SUMMARY
A preference for homologs over sister chromatids in homologous recombination is a fundamental difference in meiotic versus mitotic cells. In budding yeast, the bias for interhomolog recombination in meiosis requires the Dmc1 recombinase and the meiosis-specific kinase, Mek1, which suppresses engagement of sister chromatids by the mitotic recombinase, Rad51. Here, a combination of proteomic, biochemical and genetic approaches has identified an additional role for Mek1 in inhibiting the activity of the Rad51 recombinase through phosphorylation of its binding partner, Rad54. Rad54 phosphorylation of threonine 132 attenuates complex formation with Rad51 and a negative charge at this position reduces Rad51 function in vitro and in vivo. Thus, Mek1 phosphorylation provides a dynamic means of controlling recombination partner choice in meiosis in two ways: (1) it reduces Rad51 activity through inhibition of Rad51/Rad54 complex formation and (2) it suppresses Rad51-mediated strand invasion of sister chromatids via a Rad54-independent mechanism.
INTRODUCTION
In eukaryotic cells, double strand breaks (DSBs) on chromosomes can be catastrophic or essential, depending upon the situation. In mitotically dividing cells, DSBs may occur as a result of stalled replication forks or DNA damage. In these situations, sister chromatids are preferentially used as templates for repair, thereby keeping the DNA sequence intact (Kadyk and Hartwell, 1992). In meiosis, DSBs are necessary for generating crossovers between homologous chromosomes. In combination with sister chromatid cohesion, crossovers provide the physical connections necessary for proper segregation of homologs at the first meiotic division (Petronczki et al., 2003). In meiotic cells, therefore, the preferred templates for DSB repair are homologous chromosomes rather than sister chromatids.
During meiosis in budding yeast, DSBs are generated by a conserved, meiosis-specific transesterase called Spo11 (Keeney, 2001). Spo11 preferentially cleaves specific regions in the genome termed “hotspots”. After DSB formation, the 3′ ends of the breaks are bound by the RecA orthologs, Rad51 and Dmc1. Rad51 is the major recombinase in vegetative cells while Dmc1 is present only in meiotic cells. Rad51 loading onto the single strand ends of DSBs requires Rad52, Rad55 and Rad57 while Dmc1 is loaded by a mediator complex comprised of Mei5 and Sae3 (Hunter, 2007; Symington, 2002). Rad51 and Dmc1 co-localize to DSBs during meiosis and are required for interhomolog recombination (Bishop, 1994; Schwacha and Kleckner, 1997; Shinohara et al., 1997a). Genetic and biochemical experiments indicate that these two recombinases have over-lapping, but also non-redundant, functions in meiosis (Sheridan and Bishop, 2006).
Rad54 is a member of the Swi2/Snf2 family of DNA motor proteins and functions at several different steps during recombination: (1) stabilization of Rad51 filaments, (2) stimulation of Rad51-mediated strand invasion and (3) removal of Rad51 from DNA after joint molecules have been formed (Heyer et al., 2006; Tan et al., 2003). In meiosis, rad54Δ mutants exhibit reduced sporulation and spore viability. These defects are exacerbated when the RDH54 gene (also known as TID1) which encodes a Rad54-related protein, is deleted (Klein, 1997; Shinohara et al., 1997b). During meiosis, Dmc1 interacts with Rdh54 (Dresser et al., 1997). Although there is some functional redundancy between RAD54 and RDH54, genetic experiments indicate that Rad51/Rad54 is utilized primarily for sister chromatid recombination in vegetative and meiotic cells, while Dmc1/Rdh54 is involved primarily in interhomolog recombination during meiosis (Arbel et al., 1999; Klein, 1997; Shinohara et al., 1997b).
In budding yeast, interhomolog bias during meiosis requires not only Dmc1 but also a trio of meiosis-specific proteins, Hop1, Red1 and Mek1 (Niu et al., 2005; Wan et al., 2004). Hop1 is conserved in multicellular eukaryotes such as plants and nematodes and has been implicated in the suppression of meiotic sister chromatid repair in nematodes (Armstrong et al., 2002; Couteau et al., 2004). Mutation of these genes leads to a specific decrease in interhomolog recombination and Meiosis I non-disjunction (Hollingsworth et al., 1995). Mek1 is a serine-threonine protein kinase whose activation is dependent upon DSB formation (Niu et al., 2007). dmc1Δ cells arrest in prophase with unrepaired breaks as a result of triggering the meiotic recombination checkpoint (Bishop et al., 1992; Lydall et al., 1996). Inactivation of Mek1 after break formation in dmc1Δ mutants results in a rapid repair of DSBs by Rad51/Rad54 using sister chromatids as the template (Niu et al., 2005). The spore inviability of mek1Δ mutants indicates that the presence of Dmc1 is not sufficient to promote interhomolog recombination in the absence of Mek1 kinase activity.
Understanding the mechanisms by which Mek1 regulates meiotic recombination requires identification of Mek1 substrates. Testing candidate recombination proteins for in vitro phosphorylation by Mek1 led to the discovery that Mek1 indirectly down-regulates Rad51 recombinase activity during meiosis by phosphorylation of Rad54. This mechanism is independent of Hed1, a meiosis-specific protein that inhibits recombinase activity by binding directly to Rad51, thereby preventing Rad51/Rad54 complex formation (Busygina et al. 2008; Tsubouchi and Roeder 2006). Mek1 phosphorylation of Rad54 is separate from the mechanism that suppresses sister chromatid repair, demonstrating that Mek1 regulates meiotic recombination in at least two ways. The dynamic nature of phosphorylation as a modification suggests a mechanism by which Rad51 recombinase activity can be modulated during meiosis.
RESULTS
Rad54 and Rdh54 are substrates of Mek1 in vitro
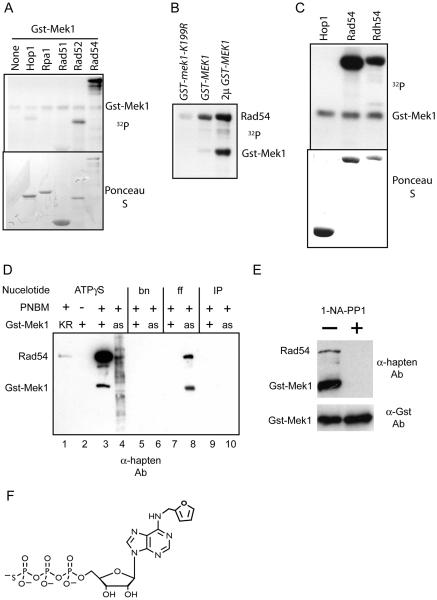
In vitro kinase assays were performed with purified proteins that mediate different steps of recombination such as RPA (single strand binding protein), Rad52, Rad51 and Rad54. Because Mek1 activation is dependent upon meiotic DSB formation, GST-Mek1 was purified from dmc1Δ-arrested cells (Niu et al., 2007). GST-Mek1 autophosphorylation was detected as has previously been observed (Niu et al., 2005). Weak labeling of Rad52 and Hop1 was detected, while Rad51 and RPA1 were not significantly phosphorylated (Figure 1A). Whether the Rad52 or Hop1 phosphorylation is biologically significant is unclear. Hop1 is a phosphoprotein in vivo, but its phosphorylation is independent of MEK1 (Niu et al 2005). In contrast, robust labeling of Rad54 was observed by Gst-Mek1 (Figure 1A), which was not seen when a catalytically inactive version of Mek1, GST-mek1-K199R, was used (Figure 1B). Kinase assays with the Rad54 paralog, Rdh54, showed that it is also a good in vitro substrate of GST-Mek1 (Figure 1C).
