Abstract
There is mounting evidence that tRNA modifications play crucial roles in the maintenance of wild type levels of several tRNA species. Here we describe a generalized framework in which to study tRNA turnover in the yeast Saccharomyces cerevisiae as a consequence of a defect in tRNA modification status. We describe several approaches for the identification of tRNA species that are reduced as a consequence of a modification defect, methods for analysis of the rate of tRNA loss and analysis of its aminoacylation, and methods for initial characterization of tRNA turnover. These approaches have been used successfully for several modification defects that result in tRNA turnover.
1. Introduction
tRNA molecules are extremely stable RNAs, with half-lives estimated in hours or days. This stability of tRNAs has been attributed in part to the ubiquitous modifications that are present on the body of tRNAs. Numerous in vitro experiments have shown that fully modified tRNAs are more thermally stable and more compactly folded than completely unmodified tRNA transcripts (Sampson and Uhlenbeck, 1988; Hall et al., 1989; Perret et al., 1990; Derrick and Horowitz, 1993; Maglott et al., 1998), although little is known about the contribution of individual modifications to folding or stability (Nobles et al., 2002). Recent experiments in the yeast Saccharomyces cerevisiae have shown that lack of individual modifications or pairs of modifications can have profound effects on levels of specific tRNAs in vivo. These results have opened the door to understanding the biological roles of modifications in maintaining tRNA levels, and the specific mechanisms by which modifications exert these effects in the cell.
Lack of specific modifications can lead to a decrease in the levels of particular tRNA species in different ways in yeast. Thus, loss of the m1A modification at position 58, due to mutation of GCD10 or GCD14, results in specific degradation of pre-tRNAiMet by the Trf4/Rrp6 nuclear pre-tRNA surveillance pathway (Anderson et al., 1998; Kadaba et al., 2004). By contrast, lack of the m7G and m5C modifications in a trm8-Δ trm4-Δ strain results in rapid degradation and deacylation of mature tRNAVal(AAC) at elevated temperatures, by a mechanism distinct from the Trf4/Rrp6 pathway (Alexandrov et al., 2006). We have also recently reported that lack of the Um44 and ac4C12 modifications in a trm44-Δ tan1-Δ strain leads to decreased levels of tRNASer(CGA) and tRNASer(UGA) in this strain by an undetermined mechanism (Kotelawala et al., 2008). In addition, mutation of certain tRNAs together with lack of particular modifications can also lead to loss of the tRNA, as has been reported for mutation of tRNASer(CGA) accompanied by lack of m5U54 (due to deletion of TRM2) or Ψ55 (PUS4) (Johansson and Bystrom, 2002) and for mutation of tRNAArg(CCG) accompanied by lack of Ψ55 (Copela et al., 2006).
There are two common features in known examples in which lack of particular tRNA modifications leads to decreased tRNA levels. First, in all cases examined so far, lack of a single modification or a combination of modifications only affects the levels of one or two tRNA species, even though the particular modifications may occur on numerous tRNAs. Thus, lack of m1A58, which is found on 23 sequenced yeast tRNAs, is primarily associated with loss of pre-tRNAiMet (Anderson et al., 1998), lack of m7G46 and m5C49, which occur together on 3 tRNAs, leads to degradation of only tRNAVal(AAC) (Alexandrov et al., 2006), and lack of Um44 and ac4C12 leads to degradation of only 2 of the 4 tRNASer species that have these modifications (Kotelawala et al., 2008). Second, although the modifications in these mutants are not present even at permissive temperatures, the levels of the affected tRNA species decrease more significantly at elevated temperatures, coinciding with the temperature sensitive growth defects of these strains.
These features have allowed us to develop a framework in which to study the turnover of tRNAs that are improperly modified. We will describe below several approaches that can be used to identify the tRNA species the levels of which may be affected by a tRNA modification defect. We will also describe methods for analysis of the levels and aminoacylation status of the affected tRNA in the mutant strain. Finally, we will discuss methods that can be used to determine if the loss of a tRNA species is due to a decrease in transcription of the tRNA or to tRNA degradation, and if the precursor or mature form of the tRNA is degraded.
2. Methodology
Identification of the tRNA species that are reduced in modification mutants.
Analysis of the levels of functional tRNA in vivo.
Characterization of the loss of tRNA to distinguish between transcription and degradation, and to identify the substrate for degradation.
3. Identification of the tRNA Species that are Reduced in Modification Mutants
To characterize how a modification defect leads to loss of tRNA, it is necessary first to identify the tRNA species that are reduced because of the modification defect. This can be accomplished by three methods: direct analysis of likely tRNA targets in non-permissive conditions; global analysis of tRNA levels in the mutant strain using microarrays; or screening for high copy suppressors of the mutant phenotype, one of which is likely to be the affected tRNA. These methods are discussed further below.
3.1. Direct Testing of Likely tRNA Targets
Direct analysis of likely target tRNAs in the modification deficient mutant strain can be successful if only a small number of tRNA species have the particular modification in question. In this case, RNA is isolated from the mutant strain and a wild type control at permissive and non-permissive conditions and analyzed by Northern blotting to detect differences in levels of candidate tRNAs. The identified target tRNA(s) can then be tested as high copy suppressors of the mutant phenotype to show that loss of the identified tRNA(s), and not other tRNA species, is responsible for the growth defect of the mutant strain.
