM dNTPs (MBI Fermentas, Vilnius, Lithuania), 2 mM MgCl2, 1X PCR buffer (MBI Fermentas, Vilnius, Lithuania), 10 pmol of primers (MWG Biotech, Martinsried, Germany) and 1.5 U Taq DNA polymerase (MBI Fermentas, Vilnius, Lithuania). PCR was performed using Eppendorf thermal cycler (Eppendorf, Hamburg, Germany). Following an initial denaturation step (5 min at 94°C), samples were subjected to 30 cycles of PCR at 94°C for 30 sec, 62°C for 1 min, and 72°C for 1.5 min with a final extension of 5 min at 72°C.
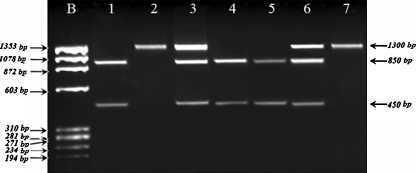
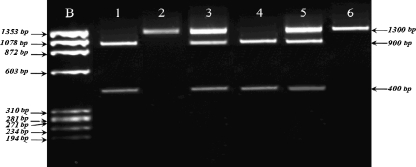
The PCR products were digested overnight with PvuII (MBI Fermentas, Vilnius, Lithuania) and XbaI (MBI Fermentas, Vilnius, Lithuania) restriction enzymes as described previously [13, 14]. After digestion with PvuII, PCR product was cut into 850 and 450 bp fragments in the presence of p allele, whereas P allele was undigested (1300 bp) (Fig. 1). After digestion with XbaI PCR product was cut into 900 and 400 bp fragments in the presence of x allele, whereas X allele was undigested (1300 bp) (Fig. 2). The PCR products and the restriction fragments were separated in 2% agarose gel stained with ethidium bromide, and were visualized by Gel Logic 100 Imaging System (GL 100) (Kodak, NY, USA). To confirm the genotypes obtained by PCR-RFLP method, DNA sequencing was carried out 5% of the samples using ABI Prism 310 Genetic Analyzer. (Applied Biosystems, Foster City, CA, USA).
Fig. 1.
RFLP analysis of the PvuII polymorphism. Lane 1, 4, 5: Two fragments of 850 bp and 450 bp for pp genotype, Lane 2: undigested PCR product of 1300 bp for PP genotype, Lane 3, 6: Three fragments of 1300 bp, 850 bp and 450 bp for Pp genotype, Lane 7: PCR product of external PCR. b: ϕX174 molecular size marker
Fig. 2.
RFLP analysis of the XBaI polymorphism. Lane 1, 4: Two fragments of 900 bp and 400 bp for xx genotype, Lane 2: undigested PCR product of 1300 bp for XX genotype, Lane 3, 5: Three fragments of 1300 bp, 900 bp and 400 bp for Xx genotype, Lane 6: PCR product of external PCR. b: ϕX174 molecular size marker
The (TA)n (rs3138774) repeat polymorphism in the ESR1 promoter region was investigated by PCR using a FAM labelled forward primer 5′ GACGCATGATATACTTCACC 3′ and reverse primer 5′ GCAGAATCAAATATCCAGATG 3′ in a 25 ml PCR reaction containing: 1X PCR buffer, 20 μM of dNTP, 2 mM MgCl2 (MBI Fermentas, Vilnius, Lithuania), 20 pmol of primers (MWG Biotech, Martinsried, Germany) and 1 U Taq DNA polymerase (MBI Fermentas, Vilnius, Lithuania). The reaction volume was made up to 25 ml using deionised water. PCR was performed using Eppendorf thermal cycler (Eppendorf, Hamburg, Germany). Following an initial denaturation step (5 min at 94°C), samples were subjected to 30 cycles of PCR at 94°C for 30 sec, 62°C for 1 min, and 72°C for 1 min with a final extension of 5 min at 72°C. The fluorescence-labelled PCR products were analysed for size using an ABI Prism 310 Genetic Analyzer. (Applied Biosystems, Foster City, CA, USA). The sizes of the PCR products were determined by Genescan v.3.0 software (Applied Biosystems, Foster City, CA, USA). ROX-500 (Applied Biosystems, Foster City, CA, USA) was used as an internal size standard.