Figure 1. In vitro kinase assays using GST-Mek1 and various recombination proteins.
A. GST-Mek1 was purified from meiotic yeast cells and incubated with 32PATP and 1 μg purified protein as indicated. RPA1, Rad51, Rad52 and Rad54 were purified from vegetative yeast cells while Hop1 was purified from bacteria. The proteins were then fractionated and transferred to a nitrocellulose membrane. The top panel shows an autoradiograph of the membrane while the bottom panel shows the filter after Ponceau S staining; B. Autoradiograph of kinase assays using bacterially purified Rad54 and GST-Mek1-K199R or GST-Mek1 purified from meiotic yeast cells expressing GST-MEK1 in single or high copy number (2μ GST-MEK1). C. Autoradiograph of kinase assays using GST-Mek1 and recombinant Rad54 and Rdh54. D. In vitro kinase assays using the semi-synthetic epitope system. Purified soluble GST-Mek1 (+) and GST-mek1-K199R (KR), as well as GST-mek1-as (as) bound glutathione-Sepharose beads were reacted with 1 μg Rad54 and either ATPγS or the indicated ATPγS analog. After alkylation with PNBM, the proteins were fractionated and transferred to nitrocellulose and probed with α-hapten antibodies. bn = benzyl-ATPγS; ff = furfuryl-ATPγS; IP = isopentyl- ATPγS. E. Kinase reactions with GST-mek1-as pulldowns in the presence or absence of 10 μM 1-NA-PP1 processed as in Panel D. In addition, part of the pulldown was probed with α-GST antibodies to detect the amount of GST-mek1-as in the reactions. F. Structure of ff-ATPγS
The direct phosphorylation of Rad54 by Mek1 was established using GST-mek1-as which contains a mutation in the ATP binding pocket that enables the kinase to utilize derivatives of ATP otherwise too bulky for Mek1 or other kinases (Wan et al., 2004). Phosphorylation was detected using the semi-synthetic epitope method (Allen et al., 2007). In the presence of ATPγS, proteins can be thiophosphorylated by kinases. Alkylation of the thiophosphates with p-nitrobenzylmesylate (PNBM) creates affinity tags that can be detected on immunoblots using the α-hapten antibody. To confirm that this method works for GST-Mek1, purified, soluble kinase was incubated with recombinant Rad54 and ATPγS, the proteins were alkylated and analyzed using the α-hapten antibody. Both autophosphorylation of GST-Mek1 and phosphorylation of Rad54 were observed (Figure 1D, lane 3). Detection of the phosphorylated proteins was dependent upon ATPγS (and not ATP), Mek1 kinase activity and alkylation by PNBM (Figure 1D, lanes 1-3)(data not shown).
To test whether Mek1 directly phosphorylates Rad54, GST-mek1-as was immobilized on glutathione Sepharose beads and the beads were incubated with Rad54 and ATPγS. Unlike purified GST-Mek1 and GST-mek1-K199R, the GST-mek1-as pulldown was contaminated with additional kinases and proteins as evidenced by the smear of proteins detected by the α-hapten antibody (Figure 1D, lane 4). GST-mek1-as autophosphorylation and robust labeling of Rad54 were not observed with ATPγS, consistent with previous observations that GST-Mek1-as does not utilize ATP efficiently in vitro (Wan et al., 2004). Kinase assays using different ATPγS analogs and the GST-mek1-as bound beads revealed specific phosphorylation of GST-mek1-as and Rad54 with furfuryl (ff)-ATPγS (Figure 1D, lane 8). GST-mek1-as has the advantage that it can be specifically inhibited by the addition of the purine analog, 1-NA-PP1, while wild-type GST-mek1 is unaffected by inhibitor (Wan et al. 2004). Addition of 1-NA-PP1 to the kinase reaction abolished phosphorylation of both GST-mek1-as and Rad54, although similar amounts of kinase were present in the reactions as determined by probing with antibodies against GST (Figure 1E). This result confirms that the observed kinase activity is due to GST-mek1-as. The loss of background labeling in the GST-mek1-as pulldowns and the absence of phosphorylation in the GST-Mek1 plus ff-ATPγS reaction, indicates that GST-mek1-as can specifically utilize ff-ATPγS in vitro to directly phosphorylate both Rad54 and itself.
Mek1 phosphorylates the N-termini of Rad54 and Rdh54
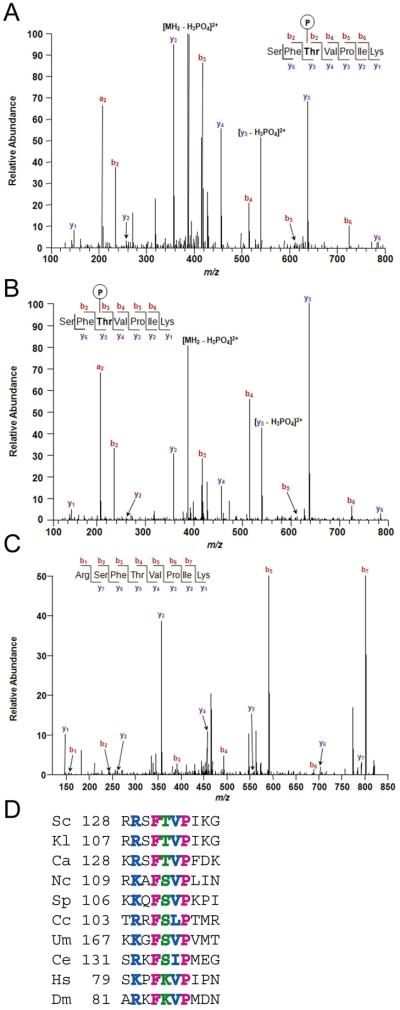
In vitro Mek1 phosphorylation sites on recombinant Rad54 and Rdh54 were mapped using mass spectrometry (MS) analysis. Because Rad54 and Rdh54 were purified from bacteria, any phosphates detected by MS should be due to Mek1. Three phosphorylation sites were detected for Rad54 (Ascore > 19): T58, T132 and T231 (Figure 2A)(Supplemental Figure 1A and B) (Beausoleil et al., 2006). Two sites with an Ascore >19 were observed for Rdh54: S85 and T89 (Supplemental Figure 1C and D). All of these sites are located in the N-terminal regions of the proteins outside of the catalytic cores. Comparing the protein sequences of Rad54 and Rdh54 reveals that their N-termini are not well conserved (Shinohara et al., 1997b) and that the Rad54 phosphorylation sites do not correspond with the Rdh54 sites.
Figure 2. MS analysis of peptides containing T132 of in vitro and in vivo phosphorylated Rad54.
Assignment of b (red) and y (blue) ion series in MS/MS scan from precursor peptide ions of Rad54 containing T132. A. Recombinant Rad54 phosphorylated in vitro by Mek1. B. Rad54-3FLAG purified from dmc1Δ-arrested meiotic yeast cells. C. Rad54-3FLAG purified from dmc1Δ mek1Δ-arrested meiotic yeast cells. In addition to the SEQUEST searches, manual inspection of the data using extracted ion chromatograms revealed no phosphorylated peptides with the sequence RSFTVPIK or SFTVPIK . Xcorr values are given in Supplementary Table 1. D. Alignment of region containing Rad54 T132 from different species. The T132 position is indicated in green, identical and similar amino acids are in pink and blue, respectively. Sc, Saccharomyces cerevisiae; Kl, Kluveryomyces lactis; Ca, Candida albicans; Nc, Neurospora crassa; Sp, Schizosaccharomyces pombe; Cc, Coprinus cinereus; Um, Ustilago maydis; Ce, Caenorhabditis elegans; Hs, Homo sapiens; Dm, Drosophila melanogaster.