This approach has been used successfully in our laboratory to show that a decrease in the levels of tRNASer(CGA) and tRNASer(UGA) occurs in a trm44-Δ tan1-Δ strain, lacking Um44 and ac4C12, and leads to the temperature sensitive phenotype of this strain (Kotelawala et al., 2008). Since the Um44 modification occurs only on the four tRNASer species in yeast, and ac4C12 occurs on tRNASer and tRNALeu species, the levels of each of these tRNAs in the trm44-Δ tan1-Δ strain were analyzed by Northern blotting. A decrease in tRNASer(CGA) and tRNASer(UGA) was observed in non-permissive conditions, whereas there was no change in the levels of any of the other species tested. Expression of both tRNAs from high copy plasmids rescued growth of the trm44-Δ tan1-Δ strain at high temperature, indicating that loss of these tRNAs is responsible for its temperature sensitive phenotype.
The primary drawback of this approach is that only 29 of the 42 tRNA species have been fully characterized in yeast (Sprinzl et al., 1999), so not all the tRNA species that have a particular modification may be known. Thus, if a target tRNA species is identified, failure to suppress the mutant phenotype by overexpression of the tRNA gene may indicate that other tRNA species also decrease due to the modification defect. However, high copy suppression of the mutant phenotype may also fail if the phenotype is not due to loss of tRNA, but is due to some other role of the modification. Nonetheless, successful suppression of the mutant phenotype by tRNA overexpression indicates that the phenotype is due to loss of tRNA, and that the identified tRNA species is(are) likely the only one(s) affected.
3.2. Microarray Analysis of tRNA levels
Global analysis of tRNA levels using microarray methods is the most general way to identify target tRNAs whose levels are affected by a modification defect. Indeed, this approach is necessary if the modification defect does not lead to a growth phenotype, and is useful if the modification affected by the mutation is present on numerous tRNA species, which would make direct analysis of likely targets more difficult. In this microarray approach, tRNA levels in the mutant strain are compared directly to those in the wild type strain when both are grown under conditions in which tRNA levels are expected to be affected. The methods used for preparation and microarray analysis of RNA have been described in detail by Hughes et al. (Hughes et al., 2006). Any observed changes in tRNA levels are then verified by Northern analysis of RNA isolated from the mutant and wild type strains under non-permissive conditions.
We used this approach to show that loss of tRNAVal(AAC) occurs in strains lacking either D47 (due to lack of DUS3), Ψ13 (PUS7), or m5C34, 40, 48, 49 (TRM4) in combination with m7G46 (TRM8/TRM82) (Alexandrov et al., 2006). RNA isolated from the trm8-Δ dus3-Δ, trm8-Δ pus7-Δ, and trm8-Δ trm4-Δ strains grown at semi-permissive conditions, and from the corresponding single mutants was compared with RNA from a wild type control by direct labeling of the RNA. Examination of relative hybridization of different tRNA species revealed that tRNAVal(AAC) levels were decreased in all three double mutant strains relative to wild type. This was then confirmed by Northern analysis of RNA isolated from these strains under non-permissive conditions. Furthermore, high copy suppression demonstrated that loss of tRNAVal(AAC) is the primary cause of the temperature sensitive phenotype of the trm8-Δ trm4-Δ strain.
An important consideration when interpreting the results of microarray analysis of tRNA levels is that certain modifications hinder hybridization of the tRNA to the microarray probes. This was initially observed for dihydrouridine (Xing et al., 2004), but has since been demonstrated for a number of other modifications that impact Watson-Crick base pairing (Hiley et al., 2005; Hughes et al., 2006). Lack of one of these modifications results in increased hybridization of the tRNA to the corresponding probe compared with the modified tRNA, which may mask a decrease in the abundance of the tRNA in the mutant strain. It is therefore important to analyze the relative hybridization of multiple probes that hybridize at different positions of a single tRNA species.
3.3. Multicopy Suppression of Mutant Phenotype
Screening for high copy suppressors of the mutant strain phenotype to identify affected tRNA species relies on two assumptions: first, that a decrease in tRNA levels is responsible for the growth phenotype of the modification mutant; and second, that only a single tRNA species is severely affected by the mutation. As in other multicopy suppression screens, the mutant strain is transformed with the genomic library of choice to obtain ~5x representation of each plasmid to ensure complete genome coverage, and transformants are replica-plated to non-permissive conditions to select for suppression of the mutant phenotype. The identity of the suppressor gene(s) is determined by sequencing of the recovered plasmid, followed by sub-cloning of the candidate tRNA genes and re-transformation to ensure plasmid dependence. A decrease in the levels of the candidate tRNA is then confirmed by analysis of tRNA levels in the original mutant strain, by Northern blotting. This approach was used to show that degradation of pre-tRNAiMet due to lack of m1A58 is responsible for the temperature sensitive phenotype of gcd10 mutants (Anderson et al., 1998).