Statistical analysis
Allele frequencies were determined by the gene counting method. Hardy-Weinberg equilibrium was tested using Genepop Version 4.0 [15]. The relationship between ESR1 genotypes and infertility was analyzed using the χ2 test. Reference genotypes/alleles were used to calculate crude odds ratios (ORs) and 95% confidence intervals (CIs). Clinical characteristics of the study and control subjects were compared using one way analysis of variance (One way-ANOVA) and Kruskall-Wallis. The Linkage Disequilibrium (LD) values for the three pairs of SNPs have been calculated using Haploview Version 4.0 (Website: http://www.broad.mit.edu/mpg/haploview) [16]. Haplotype frequencies were estimated by web based haplotype analysis tool (HAP) (Website: http://research.calit2.net/hap). The most common haplotype was used as the reference, and association between other haplotypes and infertility risk was calculated by using the χ2 test. P values of <0.05 were regarded as statistically significant. SPSS system 15.0 version was used for calculation.
Results
The ESR1 genotypes of the three polymorphisms were in Hardy-Weinberg equilibrium in the fertile group. With the exception of (TA)n microsatellite repeat, other ESR1 polymorphisms were in agreement with the Hardy-Weinberg equilibrium in the infertile group. Genotype distributions and allele frequencies of three ESR1 polymorphisms are shown in Table 1. For the PvuII polymorphism, a noteworthy association was observed between infertile and fertile groups (p < 0.001). Moreover, compared with the PP genotype, the ORs for the Pp and pp elevated 2.25 (95% Cl, 1.22–4.17) and 4.10 (95% Cl, 1.80–9.34) times, respectively. In addition, PvuII alleles, present in the infertile group had also significant difference with the control group (p < 0.001) (Table 1).
Table 1.
Incidence of PvuII, XbaI and (TA)n repeat dinucleotid genotypes in infertile and control groups
| Controls | Cases | p values | OR (95% CI) | p values | |||
|---|---|---|---|---|---|---|---|
| Genotypes | n | (%) | N | (%) | |||
| PvuII | p < 0.001 | ||||||
| PP | 53 | 49.5 | 28 | 26.9 | 1a | ||
| Pp | 42 | 39.3 | 50 | 48.1 | 2.25 (1.22–4.17) | 0.010 | |
| pp | 12 | 11.2 | 26 | 25 | 4.10 (1.80–9.34) | p < 0.001 | |
| Alleles | |||||||
| P | 148 | 69.2 | 106 | 51.0 | 1a | ||
| p | 66 | 30.8 | 102 | 49.0 | 2.16 (1.45–3.21) | p < 0.001 | |
| Genotypes | n | (%) | N | (%) | |||
| XbaI | 0.002 | ||||||
| XX | 54 | 50.5 | 36 | 34.6 | 1a | ||
| Xx | 49 | 45.8 | 50 | 48.1 | 1.53 (0.86–2.73) | 0.148 | |
| xx | 4 | 3.7 | 18 | 17.3 | 6.75(2.11–21.59) | p < 0.001 | |
| Alleles | |||||||
| X | 157 | 73.4 | 122 | 58.7 | 1a | ||
| x | 57 | 26.6 | 86 | 41.3 | 1.94 (1.29–2.93) | p < 0.001 | |
| Genotypes | .n | (%) | N | (%) | |||
| (TA)n repeat | 0.004 | ||||||
| SS | 58 | 54.2 | 35 | 33.6 | 1a | ||
| SL | 41 | 38.3 | 50 | 48.1 | 2.02 (1.12–3.64) | 0.019 | |
| LL | 8 | 7.5 | 19 | 18.3 | 3.94 (1.56–9.94) | 0.004 | |
| Alleles | |||||||
| S | 157 | 73.4 | 120 | 57.7 | 1a | ||
| L | 57 | 26.6 | 88 | 42.3 | 2.02 (1.34–3.04) | 0.001 | |
OR odds ratio, CI confidence interval
aReference genotype/allele
P values <0.05 are shown in bold
There is also an outstanding correlation revealed between XbaI polymorphism and the risk of infertility (p = 0.002). The xx genotype (OR 6.75 [95% CI: 2.11–21.59]) was an approximately sevenfold increased predisposition compared with the carriers of the homozygote common allele (XX genotype), whereas in heterozygotes (Xx genotype) no significant disposition was detected (OR 1.53 [95% CI: 0.86–2.73]). Besides, we found a considerable difference in the allele frequencies of the XbaI polymorphism among groups (p < 0.001) (Table 1).