T132 of Rad54 is phosphorylated in meiotic cells
To test whether Rad54 is phosphorylated during meiosis, Rad54-FLAG was purified from dmc1Δ-arrested cells after five hours in Spo medium at which time Mek1 is constitutively active (Wan et al., 2004). MS analysis found no evidence for in vivo phosphorylation of either T58 or T231. This finding, along with the fact that no phenotypes were observed for mutants containing non-phosphorylatable amino acids at these positions (data not shown), suggests that phosphorylation of T58 and T231 may be an in vitro artifact. In contrast, peptide ions indicative of T132 phosphorylation were observed (Figure 2B). Rad54 T132 was not phosphorylated when Rad54-Flag was purified from dmc1Δ mek1Δ meiotic cells, however. For example, the y5 ion derived from Rad54-3FLAG from the MEK1 diploid has a mass to charge (m/z) ratio of 637.32 which is 79.97 m/z larger than the y5 ion from Rad54-3FLAG purified from mek1Δ meiotic cells (Figure 2B and C). This difference is consistent with the absence of a phosphate group at T132 in the peptide represented in Figure 2C, confirming that T132 phosphorylation is dependent upon MEK1in vivo. T132 resides in a patch of amino acids that is highly conserved in the Rad54 orthologs of fungi and nematodes. This region is also conserved in humans and fruit flies, although in these species Rad54 cannot be phosphorylated at this position since threonine is replaced by lysine (Figure 2D).
RAD54-T132A partially suppresses the sporulation and spore viability defects of dmc1Δ
An allele of RAD54 that substitutes T132 with alanine fully complements the sporulation and spore viability defects of rad54Δ (Table 1A). Complementation was also observed with a mutant containing a negatively charged aspartic acid substitution that can mimic phosphorylation (RAD54-T132D) (Table 1A). Therefore these mutations do not negatively affect Rad54 function in an otherwise wild-type meiosis. Furthermore, whatever meiotic process requires RAD54 for wild-type levels of sporulation and spore viability is independent of Mek1 phosphorylation of T132.
Table 1.
Sporulation and spore viability in various RAD54 strains.
| Relevant genotypea | MEK1 | mek1Δ | ||
|---|---|---|---|---|
| A | % Spob | % s. v.c (# asci) | % Spo | % s. v. (# asci) |
| rad54Δ::pRS306 | 36.5 | 31.5 (50) | ND | ND |
| rad54Δ::RAD54 | 93.5 | 95.5 (50) | ND | ND |
| rad54Δ::RAD54-T132A | 93.8 | 100.0 (50) | ND | ND |
| rad54Δ::RAD54-T132D | 92.0 | 97.6 (51) | ND | ND |
| B | ||||
| dmc1Δ RAD54 /YEp24 | 0.8 | ND | 73.2 | <1.9 (13) |
| dmc1Δ RAD54 /2μd PHOP1-RAD51 | 36.6 | 70.0 (102) | 72.6 | 0.8 (101) |
| dmc1Δ RAD54/2μd RAD54 | 4.0 | 56.8 (12) | 80.7 | <1.9 (13) |
| dmc1Δ RAD54 /2μ RAD54-T132A | 51.2 | 57.3 (65) | 86.3 | <1.9 (13) |
| dmc1Δ RAD54 /2μ RAD54-T132D | 8.3 | 46.2 (13) | 81.0 | <1.9 (13) |
| C | ||||
| dmc1Δ rad54Δ::RAD54 | 1.1 | ND | 81.5 | 0.6 (78) |
| dmc1Δ rad54Δ::RAD54-T132A | 22.2 | 38.1 (118) | 79.0 | <1.2 (78) |
| dmc1Δ rad54Δ::RAD54-T132D | 0.2 | ND | 75.5 | 1.6 (78) |
| D | ||||
| hed1Δ dmc1Δ rad54Δ::RAD54 | 80.6 | 33.5 (182) | ND | ND |
| hed1Δ dmc1Δ rad54Δ::RAD54-T132A | 83.5 | 12.2 (78) | ND | ND |
| hed1Δ dmc1Δ rad54Δ::RAD54-T132D | 91.6 | 25.0 (78) | ND | ND |
| E | ||||
| hed1Δ dmc1Δ rad54Δ::RAD54 | 6.8 | 42.1 (104) | ND | ND |
| HED1 dmc1Δ rad54Δ::RAD54 | ||||
| hed1Δ dmc1Δ rad54Δ::RAD54-T132Ab | 66.0 | 61.2 (78) | ND | ND |
| HED1 dmc1Δ rad54Δ::RAD54-T132A | ||||
| hed1Δ dmc1Δ rad54Δ::RAD54-T132D | 5.6 | 26.9 (78) | ND | ND |
| HED1 dmc1Δ rad54Δ::RAD54-T132D | ||||
| F | ||||
| hed1Δ rad54Δ::RAD54 | 88.0 | 95.1 (40) | ND | ND |
| hed1Δ rad54Δ::RAD54-T132A | 89.8 | 96.9 (40) | ND | ND |
| hed1Δ rad54Δ::RAD54-T132D | 92.3 | 96.9 (40) | ND | ND |
All strains are diploid and homozygous unless otherwise indicated.
Sporulation was measured by counting between 400-1200 cells using phase contrast microscopy.
% s. v. = % spore viability.
2μ indicates a high copy number plasmid.
The dmc1Δ defects in DSB repair and interhomolog recombination can be partially overcome by over-expression of RAD51 and, to a lesser extent, RAD54 (Bishop et al., 1999; Tsubouchi and Roeder, 2003) (Table 1B). To test whether Mek1 phosphorylation of T132 affects the ability of RAD54 over-expression to suppress dmc1Δ, RAD54-T132A was introduced into a dmc1Δ diploid on a multi-copy plasmid and found to produce over 10 times as many asci as RAD54 (Table 1B). The majority of spores produced by these tetrads were viable, suggesting that RAD54-T132A promotes interhomolog recombination in the absence of dmc1Δ. In fact, a map distance of 28 cM for the HIS4-MAT interval was observed in 222 four-viable spore tetrads (although this distance is less than the isogenic wild-type strain which was 40 cM). Sporulation was increased ~20-fold in a dmc1Δrad54Δ diploid containing two integrated copies of RAD54-T132A compared to integrated RAD54 (Table 1C). Therefore preventing Mek1 phosphorylation of T132 appears to make the Rad54 protein more active. The idea that phosphorylation of T132 acts to suppress Rad54 activity is supported by the finding that RAD54-T132D restores the low level of sporulation observed in RAD54 dmc1Δ (Table 1B and C).
Non-phosphorylatable mutants of RDH54 (RDH54-S85A and RDH54-T89A) complemented the sporulation and spore viability defects of rdh54Δ. In contrast to RAD54, over-expression of RDH54 did not improve the sporulation of dmc1Δ, nor did either of the alanine mutants (data not shown). Therefore it is unclear whether the phosphorylation of Rdh54 observed by Mek1 in vitro is functionally important.