As with all screens using a genomic plasmid library, this type of a screen may not be successful if the gene encoding the affected tRNA is not present in the plasmid library used, especially if it is a single copy tRNA gene. This problem of gene representation has been partially resolved by creation of a tiled array genomic library, which covers 97% of the yeast genome and contains 98% of non-coding RNAs (Jones et al., 2008). It is of course entirely possible that this approach will yield other multi-copy suppressors that are not tRNA genes. Such suppressors may ultimately be very interesting in understanding the nature of the defect.
4. Analysis of the levels of functional tRNA in vivo
Identification of the target tRNA species that is(are) affected by a modification mutant allows detailed analysis of the loss of the affected tRNA, including the rate of tRNA loss, the functional status of the tRNA that remains, and eventually the mechanism by which the tRNA is lost. These questions can be addressed by first performing a detailed time course analysis of tRNA levels in the mutant strain, and by determining the aminoacylation level of the tRNA at different times, as described below.
4.1. Growth of Cells for RNA Isolation
The time course with which a target hypomodified tRNA is lost in vivo is very informative as to the nature of the tRNA loss. For example, a very rapid loss of a tRNA upon shift to non-permissive conditions indicates that loss of the tRNA is likely due to degradation of the mature tRNA species, whereas slow loss of a tRNA may be due to degradation of the precursor or the mature form, or perhaps to a decrease in tRNA transcription. The appropriate time scale in which to analyze tRNA levels in a mutant strain is dictated by the time it takes for a growth phenotype to become apparent in liquid growth medium when the strain is grown under non-permissive conditions.
We have frequently observed that growth phenotypes of mutant strains are more pronounced on solid media than in liquid culture, which makes it very important to determine growth conditions that will allow analysis of tRNA in the mutant strain under non-permissive conditions. This was the case for the trm44-Δ tan1-Δ strain, which does not grow at 35 °C when grown on YPD plates, but grows like wild type in liquid YPD media at temperatures as high as 37 °C. To circumvent this problem, this strain was grown at 36.5 °C in liquid YP media supplemented with 3% glycerol instead of 2% dextrose, which allows the temperature sensitive growth defect to be observed (Kotelawala et al., 2008). Similarly, trm8-Δ trm4-Δ mutants do not grow at 33 °C or above on YPD plates, but continue growing in liquid culture for more than 13 hours after shift to 33 °C, and require shift to 37 °C to stop growth rapidly and completely (Alexandrov et al., 2006).
Once conditions that limit growth of the mutant strain in liquid media have been determined, a preliminary growth curve is done to determine the time it takes for a growth defect to become apparent in the mutant strain. To do this, the mutant strain and a wild type control are grown under permissive conditions for at least three generations to establish exponential growth, and then shifted to non-permissive conditions. Growth of the strains is monitored by measuring OD600 of the cultures before and after shift to determine the exact time course of cessation of growth.
To analyze levels of the target tRNA species, cells are grown under permissive conditions, shifted to restrictive conditions, and sampled at appropriate times. If the phenotype of the mutant strain becomes apparent soon after shift to non-permissive conditions, as in the case of the trm8-Δ trm4-Δ strain lacking m7G and m5C, mixing of the culture with pre-heated media achieves rapid shift to non-permissive temperature, and allows detailed analysis of the rate of tRNA degradation. Typically, one volume of culture (eg., 35 ml) at 29 °C is mixed with two volumes of media (70 ml) pre-heated to 41 °C. For other initial and final desired conditions, the temperature of the pre-heated media can be calculated using the following formula: Tmedia=(Ttotal×Vtotal − Tculture×Vculture)/Vmedia, where T is temperature in degrees Kelvin, and V is volume, although the temperature of the media should not exceed 45 °C to avoid a heat shock response. After the temperature shift, cells are grown with shaking in a water bath to ensure that the appropriate temperature is maintained.
Cells (typically 20 ODs) are harvested immediately before shift to non-permissive conditions, and then at different times after temperature shift. A rapid method that minimizes changes in tRNA levels that may occur after cells are removed from growth is harvesting by filtration. The culture is filtered through a 0.45 µm vacuum filter (Nalgene, #296-3354) (requiring ~1 minute for 100 ml culture), cells are washed with 10 ml of cold water (less than 1 minute), washed off the filter by pipetting with 4 ml of cold water, split into two 2 ml microfuge tubes and immediately frozen on dry ice for storage at −70 °C. Using this procedure requires only ~2–3 minutes from the time the sample is taken until cells are frozen.
A more common method that requires significantly more time is to chill the culture on ice (or to add ice or frozen media to the culture), followed by centrifugation at 4 °C to pellet cells, washing of cells in cold water, re-centrifugation to pellet cells, and freezing of cells on dry ice for storage at −70 °C.
4.2. Preparation of RNA for Analysis of tRNA levels
To analyze loss of target tRNA species in the mutant strain under non-permissive conditions, RNA is isolated from cells using the hot phenol low molecular weight RNA isolation protocol, based on the method described by Rubin (Rubin, 1975), which allows extraction of small RNAs from cells with minimal contamination from cell debris.
Use ~10 ODs of cells to yield ~20 µg bulk RNA. If cells were harvested using the rapid filtration method, thaw cells on ice, centrifuge in a microfuge for 1 minute at maximum speed at 4 °C, and discard supernatant.