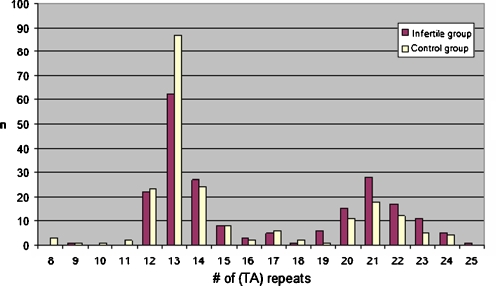
For the (TA)n repeat polymorphism, we observed 18 different alleles with (TA)n repeat number ranging between 8 and 25 (Fig. 3). This polymorphism showed a bimodal distribution, with two peaks at 13 repeats (35.3% of alleles) and 21 repeats (10.9% of alleles), and a breakpoint at 18 TA repeats as reported previously [17–19]. This cutoff point was used to divide alleles into categories of either short (S < 18 TA repeats) or long alleles (L ≥ 18 TA repeats). When divided upon this basis, for the infertile group the (TA)n repeat genotype frequencies were SS = 35 (33.6%); SL = 50 (48.1%); and LL = 19 (18.3%). For the control group same frequences were SS = 58 (54.2%); SL = 41 (38.3%); and LL = 8 (7.5%) (Table 1). A significant relationship was observed between infertile and control group in terms of (TA)n repeat microsatellite polymorphism (p < 0.001). It had been revealed that longer ESR (TA)n was linked with a higher risk for infertility.
Fig. 3.
Frequence distribution of dinucleotide TA repeats in ESR1 gene
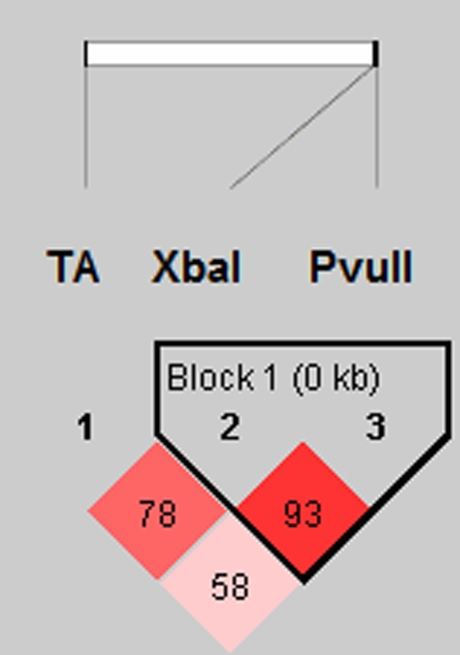
Linkage disequilibrium (LD) was only observed between PvuII and XbaI polymorphisms. No LD was detected between the (TA)n microsatellite polymorphism and PvuII-XbaI (Fig. 4). The distributions of ESR1 haplotypes with estimation of ORs in infertile women and controls are presented in Table 2.
Fig. 4.
Linkage disequilibrium comparisons of TA, PvuII and XbaI polymorphisms in settlement order in the ESR1 gene
Table 2.
Haplotype frequencies of PvuII and XbaI polymorphisms in control and study groups
| Haplotype | Control (2n = 214) (%) | Infertile (2n = 208) (%) | OR (%95 CI) | pa |
|---|---|---|---|---|
| PX | 145 (67.8) | 104 (50.0) | 1b | |
| Px | 54 (25.2) | 84 (40.4) | 2.17 (1.42–3.32) | 0.0001 |
| pX | 12 (5.6) | 18 (8.7) | 2.09 (0.97–4.53) | 0.057 |
| px | 3 (1.4) | 2 (0.9) | 0.93 (0.15–5.66) | 1.000 |
OR Odds Ratio, CI Confidence Interval
aχ2 test was used to compare the groups. Statistically significant results were shown in bold
bReference haplotype
Table 3 summarizes number of follicles and retrieved oocytes, maturation, fertilization and pregnancy rates in each PvuII, XbaI and (TA)n genotype groups. A statistically significant difference was found between maturation and the fertilization rates in women with different ESR1 PvuII genotypes. ESR1 pp genotype tended to be associated with a lower number of fertilized oocyte (p = 0.003). Each PvuII genotype was significantly related with each other in terms of maturation and fertilization rates. Both maturation and fertilization rates and also number of high quality embryos were higher for women with the ESR1 PvuII PP genotype; lower for women with the ESR1 PvuII Pp genotype and the lowest for women with the ESR1 PvuII pp genotype (p < 0.001).
