Abstract
SUMMARY
2-Oxoglutarate-dependent dioxygenases, including the EglN prolyl hydroxylases that regulate HIF, can be inhibited with drug-like molecules. EglN2 is estrogen-inducible in breast carcinoma cells and the lone Drosophila EglN interacts genetically with Cyclin D1. Although EglN2 is a non-essential gene we found that EglN2 inactivation decreases Cyclin D1 levels and suppresses mammary gland proliferation in vivo. Regulation of Cyclin D1 is a specific attribute of EglN2 among the EglN proteins and is HIF-independent. Loss of EglN2 catalytic activity inhibits estrogen-dependent breast cancer tumorigenesis and can be rescued by exogenous Cyclin D1. EglN2 depletion also impairs the fitness of lung, brain, and hematopoietic cancer lines. These findings support the exploration of EglN2 inhibitors as therapeutics for estrogen-dependent breast cancer and other malignancies.
SIGNIFICANCE
Cyclin D1 plays an important role in many cancers, including breast cancer. The observations described herein predict that inhibiting EglN2 catalytic activity will diminish Cyclin D1 levels in cancer cells and impair their ability to proliferate in vivo. Notably, EglN2 is estrogen-inducible and loss of either EglN2 or Cyclin D1 leads to mammary gland hypoproliferation. Therefore the relationship between EglN2 and Cyclin D1 might be especially relevant in hormone-sensitive breast cancer, where new therapies are needed for women who become refractory to estrogen antagonists. EglN2 appears to be an attractive drug target because EglN2 is not essential in mammals and it has already been established that enzymes of this class can be inhibited with drug-like small organic molecules.
INTRODUCTION
Most successful drugs are small organic molecules that bind to, and inhibit, specific cellular proteins. Proteins that serve as enzymes have proven to be particularly tractable as drug targets. Establishing additional classes of enzymes that can be manipulated with small organic molecules opens new avenues for drug discovery.
The 2-oxoglutarate and iron-dependent dioxygenase superfamily includes the collagen prolyl and lysyl hydroxylases, the FTO and AlkB DNA demethylases, the JmC-containing histone demethylases, the FIH1 asparaginyl hydroxylase, and the EglN family prolyl hydroxylases (Aravind and Koonin, 2001; Klose et al., 2006; Pollard et al., 2008; Taylor, 2001). These enzymes can be inhibited with drug-like small molecules that compete with 2-oxoglutarate or interfere with iron utilization, both in vitro and in vivo (Bruegge et al., 2007; Mole et al., 2003; Ozer and Bruick, 2007; Safran et al., 2006).
There are three EglN (also called PHD or HPH) family members in humans, called EglN1, EglN2, and EglN3 (Kaelin and Ratcliffe, 2008). All three enzymes are capable of hydroxylating the alpha subunit of the heterodimeric transcription factor HIF (hypoxia-inducible factor). Prolyl hydroxylated HIFα is recognized by a ubiquitin ligase complex containing the pVHL tumor suppressor protein, leading to its polyubiquitinylation and subsequent proteasomal degradation. EglN family members exhibit Km values for oxygen that exceed the oxygen concentrations found in mammalian tissues (Kaelin and Ratcliffe, 2008). Accordingly, these enzymes are highly sensitive to decrements in oxygen availability, such as might occur following an interruption in blood supply. HIF regulates a program of gene expression that facilitates survival under hypoxic conditions through cell-intrinsic changes in metabolism and cell-extrinsic changes affecting oxygen delivery. For example, HIF activates the transcription of genes such as erythropoietin that enhance red blood cell production and hence blood oxygen carrying capacity. EglN antagonists stimulate red blood cell production in mammals and are currently undergoing Phase II testing for different forms of anemia (Hsieh et al., 2007; Safran et al., 2006).
EglN1 (also called PHD2) is the primary prolyl hydroxylase responsible for HIF regulation (Berra et al., 2003; Minamishima et al., 2008; Takeda et al., 2008). EglN2 (also called PHD1) and EglN3 (also called PHD3) might also regulate HIF under certain conditions (Appelhoff et al., 2004). For example, EglN3 is itself a HIF target, is induced by hypoxia, and has a lower oxygen Km that EglN1 (Appelhoff et al., 2004; Minamishima et al., 2009). Cell culture and animal experiments support that EglN3 partially compensates for EglN1 when the latter is inactivated by hypoxia (Appelhoff et al., 2004; Minamishima et al., 2009). Whether EglN2 and EglN3 have HIF-independent functions is less clear although recent studies support a HIF-independent role for EglN3 in the control of apoptosis (Rantanen et al., 2008; Schlisio et al., 2008).
Polyak and coworkers reported that EglN2 mRNA accumulates in breast cancer cells that have been stimulated to proliferate with estrogen and that EglN2 overexpression promotes estrogen-independent growth and tamoxifen resistance (Seth et al., 2002). Frei and Edgar noted that certain phenotypes observed in flies engineered to overproduce Cyclin D1 were abrogated by concurrent inactivation of Egl9, which is the lone ancestral EglN family member in Drosophila (Frei and Edgar, 2004). Since Cyclin D1 plays an important role in many forms of cancer, including breast cancer, and is induced by estrogen in estrogen-receptor positive breast cancers (Bartkova et al., 1994; Landis et al., 2006; Roy and Thompson, 2006; Yu et al., 2001), we asked whether EglN2 activity affects Cyclin D1 activity.
RESULTS
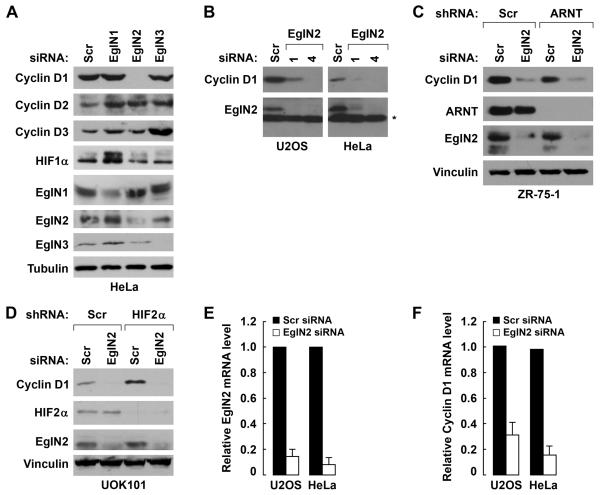
Toward this end, we transiently transfected HeLa cervical carcinoma cells, U2OS osteosarcoma cells, and both T47D and ZR-75-1 breast carcinoma cells with previously validated siRNAs that are specific for EglN1, EglN2, or EglN3 (Appelhoff et al., 2004). Downregulation of EglN2, but not EglN1 or EglN3, decreased Cyclin D1 protein levels (Fig 1A, Supplemental Fig 1A, and data not shown). Similar results were observed with a second, independent, EglN2 siRNA and downregulation of Cyclin D1 by the two different EglN2 siRNAs mirrored their ability to downregulate EglN2 (Fig 1B and Supplemental Fig 1B). In some experiments Cyclin D3 was also decreased (data not shown). As expected, suppression of EglN1, but not EglN2 or EglN3, induced HIF1α (Fig 1A). These results suggest that Cyclin D1 is specifically regulated by EglN2 amongst the EglN family members and that EglN2 regulates Cyclin D1 in a HIF-independent manner.
Fig 1. EglN2 Regulates Cyclin D1.
(A) Immunoblot analysis of Hela cells 48 hours after transfection with siRNAs targeting EglN1, EglN2, EglN3, or a scrambled control siRNA.
(B, E-F) Immunoblot (B) and qRT-PCR (E and F) analysis of U2OS and HeLa cells transfected with two independent siRNAs (#1 and #4) targeting EglN2 (* indicates nonspecific bands). Error bars = 1 SEM.
(C) Immunoblot analysis of ZR-75-1 cells infected with a lentivirus encoding an ARNT shRNA or scrambled control (Scr) followed by tranfection with siRNA against EglN2 or scrambled control.
(D) Immunoblot analysis of UOK101 cells infected with a lentivirus encoding an HIF2α shRNA or scrambled control (Scr) followed by tranfection with siRNA against EglN2 or scrambled control.