To extract RNA, add 0.5 ml of RNA Extraction Buffer (0.1 M NaOAc pH 5.2, 20 mM EDTA pH 8.0, 1% SDS) and 0.5 ml of phenol (equilibrated with 0.5 M Tris-HCl pH 7.5), and then vortex tubes for 30 seconds every 3 minutes for a total of 21 minutes, placing tubes at 60 °C between vortex treatments.
Centrifuge tubes for 5 minutes at 7,000 RPM at 4 °C in a microfuge and transfer the aqueous layer to pre-spun 2 ml Phase Lock Gel Heavy tubes (Eppendorf/5 PRIME cat #2302830).
To remove protein contamination, add 0.4 ml of 25:24:1 phenol/chloroform/iso-amyl alcohol (equilibrated with 0.5 M Tris-HCl pH 7.5) to each tube, mix, and centrifuge for 5 minutes at maximum speed in a microfuge. Repeat this step a second time in the same tube.
To precipitate nucleic acid and remove phenol contamination, transfer the aqueous layer to a new 1.5 ml microfuge tube containing 1 ml 100% EtOH, mix the sample, freeze on dry ice, and then centrifuge for 15 minutes at maximum speed at 4 °C, and discard the supernatant. An RNA pellet should be visible at the bottom of the tube.
Resuspend the pellet in 225 µl of TE 8 (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0), add 25 µl of 3M NaOAc pH 5.2 and 625 µl of 100% EtOH, and mix (solution may become cloudy). Centrifuge samples for 15 minutes at maximum speed at 4 °C and discard supernatant. Wash pellets with 100 µl of cold 70% EtOH, centrifuge for 5 minutes at maximum speed at 4 °C, and remove all supernatant by re-spinning and air drying.
Resuspend the pellet in 50 µl of TE 7.5 (10 mM Tris-HCl pH 7.5, 1 mM EDTA pH 8). Store at −70 °C.
RNA prepared using the Hot Phenol method is separated on a 25 cm long and 0.75 mm thick 10% polyacrylamide gel (19:1) containing 1X TBE (0.9 M Tris-Borate, 1 mM EDTA, pH 8.3, from a 10X stock) and 8 M urea, followed by transfer to Hybond N+ membrane (Amersham Biosciences #RPN303B) in 1X TAE (40 mM Tris-Acetate, 1mM EDTA, pH 8.1, from a 50X stock) at 4 °C in a BioRad Trans-Blot apparatus for 5 hours at 40V. The RNA is then UV crosslinked to the membrane with 120,000 µJ at 254 nm in a Stratagene UV Crosslinker (model #2400) for Northern analysis. The membrane can be stored in 1X TAE at 4 °C for up to two months.
4.3. Preparation of RNA under Acidic Conditions for Analysis of Aminoacylation
To analyze changes in the aminoacylation status of the target tRNA species, RNA is isolated under acidic conditions to preserve aminoacylation, and is resolved on an acidic gel. This method was described by Varshney et al. (Varshney et al., 1991) and applications of this approach and some related methodology are discussed in a recent review (Kohrer and Rajbhandary, 2008).
Use ~10 ODs of cells to yield ~30 µg total RNA. If cells were harvested using the rapid filtration method, thaw cells on ice, centrifuge in a tabletop microfuge for 1 minute at maximum speed at 4 °C, and discard supernatant.
To extract RNA, add 200 µl of ice cold Acidic RNA Elution Buffer (0.3 M NaOAc, 10 mM EDTA, pH 4.5) to each sample and 0.5 mm glass beads to saturate cells (volume will go up to ~0.5 ml). Then vortex tubes for 15 seconds every 3 minutes for 12 minutes, keeping samples on ice between vortex steps.
To remove contaminants, add 1 ml phenol equilibrated with 0.3 M NaOAc pH 4.5, 10 mM EDTA, vortex tubes for 15 seconds every 3 minutes for 9 minutes, keeping the samples on ice between vortex steps. Centrifuge tubes for 10 minutes at 5,000 RPM in a microfuge at 4 °C to pellet cell debris and separate phenol and aqueous phases. The aqueous phase may appear cloudy due to precipitation of sodium acetate on ice.
To remove phenol and salt contamination, carefully transfer the aqueous phase to new 1.5 ml microfuge tube containing 350 µl 100% EtOH, mix, freeze on dry ice, centrifuge for 15 minutes at maximum speed at 4 °C, and discard supernatant. An RNA pellet should be visible at bottom of tube.
Resuspend the pellet in 200 µl of cold Acidic RNA Elution Buffer, by placing tubes at 4 °C in a refrigerator, not on ice, to facilitate resuspension of RNA. This may take ~15–25 minutes. Add 400 µl of 100% EtOH, mix, freeze on dry ice, and then centrifuge the sample for 15 minutes at maximum speed at 4 °C in a microfuge, and discard the supernatant. Wash the pellet with 100 µl of cold 70% EtOH, centrifuge for 5 minutes at maximum speed at 4 °C, and discard the supernatant.
Resuspend the pellet at 4 °C in 60 µl of 10 mM NaOAc pH 4.5, 1 mM EDTA. Make 20 µl aliquots of each sample, quick-freeze on dry ice and store at −70 °C. This storage method avoids the necessity of subjecting samples to multiple freeze-thaw cycles.