Table 3.
Follicle number, oocyte number, follicle:oocyte ratio, maturation rate and fertilization rate in each of the three genotypes for PvuII, XbaI and TA dinucleotide polymorphisms
| Genotypes of ESR1, PvuII | Genotypes of ESR1, XbaI | Genotypes of ESR1,(TA)n repeat | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PP (n = 28) | Pp (n = 50) | pp (n = 26) | p value | XX (n = 36) | Xx (n = 50) | xx (n = 18) | p value | SS (n = 35) | SL (n = 50) | LL (n = 19) | p value | |
| # of follicles Median(Min-max)% | 18.5 (15.25–23.00) | 19.0 (15.00–23.25) | 19.0 (16.00–25.00) | 0.579a | 18.5 (16.00–23.75) | 19.0 (15.00–24.00) | 19.0 (16.00–23.25) | 0.972a | 18.0 (15.00–24.00) | 19.5 (16.00–23.00) | 19.0 (15.00–24.00) | 0.894a |
| # of collected oocyte Median(Min-max)% | 18.5 (14.00–22.00) | 17.5 (15.00–22.25) | 19.0 (15.00–22.25) | 0.812a | 18.0 (15.00–22.00) | 17.5 (15.00–23.00) | 18.5 (15.00–22.00) | 0.938a | 17.0 (14.00–23.00) | 18.0 (16.00–22.25) | 19.0 (14.00–22.00) | 0.836a |
| Maturation rate Median(Min-max)% | 94.0 (87.09–100.0)b | 93.5 (86.76–96.03)c | 82.0 (63.38–89.83)b,c | p < 0.001a | 94.3 (87.84–100.00)d | 91.7 (80.75–95.50)e | 86.5 (65.00–91.39)d,e | p < 0.001a | 92.9 (83.33–95.55) | 92.5 (85.80–95.59) | 86.4 (66.67–92.86) | 0.075a |
| Fertilization rate Median(Min-max)% | 86.6 (72.1–94.02)b,f | 71.0 (59.58–81.25)f | 60.0 (48.86–75.48)b | p < 0.001a | 84.0 (72.61–94.36)d,g | 68.8 (59.58–78.33)g | 59.5 (39.78–79.56)d | p < 0.001a | 80.0 (70.0–92.85)h,j | 71.1 (60.0–81.53)i,j | 58.8 (40.00–71.43)h,i,j | p < 0.001a |
| # of grade 1–2 embryos Median (Min-max)% | 10.0b (6.00–14.00) | 9.5c (4.00–16.00) | 8.0b,c (4.00–12.00) | p < 0.001 | 9.0 (4.50–17.00) | 10.0 (6.00–14.00) | 8.5 (3.50–14.00) | 0.668 | 10.0 (7.00–12.00) | 12.0 (6.00–17.00) | 10.0 (5.00–14.00) | 0.772 |
| # of grade 3–4 embryos Median(Min-max)% | 6.0b (3.00–10.00) | 7.0c (2.00–10.00) | 7.5b,c (3.00–11.00) | p < 0.001 | 7.0 (2.00–11.00) | 8.0 (2.00–7.00) | 8.0 (3.0–15.00) | 0.645 | 6.5 (3.00–10.00) | 5.5 (2.00–11.00) | 6.0 (2.00–12.00) | 0.739 |
| Frequency (%) of clinical pregnancies | 35.7 | 34.0 | 30.8 | 0.556k | 33.3 | 36.0 | 27.8 | 0.895k | 31.4 | 36.0 | 31.5 | 0.712k |
aKruskal Wallis test
bSignificant relationship was found between the groups when PP and pp genotypes were compared p < 0.001
cSignificant relationship was found between the groups when Pp and pp genotypes were compared p < 0.001
fSignificant relationship was found between the groups when PP and Pp genotypes were compared p < 0.001
dSignificant relationship was found between the groups when XX and xx genotypes were compared p < 0.001
eSignificant relationship was found between the groups when Xx and xx genotypes were compared p = 0.028
gSignificant relationship was found between the groups when XX and Xx genotypes were compared p < 0.001
hSignificant relationship was found between the groups when SS and LL genotypes were compared p < 0.001
iSignificant relationship was found between the groups when SS and SL genotypes were compared p = 0.011
jSignificant relationship was found between the groups when SL and LL genotypes were compared p = 0.013
kLogistic regression analysis
The associations between the ESR1 XbaI polymorphism and IVF parameters were assessed and a statistically significant difference was found between fertilization rate. XX genotypes showed a higher fertilization rate and difference was significant between XX and Xx or xx genotypes (p < 0.001) (Table 3).