In further support of the latter conclusion, downregulation of Cyclin D1 after EglN2 loss was not affected by concurrent inactivation of the HIFα heterodimeric partner ARNT (HIF1β) (Fig 1C and Supplemental Fig 2A). In addition, EglN2 loss decreased Cyclin D1 in UOK101 and 769-P VHL−/−renal carcinoma cells, which constitutively produce HIF2α protein due to the absence of pVHL and produce neither HIF1α mRNA nor protein (Maxwell et al., 1999) (Fig 1D and Supplemental Fig 2B and data not shown). Moreover, elimination of HIF2α in these cells with a highly effective shRNA did not prevent the loss of Cyclin D1 in cells depleted of EglN2 (Fig 1D and Supplemental Fig 2B). Collectively, these results strongly suggest that the regulation of Cyclin D1 by EglN2 is not mediated by changes in HIF activity. Note that in some experiments EglN2 protein migrated as a doublet (for example, Fig 1C), probably due to alternative translation initiation (Tian et al., 2006) .
Both EglN2 mRNA and Cyclin D1 mRNA levels were diminished in cancer cells transfected with EglN2 siRNA, but not in cells transfected with a scrambled control siRNA (Fig 1E and 1F, Supplemental Fig. 2C and 2D). Moreover, we have not detected specific binding of EglN2 to Cyclin D1 and EglN2 failed to hydroxylate Cyclin D1 in vitro (data not shown). Collectively, these results suggest that the regulation of Cyclin D1 by EglN2 is indirect and involves changes in Cyclin D1 transcription or mRNA stability.
To examine this further, we measured the levels of heterogenous nuclear Cyclin D1 RNA, indicative of newly transcribed mRNA precursors, and recruitment of RNA Polymerase II to the Cyclin D1 promoter, indicative of on-going transcription, in cells after EglN2 depletion. In both T47D and ZR-75-1 breast carcinoma cells depletion of EglN2 with an effective shRNA decreased heterogenous nuclear Cyclin D1 RNA levels (Supplemental Fig. 3A-D) and decreased loading of RNA Polymerase II onto the Cyclin D1 promoter (Supplemental Fig. 3E and 3F) relative to cells treated with a scrambled control shRNA. In contrast, we did not detect a difference in Cyclin D1 mRNA stability in cells that were infected to produce either the EglN2 or control shRNA and then treated with actinomycin to prevent new mRNA synthesis (data not shown). Therefore the regulation of Cyclin D1 by EglN2 is at least partly at the level of transcription.
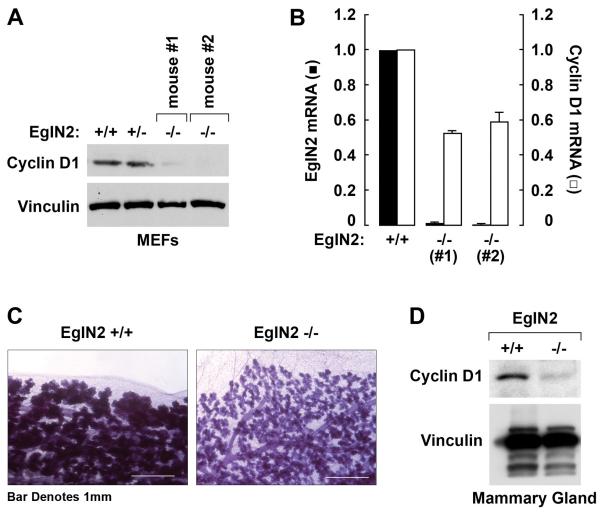
EglN2−/− mice are viable and grossly normal (Aragones et al., 2008; Takeda et al., 2006). In keeping with the siRNA-based experiments described above, we found that Cyclin D1 mRNA and protein levels are diminished in EglN2−/− mouse embryo fibroblasts (MEFs) (Fig 2A and 2B). Moreover, we observed that older, pregnant, EglN2−/− mice do not breastfeed their pups properly compared to littermate controls, a phenotype previously observed in Cyclin D1−/− mice (Sicinski et al., 1995). Moreover, mammary glands from older, lactating, EglN2−/− mice revealed evidence of hypoproliferation reminiscent of, but not as severe as, seen in Cyclin D1−/− mice (Sicinski et al., 1995) (Fig 2C) and exhibited lower levels of Cyclin D1 protein (Fig 2D). Therefore EglN2 regulates Cyclin D1 in vivo, with loss of EglN2 leading to a hypomorphic Cyclin D1 phenotype.
Fig 2. Decreased Cyclin D1 Levels in EglN2−/−Mice.
(A and B) Immunoblot (A) and qRT-PCR analysis (B) of MEFs prepared from littermates with the indicated genotypes. Error bars = 1 SEM.
(C) Whole mounts of mammary glands from wild-type and EglN2−/− mice 1 day postpartum. Magnification= 6X.
(D) Immunoblot analysis of mammary glands as in (C).
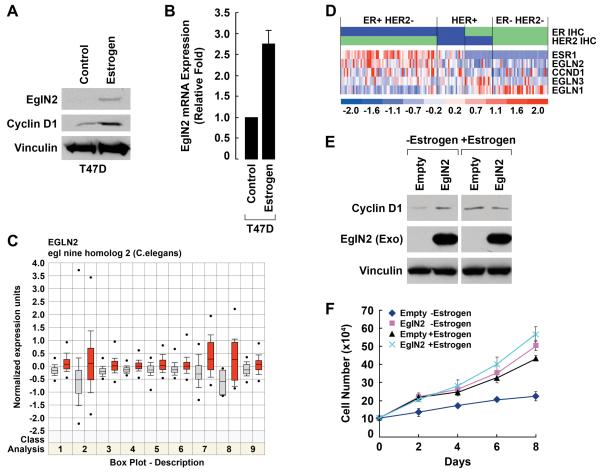
Since EglN2 mRNA is induced by estrogen in human breast cancer cells (Appelhoff et al., 2004; Seth et al., 2002), and EglN2 loss affects mammary gland proliferation, we next focused our attention on the role of EglN2 in human breast cancer. We first confirmed that EglN2 protein levels, like EglN2 mRNA levels, are induced by estrogen in human (T47D) breast cancer cells (Fig 3A and B). Moreover, EglN2 mRNA levels are increased in estrogen receptor (ER) positive breast cancers compared to ER negative breast cancers (Fig 3C and D) and Cyclin D1 mRNA and EglN2 mRNA levels are positively correlated with one another across breast cancers (Supplemental Fig 4). By contrast, EglN1 mRNA levels appear to be highest in ER negative, Her2 negative, breast cancers and EglN3 mRNA levels highest in ER negative, Her2 positive, breast cancers (Fig 3D). Notably, both EglN1 and EglN3 are HIF targets. Although Her2 activation has been reported to activate HIF (Laughner et al., 2001; Li et al., 2005) we observed the clearest evidence of HIF activation, as determined by accumulation of canonical HIF-responsive mRNAs, in the ER negative, Her2 negative breast cancers (A.L.R. and W.G.K.-data not shown).
Fig 3. EglN2 is Estrogen Inducible.
(A and B) Immunoblot (A) and qRT-PCR analysis (B) of T47D cells treated with estrogen or vehicle. Error bars = 1 SEM.
(C) Normalized EglN2 mRNA levels in 9 publically available mRNA expression profile datasets. Red boxes = ER positive breast cancers. Blue Boxes = ER negative breast cancers. P values for these 9 datasets are: 1.2E-9; 3.4E-8; 6.2E-8; 1.1E-7; 2.1E-7; 6.3E-7; 1.4E-6; 5.6E-5 and 9.6E-5, respectively.
(D) Relative expression of EglN2 and other genes of interest in subsets of breast cancers as analyzed by gene expression array. Samples are arranged into subsets according to immunohistochemistry (IHC) staining results for estrogen receptor (Blue indicates ≥1% positive nuclei and green indicates <1% positive nuclei) and HER-2/neu [blue indicates HER-2/neu score of 3+ (strong complete membrane staining in >10% of cells) and green a HER-2/neu score of 0, 1 or 2+]. The middle panel is a display of the relative gene expression (red indicates high expression and blue indicates low expression) with each column representing an individual tumor sample and each row representing the results for the indicated genes. Cyclin D1 (CCND1) mRNA levels are higher in ER positive tumors compared to ER negative tumors [mean levels 500 vs 330 (arbitrary units), p < 2E-8].
