Abstract
During fin regeneration, osteoblasts must continually differentiate for outgrowth of the bony fin rays. Bone maturity increases in a distal-proximal manner, and osteoblast maturation can be detected similarly when following gene expression. We find that early markers for osteoblast differentiation are expressed in a discrete domain at the distal end of the fin, just proximal to the adjacent germinal compartment of dividing cells. Matrix genes, required at later stages developmentally, are expressed in a population of cells proximally to the early genes. A marker for mature osteoblasts is expressed in cells further proximal. These domains of gene expression are partially overlapping, perhaps revealing additional levels of osteoblast maturity. We suggest a model for growth where new cells are continually added to the distal-most osteoblast compartment, while osteoblasts in more proximal locations differentiate, thus translating developmental time to location on the proximal-distal axis.
Keywords: bone growth, osteoblast maturation, fin regeneration, zebrafish, cx43
Introduction
We utilize growth of the zebrafish fin to reveal underlying cellular and molecular mechanisms regulating bone growth. Fins are comprised of segmented bony fin rays surrounded by a multilayered epithelium (Goss and Stagg, 1957; Haas, 1962). Each fin ray is comprised of two hemirays of bone matrix that extend longitudially along the proximal-distal axis. The hemirays surround a loose mesenchyme of undifferentiated cells as well as blood vessels and nerves. Distally, the hemirays are lined with collagen-like fibrils called actinotrichia (Becerra et al., 1983), which serve as the substrate for osteoblasts to align and secrete bone matrix directly via intramembraneous ossification (Landis and Geraudie, 1990). Thus, osteoblasts are found laterally in association with the bone matrix, while the mesenchyme is located medially. Cell proliferation contributing to new fin growth occurs medially and in the distal mesenchyme (Goldsmith et al., 2003). Thus, growth occurs at the distal end of the fin.
During fin regeneration all of the tissues of the zebrafish fin are restored in form and function. In particular, osteoblasts must continuously differentiate for outgrowth of the bony fin rays. Regeneration begins with wound healing by the migration of epithelial cells to cover the wound (Poleo et al., 2001; Santos-Ruiz et al., 2002). The mesenchymal cells beneath the amputation plane become disorganized and migrate distally to establish the proliferating cells of the regeneration blastema (Poleo et al., 2001; Santos-Ruiz et al., 2002). It is not clear if these cells represent stem cell populations or if they are the result of de-differentiation (Akimenko et al., 2003; Poss et al., 2003). Next, cells of the blastema compartmentalize into a non-proliferative, msxb-positive population in the distal-most region and a more proximal region of highly proliferative (msxb-negative) cells (Nechiporuk et al., 2003). Blastemal organization is complete by around 3 days post amputation (dpa) and outgrowth proceeds for approximately 2 weeks by coordinating cell proliferation with differentiation to replace lost tissue. Indeed, there is evidence from studies completed in Carassius auratus that dividing cells of the blastema cross the rows of actinotrichia and contribute to the population of osteoblasts (Santamaria et al., 1996).
One gene shown to be expressed in the proliferating compartment of the blastema is connexin43 (cx43, Iovine et al., 2005). Interestingly, mutations in cx43 cause the phenotypes of the short fin mutant, including short bony fin ray segments and reduced cell proliferation (Iovine et al., 2005; Hoptak-Solga et al., 2008). Connexins are the subunits of gap junctions, proteinaceous channels that permit the exchange of small molecules (< 1200 Da) among neighboring cells. It is not understood how defects in this seemingly simple mode of communication might lead to abnormalities in bone growth. Recent work from our lab indicates that Cx43 functions autonomously to establish the population of dividing cells (Hoptak-Solga et al., 2008), but may not contribute to later events such as osteoblast differentiation. Still, it is of interest to pursue this question further in order to appreciate how proliferation and differentiation are coordinated during fin regeneration. Therefore, here we evaluate osteoblast differentiation with respect to cx43 expression and cell proliferation. We find that osteoblast differentiation occurs in overlapping compartments of increasing maturity along the distal-proximal axis coincident with new bone growth. The population of cx43-positive proliferating cells resides in close proximity to the differentiating osteoblasts, but these proliferative cells down-regulate cx43 as they leave the blastema. This study therefore supports our previous findings and extends our understanding of osteoblast maturation during fin regeneration.