Differences in the outcomes of IVF according to ESR1 (TA)n genotypes were also observed. Three ESR1 (TA)n genotypes were also differed with each other significantly in terms of fertilization rates (p = 0.011 for SS and SL genotypes, p < 0.001 for SS and LL genotypes and p = 0.013 for SL and LL genotypes). The mean fertilization rates were higher in SS genotypes (%80.0), lower in SL genotypes (%71.1) and the lowest in LL genotypes (%58.8) (Table 3).
The mean clinical pregnancy rate for all study patients was 33.7% (35/104). The associations between the ESR1 genotypes and the occurence of clinical pregnancy were examined but none of the ESR1 variants included in this study predicted the probability for clinical pregnancy per embryo transfer.
Discussion
Since the first application of assited reproduction techniques, many factors have been associated with the outcome of IVF treatment [20]. Although all the patients were exposed to the same IVF protocol in follicular stimulation, follicular responses among the patients differed significantly. The alterations in the genotype of the estrogen and/or its receptor may be one of the factors that contribute toward such observed variability. Since the estrogen hormone affects maturation of oocytes and provides an optimal oocyte cytoplasm and oolemma maturation [3], it counts as an important factor that determines the quality of oocyte. A good quality oocyte is essential for maturation, fertilization and post-embryonic development. Edwards et al. (1984) reported that fertilization rates were higher with mature oocytes and Tesarik et al. (1995) found that addition of estradiol to human oocyte maturation medium increased the fertilization and cleavage rates of in-vitro matured oocytes [20, 21].
In the present study we have analysed three polymorphisms of the ESR1 gene in patients undergoing ovarian stimulation for in-vitro fertilization and embryo transfer in order to assess the numbers of follicles and oocytes produced by individuals having different ESR1 genotypes as well as the maturation, fertilization and pregnancy rates obtained from these patients. Throughout the study we investigated whether these polymorphisms affect the response and the incidence of a polymorphism is different between women undergoing IVF and controls.
PvuII and XbaI polymorphisms located in intron 1 are approximately 50 bp apart from each other. Their location in the intron makes it unlikely that the polymorphisms may affect ESR expression. Neither PvuII nor XbaI polymorphism cause amino acid substitutions. However they may be in linkage disequilibrium with other ESR1 mutations which may affect both the estrogen receptor gene expression and function [13].
The ESR1 gene promoter has a very complex genomic organization. It contains multiple promoter regions with alternative splice sites, resulting in expression of alternative first exons and different estrogen protein transcripts [22]. Thus (TA)n dinucleotide repeat lenght may affect alternative promoter and first exon usage resulting in different expression patterns [17]. Different expression patterns may affect the function of ESR protein and also the amounts of ESR1 protein that is produced.
We first evaluated the genotype distribution and allele frequencies of the three polymorphisms in Turkish women both in the controls as well as in patients undergoing IVF program. For the PvuII, XbaI and (TA)n polymorphisms, significant relationship was observed between infertile and fertile groups (p = 0.001, p = 0.002 and p = 0.004 respectively). Furthermore we found a considerable difference in the allele frequencies in every three polymorphisms among the groups (p = 0.001, for each three). These results indicate that these polymorphisms may have impact on infertility.
Role of ESR1 polymorphisms on human fertility was indicated previously [23] and it has been reported that ESR1 genotype manifest in modern societies as successful outcome in women undergoing IVF [7, 8]. According to some previous findings polymorphism can affect the outcome of IVF by affecting folliculogenesis, oocyte maturation, embryo quality and endometrial receptivity [8]. In the present study strong negative associations were found between severity of PvuII polymorphism in the ESR1 gene with embryo quality (p < 0.001).
Sundarrajan et al. examined the relationship of PvuII and a rare BstUI polymorphism in the ESR1 gene to the mean numbers of follicles and oocytes, their mean ratios, mean number of embryos and pregnancy rates [8]. They reported that the mean follicular number, oocyte number, embryo number, follicular size and prengancy rate were significantly smaller in patients homozygous for PvuII polymorphism.