(E and F) Immunoblot analysis (E) and proliferation assay (F) of T47D cells infected with retrovirus encoding HA-EglN2 or with the empty vector in the presence or absence of estrogen (10nM) treatment. In (E) estrogen exposure was for 48 hours prior to cell harvest and anti-HA antibody was used to detect exogenous (Exo) EglN2. Error bars = 1 SEM.
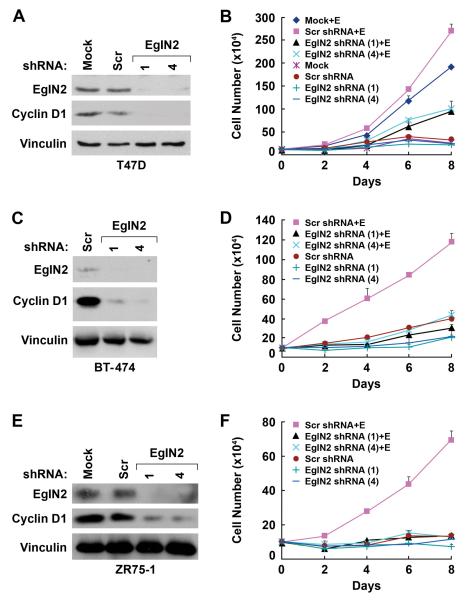
Forced overexpression of EglN2 promotes colony formation by T47D cells (Seth et al., 2002). Similarly, we found that overexpression of EglN2 was sufficient to promote the proliferation of T47D cells in the absence of estrogen (Fig 3E and F). Exogenous EglN2 had only minimal effects on Cyclin D1 and proliferation in the presence of estrogen, however, presumably because the endogenous EglN2 is no longer limiting under these conditions (Fig 3E and F). To ask if EglN2 loss would inhibit breast cancer cell proliferation we infected T47D cells with retroviral vectors encoding short hairpin RNAs corresponding to the two EglN2 siRNAs used above. Downregulation of both EglN2 and Cyclin D1 by the EglN2 shRNAs, but not control (scrambled GFP) shRNA, was confirmed by immunoblot analysis (Fig 4A).
Fig 4. Downregulation of EglN2 Suppresses Cancer Cell Proliferation.
(A and B). Immunoblot (A) and cell proliferation assay (B) of T47D cells infected with retrovirus encoding shRNA against EglN2 (sequence 1 or 4) or a scrambled control shRNA. In (B) cells were grown in Phenol Red-free RPMI medium supplemented with 5% charcoal/dextran treated FBS in the presence or absence of estrogen (10 nM) as indicated.
(C - F). Immunoblot (C and E) and cell proliferation (D and F) assay of BT474 (C and D) and ZR75-1 cells (E and F). Error bars = 1 SEM.
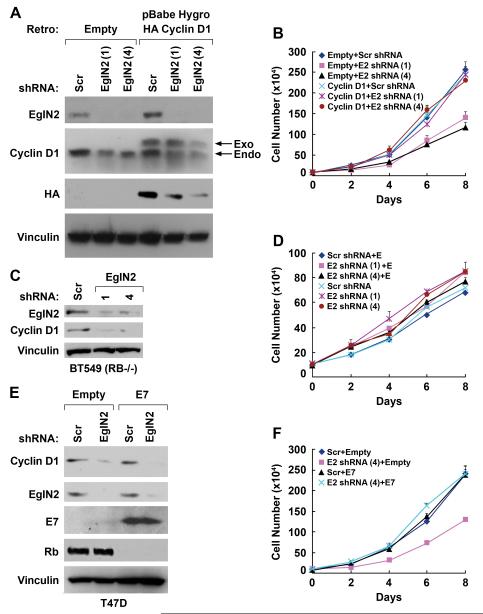
As expected, T47D cells infected with the control shRNA, like parental T47D cells, proliferated in the presence of estrogen but not in its absence (Fig 4B). Proliferation in the presence of estrogen was markedly reduced, however, in T47D cells infected with either of the two EglN2 shRNAs (Fig 4B). The effects of EglN2 reduction on proliferation were even more striking in the estrogen-dependent cell lines BT-474 (Fig 4C and 4D) and ZR75-1 (Fig 4E and 4F).
The observation that two independent EglN2 shRNAs, but not the control shRNA, inhibited cell proliferation argues that this phenotype is due to effects on EglN2 activity (“on-target”). To test this further, we performed rescue experiments using T47D cells that were infected with a retrovirus encoding a non-natural EglN2 mRNA in which translationally silent mutations were introduced into the sequence targeted by shRNA #4. These cells, but not cells infected with an empty retrovirus, were now insensitive to the Cyclin D1 suppressive (Fig 5A) and antiproliferative effects of the EglN2 shRNA #4 (Fig 5B). These findings, together with our analysis of EglN2−/− cells, support that EglN2 regulates Cyclin D1-dependent cell proliferation.
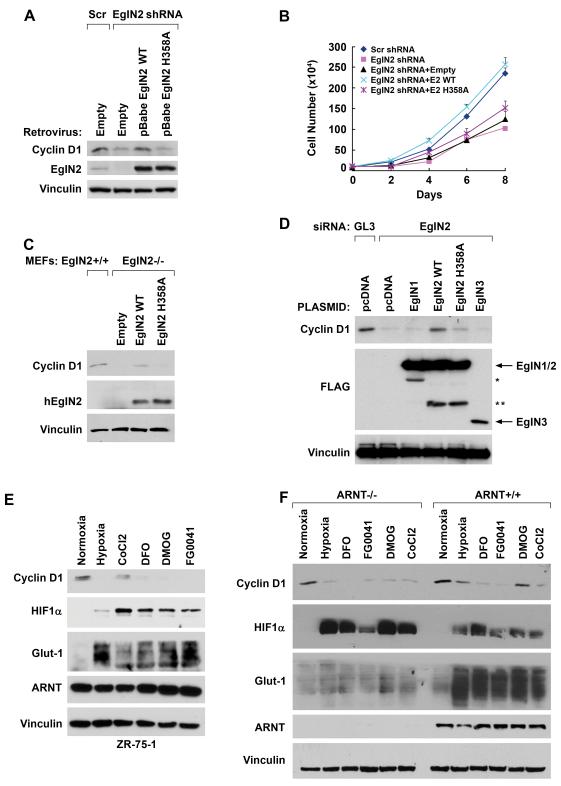
Fig 5. Control of Cyclin D1 and Proliferation by EglN2 is Hydroxylase-Dependent.
(A and B). Immunoblot (A) and cell proliferation assay (B) of T47D cells that were first infected with a retrovirus expressing an shRNA-resistant mRNA encoding wild-type or catalytic-dead (H358) EglN2 (or with empty vector) and then infected with a EglN2 shRNA retrovirus (or scrambled shRNA vector).
(C) Immunoblot analysis of EglN2−/− MEFs infected with retroviruses encoding EglN2 (wild-type), EglN2 H358A, or with the empty vector. An EglN2+/+ extract was included in lane 1 as a control. Note that the EglN2 antibody does not recognize murine EglN2.
(D) Immunoblot analysis of Hela cells transiently transfected with plasmids encoding the indicated EglN proteins (or the empty vector) and siRNAs against EglN2 or luciferase (GL3). The molecular bases for the bands indicated by the asterisks are unknown.
(E and F) Immunoblot analysis of ZR-75-1 cell (E) and isogenic murine hepatoma cells (ARNT −/− or ARNT +/+)(F) treated overnight with hypoxia (0.2% O2), CoCl2 (200 μM), DFO (200 μM), DMOG (1 mM) or FG0041 (40 μM).
Error bars = 1 SEM.