Results and Discussion
Osteoblast differentiation occurs in compartments of increasing maturity
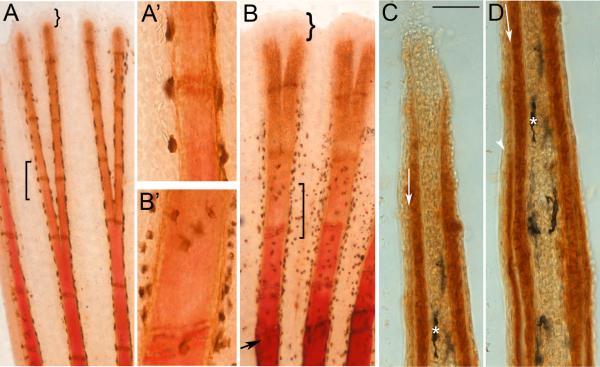
We begin by revealing differences in bone maturity along the proximal-distal axis in order to demonstrate that newer and younger tissue is located in the more distal locations. We evaluated alizarin red staining, which detects calcified bone matrix, in combination with a marker for mature osteoblasts, ZNS5 (Johnson and Weston, 1995). Alizarin red strongly stains the most proximal bony elements of ontogenetic and regenerating fins, but fails to stain the distal-most 0.75–1.2 mm of fin tissue (Figure 1 A, B). Mature ZNS5-positive osteoblasts are located along most of the length of the fin rays, including the alizarin red negative portions of the fin rays (Figure 1). Osteoblasts are limited to the medial surface of the bone matrix distally, and surrounding the bone matrix proximally (Santamaria and Becerra, 1991). Indeed, cryosectioning of ZNS5-positive fins revealed that ZNS5-positive cells are also located medial to the deposited bone matrix in more distal, younger tissue (Figure 1 C), and surrounding the matrix in the more mature and proximal tissue (Figure 1 D, and also observed in Smith et al., 2006; Smith et al., 2008). Therefore, the failure to detect alizarin red in the distal fin rays is not due to the absence of matrix-secreting osteoblasts. Further, the vital dye calcein can detect bone matrix in the alizarin red-negative region of the fin rays (Du et al., 2001), revealing that bone matrix is present and can be visualized by more sensitive methods. Thus, differences in alizarin red reactivity in fins reveals that the maturity of bone matrix increases from distal to proximal locations.
Figure 1.
Differences in the maturity of bone matrix can be detected using alizarin red. (A) Ontogenetic fin stained for both alizarin red and ZNS5. Square bracket identifies transition from the alizarin red positive staining (proximally) to the alizarin red negative region (A’ shows this in higher magnification). (B) Regenerating fin (5 dpa) stained for both alizarin red and ZNS5. Arrow points to amputation plane. Square bracket identifies transition from the alizarin red positive staining (proximally) to the alizarin red negative region (B’ shows this in higher magnification). Curved brackets in (A) and (B) identifies the ZNS5-negative region (the remainder of the fin ray is ZNS5-positive). (C) and (D) Sequential images of a single longitudinal cryosection through a ZNS5-stained fin ray (i.e. staining was completed on whole mount fins prior to sectioning). Arrow points to unstained bone matrix. Asterisk identifies a landmark melanocyte. Arrowhead in (D) points to ZNS5-positive signal on the lateral surface of the bone matrix (signal can also be observed medial to the bone matrix). Scale bar for C and D is 50 μm.
We and others have observed differences in the maturity of bone matrix along the proximal-distal axis of fin rays in teleosts (Santamaria et al., 1992; Mari-Beffa et al., 1996). We next wished to determine if similar differences in osteoblast maturation could be detected molecularly. Therefore, we evaluated the expression of genes representing different stages of osteoblast differentiation. In addition to ZNS5 shown in Figure 1, our analysis included the osteoblast markers for the early transcription factors runx2a and runx2b (i.e. runx2 is duplicated in zebrafish, Flores et al., 2004) and osterix (osx), and the matrix gene bone sialoprotein (spp-1). These genes were chosen because they likely represent markers for osteoblasts at different levels of maturity. The runx2 genes are believed to be expressed earliest in the osteoblast lineage, and may directly regulate the expression of osx (Nishio et al., 2006). Bone matrix genes, such as spp-1, are expressed later.