RAD54-T132A suppression of dmc1Δ is dependent on MEK1
For over-expression of RAD51 to restore interhomolog recombination and spore viability to dmc1Δ strains, Mek1 must be active (Niu et al., 2005). Deletion of MEK1 in dmc1Δ strains expressing RAD54-T132A results in high levels of sporulation, similar to dmc1Δmek1Δ diploids (Table 1B and C). Spore viability is less than 1%, however, presumably because DSB repair between sister chromatids fails to create the interhomolog connections needed for accurate Meiosis I segregation (Niu et al., 2005) (Table 1B and C). mek1Δ diploids initiate wild-type levels of DSBs, ruling out the idea that improved sporulation is due simply to a reduction in DSBs (Pecina et al. 2002). The finding that Mek1 is required for interhomolog recombination to occur in the RAD54-T132A strain indicates that Mek1 plays a role in the suppression of meiotic inter-sister DSB repair that is distinct from its role in the phosphorylation of Rad54.
A negative charge at Rad54 T132 reduces Rad51/Rad54 complex formation in vitro
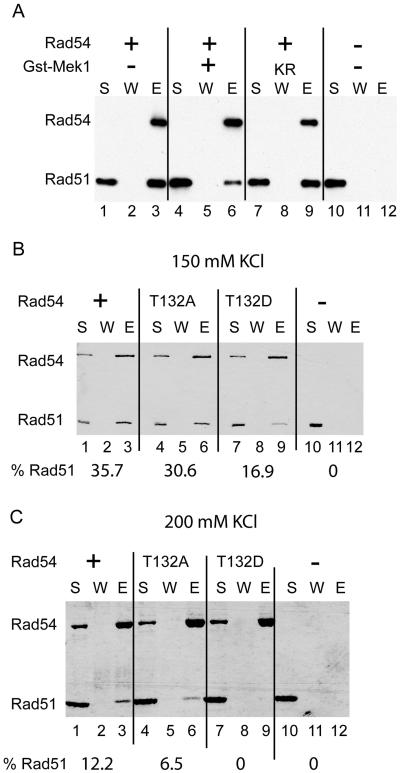
Rad51 and Rad54 physically interact in vegetative yeast cells (Clever et al., 1997). In vitro pulldown experiments using recombinant proteins have shown that deletion of the N-terminal 129 amino acid residues of Rad54 impairs interaction with Rad51 (Raschle et al., 2004). Given the known involvement of the Rad54 N-terminus in Rad51 interaction we reasoned that phosphorylation of T132 by Mek1 could regulate the affinity of Rad54 for Rad51. To test this hypothesis, recombinant His6-S-Rad54 was phosphorylated in vitro by GST-Mek1 and then examined for interaction with Rad51. Protein complexes were captured on nickel beads that bound the His6-tag on Rad54. The ratio of Rad51/Rad54 eluted from the beads was reduced when phosphorylated Rad54 was used (Figure 3A, compare lanes 3 and 6). As expected, reaction with the catalytically inactive GST-mek1-K199R kinase did not affect Rad51/Rad54 complex formation (Figure 3A, compare lanes 3 and 9). Therefore, Mek1 phosphorylation decreases Rad54's ability to bind Rad51.
Figure 3. In vitro pulldown experiments using Rad51 and modified forms of Rad54.
A. Purified His6-S-Rad54 (500 ng) (indicated at Rad54) was reacted with 200 ng GST-Mek1 (+) or GST-Mek1-K199R (KR) and ATP for 30 min at room temperature then incubated with 500 ng of Rad51 at 180 mM KCl. After capturing the protein complexes with Ni-NTA-agarose, the beads were washed with SDS to elute bound proteins. The supernatant containing unbound proteins (S), wash (W) and SDS eluate (E) were probed on an immunoblot with α-His6 and α-Rad51 antibodies. B. Rad51 (5 μg) was incubated with His6-S-Rad54 (5 μg), His6-S-Rad54-T132A (5 μg), or His6-S-Rad54-T132D (5 μg) in 150 mM KCl (Panel B) or 200 mM KCl (Panel C). The supernatant (S), wash (W), and SDS eluate (E) were analyzed by Coomassie staining of SDS polyacrylamide gels. % Rad51 indicates the fraction of Rad51 that bound to Rad54.
Some residual Rad51 binding was observed with phosphorylated Rad54, perhaps because the kinase reaction was not complete. In addition, the phosphorylation experiment does not address whether a negative charge specifically at T132 is deleterious for Rad51/Rad54 complex formation. Genetic experiments indicate that the T132D mutant behaves like a constitutively phosphorylated protein (Table 1), making it a useful phosphomimic for in vitro experiments. His6-S-tagged Rad54, Rad54-T132A and Rad54-T132D were purified to homogeneity from E. coli (Supplemental Figure 3), incubated with Rad51 and precipitated with S-protein agarose beads. At 150 mM salt, a two-fold reduction in Rad51 binding was observed with Rad54-T132A compared to Rad54, in contrast to Rad54-T132A which bound Rad51 nearly as well as wild type (Figure 3B, lanes 3, 6 and 9). Increasing the salt to concentration to 200 mM KCl eliminated Rad51 binding to Rad54-T132D but not Rad54-T132A (Figure 3B, lanes 3, 6 and 9). Efficient Rad51 binding was observed with Rad54-T132D at lower salt concentrations, indicating that a negative charge at T132 decreases, but does not abolish, the affinity of Rad54 for Rad51 (data not shown).
The T132D mutation impairs functional synergy of the Rad51/Rad54 pair
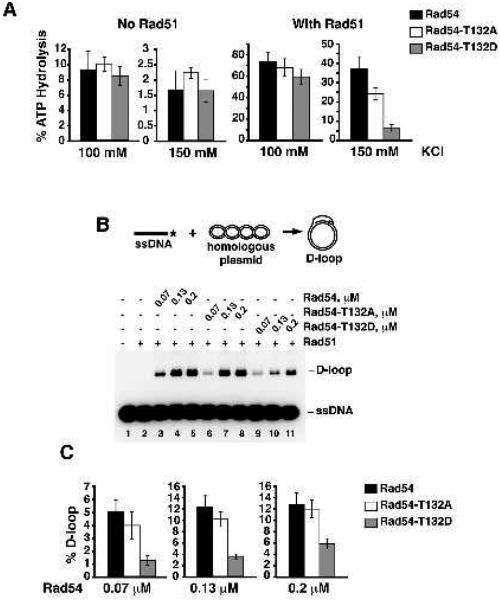
The ATPase activity of Rad54 is strongly stimulated by Rad51 (Mazin et al., 2000; Van Komen et al., 2000) Conversely, the recombinase activity of Rad51 is greatly enhanced by Rad54 (Petukhova et al., 1999). Given that the T132D mutation reduces the affinity of Rad54 for Rad51, the functional synergy of Rad51/Rad54 should be attenuated as well. Rad54, Rad54-T132A and Rad54-T132D possess similar levels of ATPase activity at 100 and 150 mM KCl concentrations (Figure 4A). In contrast, increasing KCl concentration markedly reduced Rad51-stimulated ATP hydrolysis by Rad54-T132D (Figure 4A). Similarly, D-loop reactions carried out at 150 mM salt produced less product with Rad54-T132D (Figure 4B and C). For both assays, the Rad54-T132D mutant exhibited comparable levels of activity as wild type under low salt conditions (Supplemental Figure 4), further supporting the idea that the negative charge is affecting the affinity of Rad54 for Rad51.
Figure 4. Effect of Rad54-T132D on various enzymatic activities.