In parallel, we tested cells producing an shRNA #4-resistant EglN2 mRNA encoding a hydroxylase-defective EglN2 mutant (EglN2 H358A) [(Epstein et al., 2001) and Supplemental Fig 5]. This mutant, in which a canonical histidine residue within the EglN2 catalytic domain has been replaced by alanine, did not rescue cyclin D1 levels (Fig 5A) and did not rescue proliferation in T47D cells infected to produce EglN2 shRNA #4 (Fig 5B). Similarly, the retrovirus encoding wild-type EglN2, but not EglN2 H358A, restored Cyclin D1 in EglN2−/− MEFs (Fig 5C). It should be noted that the EglN2 H358A has a shorter half-life than wild-type EglN2 (Supplemental Fig 6 and data not shown). Therefore a higher titer of the EglN2 H358A virus was used to achieve comparable levels of wild-type and mutant EglN2 in Fig 5A and 5C. In viral titration experiments wild-type EglN2 rescued Cyclin D1 levels over a wide range of titers whereas mutant EglN2 did not rescue at any titer tested (Supplemental Fig 6 and data not shown).
Similarly, wild-type EglN2, but not EglN2 H358A, rescued Cyclin D1 production in HeLa cells transiently transfected with EglN2 siRNA #4 (Fig 5D). This effect was specific because neither EglN1 nor EglN3 rescued Cyclin D1 levels when tested in parallel (Fig 5D). In these experiments EglN3, however, appeared to be unstable, possibly due to polyubiquitinylation by SIAH (Nakayama et al., 2004). Finally, endogenous Cyclin D1 levels were decreased in cells exposed to either hypoxia or small molecule hydroxylase inhibitors (Fig 5E and 5F). Downregulation of Cyclin D1 by hypoxia and hydroxylase inhibitors was due, at least in part, to decreased Cyclin D1 mRNA levels and was not a indirect consequence of activating the HIF transcriptional response as it did not depend upon the canonical HIFα partner ARNT (HIF1β) (Fig 5F and Supplemental Fig. 7). These results strongly suggest that regulation of Cyclin D1 by EglN2 is linked to the ability of EglN2 to hydroxylate one or more substrates other than HIF.
Cyclin D1 stimulates proliferation by promoting the phosphorylation of the retinoblastoma protein (pRB) and cells lacking pRB are inured to inhibitors of Cyclin D1-associated kinase activity. Overexpression of Cyclin D1, like overexpression of EglN2 itself, rescued cell proliferation in T47D cells expressing an EglN2 shRNA (Fig 6A and 6B). Moreover, the proliferation of breast cancer cells lacking pRB function due to RB1 mutation (Fig 6C and 6D), exogenous expression of the E7 oncoprotein (Fig 6E and 6F), or exogenous expression of RB1 shRNAs (Supplemental Fig 8) was not impaired by EglN2 loss despite diminished Cyclin D1 levels. These results support that impaired proliferation in cells lacking EglN2 is due, at least partly, to loss of Cyclin D1. We also noted that exogenous Cyclin D levels were diminished by EglN2 in some experiments (Fig 6A). The significance of this finding is not yet clear but could reflect a postranscriptional link between EglN2 and Cyclin D1.
Fig 6. Impaired Proliferation of Cells Lacking EglN2 is Due to Cyclin D1 Loss.
(A and B). Immunoblot (A) and cell proliferation (B) assay of T47D cells that were infected with retroviruses encoding shRNAs against EglN2 [sequence 1 or 4](or scrambled control) and then infected with a retrovirus encoding HA-Cyclin D1 (or empty vector).
(C and D). Immunoblot (C) and cell proliferation assay (D) of BT549 RB−/− breast carcinoma cells infected with retroviruses encoding shRNAs against EglN2 (sequence 1 or 4) or scrambled control shRNA. In (D) cells were grown in Phenol Red-free RPMI medium supplemented with 5% charcoal/dextran treated FBS in the presence or absence of estrogen (10 nM) as indicated.
(E and F). Immunoblot (E) and cell proliferation (F) assay of T47D cells that were infected with a retrovirus encoding E7 (or empty retrovirus) and then superinfected retroviruses encoding shRNAs against EglN2 [sequence 4](or scrambled control). Note that E7 promotes the degradation of pRB (Boyer et al., 1996) and that hyperphosphorylated and hypophosphorylated pRB are not resolved under these electrophoretic conditions.
Error bars = 1 SEM.
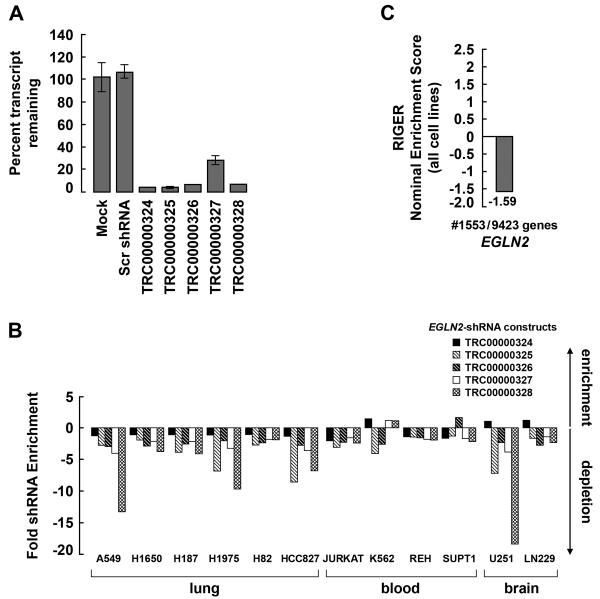
To ask whether EglN2 ablation would diminish cancer cell proliferation or survival in other cell lineages, we obtained multiple shRNA constructs targeting EglN2 from The RNAi Consortium (TRC) at the Broad Institute. We confirmed that each of these constructs effectively suppressed EglN2 (Fig 7A). We next examined data from a set of 12 cancer cell lines that were screened with a pooled version of the TRC shRNA library (Luo et al., 2008) to determine if cells harboring these EglN2-specific shRNAs were depleted from a population of cells during 28 days. Indeed, we observed that cells expressing EglN2-specific shRNAs were depleted from the population for each of the cell lines tested. Specifically, we observed that these constructs were depleted 2.8 fold on average with a maximum depletion of 18.4 fold for construct #5 in the U251 cell line (Fig 7B). EglN2 ranks in the top 16.5% (rank #1553/9423 genes) of depleted genes across all 12 cell lines as compared to all genes examined in this pooled screen (Fig 7C). Together, these observations support that EglN2 is essential for proliferation of cancer cell lines derived from multiple lineages.
Figure 7. Impaired Fitness of Diverse Cancer Cell Lines Lacking EglN2.
(A). EglN2 mRNA abundance, as determined by real-time PCR, in U2OS cells infected with the indicated lentiviruses.
(B). Normalized abundance of the indicated EglN2 shRNA vectors, determined using microarray hybridization of genomic DNA, 28 days after initial infection with a pool containing ~45,000 lentiviral shRNA vectors and subsequent passage in vitro.
(C). EglN2 is an essential gene as determined by the RIGER algorithm (Luo et al 2008). The 5 shRNA constructs targeting EglN2 were treated as a set which was compared to the sets derived from shRNA constructs targeting each of the other ~9,500 genes within each of the 12 cell lines shown in (B). A KS statistic was used to assess bias of the EglN2 shRNA set as showing evidence of depletion during the experiment in (B). A second application of RIGER was then used to identify genes commonly essential among the 12 cell lines. The score and rank of EglN2 from this analysis are shown.
Error bars = 1 SEM.
To ask whether downregulation of EglN2 would affect tumor growth in vivo, ZR75-1 breast carcinoma cells were next infected with lentiviruses encoding EglN2 shRNAs (#1 or #4) under the control of a doxycycline-inducible promoter. Cells infected with an analogous lentivirus encoding a scrambled GFP shRNA served as a control. As expected, doxycycline treatment of cells infected with the EglN2 shRNA lentiviruses led to decreased EglN2 and Cyclin D1 protein levels, and decreased cell proliferation compared to cells grown in the absence of doxycycline (Fig 8A and 8B). Cessation of proliferation in this model was associated with an apparent G1/S block, consistent with loss of Cyclin D1 function (Supplemental Fig 9). These effects of doxycycline were specific because they were not observed in cells infected with the control shRNA lentivirus. Gene expression profiling, combined with gene set enrichment analysis, confirmed that EglN2 depletion led to downregulation genes linked to cell-cycle progression, estrogen-dependent signaling, and tamoxifen-resistance (Supplemental Fig 10 and Supplemental Tables 1, 2, 3).