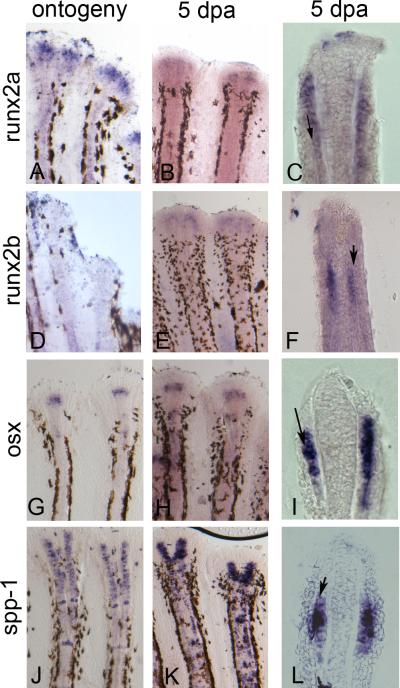
We completed whole mount in situ hybridization during ontogeny and regeneration for runx2a, runx2b, osx, and spp-1 (Figure 2). We find that the expression patterns are largely similar during ontogeny and regeneration, except that we could not detect runx2b during ontogeny. All of the genes except spp-1 appeared to exhibit discrete domains of expression towards the distal ends of the fin rays. In contrast, spp-1 has not only a strong distal expression domain but also strong sporadic expression along the entire length of the regenerate, including some staining in mature and developing joints. Cryosectioning of regenerating fins was completed to determine the tissue-specific location. All genes are expressed in the lateral compartment (and medial of the deposited bone matrix), consistent with the location of osteoblasts (Figure 2).
Figure 2.
Expression of osteoblast markers during ontogeny and regeneration is in the lateral osteoblast compartment. Ontogeny is on the left, 5 dpa regenerating fin is in the middle, cryosection of 5 dpa regenerating fins is on the right. (A,B,C) runx2a; (D,E,F) runx2b; (G,H,I) osx; (J,K,L) spp-1. Cells expressing all genes are located medial to the deposited bone matrix (arrows).
While evaluating these expression patterns, we noticed apparent differences in the proximal-distal locations of early (i.e. runx2a) and late (ZNS5) expression domains. We evaluated these differences more carefully by measuring the distal-most and proximal-most boundaries of expression (Table 1). Using this method, we identified domains of expression where the early osteoblast markers are located more distal than the later osteoblast markers. Indeed, we found that all of the early transcription factors (runx2a, runx2b, osx) are located in the distal-most 36–98 μm of the fin ray; spp-1 is found from 64– 200 μm; ZNS5-positive cells are first identified at about 100 μm (and along the entire length of the fin rays). Thus, we find three partially overlapping domains representing different stages of osteoblast maturation, consistent with the notion that more distal cells represent the least mature osteoblasts. Our results are strikingly similar to a recent report on osteoblast differentiation in zebrafish embryonic bone (Li et al., 2009). Based on the relative timing of gene expression of runx2a, runx2b, osterix, and other matrix genes, the authors describe three overlapping stages of bone development (runx2a and runx2b during an early stage; osx during an intermediate stage; matrix genes during the mature stage). Our results do not distinguish the timing of osx expression from runx2a and runx2b, but it remains possible that the runx2 genes may regulate osx during fin growth. Thus, from both studies the relative order of gene expression during osteoblast differentiation appears to be largely conserved.
Table 1.
Expression domains of genes expressed during fin regeneration
| Marker | Type | Distal (μm) | Proximal (μm) | Range (μm) |
|---|---|---|---|---|
| cx43 | proliferating | 14 +/− 3.5 | 96 +/−11.4 | 10.5 – 107.4 |
| runx2a | early ost. | 48 +/− 10.2 | 83 +/−13.7 | 37.8 – 96.7 |
| runx2b | early ost. | 47 +/− 10.9 | 83.2 +/− 15.1 | 36.1 – 98.3 |
| osx | early ost. | 55 +/− 8.1 | 79.5 +/− 4.6 | 46.9 – 84.1 |
| spp-1 | late ost. | 79 +/− 15.0 | 198 +/− 22.7 | 64 – 220.7 |
| ZNS5 | mature ost. | 114 +/− 11 | N/A | 103 - |
The distance between the distal or proximal staining boarder and the distal tip of the fin was measured from the D + 3 or V + 3 fin rays from five regenerating fins (5 dpa). The range is from the distal-most to the proximal-most staining. ZNS5 is expressed along the entire length of the fin rays (and does not have a proximal boarder).