Aminoacylated RNA prepared using the Acidic RNA prep is resolved on a 0.75 mm thick 6.5 % polyacrylamide gel (19:1) containing 8 M urea and 0.1 M NaAc pH 4.5 at 4 °C. Prior to loading, each sample is mixed with an equal volume of Acidic RNA dye (0.1 M NaOAc pH 4.5, 8M urea, 0.05% bromophenol blue, 0.05% xylene cyanol). To obtain maximum resolution between the aminoacylated and de-acylated tRNA species, RNA is separated on a 40 cm gel by electrophoresis until the bromophenol blue is just at the bottom of the gel. We typically run the gel at 450V for ~20 hrs and make sure that the gel stays cold to the touch. In this system, the aminoacylated tRNA migrates slower than its corresponding deacylated species, and tRNA migrates below the xylene cyanol dye, about three-fifths of the way down the gel. A deacylated RNA control is obtained by incubation in buffer containing 0.1 M Tris-HCl (pH 9.0) and 1 mM EDTA for 30 min at 37 °C, followed by ethanol precipitation and resuspension in 10 mM NaOAc, 1 mM EDTA, pH 4.5. After separation on the gel, RNA is transferred to a Hybond N+ membrane and crosslinked as described above. The membrane can be stored in 1X TAE at 4 °C for up to two months.
4.4. Northern Blot Hybridization and Quantification
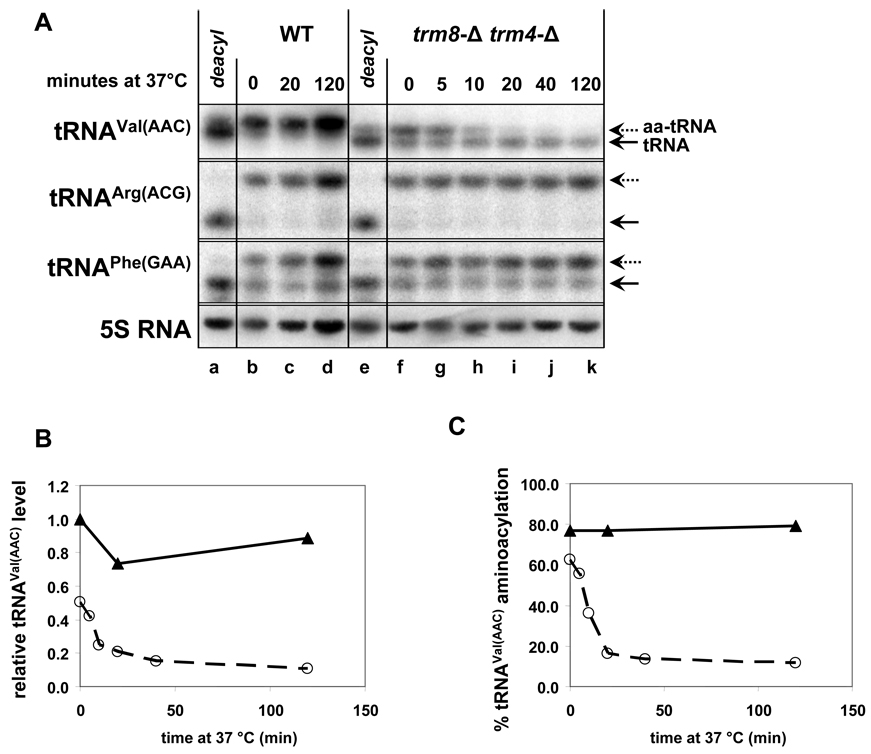
RNA isolated and separated by either of the described methods is visualized with oligonucleotide DNA probes labeled with 32P at the 5’ end using standard Northern blotting techniques. Between 2 and 20 µg of bulk RNA has been used to accurately determine changes in tRNA levels, although using less bulk RNA (2 µg) results in a cleaner blot with sharper bands. The membrane is hybridized with 20 pmol of 32P-labeled probe in 50 ml of hybridization buffer (6X SSC [from a 20X stock of 3 M NaCl, 0.3 M. Sodium Citrate, pH 7.0], 5X Denhardt’s solution [from a 50X stock of 1% ficoll, 1% BSA, 1% polyvinylpyrrolidone], 25 mM sodium phosphate buffer, pH 6.5, 0.5% SDS) at 37 °C for 1 hour or longer using 30 cm hybridization bottles (VWR #KT736500-3530) and a hybridization oven (Hybaid). The membrane is then washed twice with 30 ml of 1X SSC, 0.1% SDS at 37 °C for 15 minutes. Since tRNAs are highly expressed, even single copy tRNA species can be easily detected after an overnight exposure. The probe is stripped off the membrane by two successive incubations in 0.1X SSC, 0.5% SDS at 90 °C for 20 minutes before hybridization with a new probe. A typical analysis is shown in Figure 1, which details the loss of tRNAVal(AAC) in a trm8-Δ trm4-Δ strain after shift to 37 °C.
Figure 1. tRNAVal(AAC) levels and aminoacylation levels both decrease at 37 °C in a trm8-Δ trm4-Δ strain.