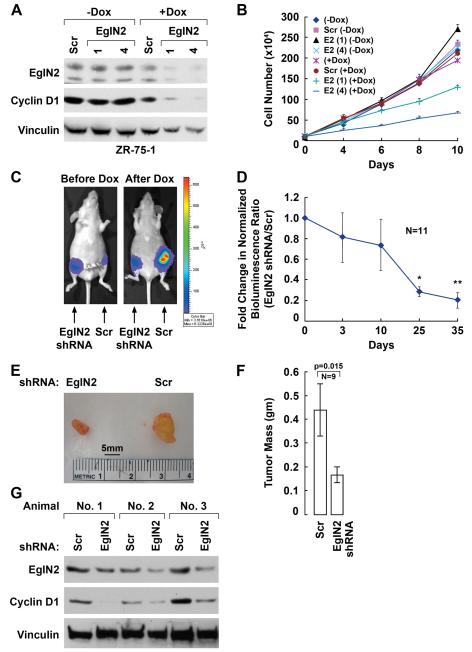
Fig 8. Downregulation of EglN2 Suppresses Tumorigenesis.
(A) and (B). Immunoblot (A) and cell proliferation assay (B) of ZR75-1 cells infected with doxycycline (DOX)-inducible lentiviruses encoding shRNAs against EglN2 [sequence 1 or 4](or scrambled control). Cells were grown in RPMI supplemented with 10% fetal bovine serum in the presence or absence of doxycycline.
(C). Representative bioluminescent images of orthotopic tumors formed by ZR75-1 cells as in (A) that were then superinfected with a retrovirus encoding firefly luciferase. 8 × 106 cells were injected into the 4th mammary glands of nude mice implanted with estrogen pellets. Bioluminescent images were obtained one week later (day 0) and serially after mice were begun on chow containing doxycycline (day 3). Shown in (C) are day 0 image (Before Dox) and day 35 (After Dox).
(D). Quantitation of imaging studies as in (C). *p<0.01 for comparison between Day 25 and Day 0, **p<0.01 for comparison between Day 35 and Day 0. Error bars = 1 standard error of the mean. See Methods for normalization.
(E). Representative gross appearance of tumors at necropsy.
(F). Mean tumor weight at necropsy. Error bars = 1 SEM.
(G). Immunoblot analysis of tumors removed from 3 mice at necropsy.
Next the ZR75-1 cells infected with the inducible shRNA lentiviruses were infected with a retrovirus encoding luciferase and grown orthotopically in the mammary glands of immuncompromised mice. The luciferase activity of the EglN2 shRNA cells was comparable to the luciferase activity of the control shRNA cells in vitro (data not shown). One mammary gland was injected with the inducible EglN2 shRNA cells and the contralateral mammary gland was injected with the inducible control shRNA. Mice were treated with a depot form of estrogen to promote the growth of the breast cancer cells and live tumor cell burden was monitored non-invasively with bioluminescent imaging beginning 1 week after cell implantation. At this timepoint, the EglN2 shRNA tumors usually exhibited stronger luciferase signals than the control shRNA cells, arguing that the EglN2 shRNA cells were at least as tumorigenic as the control cells in vivo prior to the administration of doxycycline. Three days later imaging was repeated. Mice in which both tumors had increased in signal intensity, indicative of tumor cell expansion, were then fed chow containing doxycycline and serially monitored using bioluminescence. Over time there was a progressive decline in the EglN2 shRNA tumor signal relative to the control shRNA tumor signal, largely due to continued expansion of the tumors formed by the control shRNA cells and an apparent arrest of the EglN2 shRNA cells (Fig 8C and 8D). After 5–6 weeks of doxycycline treatment the mice were sacrificed and the tumors were excised and weighed. Consistent with the bioluminescent images, the tumors formed by the EglN2 shRNA cells were smaller than the tumors formed by the control shRNA cells (Fig 8E and 8F). It should be noted that bioluminescent imaging detects viable tumor cells, whereas tumor mass includes contributions from stroma, host cells, and non-viable tumor cells. Similar results were observed with T47D cells (Supplemental Fig 11) and MCF7 cells (Supplemental Fig 12). Immunoblot analysis of ZR75-1 tumor extracts prepared at necropsy confirmed that Cyclin D1 levels were diminished in tumors following induction of the EglN2 shRNAs in vivo (Fig 8G and data not shown). Conversely, expression of Cyclin D1 under the control of a constitutively active promoter restored the ability of EglN2-depleted ZR75-1 cells to proliferate in vivo (Supplemental Fig 13). Therefore loss of EglN2 decreases Cyclin D1 levels and inhibits tumor growth in vivo.
DISCUSSION
We found that loss of EglN2, but not loss of the paralogous proteins EglN1 and EglN3, decreases Cyclin D1 mRNA and protein levels, decreases cell proliferation, and decreases tumor formation. Impaired proliferation in cells lacking EglN2 could be rescued by restoring Cyclin D1 protein production and was not observed in cells lacking the pRB tumor suppressor protein, which is required for growth inhibition in cells deprived of Cyclin D1-associated kinase activity. Therefore loss of Cyclin D1 causes, and does not merely correlate with, impaired proliferation in EglN2-defective cells. An intimate, causal, connection between EglN2, Cyclin D1, and cell proliferation is also suggested by similarities between EglN2−/− mice and Cyclin D1−/− mice, both of which display impaired mammary gland proliferation in response to pregnancy. Regulation of Cyclin D1 and cell proliferation by EglN2 depends on EglN2 catalytic activity, suggesting that small molecule EglN2 inhibitors would have anticancer activity.
Such inhibitors, were they to be developed, might be particularly useful for the treatment of estrogen-dependent breast cancer. Polyak and coworkers reported previously that EglN2 is estrogen-inducible and that EglN2 overexpression promotes breast cancer cell proliferation (Seth et al., 2002). The latter observation, together with our EglN2 loss of function studies, indicates that EglN2 activity regulates breast cancer proliferation in response to estrogen. Cyclin D1, which is also induced by estrogen, is an important regulator of breast cancer proliferation and is frequently amplified or otherwise overexpressed in this disease (Roy and Thompson, 2006; Steeg and Zhou, 1998). Loss of Cyclin D1-associated kinase activity is sufficient to prevent or delay the development of breast cancer in mouse models (Landis et al., 2006; Yu et al., 2001; Yu et al., 2006). Cyclin D1 also has kinase-independent functions related to estrogen receptor signaling and mammary epithelial proliferation (Landis et al., 2006; Neuman et al., 1997; Yu et al., 2006; Zwijsen et al., 1997). Therefore EglN2 inhibitors might prove more efficacious than small molecule inhibitors of cdk4 and cdk6, which are the catalytic partners of Cyclin D1. Moreover, we have observed loss of the estrogen receptor in breast cancer cells deprived of EglN2 (Supplemental Table 2 and Q.Z. and W.G.K.-unpublished data). Therefore EglN2 antagonists and ER antagonists might be additive or synergistic when used to treat ER positive breast cancers.
Downregulation of Cyclin D1, and impaired proliferation, following EglN2 loss was not restricted to breast cancer cells, however, but appears to be common across a variety of tumor types. Therefore EglN2 inhibitors might be useful beyond the treatment of ER positive breast cancer. It will also be of interest to see if EglN2, which maps to chromosome 19q13.2, is mutationally activated in any human cancers.
Downregulation of EglN1, the primary HIF prolyl hydroxylase, led to increased HIFα, as expected, but did not decrease Cyclin D1 levels. Conversely, downregulation of EglN2 decreased Cyclin D1 without appreciably affecting HIFα protein levels. Moreover, downregulation of EglN2 decreased Cyclin D1 in cells lacking ARNT (HIF1β), the heterodimeric partner for HIFα and in cells lacking pVHL, which targets hydroxylated HIFα for destruction. These observations strongly suggest that loss of Cyclin D1 upon EglN2 inactivation is not a secondary consequence of changes in HIF activity. Nonetheless, there is the potential for crosstalk between HIF and Cyclin D1. For example, HIF can transcriptionally activate REDD1, which inhibits mTOR activity (Brugarolas et al., 2004; Reiling and Hafen, 2004) and thereby decreases Cyclin D1 translation. Moreover, our findings do not in any way preclude a role for EglN2 in the regulation of HIF activity, such as has been inferred from studies of EglN2 null animals subjected to regional ischemia (Aragones et al., 2008).