Sundarrajan et al. (1999) also investigated 72 pregnant patients and found that the number of obtained and replaced embryos in each of the three PvuII genotypes showed a highly significant negative correlation with the severity of the polymorphism [8]. In our study fertilization rate was found to have a strong negative correlation with the severity of each PvuII, XbaI and (TA)n repeat polymorphisms. Moreover a significant relationship was observed between maturation rates and PvuII & XbaI genotypes (p < 0.001).
Recently, Altmae et al. (2007) evaluated the impacts of ESR1 PvuII, XbaI and (TA)n genotypes on the the etiology of female infertility, as well as their contributions to the COH and pregnancy outcome of IVF in 159 infertile women undergoing IVF-ET [10]. They concluded that ESR1 variants predict the chance for clinical pregnancy rate per COH rather than per single embryo transfer. Contrary to findings in this study, Georgiou et al. (1997) and Sundarrajan et al. (1999) showed that there was a relationship between some ESR1 variants and clinical pregnancy rate per embryo transfer [7, 8]. In our study the impact of polymorphism on maturation rate, fertilizaton rate and post-embryonic development was shown nevertheless no statistical relationship was detected between any of the ESR1 variants and pregnancy rate.
When a haplotype analysis was made in order to interrogate the linkage between these three polymorphisms, linkage disequilibrium was only detected between PvuII and XbaI polymorphisms. For the infertile and fertile group four haplotypes were observed and the most frequent haplotype; PX in the control group was assigned as the reference. Infertility risk was estimated by comparing other haplotype frequencies against the reference and infertility risk for women having Px genotype was found to be approximately 2 times higher than that for the women having PX genotype (p < 0.001).
Conclusions
In conclusion, this study showed a significant association between ESR1 genotypes and risk for infertility and some IVF parameters. Still, further studies are needed to confirm our findings in larger scale studies, which will probably reveal more significant results. The expression of ESR1 could be regulated depending on the ESR1 genotypes and PvuII, XbaI and (TA)n polymorphisms may serve as markers in predicting the risk for infertility, ovarian response of IVF patients and success rates of IVF treatment. Nevertheless further studies are necessary to determine whether it is possible to apply this relationship to the pre-cycle evaluation of individual genetic predisposition.
Acknowledgments
This work was supported by the Gazi University Research Fund as a project with code number 01/2007–89.
Footnotes
Capsule
In this study role of ESR1 gene polymorphisms on infertility was focused and a significant association between ESR1 genotypes and some IVF parameters was reported.
References
- 1.Goldenberg RL, Vaitukaitis JL, Ross GT. Estrogen and follicle- stimulating hormone interactions on follicle growth in rats. Endocrinology. 1972;90:1492–8. [DOI] [PubMed]
- 2.Ireland JJ, Richards JS. Acute effects of estradiol and follicle-stimulating hormone on specific binding of human [125I] iodofollicle-stimulating hormone to rat ovarian granulosa cells in vivo and in vitro. Endocrinology. 1978;102:876–83. [DOI] [PubMed]
- 3.Filicori M, Cognigni GE, Taraborrelli S, Spettoli D, Ciampaglia W, de Fatis CT, et al. Luteinizing hormone activity supplementation enhances follicle-stimulating hormone efficacy and improves ovulation induction outcome. J Clin Endocrinol Metab. 1999;84(8):2659–63. [DOI] [PubMed]
- 4.Walter P, Green S, Greene G, Krust A, Bornert JM, Jeltsch JM, et al. Cloning of the human estrogen receptor cDNA. Proc Natl Acad Sci U S A. 1985;82(23):7889–93. [DOI] [PMC free article] [PubMed]
- 5.Mosselman S, Polman J, Dijkema R. ER beta: identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996;392(1):49–53. [DOI] [PubMed]