A. ATP hydrolysis by Rad54, Rad54-T132A, or Rad54-T132D (40 nM) with or without Rad51 (890 nM) was examined in the presence of 100 or 150 mM KCl. Error bars represent SEM. B. Schematic of the D-loop reaction (top). The reaction was carried out in the presence of 150 mM KCl. Rad51 (0.1 μM) was incubated with radiolabeled ss 90-mer oligo (3 μM nucleotides) followed by the addition of the indicated amount of Rad54, Rad54-T132A, or Rad54-T132D. The reaction was initiated by the addition of the pBluescript SK replicative form I DNA (45 μM base pairs). C. Quantification of the D-loop product based on three independent experiments. Error bars represent SEM.
The T132D mutation affects Rad54 activity in vivo
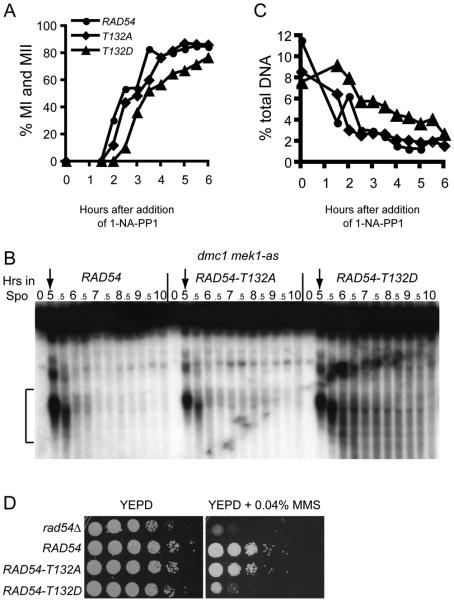
Mek1-as kinase activity can be abolished in vivo by the addition of the 1-NA-PP1 inhibitor directly to the sporulation medium (Wan et al. 2004). When Mek1-as is inactivated by addition of inhibitor to dmc1Δ-arrested cells, DSBs rapidly disappear due to RAD54-dependent repair using sister chromatids as templates (Niu et al., 2005). If Rad51/Rad54 complexes are required under these conditions, then dmc1Δ RAD54-T132D mek1-as cells should exhibit a delay in this repair. dmc1Δ mek1-as diploids containing RAD54, RAD54-T132A or RAD54-T132D were incubated for five hours in sporulation medium. At this time, cells arrested in prophase and exhibited DSBs at the YCR048w and HIS2 hotspots (Figure 5 A and B) (data not shown). Within 90 minutes after addition of inhibitor, the RAD54 and RAD54-T132A diploids began proceeding through Meiosis I. RAD54-T132D was delayed, however, by ~1 hour, likely caused by retarded repair of DSBs. Whereas the bulk of DSBs have disappeared by 2 hours in the RAD54 and RAD54-T132A diploids, DSBs fragments at YCR048w and HIS2 persisted in the RAD54-T132D strain for several hours (Figure 5 B and C)(data not shown).
Figure 5. Effects of RAD54-T132D on meiotic inter-sister DSB repair and MMS sensitivity in vegetative cells.
A. Meiotic progression in dmc1Δ mek1-as diploids homozygous for RAD54, RAD54-T132A or RAD54-T132D. After incubation in Spo medium for 5 hours 1 μM of the Mek1-as kinase inhibitor, 1-NA-PP1, was added to initiate sister chromatid repair. The cells were fixed, stained with DAPI and examined by fluorescence microscopy. Binucleate and tetranucleate cells have completed Meiosis I (MI) and II (MII), respectively. 200 cells were counted for each timepoint. B. DSBs at the YCR048w hotspot from the timecourse in Panel A. Brackets indicate the region of the gel used to quantify DSBs. The value of the 0 timepoint was subtracted as the background. Numbers indicate the hours in Spo medium. Arrows indicate the time at which inhibitor was added. C. Quantitation of DSB fragments as a fraction of the total DNA from the experiment shown in B. D. MMS sensitivity of RAD54 mutants. Overnight cultures of haploid rad54Δ, RAD54, RAD54-T132A or RAD54-T132D strains were serially diluted 10-fold, spotted onto YEPD medium without or with 0.04% MMS and grown for two days at 30°.
In the absence of inhibitor, dmc1Δ RAD54 mek1-as exhibited 0.5% sporulation, whereas addition of inhibitor resulted in >70% sporulation for all three strains. No viable spores were observed out of 26 tetrads for each strain, as expected if repair was occurring between sister chromatids. The finding that DSB repair eventually occurs in RAD54-T132D even though Mek1 is inactivated supports the idea that a negative charge at T132 is not sufficient to completely suppress DSB repair between sister chromatids.
The T132D mutation also impairs Rad54 activity in vegetative cells, as a RAD54-T132D haploid is at least 10-fold more sensitive to 0.04% MMS than RAD54 or RAD54-T132A (Figure 5D). The RAD54-T132D mutant therefore has phenotypes consistent with a reduced ability to form Rad51/Rad54 complexes as indicated by the in vitro experiments.
Rad54 phosphorylation acts synergistically with HED1 to suppress DMC1- independent DSB repair
The meiosis-specific protein Hed1 binds to Rad51 and interferes with Rad51's ability to interact with Rad54 (Busygina et al., 2008). Consistent with this fact, deletion of HED1 suppresses the sporulation defect of a dmc1Δ diploid and produces some viable spores (Tsubouchi and Roeder, 2006)(Table 1D). Neither dmc1Δ RAD54-T132A nor hed1Δ dmc1Δ exhibits wild-type spore viability (38.1% and 33.5%, respectively), however, suggesting that Hed1 and Rad54 T132 phosphorylation may function in parallel pathways to restrain Rad51 activity. If so, then combining RAD54-T132A and hed1Δ should increase interhomolog recombination in a dmc1Δ diploid compared to either single mutant.
To test this hypothesis, a diploid homozygous for dmc1Δ and RAD54-T132A but heterozygous for hed1Δ was constructed. Decreasing the dosage of HED1 by half may make more Rad51 available for interaction with Rad54, thereby promoting complex formation. This idea is supported by the observation that hed1Δ is semi-dominant in the dmc1Δ RAD54 and dmc1Δ RAD54-T132D strains, increasing sporulation from 1.1% and 0.2% in the homozygous HED1 diploids to 6.8% and 5.6%, respectively, in the heterozygotes (compare Table 1C and E). In the HED1/hed1Δ dmc1Δ background, the RAD54-T132A strain sporulated 10-fold better and exhibited a higher level of viable spores than the comparable RAD54 and RAD54-T132D diploids, consistent with the parallel pathways hypothesis (Table 1E). In contrast, when homozygous, the hed1Δ dmc1Δ RAD54-T132A diploid exhibited a three-fold reduction in spore viability compared to the HED1 dmc1Δ RAD54-T132A diploid (compare Table 1C and D), as well as producing fewer viable spores than either hed1Δ dmc1Δ RAD54 or hed1Δ dmc1Δ RAD54-T132D (Table 1D). This decrease in spore viability may result from increased sister chromatid repair (see Discussion). It should be noted that in the presence of DMC1, sporulation and spore viability of the hed1Δ RAD54-T132A diploid is wild type, indicating that the combination of Mek1 and Dmc1 is sufficient to promote interhomolog recombination even when Rad51/Rad54 complexes are free to form (Table 1F).
DISCUSSION
Rad51 is the major recombinase in mitotically dividing cells and, in some organisms such as nematodes and fruit flies, it also serves as the recombinase for meiotic recombination (Villeneuve and Hillers, 2001). For most eukaryotes, however, including budding yeast and mammals, the meiosis-specific recombinase, Dmc1 is required. How these two recombinases function together to promote interhomolog recombination during meiosis remains an important, yet unanswered question.