The population of dividing cells is in close proximity to the cells expressing early osteoblast markers
It has been firmly established that the population of dividing cells during fin regeneration is confined to blastema (Santamaria et al., 1996; Nechiporuk and Keating, 2002). We have shown that cx43 mRNA is expressed in the regeneration blastema (Iovine et al., 2005) and that cx43 is coincident with mitotic cells (Hoptak-Solga et al., 2008). Since this population of cells may contribute to the population of differentiating osteoblasts (Santamaria et al., 1996), understanding the physical relationship of the blastema and the lateral compartments is important. Therefore, we measured the cx43-positive expression domain in 5 dpa regenerating fins and found that the cx43-positive domain spans about 14–100 μm (Table 1). This region largely overlaps the earliest osteoblast genes, and extends slightly more distal.
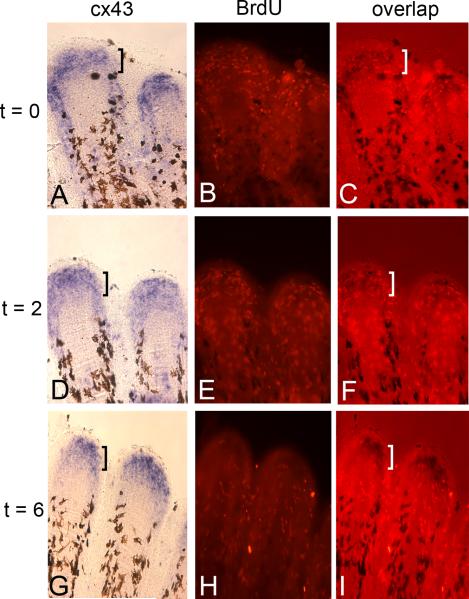
To identify the potential migration pattern of the cx43-positive dividing cells, we completed a pulse-chase experiment using bromodeoxyuridine (BrdU). BrdU is a thymidine analog that is incorporated into DNA during S-phase. Therefore, BrdU labels cells during the cell division cycle and persists as a label for a small number of subsequent cell divisions. Fins were amputated and permitted to regenerate for 5 days (i.e. 5 dpa) so that we could be sure that regeneration was in the growth stage. Fish were exposed to BrdU for a 10 minute pulse and divided into three groups. One group was harvested immediately following the BrdU exposure, which identifies the cells labeled during this time period (i.e. t = 0 or “pulse”). A second group was allowed to swim in fresh water for 2 hours following the BrdU pulse (t = 2), while a third group swam in fresh water for 6 hours following the BrdU pulse (t = 6). After harvesting, the fins were processed for both cx43 expression and BrdU. As expected, the BrdU-positive cells were almost entirely co-localized with the cx43 expression domain at the t = 0 time point (Figure 3A-C), consistent with previous findings that the cx43-positive region identifies the population of dividing cells (Hoptak-Solga et al., 2008). BrdU-positive cells begin to appear outside the cx43-positive compartment as early as at the 2 hour chase, although many BrdU-positive cells continue to overlap with the cx43-positive region (see bracket in Figure 3F). This is in contrast to the 6 hour chase, where the BrdU positive cells are located almost entirely outside the cx43-positive domain, in regions both proximal and lateral (Figure 3G-I). Thus, actively dividing cells co-express cx43 and BrdU (i.e. at the pulse), and at late time points cx43-positive cells and BrdU positive cells no longer overlap. We suggest that dividing cells migrate away from the blastemal compartment of proliferating cells, down-regulate cx43, and contribute to differentiating cell types of the regenerating fins. Indeed, this conclusion is consistent with previous findings demonstrating that proliferating cells do not populate the lateral osteoblast compartment, but rather, proliferating blastemal cells exit the cell cycle and migrate laterally before differentiating as osteoblasts (Santamaria et al., 1996; Johnson and Bennett, 1999).
Figure 3.