Hypoxia generally inhibits cell proliferation, associated with a loss of cyclin D1 and pRB hypophosphorylation, presumably as a means to conserve ATP. Our findings suggest that cessation of proliferation under these conditions is due, at least partly, to impaired EglN2 activity. EglN inhibitors that are capable of activating the HIF transcriptional program in vivo are currently being tested in the clinic for the treatment of anemia and ischemic diseases. A theoretical concern with such agents relates to the ability of HIF to promote tumor growth in some preclinical models (Semenza, 2003). Our findings suggest that such protumorigenic effects might be mitigated by antitumor effects stemming from downregulation of Cyclin D1.
Clearly it will be important to determine the EglN2 substrate that links EglN2 to Cyclin D1. It should be noted that EglN2 is a nuclear protein (Metzen et al., 2003) and regulation of Cyclin D1 appears to be largely at the level of Cyclin D1 transcription. The latter was unexpected because the study by Edgar and Frei, linking Egl9 to Cyclin D1, employed a transgenic Drosophila overexpressing Cyclin D1 under the control of a heterologous promoter (Frei and Edgar, 2004). It is possible that EglN2 also influences Cyclin D1 posttranscriptionally or that the findings of Edgar and Frei were unrelated to the EglN2 biology described here and therefore fortuitous. Regardless, higher metazoans, in contrast to Drosophila and Caenorhabditis, have three EglN family members. Our findings, together with previously published work, indicate that EglN1 has been retained as the primary HIF regulator under normal conditions, and that EglN2 and EglN3, in addition to regulating HIF, have assumed HIF-independent roles in the control of proliferation and apoptosis, respectively.
EXPERIMENTAL PROCEDURES
Cell culture
HeLa, U2OS, MEFs, UOK101, Phoenix, 293T, MCF7, ARNT−/− and ARNT+/+ cells were maintained in DMEM containing 10% fetal bovine serum (FBS) (Hyclone, Logan, Utah) except where indicated. The MCF7 media was supplemented with 10 μg/ml insulin. 769-P, ZR-75-1, BT-474, T47D and BT-549 cells were maintained in RPMI medium containing 10% FBS except where indicated. Following retroviral or lentiviral infection cells were maintained in the presence of hygromycin (200 μg/ml) or puromycin (2 μg/ml) depending upon the vector. All cells were maintained at 37 °C in 10% CO2.
Mice
EglN2 −/− mice were obtained from Regeneron Pharmaceuticals (Tarrytown, NY). Inguinal mammary glands were removed one day postpartum and whole mounts were prepared as described (Geng et al., 1999). MEFs were isolated from E13.5 day embryos as described (Kozar et al., 2004). All mouse experiments complied with NIH guidelines and were approved by the Dana-Farber Cancer Institute Animal Care and Use Committee.
siRNA
Cells grown in six-well plates were transfected with 200 nM siRNA using Lipofectamine2000 (for mixtures of plasmids and siRNA oligos) or Oligofectamine (for siRNA alone). siRNAs were purchased from Dharmacon, Inc. (Lafayette, CO). Sense strands were as follows: GFP 5′-GGCTACGTCCAGGAGCGCACC-3′; GL3 5′-CTTACGCTGAGTACTTCGATT-3′; GFP Scramble 5′-AACAGTCGCGTTTGCGACTGG-3′; EglN2-A 5′-GACTATATCGTGCCCTGCATG-3′; EglN2-4 5′-GCCACTCTTTGACCGGTTGCT-3′; EGLN1 5′-AGCTCCTTCTACTGCTGCA-3′; EGLN3 5′-CAGGTTATGTTCGCCACGT-3′ (Appelhoff et al., 2004; Schlisio et al., 2008).
Immunoblot analysis
Whole cell extracts were prepared in EBC buffer (50mM Tris [ph 8.0], 120 mM NaCl, 0.5 % NP40) containing protease inhibitors. Mouse mammary glands were lysed in NP-40 lysis buffer (10% glycerol, 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml leupeptin and aprotinin). Equal amounts of protein, as determined by the Bradford assay, were resolved by SDS-PAGE and western blot analysis was performed as previously described (Schlisio et al., 2008). Rabbit polyclonal anti-EglN1, EglN2, EglN3, HIF1α and Glut1 antibodies were from Novus Biological (Littleton, CO). Anti-Cyclin D1, D2, and D3 antibodies were from Neomarker (Fremont, CA). ARNT antibody was from BD biosciences (San Jose, CA). Anti-HA antibody was from Covance. Antibodies against Vinculin, Tubulin and FLAG (M2) were from Sigma (St. Louis, MO).
Real Time RT-PCR
Total RNA was isolated using RNeasy mini kit with on-column DNase digestion (Qiagen). First-strand cDNA was generated using StrataScript First-Stand Synthesis System (Statagene). Real-time PCR was performed in duplicate using QuantiTect SYBR Green PCR master mix (Qiagen) and the Mx3000P QPCR system (Stratagene). All values were normalized to the level of 18S rRNA (F: AAGACGATCAGATACCGTCGTAG R: GTTTCAGCTTTGCAACCATACTC) or beta-actin abundance. Real time PCR primer sequences are as follows: mouse EglN2 (F: CTGGGCAACTACGTCATCAAT R: TGCACCTTAACATCCCAGTTC ); mouse Cyclin D1 (F: CCAACAACTTCCTCTCCTGCT R: GACTCCAGAAGGGCTTCAATC); Mouse beta-actin (F: ACCAACTGGGACGACATGGA R: GGTCTCAAACATGATCTGGGTCAT ); human EglN2 (F: AACATCGAGCCACTCTTTGAC R: TCCTTGGCATCAAAATACCAG); human Cyclin D1 ( F: CCG TCC ATG CGG AAG ATC R: ATG GCC AGC GGG AAG AC ); human beta-actin (F: AGA AAA TCT GGC ACC ACA CC R: GGG GTG TTG AAG GTC TCA AA ).
Plasmids
The EglN2 open reading frame cDNA was PCR amplified with a 5′ primer that introduced a BamH1 site and a Flag epitope and a 3′ primer that introduced an EcoRI site. The product was digested with BamHI and EcoRI and cloned into pBabe-Puro vector cut with these two enzymes. pBABE-EglN2-H358A was made using a site-directed mutagenesis kit (Quick-change; Stratagene). pBabeHygro-Cyclin D1 was constructed by ligating the BamHI-SalI Cyclin D1 cDNA insert from pBabePuro-Cyclin D1 (kindly provided by Dr. P. Sicinski) into pBabeHygro vector cut with these two enzymes. pLXSN and pLXSN E7 retrovial vectors (Halbert et al., 1991) were kindly given by Dr. Karl Munger.
shRNA expression vectors corresponding to the siRNAs described above were created by ligating synthetic, duplex, oligonucleotides into pMKO.1 retroviral vector (Boehm et al., 2005) or pCCLsin.PPT.hPGK.GFP.Wpre lentiviral vector (Corso et al., 2008) (kindly provided by Dr. S. Giordano). All plasmids were confirmed by DNA sequencing.
Lentiviral Rb shRNA vectors, ARNT shRNA vectors, and lentiviral EglN2 shRNA (TRC00000324-328) were obtained from the Broad Institute RNAi consortium shRNA library. Rb #1 shRNA target sequence: CCACATTATTTCTAGTCCAAA; Rb #2 shRNA target sequence: CAGAGATCGTGTATTGAGATT; ARNT target sequences were CCTTTGTCTTTCTGTGTACTT (Fig 1) and GAGAAGTCAGATGGTTTATTT (Supplemental Fig 5); TRC00000324-328 shRNA target sequences: CGCATGGCAGACAGCTTAAAT;GCTGCATCACCTGTATCTATT;GCCACTCTTT GACCGGTTGCT;ACTGGGACGTTAAGGTGCATG;CTGGGACGTTAAGGTGCAT GG, respectively. The lentivirus encoding the HIF2α was a gift of Dr. Sungwoo Lee.