There is a large body of work indicating that Rad51 and Dmc1 have both overlapping and non-overlapping functions in meiosis (for a review, see Sheridan and Bishop, 2006). That the two recombinases are functionally distinct is clear from studies showing that filaments comprised solely of Rad51 or Dmc1 behave differently during meiosis. In dmc1Δ strains, Rad51 is efficiently recruited to DSBs, but no strand invasion of either sister chromatids or homologous chromosomes is observed (Bishop, 1994; Hunter and Kleckner, 2001; Schwacha and Kleckner, 1997). The failure to invade sister chromatids is dependent upon Mek1 kinase activity (Wan et al., 2004). In rad51Δ diploids, Dmc1 recruitment to DSBs is greatly reduced, complicating the interpretation of the recombination phenotypes of this mutant (Shinohara et al., 1997a). However, in the absence of RAD52, Dmc1 is loaded efficiently by the Mei5-Sae3 mediator complex without Rad51 (Hayase et al., 2004; Lao et al., 2008). The Dmc1 filaments formed in rad52Δ and rad51Δ mutants allow some progression beyond resected DSBs, but the joint molecules no longer exhibit interhomolog bias, despite the fact that Mek1 kinase is active (Lao et al., 2008; Schwacha and Kleckner, 1997). These results led to the proposal that Rad51 may play a structural role in creating a Rad51/Dmc1 filament, which in combination with the Mek1 phosphorylation of an as yet undetermined substrate, directs the filament away from sister chromatids and towards homologs (Sheridan and Bishop, 2006) (Figure 6A). Our work supports the idea that Rad51 recombinase activity is actively suppressed during meiosis and reveals a dynamic pathway by which this regulation can occur via the phosphorylation of Rad54 by Mek1. In addition we have shown that this down-regulation of Rad51 activity is distinct from the mechanism by which Mek1 phosphorylation inhibits Rad51-mediated strand invasion of sister chromatids.
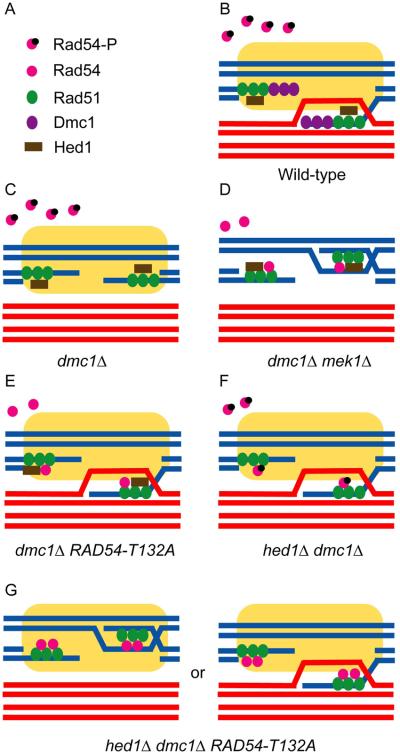
Figure 6. Model for the regulation of meiotic recombination under different genetic conditions.
A. Key for different proteins. Yellow boxes indicate the Mek1-dependent “barrier to sister chromatid repair” (BSCR). Each pair of lines indicates the DNA duplex of a sister chromatid. Blue and red lines indicate homologous chromosomes. B. Interhomolog bias is ensured in wild-type cells by (1) Dmc1, (2) prevention of Rad51/Rad54 complex formation by Hed1 binding to Rad51 and Rad54 T132 phosphorylation and (3) the BSCR. C. In dmc1Δ, DSBs are not repaired due to impaired Rad51/Rad54 complex formation and the BSCR. D. Deletion of MEK1 removes the BSCR and allows unphosphorylated Rad54 to compete with Hed1 for binding to Rad51, thereby enabling intersister DSB repair. E. Unphosphorylated Rad54-T132A can partially compete with Hed1 to form active Rad51/Rad54 complexes and the presence of the BSCR results in some of these filaments invading the non-sister chromatids of homologous chromosomes. F. Deletion of HED1 increases the amount of Rad51 available for binding, thereby allowing Rad51/Rad54 complex formation even though Rad54 is phosphorylated. Once again the BSCR promotes interhomolog strand invasion. G. The combination of hed1Δ and RAD54-T132A removes constraints on Rad51/Rad54 complex formation resulting in active filaments which are able to overcome the BSCR some fraction of the time so that DSB repair uses sister chromatids as templates even though Mek1 is active. The fact that spore viability in this situation is not 0, unlike dmc1Δ mek1Δ, indicates that the BSCR is still functioning.
Formation of Rad51/Rad54 complexes is a key regulatory step in meiotic recombination
Rad54 acts at several different steps during recombination, including stimulation of Rad51-mediated DNA strand invasion (Heyer et al., 2006; Tan et al., 2003). Rad54 performs these functions by binding to Rad51 via the Rad54 N terminus (Golub et al., 1997; Jiang et al., 1996; Raschle et al., 2004). It has previously been shown that the requirement for dmc1Δ for meiotic DSB repair and interhomolog recombination can be bypassed by: 1) over-expression of RAD51, (2) over-expression of RAD54, or (3) deletion of HED1 (Bishop et al., 1999; Tsubouchi and Roeder, 2003; Tsubouchi and Roeder, 2006). What each of these genetic conditions has in common is the potential to increase the number of Rad51/Rad54 complexes. For example, Rad51 binding to the meiosis-specific Hed1 protein does not interfere with Rad51 filament formation but does prevent Rad54 from binding to Rad51 (Busygina et al., 2008; Tsubouchi and Roeder, 2006). Therefore Hed1 likely competes with Rad54 for binding to Rad51 in vivo. Hed1 is limiting in meiotic cells, as evidenced by the observation that hed1Δ is semi-dominant in dmc1Δ strains. These results suggest that deletion of HED1 makes more Rad51 available for binding to Rad54 and that Rad51/Rad54 complex formation is the limiting factor in Rad51-mediated strand invasion during dmc1Δ meiosis. This model assumes that Rad54 binds to Rad51 filaments that have already formed at DSB ends, in idea supported by in vitro experiments showing that Rad54 interacts specifically with Rad51 nucleoprotein filaments prior to the homology search (Solinger et al., 2001).
We have discovered that, in addition to Hed1, Rad51/Rad54 complex formation is regulated by phosphorylation of a specific threonine, T132, in the N-terminus of Rad54 by the meiosis-specific kinase, Mek1. The N-terminus of Rad54 has been shown to bind to Rad51 by both two-hybrid and biochemical pulldown experiments (Golub et al., 1997; Jiang et al., 1996; Raschle et al., 2004). We show that a negative charge at position 132 of Rad54 decreases the affinity of Rad54 for Rad51, thereby attenuating the functional synergy of this protein pair. Furthermore, amino acid substitutions at position 132 that either prevent or mimic phosphorylation have opposite phenotypes. In situations where Rad54 would normally not be phosphorylated, the T132A mutant appears like wild type, while T132D is defective (i.e., MMS-sensitivity of RAD54-T132D in vegetative cells and the delay in DSB repair observed in dmc1Δ mek1-as cells after addition of inhibitor). In contrast, under meiotic conditions where Rad54 would normally be phosphorylated, RAD54-T132D looks more like wild type, while RAD54-T132A exhibits a dominant gain of function phenotype—the suppression of the interhomolog recombination and sporulation defects of dmc1Δ. RAD54-T132A is presumably dominant because the increased affinity for Rad51 allows Rad54-T132A to bind the recombinase regardless of the presence of phosphorylated Rad54. The RAD54-T132A suppression of dmc1Δ is observed even when the former is expressed at normal levels, providing further evidence that the mutant represents a more “active” version of Rad54 during meiosis.