Proliferating cells leave the blastema within 6 hours of dividing. Regenerating fins were treated with BrdU and harvested immediately (A,B,C), following a 2 hour chase in fresh water (D,E,F), or following a 6 hour chase in fresh water (G,H,I). Harvested fins were processed for cx43 in situ hybridization (left panels) and BrdU detection (middle panels). Rightward panels show overlap of bright field and fluorescence. Brackets indicate the cx43-positive domain, revealing that the majority of BrdU-labeled cells overlap with cx43. By the 6 hour chase, very few BrdU positive cells remain in the cx43-positive region.
Model of osteoblast maturation during fin regeneration
This analysis of osteoblast gene expression and cell proliferation suggests a model for osteoblast differentiation and maturation (Figure 4). Proliferating cells, identified by cx43 expression and BrdU incorporation, are found medially in the blastema partially adjacent to the population of early osteoblast differentiation (i.e. the runx2a, runx2b, and osx positive domain). More proximally, cells begin to express spp-1 (intermediate) and fully mature ZNS5-positive osteoblasts are located at an even more proximal location. These domains for early, intermediate, and mature osteoblast markers partially overlap, providing a minimum of four zones of osteoblast maturation with unique expression profiles (Figure 4). Zone 1 contains cells expressing only the early markers (i.e. runx2a, runx2b, osx), zone 2 cells express early plus intermediate markers (i.e. runx2a, runx2b, osx in addition to spp-1), zone 3 cells express intermediate plus mature markers (i.e. spp-1 and ZNS5), and zone 4 cells express only mature markers (ZNS5). It is not clear if the cells found in these zones begin to express more mature markers concurrent with expressing the less mature markers, or if there are subpopulations of cells within each zone with different expression profiles. Double-label in situ hybridization would distinguish these possibilities (i.e. co-expression of genes or independent populations of cells expressing different markers). Unfortunately, this has not yet been possible in fin tissue. This limitation does not preclude the conclusion that zones reflecting osteoblast maturity exist, although the mechanism of transitioning between zones is in question.
Figure 4.
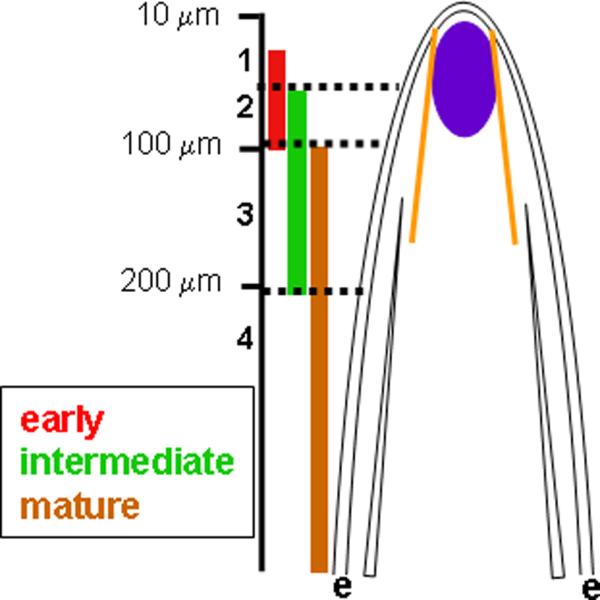
Model for osteoblast maturation in the fin ray. Early (runx2a, runx2b, osx), intermediate (spp-1), and mature (ZNS5) osteoblast domains are represented on the left. These genes are expressed in partially overlapping domains in the lateral osteoblast compartment (between the basal layer of the epidermis,e, and the actintrichia, yellow). The region of cx43-positive cells is indicated in purple in the cartoon of the fin ray. We suggest that the cx43-positive proliferating cells cross actinotrichia and contribute to the cells in zone 1.
This report provides molecular evidence that important events in fin ray maturation can be followed by examination of discrete distal-proximal compartments. Together with the apparent migration of proliferating cells away from the blastema, our data suggest that bone growth proceeds by new cells entering the lateral compartment at the level of (or distal to) zone 1. Indeed, proliferating cells of the regeneration blastema in goldfish fins were shown to contribute to the lateral population of osteoblasts (Santamaria et al., 1996). These findings do not negate the possibility that cells located in the lateral proximal mesenchyme may independently differentiate as osteoblasts, however there is no evidence that this population of cells is renewed by proliferation (Johnson and Bennett, 1999; Santamaria et al., 1996). Therefore, the finding that the earliest markers for osteoblasts are located in a discrete, distally-positioned compartment closely associated with the blastema strongly suggests that proliferating blastemal cells contribute directly to differentiating osteoblasts. Growth and differentiation therefore occur concurrently, thus translating developmental time to location on the proximal-distal axis.