(A) Northern analysis of tRNA levels and aminoacylation. Strains were grown in YPD at 28 °C to OD600 ~1.3, rapidly shifted to 37 °C, and cells were harvested by filtration at the indicated times. 2 µg of RNA isolated under acidic conditions was separated on an acidic gel and analyzed by Northern blotting. For each strain, one sample was deacylated prior to gel electrophoresis. Dashed and solid arrows indicate aminoacylated and deacylated tRNA species, respectively. Note that tRNAVal(AAC) from strains lacking TRM8 migrates faster than wild type. (B) Quantification of the levels of tRNAVal(AAC). The ordinate shows the ratio of the levels of tRNAVal(AAC) at each time point relative to its level in the wild type strain immediately before temperature shift (each value itself first normalized to 5S RNA). (C) Quantification of the aminoacylation levels of tRNAVal(AAC).
To ensure accurate quantification, and to compensate for loading errors, the level of each tRNA is normalized to a reference RNA species that should be unaffected in the mutant strain, such as 5S RNA. Aminoacylation is quantified as a ratio of the aminoacylated tRNA fraction relative to the total tRNA. Although each membrane can be probed numerous times, it is sometimes important to probe less abundant tRNA species before probing the more abundant ones, since it may be difficult to completely strip some probes off the membrane, and 5S RNA should always be probed last. We have probed a single blot with as many as fifteen different probes.
Care should be taken to design probes that do not hybridize to the site of the affected tRNA modification if it is known to affect base pairing. Since some tRNA species have very similar sequences, care must be taken to design probes that will differentiate between two similar species, as was done for tRNASer(CGA) and tRNASer(UGA), which differ at only three positions (Kotelawala et al., 2008).
4.5. Experimental Considerations
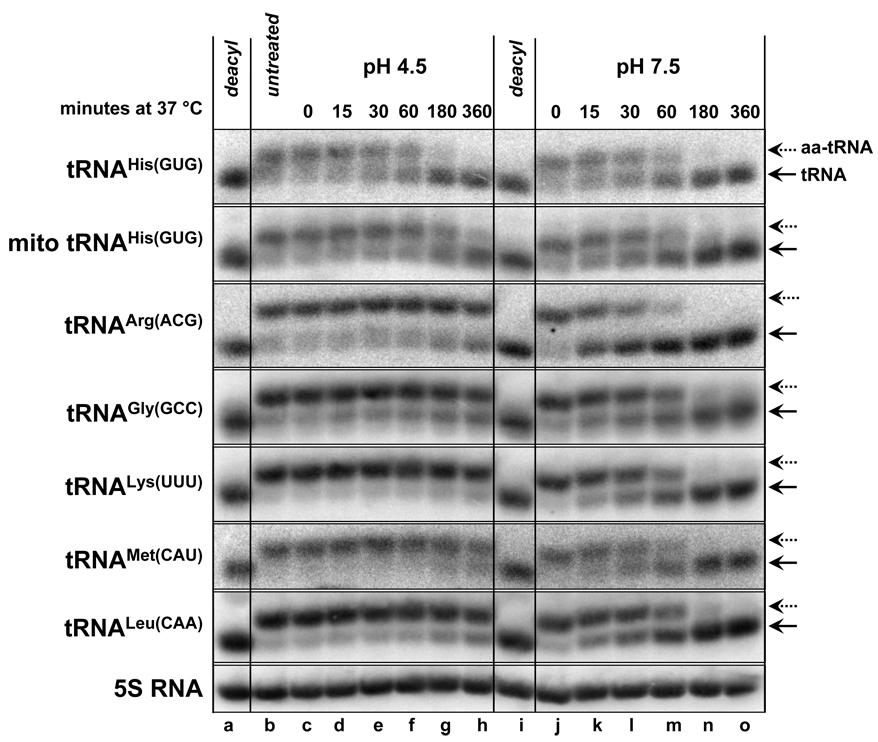
The described method for analysis of the aminoacylation status of different tRNA species has been used to analyze aminoacylation of at least 17 different tRNA species, listed in Table 1. We find that RNA isolated under acidic conditions can be stored at −70 °C for at least 2 months without loss of tRNA aminoacylation. We also find that aminoacylation of almost all tRNAs tested is stable in pH 4.5 buffer even at 37 °C, with half-lives estimated to be greater than 12 hours (Fig. 2), and is at least several-fold more stable at 4 °C. The only exceptions are the cytoplasmic and mitochondrial his-tRNAHis, which have half-lives of 3 hours and 5 hours, respectively, at pH 4.5 and 37 °C (Fig. 2), and these species are stabilized substantially at pH 3.5. However, aminoacylation of all examined tRNAs is destabilized at pH 7.5 and 37 °C, with half-lives of less than two hours, underscoring the importance of the acidic conditions and cold temperature described in the protocol (Fig. 2).
Table 1.