Virus Production and Infection
Phoenix packaging cell line was used for the generation of ecotropic retroviruses and all retroviral infections were carried out as described (Boehm et al., 2005). 293T packaging cell line was used for lentiviral amplification and all lentiviral infections were carried out as previously described (Moffat et al., 2006). Briefly, viruses were collected 48 and 72 h after transfection, filtered, and used to infect cells in the presence of 8 μg/ml polybrene prior to drug selection.
Microarray Analysis
The gene expression dataset [www.ncbi.nlm.nih.gov/geo/ (Accession number GSE5460)] and immunohistochemical analysis of primary human breast tumors used for Fig 3D was as described in (Lu et al., 2008). Raw expression data obtained using Affymetrix GENECHIP software was normalized, analyzed, and displayed using DNA-Chip Analyzer (dChip) custom software (W. H. Wong and C. Li, http://www.dChip.org/). Array probe data were normalized to the mean expression level of each probe across the sample set. The analysis in Fig 3C and Supplemental Fig 4 was performed using data and software available at www.oncomine.org and www.genesapiens.org, respectively.
For supplemental Fig 10 and Supplementary Tables 1, 2, 3 T47D breast carcinoma cells that were infected with lentivirus encoding either inducible EglN2 shRNA or Scrambled control (Scr) shRNA were treated with doxycycline (1 μg/ml) for 48 hours or left untreated. Total RNA was extracted by using RNeasy mini-kit with on-column DNase digestion (Qiagen). Biotin labeled cRNA was prepared from 1 μg of total RNA, fragmented, and hybridized to a Human Gene 1.0ST array (Affymetrix). The arrays were scanned and the data, as CEL files, were analyzed with Affymetrix® Expression Console™. The data were normalized using RMA (Robust Multi-Array) normalization (Bolstad et al., 2003). All samples successfully underwent a series of quality control tests and results from duplicate samples were highly comparable (R>0.967). Gene expression values less than a minimum threshold of 20 or a maximum threshold of 16 000 were set to 20 and 16 000, respectively. Genes with minimal variation across the data set were discarded (maximum/minimum <3 or maximum - minimum <100). GSEA was performed as described (Subramanian et al., 2005).
Cell Proliferation Assays
T47D, BT-474 and ZR-75-1 cells were plated, in triplicate, in 6 well plates (105 cells/well) in RPMI supplemented with 10% FBS or in phenol red-free RPMI containing 5% charcoal stripped serum supplemented, as indicated, with 10 nM estrogen. At the indicated time points cells were trypsinized, pelleted by centrifugation, and resuspended in RPMI supplemented with 0.2% trypan blue. The number of viable cells, as determined by Typan blue exclusion, was determined using a hemocytometer.
shRNA pooled screening
Pooled RNAi screens consisting of 45,000 shRNAs were conducted as described by Luo and coworkers (Luo et al., 2008). Briefly, an shRNA pool consisting of 45,182 individual constructs was infected into the indicated cancer cell lines. For each screen, 3.6 × 107 cells were infected at a multiplicity of infection of 0.3 to ensure that each shRNA was introduced into 200 independent cells. Early timepoint (3–4 day) samples (n = 10) and DNA control samples (n = 10) were compared with end-timepoint (4 week) samples derived from replicate (n = 10) infections of each cell line. Genomic DNA was prepared from these timepoints, the hairpin region of shRNA constructs was amplified and digested to create half-hairpin barcodes which were hybridized to a custom Affymetrix microarray. After microarray normalization, fold-depletion scores for EglN2 were calculated on a construct-by-construct basis by comparing late timepoint hybridization values to early timepoint hybridization values.
RIGER algorithm
RIGER (RNAi Gene Enrichment Ranking), a statistical approach that considers the phenotypic results for the multiple shRNA constructs targeting the same gene was deployed as described (Luo et al., 2008). Briefly, this approach is based on the GSEA methodology (Subramanian et al., 2005) and uses similar Kolmogorov-Smirnov (KS)-based statistics to calculate gene scores from a dataset of shRNA construct profiles. First, shRNA constructs targeting EglN2 were scored according to their differential effects between late and early timepoints for each cell line. An Enrichment Score was calculated for each cell line, indicating the enrichment or depletion for the 5 EglN2 hairpins treated as a set. Finally, to find genes frequently essential across multiple cell lines, a second application of RIGER was used to find genes for which the hairpins targeting the gene were depleted in at least 8/12 cell lines.
Orthotopic tumor growth assays
6-week old female nude mice (Taconic, Hudson, NY) were used for xenograft studies. Approximately 8 × 106 viable tumor cells were resuspended in 40 μl growth factor-reduced Matrigel (BD Biosciences) and injected orthotopically into mammary gland four as previously described (Minn et al., 2005). Mice were supplied with chow containing 6 g doxycycline/kg (Bioserv, Frenchtown, NJ) for a treatment period of 5–6 weeks.
For bioluminescent detection and quantification of cancer cells, mice were given a single i.p. injection of a mixture of luciferin (50 mg/kg) ketamine (150 mg/kg) and xylazine (12 mg/kg) in sterile water. Five minutes later, mice were placed in a light-tight chamber equipped with a charge-coupled device IVIS imaging camera (Xenogen, Alameda, CA). Photons were collected for a period of 1–60 s, and images were obtained by using LIVING IMAGE 2.60.1 software (Xenogen) and quantified using IGOR Pro 4.09A image analysis software (WaveMatrics, Lake Oswego, OR). The total photons from the EglN2 shRNA tumor region of interest (ROI) were divided by the total photons from Scrambled shRNA tumor ROI and, for each mouse, normalized based on the ratio prior to the onset of doxycycline treatment for that mouse. Results were presented as mean ± standard error of the mean (SEM).
Supplementary Material
ACKNOWLEDGMENTS
We thank Regeneron Pharmaceuticals for the EglN2−/− mice, Oliver Hankinson for the ARNT−/− cells, Silvia Giordano for the inducible shRNA vector, Yan Geng for help with mammary gland analysis, Margaret McLaughlin-Drubin for help with E7 immunoblots, Mariela Jaskelioff for help with xenograft studies and members of the Kaelin Laboratory for useful discussions. Supported by NCI Dana-Farber/Harvard SPORE in Breast Cancer (A.L.R.) and the Breast Cancer Research Foundation (A.L.R. and W.G.K.). QZ is supported by a postdoctoral fellowship from Terri Brodeur Breast Cancer Foundation. WGK is a Howard Hughes Medical Institute Investigator and a Doris Duke Distinguished Clinical Investigator. This work is supported in part by an NIH grant (5R01CA068490-14) to WGK. WGK owns equity in Fibrogen, Inc., which is developing prolyl hydroxylase inhibitors as potential therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Number
Microarray data from T47D cells harvested 0 or 48 hours after induction of an shRNA against EglN2, or a scrambled control shRNA, were deposited in the NIH Gene Expression Omnibus database (accession number GSE18171).