Conclusions
Multiple studies demonstrate that tissue compartmentalization is important in the growth and development of the regenerating fin. First, the blastema has been shown to be compartmentalized into a distal-most non-proliferative region and a proximal proliferative region (Nechiporuk and Keating, 2002). A more recent study reveals that the basal layer of the epidermis contains discrete compartments required to confine Shh signaling, perhaps regulating the bone patterning in the underlying osteoblast compartment (Lee et al., 2009). Here, we identify compartments reflecting osteoblast maturation. Future analyses will bring these independent studies together so that a comprehensive understanding of the molecular and cellular events regulating bone growth and regeneration can be achieved.
Experimental Procedures
Fish rearing
Wild-type fish stocks used for this study were from the C32 strain (Rawls et al., 2003). Zebrafish were raised at a constant temperature of 25°C with 14 light: 10 dark photoperiod (Westerfield, 1993).
Detection ZNS5 and alizarin red
Fins were harvested at 5 dpa and fixed in 4% paraformaldehyde in PBS O/N and dehydrated in 100% methanol at −20°C. The fins were then rehydrated in a methanol series containing PBS. Fins were treated with 1 mg/ml Collagenase in PBS for 45 min at room temperature and then blocked with 0.25% BSA in PBTx for 3×5 min. A mouse antibody against ZNS5 was diluted to 1:200 in blocking solution and incubated with fins overnight at 4°C. Fins were then washed 3×5 min in blocking solution and then treated with a 1:100 diluted anti-mouse antibody for 2 hrs at room temperature. After 3×10 min of treatment in blocking solution, the fins were treated in mouse PAP (peroxidase antiperoxidase) diluted to 1:200 at 4°C overnight. Following 3×5 min washes in blocking solution, fins were placed in DAB (DAB; 0.03% DAB in 0.1 M PBS) solution and 0.01% H2O2 was added for 10 min. The fins were then washed in PBS to stop the reaction and then mounted in glycerol. Labeled fins were examined on a Nikon Eclipse 80i microscope or processed for cryosectioning as for fins labeled by in situ hybridization (described below).
For double-staining with alizarin red, stained fins were treated with alizarin red in 0.5 % potassium hydroxide overnight, followed by multiple washes in potassium hydroxide before mounting in 50 % glycerol.
In situ hybridization
In situ hybridization was performed to detect cx43 (Iovine et al., 2005), runx2a (Flores et al., 2004), runx2b (Flores et al., 2004), osx (600 bp of coding sequence from exon 2), and spp-1 (EST fz39b11, see accession number BQ092246). Fins (5 dpa or ontogenetic fins) were fixed overnight with 4% paraformaldehyde in PBS and dehydrated in 100% methanol at −20°C. Gradual aqueous washes were completed in methanol/PBST. Tissue was treated with 5 μg/ml proteinase K (5 min for embryos; 45 min for fins) and re-fixed for 20 min. Prehybridization (50 % formamide, 5X SSC, 10 mM citric acid, 0.1 % Tween20) occurred for 1 hour at 65°C, and hybridization in the presence of digoxigenin-labeled antisense probes was completed overnight. Gradual washes into 0.2X SSC were followed by gradual washes into PBST. Anti-digoxigenin Fab fragments (pre-absorbed against zebrafish tissue) were used at 1:5000 overnight. Following extensive washes in PBST followed by three short washes in staining buffer (100 mM Tris, 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1 % Tween20, pH 9.0). Tissue was next transferred to staining solution (staining buffer plus 0.22 mg/ml NBT and 0.175 mg/ml BCIP) and development proceeded until purple color was observed.
Measurements of the distal-most and proximal-most boundaries of expression were taken only from the longest fin rays (V +3) since that was established as a standard (Iovine and Johnson, 2000). Images were collected using a digital Nikon camera and distances were determined using ImagePro software. A minimum of 5 fins (up to 10 fin rays) per marker were utilized for measurements. Standard deviation was calculated and student’s t-tests were performed to determine if data sets were statistically different (i.e. p < 0.05 indicates that data sets are different).