Summary of tRNA species probed for aminoacylation.
| tRNA probed | Probe sequence | Targeted region | Separation on acidic gel | Reference |
|---|---|---|---|---|
| Arg(ICG) | TAGCCAGACGCCGTGAC | 18–34 | excellent | (Gu et al., 2005) |
| Ala(AGC) | CATGCTAAGGGAGCGCG | 23–39 | good | Baker, M., unpublished |
| Gly(GCC) | AAGCCCGGAATCGAACCGG | 50–68 | good | (Gu et al., 2005) |
| His(GUG) | ACTAACCACTATACTAAGA | 5–23 | good | (Gu et al., 2005) |
| mito-His(GUG) | CGAACTCAGATTTAACGCA | 38–57 | good | (Gu et al., 2005) |
| Ile(IAU) | GCACGGTGCCTTAACCA | 17–32 | good | Baker, M., unpublished |
| Leu(CAA) | AGATTCGAACTCTTGCATCTT | e3–62 | good | (Alexandrov et al., 2006) |
| Lys(UUU) | TAAAAGCCGAACGCTCTACC | 18–37 | good | (Gu et al., 2005) |
| iMet(CAU) | GCCCTGCGCGCTTCCA | 16–32 | good | (Alexandrov et al., 2006) |
| Met(CAU) | TTCAGATTATGAGACTGACGC | 24–44 | good | (Alexandrov et al., 2006) |
| Phe(GAA) | TGTGGATCGAACACAGGACC | 45–64 | good | Chernyakov, I., unpublished |
| Thr(IGU) | GGATTTGAACCGATGATCT | 44–62 | good | Baker, M., unpublished |
| Trp(CCA) | AGTCGAAAGCTCTACCAT | 15–33 | poor | Baker, M., unpublished |
| Val(AAC) | TGGTGATTTCGCCCAGGA | 59–76 | poor | (Alexandrov et al., 2006) |
| Tyr(GUA) | CGAGTCGAACGCCCGAT | 46–62 | poor | (Gu et al., 2005) |
| Pro(UGG) | GGAATTGAACCCAGGGCCTCTCGCA | 38–62 | very poor | Chernyakov, I., unpublished |
| Ser(IGA) | CGCCTTAACCACTCGGCC | 9–26 | very poor | Baker, M., unpublished |
| Cys(GCA) | AATCTGCTGCGCTACCACT | 14–33 | not detected | Chernyakov, I., unpublished |
Figure 2. pH dependence of aminoacyl-tRNA stability at 37 °C.
RNA isolated from a wild type strain under acidic conditions was incubated at 37 °C in pH 4.5 buffer (10 mM NaOAc pH 4.5, 10 mM MgCl2, 20 mM KCl) or pH 7.5 buffer (50 mM HEPES pH 7.5, 10 mM MgCl2, 20 mM KCl) for the indicated times, followed by ethanol precipitation and resuspension in 10 mM NaOAc pH 4.5, 1 mM EDTA. The RNA was then separated on an acidic gel and analyzed by Northern blotting.
Certain aminoacyl-tRNAs may be less amenable to such analysis for several reasons. As listed in Table 1, separation between the aminoacylated and de-acylated tRNA differs for different tRNA species, and may depend on the nature of the amino acid. We note that for tRNACys, unlike all other tRNAs examined, there is no observed difference in mobility between the aminoacylated and deacylated samples, which might mean that the cys-tRNACys does not survive the treatment prior to analysis. We also note that certain modifications, such as m7G, can affect tRNA migration in this system (Fig. 1) and may make quantification more difficult.
An alternative approach has been used to quantify aminoacylation levels of tRNA species using a new microarray analysis method (Dittmar et al., 2005). This method employs a trichloroacetic method for tRNA isolation (Kruger and Sorensen, 1998) and relies on the resistance of aminoacylated tRNA to periodate oxidation of the 3’ end of the tRNA. In one aliquot of a sample, RNA is treated with periodate to destroy the 3' end of tRNA that is not aminoacylated, and the remaining aminoacylated tRNA is then deacylated and ligated to a fluorescently labeled oligonucleotide. In a second aliquot of the sample, all of the tRNA is deacylated and then labeled with a different fluorescent probe. Then both samples are compared on microarrays to evaluate aminoacylation levels (Dittmar et al., 2005).
5. Characterization of the Loss of tRNA
Since tRNA degradation in response to absence of modification has been shown to occur at both the pre-tRNA and the mature tRNA level (Anderson et al., 1998; Alexandrov et al., 2006), it is important to determine whether the precursor or mature target tRNA species is degraded due to lack of the modification of interest. Degradation of mature tRNA is easily assessed by analysis of tRNA from cells grown in the presence of a transcription inhibitor, such as thiolutin (Jimenez et al., 1973). Addition of 5 µg/ml thiolutin effectively inhibits ribonucleotide incorporation within ~5 minutes, and growth stops within ~25 minutes after addition of the drug. Since processing of pre-tRNA to mature tRNA continues after thiolutin treatment, disappearance of pre-tRNA is expected whether or not the pre-tRNA is degraded. Disappearance of pre-tRNA is also a convenient indication that thiolutin treatment is effective. If loss of the mature tRNA is still observed when the mutant strain is treated with thiolutin, or occurs to a greater extent after treatment, then mature tRNA itself is the degradation substrate. However, this method cannot directly distinguish between degradation of pre-tRNA or a decrease in transcription of the tRNA.