REFERENCES
- Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- Aragones J, Schneider M, Van Geyte K, Fraisl P, Dresselaers T, Mazzone M, Dirkx R, Zacchigna S, Lemieux H, Jeoung NH, Lambrechts D, Bishop T, Lafuste P, Diez-Juan A, Harten SK, Van Noten P, De Bock K, Willam C, Tjwa M, Grosfeld A, Navet R, Moons L, Vandendriessche T, Deroose C, Wijeyekoon B, Nuyts J, Jordan B, Silasi-Mansat R, Lupu F, Dewerchin M, Pugh C, Salmon P, Mortelmans L, Gallez B, Gorus F, Buyse J, Sluse F, Harris RA, Gnaiger E, Hespel P, Van Hecke P, Schuit F, Van Veldhoven P, Ratcliffe P, Baes M, Maxwell P, Carmeliet P. Deficiency or inhibition of oxygen sensor Phd1 induces hypoxia tolerance by reprogramming basal metabolism. Nat Genet. 2008;40:170–180. doi: 10.1038/ng.2007.62. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Bio. 2001;2 doi: 10.1186/gb-2001-2-3-research0007. research0007.0001-0007.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Lukas J, Muller H, Lutzhoft D, Strauss M, Bartek J. Cyclin D1 protein expression and function in human breast cancer. Int J Cancer. 1994;57:353–361. doi: 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. Embo J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Hession MT, Bulmer SE, Hahn WC. Transformation of human and murine fibroblasts without viral oncoproteins. Mol Cell Biol. 2005;25:6464–6474. doi: 10.1128/MCB.25.15.6464-6474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- Bruegge K, Jelkmann W, Metzen E. Hydroxylation of hypoxia-inducible transcription factors and chemical compounds targeting the HIF-alpha hydroxylases. Curr Med Chem. 2007;14:1853–1862. doi: 10.2174/092986707781058850. [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corso S, Migliore C, Ghiso E, De Rosa G, Comoglio PM, Giordano S. Silencing the MET oncogene leads to regression of experimental tumors and metastases. Oncogene. 2008;27:684–693. doi: 10.1038/sj.onc.1210697. [DOI] [PubMed] [Google Scholar]
- Epstein A, Gleadle J, McNeill L, Hewitson K, O’Rourke J, Mole D, Mukherji M, Metzen E, Wilson M, Dhanda A, Tian Y, Masson N, Hamilton D, Jaakkola P, Barstead R, Hodgkin J, Maxwell P, Pugh C, Schofield C, Ratcliffe P. C. elegans EGL-9 and Mammalian Homologs Define a Family of Dioxygenases that Regulate HIF by Prolyl Hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Frei C, Edgar BA. Drosophila cyclin D/Cdk4 requires Hif-1 prolyl hydroxylase to drive cell growth. Dev Cell. 2004;6:241–251. doi: 10.1016/s1534-5807(03)00409-x. [DOI] [PubMed] [Google Scholar]
- Geng Y, Whoriskey W, Park MY, Bronson RT, Medema RH, Li T, Weinberg RA, Sicinski P. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell. 1999;97:767–777. doi: 10.1016/s0092-8674(00)80788-6. [DOI] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MM, Linde NS, Wynter A, Metzger M, Wong C, Langsetmo I, Lin A, Smith R, Rodgers GP, Donahue RE, Klaus SJ, Tisdale JF. HIF-prolyl hydroxylase inhibition results in endogenous erythropoietin induction, erythrocytosis, and fetal hemoglobin expression in rhesus macaques. Blood. 2007;110:2140–2147. doi: 10.1182/blood-2007-02-073254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya S, Bronson RT, Akashi K, Sicinski P. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–491. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer Cell. 2006;9:13–22. doi: 10.1016/j.ccr.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YM, Zhou BP, Deng J, Pan Y, Hay N, Hung MC. A hypoxia-independent hypoxia-inducible factor-1 activation pathway induced by phosphatidylinositol-3 kinase/Akt in HER2 overexpressing cells. Cancer Res. 2005;65:3257–3263. doi: 10.1158/0008-5472.CAN-04-1284. [DOI] [PubMed] [Google Scholar]
- Lu X, Lu X, Wang ZC, Iglehart JD, Zhang X, Richardson AL. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat. 2008;108:191–201. doi: 10.1007/s10549-007-9596-6. [DOI] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P, Weisner M, Chang G-W, Clifford S, Vaux E, Pugh C, Maher E, Ratcliffe P. The von Hippel-Lindau gene product is necessary for oxgyen-dependent proteolysis of hypoxia-inducible factor α subunits. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Metzen E, Berchner-Pfannschmidt U, Stengel P, Marxsen JH, Stolze I, Klinger M, Huang WQ, Wotzlaw C, Hellwig-Burgel T, Jelkmann W, Acker H, Fandrey J. Intracellular localisation of human HIF-1 alpha hydroxylases: implications for oxygen sensing. J Cell Sci. 2003;116:1319–1326. doi: 10.1242/jcs.00318. [DOI] [PubMed] [Google Scholar]
- Minamishima YA, Moslehi J, Bardeesy N, Cullen D, Bronson RT, Kaelin WG., Jr. Somatic inactivation of the PHD2 prolyl hydroxylase causes polycythemia and congestive heart failure. Blood. 2008;111:3236–3244. doi: 10.1182/blood-2007-10-117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamishima YA, Moslehi J, Padera RF, Bronson RT, Liao R, Kaelin WG., Jr. A Feedback Loop Involving the Phd3 Prolyl Hydroxylase Tunes the Mammalian Hypoxic Response In Vivo. Mol Cell Biol. 2009 doi: 10.1128/MCB.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Mole DR, Schlemminger I, McNeill LA, Hewitson KS, Pugh CW, Ratcliffe PJ, Schofield CJ. 2-oxoglutarate analogue inhibitors of HIF prolyl hydroxylase. Bioorg Med Chem Lett. 2003;13:2677–2680. doi: 10.1016/s0960-894x(03)00539-0. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB, Bowtell DD, Ronai Z. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117:941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Neuman E, Ladha MH, Lin N, Upton TM, Miller SJ, DiRenzo J, Pestell RG, Hinds PW, Dowdy SF, Brown M, Ewen ME. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol. 1997;17:5338–5347. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer A, Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat Chem Biol. 2007;3:144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- Pollard P, Loenarz C, Mole D, McDonough M, Gleadle J, Schofield C, Ratcliffe P. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem J. 2008;416:387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- Rantanen K, Pursiheimo J, Hogel H, Himanen V, Metzen E, Jaakkola PM. Prolyl Hydroxylase PHD3 Activates Oxygen-dependent Protein Aggregation. Mol Biol Cell. 2008;19:2231–2240. doi: 10.1091/mbc.E07-11-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling JH, Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 2004;18:2879–2892. doi: 10.1101/gad.322704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy PG, Thompson AM. Cyclin D1 and breast cancer. Breast. 2006;15:718–727. doi: 10.1016/j.breast.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Safran M, Kim WY, O’Connell F, Flippin L, Gunzler V, Horner JW, Depinho RA, Kaelin WG., Jr. Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: assessment of an oral agent that stimulates erythropoietin production. Proc Natl Acad Sci U S A. 2006;103:105–110. doi: 10.1073/pnas.0509459103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlisio S, Kenchappa RS, Vredeveld LC, George RE, Stewart R, Greulich H, Shahriari K, Nguyen NV, Pigny P, Dahia PL, Pomeroy SL, Maris JM, Look AT, Meyerson M, Peeper DS, Carter BD, Kaelin WG., Jr. The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008;22:884–893. doi: 10.1101/gad.1648608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Seth P, Krop I, Porter D, Polyak K. Novel estrogen and tamoxifen induced genes identified by SAGE (Serial Analysis of Gene Expression) Oncogene. 2002;21:836–843. doi: 10.1038/sj.onc.1205113. [DOI] [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Parker SB, Li T, Fazeli A, Gardner H, Haslam SZ, Bronson RT, Elledge SJ, Weinberg RA. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- Steeg PS, Zhou Q. Cyclins and breast cancer. Breast Cancer Res Treat. 1998;52:17–28. doi: 10.1023/a:1006102916060. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Aguila HL, Parikh NS, Li X, Lamothe K, Duan LJ, Takeda H, Lee FS, Fong GH. Regulation of adult erythropoiesis by prolyl hydroxylase domain proteins. Blood. 2008;111:3229–3235. doi: 10.1182/blood-2007-09-114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Ho VC, Takeda H, Duan LJ, Nagy A, Fong GH. Placental but not heart defects are associated with elevated hypoxia-inducible factor alpha levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol. 2006;26:8336–8346. doi: 10.1128/MCB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS. Characterization and comparative analysis of the EGLN gene family. Gene. 2001;275:125–132. doi: 10.1016/s0378-1119(01)00633-3. [DOI] [PubMed] [Google Scholar]
- Tian YM, Mole DR, Ratcliffe PJ, Gleadle JM. Characterization of different isoforms of the HIF prolyl hydroxylase PHD1 generated by alternative initiation. Biochem J. 2006;397:179–186. doi: 10.1042/BJ20051996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- Yu Q, Sicinska E, Geng Y, Ahnstrom M, Zagozdzon A, Kong Y, Gardner H, Kiyokawa H, Harris LN, Stal O, Sicinski P. Requirement for CDK4 kinase function in breast cancer. Cancer Cell. 2006;9:23–32. doi: 10.1016/j.ccr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Zwijsen RM, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJ. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.