Following whole mount in situ hybridization (or ZNS5-labeling), fins were embedded in 1.5 % agarose/5% sucrose, and equilibrated overnight in 20 % sucrose. Fins were mounted in OCT and cryosectioned (18–20 μm sections) using a Reichert-Jung 2800 Frigocut cryostat. Sections were collected on Superfrost Plus slides (Fisher) and mounted in 100 % glycerol.
Detection of proliferating cells using BrdU on fins processed for in situ hybridization
Fins were amputated and permitted to regenerate for 5 days. Individuals were moved to water containing 50 μg/ml BrdU water for 10 min. Fins were harvested immediately (pulse, t = 0), or fish were moved to fresh water without BrdU (chase). Fins were harvested after 2 hours (t = 2) or after 6 hours (t = 6). Harvested fins were fixed in 4% paraformaldehyde in PBS, and dehydrated in methanol before processing for in situ hybridization for cx43. Following in situ hybridization (above), detection for BrdU on whole mount fins was completed as described (Nechiporuk and Keating, 2002). Briefly, fins were washed in PBTx, treated with 2N HCl in PBTx for 30 min at room temperature and then blocked with 0.25% BSA in PBTx for 30 min. A rat monoclonal antibody against BrdU (Roche) was diluted to 1:50 in blocking solution and incubated with fins overnight at 4°C. Extensive washes were completed in PBTx (last wash in blocking solution). A goat anti-rat Alexa546 antibody (Molecular Probes) was diluted to 1:200 in blocking solution and incubated with fins overnight at 4°C. Extensive washes were completed prior to mounting in 50 % glycerol. Labeled cells were visualized on a Nikon Eclipse 80i microscope.
Acknowledgements
The authors wish to thank Jake Fugazzotto for fish rearing and for cryosectioning fin tissue. We thank M. V. Flores for sharing the runx2a and runx2b plasmids. This work was supported by the NICHD (HD047737 to MKI) and the NIAMS (AR048101 to SF).
Footnotes
NICHD: R01HD047737
NIAMS: AR048101
Bibliography
- Akimenko MA, Mari-Beffa M, Becerra J, Geraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. 2003;226:190–201. doi: 10.1002/dvdy.10248. [DOI] [PubMed] [Google Scholar]
- Becerra J, Montes GS, Bexiga SR, Junqueira LC. Structure of the tail fin in teleosts. Cell Tissue Res. 1983;230:127–137. doi: 10.1007/BF00216033. [DOI] [PubMed] [Google Scholar]
- Du SJ, Frenkel V, Kindschi G, Zohar Y. Visualizing normal and defective bone development in zebrafish embryos using the fluorescent chromophore calcein. Dev Biol. 2001;238:239–246. doi: 10.1006/dbio.2001.0390. [DOI] [PubMed] [Google Scholar]
- Flores MV, Tsang VW, Hu W, Kalev-Zylinska M, Postlethwait J, Crosier P, Crosier K, Fisher S. Duplicate zebrafish runx2 orthologues are expressed in developing skeletal elements. Gene Expr Patterns. 2004;4:573–581. doi: 10.1016/j.modgep.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Goldsmith MI, Fisher S, Waterman R, Johnson SL. Saltatory control of isometric growth in the zebrafish caudal fin is disrupted in long fin and rapunzel mutants. Dev Biol. 2003;259:303–317. doi: 10.1016/s0012-1606(03)00186-6. [DOI] [PubMed] [Google Scholar]
- Goss RJ, Stagg MW. The regeneration of fins and fin rays in Fundulus heteroclitus. J Exp Zool. 1957;136:487–507. doi: 10.1002/jez.1401360306. [DOI] [PubMed] [Google Scholar]
- Haas HJ. Studies on mechanisms of joint and bone formation in the skeleton rays of fish fins. Dev Biol. 1962;5:1–34. doi: 10.1016/0012-1606(62)90002-7. [DOI] [PubMed] [Google Scholar]
- Hoptak-Solga AD, Nielsen S, Jain I, Thummel R, Hyde DR, Iovine MK. Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration. Dev Biol. 2008;317:541–548. doi: 10.1016/j.ydbio.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Higgins EP, Hindes A, Coblitz B, Johnson SL. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev Biol. 2005;278:208–219. doi: 10.1016/j.ydbio.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Iovine MK, Johnson SL. Genetic analysis of isometric growth control mechanisms in the zebrafish caudal Fin. Genetics. 2000;155:1321–1329. doi: 10.1093/genetics/155.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Bennett P. Growth control in the ontogenetic and regenerating zebrafish fin. Methods Cell Biol. 1999;59:301–311. doi: 10.1016/s0091-679x(08)61831-2. [DOI] [PubMed] [Google Scholar]
- Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in Zebrafish fin regeneration. Genetics. 1995;141:1583–1595. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis WJ, Geraudie J. Organization and development of the mineral phase during early ontogenesis of the bony fin rays of the trout Oncorhynchus mykiss. Anat Rec. 1990;228:383–391. doi: 10.1002/ar.1092280404. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hami D, De Val S, Kagermeier-Schenk B, Wills AA, Black BL, Weidinger G, Poss KD. Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev Biol. 2009;331:270–280. doi: 10.1016/j.ydbio.2009.05.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Felber K, Elks P, Croucher P, Roehl HH. Tracking gene expression during zebrafish osteoblast differentiation. Dev Dyn. 2009;238:459–466. doi: 10.1002/dvdy.21838. [DOI] [PubMed] [Google Scholar]
- Mari-Beffa M, Mateos I, Palmqvist P, Becerra J. Cell to cell interactions during teleosts fin regeneration. Int J Dev Biol Suppl. 1996;1:179S–180S. [PubMed] [Google Scholar]
- Nechiporuk A, Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–2617. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- Nechiporuk A, Poss KD, Johnson SL, Keating MT. Positional cloning of a temperature-sensitive mutant emmental reveals a role for sly1 during cell proliferation in zebrafish fin regeneration. Dev Biol. 2003;258:291–306. doi: 10.1016/s0012-1606(03)00129-5. [DOI] [PubMed] [Google Scholar]
- Nishio Y, Dong Y, Paris M, O’Keefe RJ, Schwarz EM, Drissi H. Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene. 2006;372:62–70. doi: 10.1016/j.gene.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Poleo G, Brown CW, Laforest L, Akimenko MA. Cell proliferation and movement during early fin regeneration in zebrafish. Dev Dyn. 2001;221:380–390. doi: 10.1002/dvdy.1152. [DOI] [PubMed] [Google Scholar]
- Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. 2003;226:202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Frieda MR, McAdow AR, Gross JP, Clayton CM, Heyen CK, Johnson SL. Coupled mutagenesis screens and genetic mapping in zebrafish. Genetics. 2003;163:997–1009. doi: 10.1093/genetics/163.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria JA, Becerra J. Tail fin regeneration in teleosts: cell-extracellular matrix interaction in blastemal differentiation. J Anat. 1991;176:9–21. [PMC free article] [PubMed] [Google Scholar]
- Santamaria JA, Mari-Beffa M, Becerra J. Interactions of the lepidotrichial matrix components during tail fin regeneration in teleosts. Differentiation. 1992;49:143–150. doi: 10.1111/j.1432-0436.1992.tb00662.x. [DOI] [PubMed] [Google Scholar]
- Santamaria JA, Mari-Beffa M, Santos-Ruiz L, Becerra J. Incorporation of bromodeoxyuridine in regenerating fin tissue of the goldfish Carassius auratus. J Exp Zool. 1996;275:300–307. doi: 10.1002/(SICI)1097-010X(19960701)275:4<300::AID-JEZ8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Santos-Ruiz L, Santamaria JA, Ruiz-Sanchez J, Becerra J. Cell proliferation during blastema formation in the regenerating teleost fin. Dev Dyn. 2002;223:262–272. doi: 10.1002/dvdy.10055. [DOI] [PubMed] [Google Scholar]
- Smith A, Avaron F, Guay D, Padhi BK, Akimenko MA. Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblast differentiation and function. Dev Biol. 2006;299:438–454. doi: 10.1016/j.ydbio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Smith A, Zhang J, Guay D, Quint E, Johnson A, Akimenko MA. Gene expression analysis on sections of zebrafish regenerating fins reveals limitations in the whole-mount in situ hybridization method. Dev Dyn. 2008;237:417–425. doi: 10.1002/dvdy.21417. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A guide for the laboratory use of zebrafish (Brachydanio rerio) University of Oregon Press; Eugene, OR: 1993. [Google Scholar]