A pulse-chase experiment can be used to distinguish between degradation of tRNA and transcription effects. In this approach, nascent RNA is labeled by addition of [3H]uracil, followed by a chase with an excess of unlabeled nucleotide. RNA is then isolated at different times during the chase and the relative amounts of the labeled tRNA are analyzed by hybridization to an oligonucleotide probe. A decrease in the amount of labeled tRNA that hybridizes to the corresponding probe during the chase time course indicates that the tRNA is subject to degradation over this time course. A pulse-chase experiment and a thiolutin experiment were both used to demonstrate that pre-tRNAiMet is degraded in cells lacking m1A58 (Anderson et al., 1998).
6. Conclusions
The methods described here allow identification and initial characterization of tRNA turnover as a consequence of a modification defect. A more complete analysis of tRNA turnover requires characterization of the mechanism by which tRNA is turned over, which can be done by biochemical methods, or by analysis of genetic suppressors of the modification mutant strain. This was done by Anderson and coworkers to show that degradation of pre-tRNAiMet lacking m1A58 requires polyadenylation by the TRAMP complex, followed by degradation by Rrp6 and the nuclear exosome (Kadaba et al., 2004; Kadaba et al., 2006). Further analysis of the mechanism of tRNA turnover in this and other modification mutants will shed more light on the precise biological role of tRNA modifications and their effect on tRNA function in the cell.
ACKNOWLEDGEMENTS
We thank Andrei Alexandrov, Weifeng Gu, and Lakmal Kotelawala for earlier contributions. This research is supported by NIH grant GM52347 to EMP. ISC and MB were supported by Training in Cellular, Biochemical and Molecular Sciences Training Grant 5T32 GM068411.
REFERENCES
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1- methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copela LA, Chakshusmathi G, Sherrer RL, Wolin SL. The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability. RNA. 2006;12:644–654. doi: 10.1261/rna.2307206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrick WB, Horowitz J. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: evidence for stable base modification-dependent conformers. Nucleic Acids Res. 1993;21:4948–4953. doi: 10.1093/nar/21.21.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol. Cell. Biol. 2005;25:8191–8201. doi: 10.1128/MCB.25.18.8191-8201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall KB, Sampson JR, Uhlenbeck OC, Redfield AG. Structure of an unmodified tRNA molecule. Biochemistry. 1989;28:5794–5801. doi: 10.1021/bi00440a014. [DOI] [PubMed] [Google Scholar]
- Hiley SL, Jackman J, Babak T, Trochesset M, Morris QD, Phizicky E, Hughes TR. Detection and discovery of RNA modifications using microarrays. Nucleic Acids Res. 2005;33:e2. doi: 10.1093/nar/gni002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Hiley SL, Saltzman AL, Babak T, Blencowe BJ. Microarray analysis of RNA processing and modification. Methods Enzymol. 2006;410:300–316. doi: 10.1016/S0076-6879(06)10014-2. [DOI] [PubMed] [Google Scholar]
- Jimenez A, Tipper DJ, Davies J. Mode of action of thiolutin, an inhibitor of macromolecular synthesis in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 1973;3:729–738. doi: 10.1128/aac.3.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, Bystrom AS. Dual function of the tRNA(m(5)U54)methyltransferase in tRNA maturation. RNA. 2002;8:324–335. doi: 10.1017/s1355838202027851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GM, Stalker J, Humphray S, West A, Cox T, Rogers J, Dunham I, Prelich G. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nature Methods. 2008 doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer C, Rajbhandary UL. The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyl-tRNA synthetases. Methods. 2008;44:129–138. doi: 10.1016/j.ymeth.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelawala L, Grayhack EJ, Phizicky EM. Identification of yeast tRNA Um44 2'-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNASer species. RNA. 2008;14:158–169. doi: 10.1261/rna.811008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger MK, Sorensen MA. Aminoacylation of hypomodified tRNAGlu in vivo. J. Mol. Biol. 1998;284:609–620. doi: 10.1006/jmbi.1998.2197. [DOI] [PubMed] [Google Scholar]
- Maglott EJ, Deo SS, Przykorska A, Glick GD. Conformational transitions of an unmodified tRNA: implications for RNA folding. Biochemistry. 1998;37:16349–16359. doi: 10.1021/bi981722u. [DOI] [PubMed] [Google Scholar]
- Nobles KN, Yarian CS, Liu G, Guenther RH, Agris PF. Highly conserved modified nucleosides influence Mg2+dependent tRNA folding. Nucleic Acids Res. 2002;30:4751–4760. doi: 10.1093/nar/gkf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret V, Garcia A, Puglisi J, Grosjean H, Ebel JP, Florentz C, Giege R. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie. 1990;72:735–743. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- Rubin GM. Preparation of RNA and ribosomes from yeast. Methods Cell Biol. 1975;12:45–64. doi: 10.1016/s0091-679x(08)60951-6. [DOI] [PubMed] [Google Scholar]
- Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. U S A. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS, Emmerich J, Bauer F. Compilation of tRNA sequences and sequences of tRNA genes. 1999 doi: 10.1093/nar/gki012. pp http://www.unibayreuth.de/departments/biochemie/trna/ [DOI] [PMC free article] [PubMed]
- Varshney U, Lee C, RajBhandary U. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- Xing F, Hiley SL, Hughes TR, Phizicky EM. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 2004;279:17850–17860. doi: 10.1074/jbc.M401221200. [DOI] [PubMed] [Google Scholar]