- 6.Aléssio AM, Siqueira LH. de Carvalho ECCouto, Barini R, Mansur AP, Hoehr NF et al. Estrogen Receptor Alpha and Beta Gene Polymorphisms Are Not Risk Factors for Recurrent Miscarriage in a Brazilian Population. Clin Appl Thromb/Hemost. 2008;14:180–5. [DOI] [PubMed]
- 7.Georgiou I, Konstantelli M, Syrrou M, Messinis IE, Lolis DE. Oestrogen receptor gene polymorphisms and ovarian stimulation for in-vitro fertilization. Hum Reprod. 1997;12(7):1430–3. [DOI] [PubMed]
- 8.Sundarrajan C, Liao W, Roy AC, Ng SC. Association of oestrogen receptor gene polymorphisms with outcome of ovarian stimulation in patients undergoing IVF. Mol Hum Reprod. 1999;5:797–802. [DOI] [PubMed]
- 9.Kukuvitis A, Georgiou I, Bouba I, Tsirka A, Giannouli CH, Yapijakis C, et al. Association of oestrogen receptor alpha polymorphisms and androgen receptor CAG trinucleotide repeats with male infertility: a study in 109 Greek infertile men. Int J Androl. 2002;25(3):149–52. [DOI] [PubMed]
- 10.Altmäe S, Haller K, Peters M, Hovatta O, Stavreus-Evers A, Karro H, et al. Allelic estrogen receptor 1 (ESR1) gene variants predict the outcome of ovarian stimulation in in vitro fertilization. Mol Hum Reprod. 2007;13(8):521–6. [DOI] [PubMed]
- 11.Hsieh YY, Wang YK, Chang CC, Lin CS. Estrogen receptor alpha-351 XbaI*G and -397 PvuII*C-related genotypes and alleles are associated with higher susceptibilities of endometriosis and leiomyoma. Mol Hum Reprod. 2007;13(2):117–22. [DOI] [PubMed]
- 12.Baltaci V, Şatıroğlu H, Kabukçu C, Ünsal E, Aydınuraz B, Üner Ö, et al. Relationship Between Embryo Quality and Aneuploidies. Reprod BioMed Online. 2006;12:77–82. [DOI] [PubMed]
- 13.Yaich L, Dupont WD, Cavener DR, Parl FF. Analysis of the PvuII restriction fragment length polymorphism and exon structure of the estrogen receptor gene in breast cancer and peripheral blood. Cancer Res. 1992;52:77–83. [PubMed]
- 14.Kobayashi S, Inoue S, Hosoi T, Ouchi Y, Shiraki M, Orimo H. Association of bone mineral density with polymorphism of the estrogen receptor gene. J Bone Miner Res. 1996;11:306–11. [DOI] [PubMed]
- 15.Raymond M, Rousset F. GENEPOP (version 4.0): an updated version of GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Heredity. 1997;86:248–9.
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. [DOI] [PubMed]
- 17.Becherini L, Gennari L, Masi L, Mansani R, Massart F, Morelli A, et al. Evidence of a linkage disequilibrium between polymorphisms in the human estrogen receptor alpha gene and their relationship to bone mass variation in postmenopausal Italian women. Hum Mol Genet. 2000;9(13):2043–50. [DOI] [PubMed]
- 18.Albagha OM, Pettersson U, Stewart A, McGuigan FE, MacDonald HM, Reid DM, et al. Association of oestrogen receptor alpha gene polymorphisms with postmenopausal bone loss, bone mass, and quantitative ultrasound properties of bone. J Med Genet. 2005;42:240–6. [DOI] [PMC free article] [PubMed]
- 19.Van Meurs JB, Schuit SC, Weel AE, van der Klift M, Bergink AP, Arp PP, et al. Association of 5′ estrogen receptor alpha gene polymorphisms with bone mineral density, vertebral bone area and fracture risk. Hum Mol Genet. 2003;12(14):1745–54. [DOI] [PubMed]
- 20.Edwards RG, Fishel SB, Cohen J, Fehilly CB, Purdy JM, Slater JM, et al. Factors influencing the success of in vitro fertilization for alleviating human infertility. J In Vitro Fert Embryo Transf. 1984;1(1):3–23. [DOI] [PubMed]
- 21.Tesarik J, Mendoza C. Nongenomic effects of 17 beta-estradiol on maturing human oocytes: relationship to oocyte developmental potential. J Clin Endocrinol Metab. 1995;80(4):1438–43. [DOI] [PubMed]
- 22.Kos M, Reid G, Denger S, Gannon F. Genomic organization of the human ERα gene promoter region. Mol Endocrinol. 2001;15:2057–63. [DOI] [PubMed]
- 23.Corbo RM, Ulizzi L, Piombo L, Martinez-Labarga C, De Stefano GF, Scacchi R. Estrogen receptor alpha polymorphisms and fertility in populations with different reproductive patterns. Mol Hum Reprod. 2007;13(8):537–40. [DOI] [PubMed]