The negative charge conferred by T132 phosphorylation decreases the affinity of Rad54 for Rad51, but does not abolish the interaction. Therefore any situation that increases the amount of Rad51 binding sites, either over-expression of RAD51 or exposure of Rad51 by removal of Hed1 may allow some fraction of phosphorylated Rad54 to bind. In contrast, preventing Rad54 phosphorylation enables some fraction of Rad54 to bind Rad51 even though Hed1 is present. Therefore phosphorylation of T132 presents a way for the cell to dynamically regulate Rad51 activity during meiosis by controlling the ability of Rad51/Rad54 complexes to form.
Down-regulation of Rad51 activity during meiosis occurs by at least two independent pathways
Neither hed1Δ nor RAD54-T132A completely rescues the spore inviability of dmc1Δ, indicating that interhomolog recombination is not occurring at wild-type levels. One explanation is that Dmc1 is simply better than Rad51 at generating interhomolog crossovers (Tsubouchi and Roeder, 2006). Another possibility is that Dmc1 is necessary for the establishment of interference, the process by which crossovers are distributed throughout the genome such that each pair of homologous chromosomes receives at least one. There is a discrepancy in the literature on this point—while a reduction in interference was observed for the DMC1-independent crossovers produced by over-expression of RAD54, no such reduction was observed when RAD51 was over-expressed (Shinohara et al., 2003; Tsubouchi and Roeder, 2003). Since it seems likely that the mechanism of dmc1Δ suppression is the same in both cases (assembly of Rad51/Rad54 complexes), the differences between these two studies remain to be resolved.
We propose an alternative explanation—that Hed1 binding to Rad51 and Mek1 phosphorylation of Rad54 act independently to prevent Rad51/Rad54 complex formation (Figure 6). Thus maximal levels of Rad51 activity cannot be achieved in strains in which Rad54 is not phosphorylated, because most of the Rad51 is bound by Hed1 (Figure 6E) nor in hed1Δ diploids because Rad54 is still phosphorylated (Figure 6F). This hypothesis is supported by our discovery that combining RAD54-T132A with half the amount of HED1 increases both sporulation and spore viability in the dmc1Δ background compared to either single mutant. An unexpected result was observed, however, in the hed1Δ dmc1Δ RAD54-T132A homozygous diploid. In this diploid, the two meiosis-specific factors that normally restrain Rad51 activity are eliminated, potentially allowing Rad51/Rad54 complexes to form as efficiently in mitotic cells. Although sporulation was increased relative to the comparable HED1/hed1Δ heterozygote, spore viability decreased. This reduction in spore viability could be explained if more of the DSB repair occurring in the hed1Δ RAD54-T132A dmc1Δ diploid was directed between sister chromatids than for the RAD54 or RAD54-T132D strains, even though Mek1 is active (Figure 6G).
Taken together, these results suggest that different outcomes may result depending on the level of recombinase activity. In dmc1Δ strains endogenous levels of phosphorylated Rad54, Hed1 and Rad51 are insufficient for interhomolog recombination, consistent with the finding that recombination proteins are limiting in dmc1Δ cells (Johnson et al., 2007) (Figure 6C). Increasing the number of Rad51/Rad54 complexes promotes interhomolog recombination up to a certain point. However, if too many Rad51/Rad54 complexes are made, the Mek1 imposed barrier to sister chromatid repair may be overcome, resulting in fewer interhomolog connections and a reduction in spore viability. Modulating Rad51 activity may be especially important in organisms such nematodes and fruit flies which lack Dmc1.
Mek1 suppresses inter-sister DSB repair independently of Rad54 phosphorylation
Given that Mek1 can indirectly down-regulate Rad51 activity by phosphorylating Rad54, it is reasonable to suppose that this could be the mechanism by which Mek1 suppresses inter-sister DSB repair. If this were true, however, then situations that result in activated Rad51 (i.e. Rad51/Rad54 complex formation) should overcome the Mek1 barrier and repair via sister chromatids. In other words, if Mek1 phosphorylation of Rad54 was solely responsible for suppressing meiotic sister chromatid repair, the phenotype of RAD54-T132A dmc1Δ should be the same as dmc1Δ mek1Δ, but this is not the case. Whereas dmc1Δ mek1Δ mutants sporulate at levels >80% but produce <1% viable spores, dmc1Δ RAD54-T132A exhibits only 22% sporulation with ~40% of the spores being viable. Furthermore, inactivation of Mek1-as in dmc1Δ RAD54-T132A after DSB formation results in rapid repair of DSBs off sister chromatids. Conversely, although aspartic acid at position 132 acts as a good phosphomimic by preventing repair in dmc1Δ MEK1 diploids, it does not prevent the sister repair observed when Mek1-as is inactivated. Therefore the ability of Rad51/Rad54 complexes to mediate interhomolog strand invasion requires that Mek1 phosphorylate some other substrate that then suppresses strand invasion of sister chromatids (Figure 6).
Phosphorylation provides a dynamic way of regulating Rad51-strand exchange activity during meiosis
In the hed1Δ RAD54-T132A double mutant, the meiosis-specific barriers to Rad51/Rad54 complex formation are removed, yet there are no obvious deleterious phenotypes indicating that down-regulation of Rad51 is unnecessary if Dmc1 is present. This raises the important question of why redundant mechanisms have evolved for ensuring that Rad51 activity is constrained during meiosis. One explanation proposed by Tsubouchi and Roeder (2006) is that Hed1 ensures coordination between Rad51 and Dmc1 by inhibiting Rad51 activity only if Dmc1 is absent. Because Hed1 co-localizes with Rad51 in DMC1 cells, these authors suggested that there may be a conformational change in the Rad51/Rad54 complex induced by Dmc1 that prevents Hed1 repression of Rad51 activity. The discovery of a second pathway for Rad51 inhibition makes this explanation less likely, given that Dmc1 would now have to overcome the lack of affinity resulting from Rad54 phosphorylation in addition to inactivating Hed1. We prefer an alternative explanation that the presence of Rad51, but not its activity, is important for making recombinase filaments that are efficient in interhomolog recombination (Hunter, 2007; Sheridan and Bishop, 2006; Tsubouchi and Roeder, 2006).
We propose that the combination of Hed1 and Rad54 T132 phosphorylation suppresses Rad51 recombinase activity by preventing formation of Rad51/Rad54 complexes (Figure 6B). This idea is supported by the fact that RAD54 is dispensable for interhomolog recombination during meiosis (Shinohara et al., 1997b). If RAD54 is not required for interhomolog recombination and if it is normally prevented from interacting with Rad51, why do rad54Δ mutants exhibit reduced levels of sporulation and spore viability? The RAD54-T132A mutant completely complements the meiotic defects of rad54Δ, arguing that phosphorylation of Rad54 is not required for wild-type levels of sporulation and spore viability. One possibility is that intersister recombination is used to repair any DSBs that remain after all sixteen homologous chromosomes have been connected by crossovers (Hunter, 2007). The fact that the barrier to sister chromatid repair is mediated by a kinase makes elimination of this constraint possible simply by inactivation of Mek1. Inhibition of Mek1 kinase activity could serve to simultaneously allow Rad51/Rad54 complex formation as well as eliminate the barrier to sister chromatid repair (Figure 6D). The rad54Δ phenotypes can therefore be explained by the inability to repair these residual DSBs in late prophase after interhomolog recombination has been completed. Evidence for two rounds of Rad51-mediated recombination exists in nematodes (Hayashi et al., 2007). In this organism, Rad51 is localized to DSBs early in meiotic prophase in a Rad50-dependent manner and these breaks have the ability to form interhomolog crossovers. At mid to late pachytene there is a switch such that Rad51 is loaded onto breaks independently of Rad50. These breaks are not competent for formation of interhomolog crossovers but may be repaired by sister chromatids as way of maintaining genome integrity.
In summary, this work demonstrates a newly discovered regulatory mechanism for controlling Rad51 recombinase activity during meiosis—suppression Rad51/Rad54 complex formation by Mek1-mediated phosphorylation of Rad54. This mode of regulation allows for Rad51 activity to be rapidly modulated up and down and may be critical for ensuring that all breaks are repaired before the onset of the meiotic divisions.
EXPERIMENTAL PROCEDURES
Yeast strains and timecourses
All yeast strains were derived from the SK1 background. Complete genotypes are presented in Supplemental Table 2. Strain constructions are described in the Supplemental Methods. Timecourses and DSB analysis were carried out at 30°C as described in (Niu et al., 2005). The mek1-as inhibitor, 1-NA-PP1, is described in Wan et al. (2004).
Plasmids
Plasmid constructions are described in the Supplemental Methods.
Protein purification and MS
Purification of RPA1, Rad51, Rad52 and Rad54 out of yeast, as well as purification of recombinant Rad54 and Rdh54 from bacteria, have been published (Chi et al., 2006; Raschle et al., 2004; Song and Sung, 2000; Sung and Stratton, 1996; Van Komen et al., 2006). Purification of GST-Mek1 and Rad54-3Flag from dmc1Δ arrested yeast cells, as well as recombinant Hop1, is described in the Supplemental Methods. MS analysis is also described in the Supplemental Methods.
In vitro kinase assays
Detailed protocols for kinase assays using both ATP and ATPγS analogs are described in the Supplemental Methods. ATPγS was purchased from Sigma. The syntheses of N6-benzyl- and N6-isopentyl-ATPγS are described in Allen et al. (2007). The synthesis of N6-furfuryl-ATPγS is described in the Supplemental Methods.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Aaron Neiman for comments on the manuscript and Doug Bishop, Scott Keeney and Michael Lichten for helpful discussions. Doug Bishop, Mike Dresser, Kurt Runge and Toshi Tsukiyama generously supplied plasmids or strains. We thank Anglina Kataria for making the rdh54 mutants. This work was supported by NIH grants GM50717 to N. M. H., GM57814 to P. S., HG3456 to S. P. G. and AI44009 to K. M. S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental data
Supplemental data include two tables, three figures and one text file.
REFERENCES
- Allen JA, Li M, Brinkworth CS, Paulson JL, Wang D, Hubner A, Chou W-H, Davis RJ, Burlingame AL, Messing RO, et al. A semisynthetic epitope for kinase substrates. Nature Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbel A, Zenvirth D, Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. Embo J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong SJ, Caryl AP, Jones GH, Franklin FC. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci. 2002;115:3645–3655. doi: 10.1242/jcs.00048. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Nikolski Y, Oshiro J, Chon J, Shinohara M, Chen X. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells. 1999;4:425–443. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 2008;22:786–795. doi: 10.1101/gad.1638708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Kwon Y, Seong C, Epshtein A, Lam I, Sung P, Klein HL. Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J Biol Chem. 2006;281:26268–26279. doi: 10.1074/jbc.M602983200. [DOI] [PubMed] [Google Scholar]
- Clever B, Interthal H, Schmuckli-Maurer J, King J, Sigrist M, Heyer W-D. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. The EMBO J. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F, Nabeshima K, Villeneuve A, Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr Biol. 2004;14:585–592. doi: 10.1016/j.cub.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Dresser ME, Ewing DJ, Conrad MN, Dominguez AM, Barstead R, Jiang H, Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub EI, Kovalenko OV, Gupta RC, Ward DC, Radding CM. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 1997;25:4106–4110. doi: 10.1093/nar/25.20.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayase A, Takagi M, Miyazaki T, Oshiumi H, Shinohara M, Shinohara A. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004;119:927–940. doi: 10.1016/j.cell.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Chin GM, Villeneuve AM. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 2007;3:e191. doi: 10.1371/journal.pgen.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W-D, Li X, Rolfsmeier M, Zhang X-P. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- Hunter N. Meiotic Recombination. Springer-Verlag; Heidelberg: 2007. [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double- strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Jiang H, Xie Y, Houston P, Stemke-Hale K, Mortensen UH, Rothstein R, Kodadek T. Direct association between the yeast Rad51 and Rad54 recombination proteins. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- Johnson R, Borde V, Neale MJ, Bishop-Bailey A, North M, Harris S, Nicolas A, Goldman AS. Excess single-stranded DNA inhibits meiotic double strand repair. PLoS Genet. 2007;3:e223. doi: 10.1371/journal.pgen.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Klein HL. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol Cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- Mazin AV, Bornarth CJ, Solinger JA, Heyer WD, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- Niu H, Li X, Job E, Park C, Moazed D, Gygi SP, Hollingsworth NM. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol Cell Biol. 2007;27:5456–5467. doi: 10.1128/MCB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- Raschle M, Van Komen S, Chi P, Ellenberger T, Sung P. Multiple interactions with the Rad51 recombinase govern the homologous recombination function of Rad54. J Biol Chem. 2004;279:51973–51780. doi: 10.1074/jbc.M410101200. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Sheridan S, Bishop DK. Red-Hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 2006;20:1685–1691. doi: 10.1101/gad.1447606. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK. Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes to Cells. 1997a;2 doi: 10.1046/j.1365-2443.1997.1480347.x. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Sakai K, Shinohara A, Bishop DK. Crossover interference in Saccharomyces cerevisiae requires a TID1/RDH54- and DMC1-dependent pathway. Genetics. 2003;163:1273–1286. doi: 10.1093/genetics/163.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Shita-Yamaguchi E, Buerstedde J-M, Shinagawa H, Ogawa H, Shinohara A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homolog of RAD54, RDH54/TID1 in mitosis and meiosis. Genetics. 1997b;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Lutz G, Sugiyama T, Kowalczykowski SC, Heyer WD. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J Mol Biol. 2001;307:1207–1221. doi: 10.1006/jmbi.2001.4555. [DOI] [PubMed] [Google Scholar]
- Song B, Sung P. Functional interactions among yeast Rad51 recombinase, Rad52 mediator and replication protein A in DNA strand exchange. J Biol Chem. 2000;275:15985–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- Sung P, Stratton S. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J Biol Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TLR, Kanaar R, Wyman C. Rad54, a jack of all trades in homologous recombination. DNA Repair. 2003;2:787–794. doi: 10.1016/s1568-7864(03)00070-3. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–1775. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Komen S, Macris M, Sehorn MG, Sung P. Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol. 2006;408:445–463. doi: 10.1016/S0076-6879(06)08028-1. [DOI] [PubMed] [Google Scholar]
- Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106:647–650. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- Wan L, de los Santos T, Zhang C, Shokat K, Hollingsworth NM. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic DSB repair in budding yeast. Mol Biol Cell. 2004;15:11–23. doi: 10.1091/mbc.E03-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.