SUMMARY
Fibrinogen (Fbg) mediates platelet aggregation through its binding to the αIIbβ3 integrin receptor. Despite the many studies of platelet aggregation and blood clotting, the interaction of Fbg with the platelet integrin has remained unresolved. This paper reports on the use of self-assembled monolayers (SAMs) of alkanethiolates on gold to study the adhesion of αIIbβ3 CHO K1 cells to the GRGDS and HHLGGAKQAGDV motifs within Fbg. The peptides were immobilized to a monolayer that otherwise prevented nonspecific interactions of the cells with the substrate. Monolayers presenting GRGDSC or HHLGGAKQAGDVC were effective at mediating αIIbβ3 CHO K1 cell adhesion and spreading and were comparable to the use of Fbg-coated substrates, suggesting that both sequences can bind the receptor independently. A comparison of cell adhesion to several peptide truncations revealed that AGD was the minimal binding sequence in HHLGGAKQAGDV, and inhibition experiments showed that AGD and RGD were competitive ligands for the receptor. A peptide array of GXGDSC peptides revealed that αIIbβ3 CHO K1 cells adhered to peptides containing basic or hydrophobic residues in the X position, revealing the relaxed specificity with which αIIbβ3 recognizes its ligands. This work therefore suggests that AGD and RGD interact with Fbg in a functionally similar manner and that the use of AGD peptides may lead to a new generation of anti-thrombotic agents.
INTRODUCTION
Fibrinogen (Fbg) is an abundant plasma protein that is essential for homeostasis. This protein is a disulfide-linked homodimeric complex assembled fromα, β and γ subunits and presents multiple peptide motifs that bind the αIIbβ3 integrin receptor present on platelets and αvβ3 integrin on endothelial cells. This way, Fbg can aggregate platelets and localize the clot to activated endothelium. Fbg also serves as an extracellular matrix protein to mediate cell adhesion following its conversion to insoluble fibrin by the protease thrombin (Bini et al., 2000). Consequently, a substantial effort has been directed towards identifying the binding sequences in Fbg that mediate platelet aggregation and adhesion, and in understanding the differential roles of these ligands. This past work has implicated two sequences for platelet aggregation—the RGD site on the α subunit and a carboxy-terminal peptide on the γ subunit—yet the mechanistic roles of the two peptides remain controversial. Here, we report a study that uses model substrates that present defined peptide ligands to show that both the RGD and the γ-derived AGD sequences serve as competitive ligands for the αIIbβ3 receptor, and we show that the platelet receptor has a relaxed specificity for its ligands and recognizes peptides having a hydrophobic residue in the first position of the canonical RGD motif.
Fbg contains two peptide motifs that are important to its ability to aggregate platelet receptors: an RGD sequence at position 572-4 on the Aα chain and a HHLGGAKQAGDV sequence at position 400-11 of the γ chain. There is a second RGD site at position 95 in the Aα chain, but this ligand is likely conformationally masked within a coiled-coil domain and does not participate in the initial aggregation of platelets (Doolittle et al., 1978, Ugarova et al., 1993). A consensus has emerged that the RGD sequence is important for binding to the αvβ3 receptor on endothelial cells and thereby serves to localize a thrombus to regions of activated endothelium. Further, a series of studies has established that the γ peptide interacts with the platelet receptor and is necessary for fibrinogen-mediated aggregation of platelets (Hawiger atl al., 1982, Kloczewiak, 1984, Farrell et al., 1992). What has been less clear is whether the RGD motif is also necessary in platelet aggregation and whether the γ and RGD peptides bind to common or separate sites on the receptor.
Bennett and coworkers reported studies that supported a model wherein the two peptides bind to non-overlapping sites on the receptor. That study used two monoclonal antibodies to probe the interaction of the receptor with the ligands: PAC-1, which competes with Fbg in binding to αIIbβ3, and A2A9, which binds the integrin at a different site than does PAC-1 and sterically blocks the binding of Fbg to the receptor. The peptide RGDS blocked the binding of both PAC-1 and Fbg to platelets with equal potency. The γ-derived peptide LGGAKQAGDV also inhibited Fbg binding to platelets with an affinity comparable to that of RGDS, but was 2.5-fold less potent in inhibiting PAC-1 binding to αIIbβ3. Finally, LGGAKQAGDV, but not RGD, could inhibit the binding of A2A9 to platelets. These results suggest that the two peptides interact with the integrin at two different sites (Bennett et al., 1988). Another cross-linking study of the complex suggested that GRGDS interacts with the β3 subunit (D’ Souza et al., 1988) while HHLGGAKQAGDV interacts with the heavy chain in the αIIb subunit, giving further evidence in support of two-binding site model (D’ Souza et al., 1990). Further support for this model came from studies that used surface plasmon resonance experiments to show that the two binding sites in αIIbβ3 are allosterically related (Hu et al., 1999) and a report that the peptides served two distinct functions, with the initial cell adhesion mediated by HHLGGAKQAGDV, and subsequent cell spreading mediated by GRGDS (Salsmann et al., 2005).
Other studies have supported a model wherein the two peptides bind a common site on the platelet receptor. For example, affinity chromatography experiments found that αIIbβ3 bound to a resin presenting either RGD or γ chain peptides and could then be eluted with either peptide (Lam et al., 1987). A recent report by Springer and coworkers of the crystal structure for the complex provides the strongest evidence that the two peptides bind a common site. The structure reveals that the HHLGGAKQAGDV peptide binds to an extended site on the receptor that includes the RGD-binding site. Further, the structure revealed that the peptide adopts a conformation that allows the lysine residue of the KQAGD sequence to interact with the Arg-binding pocket on the receptor and was interpreted to suggest that the minimal sequence for the ligand is KQAGD.
In this paper, we use model substrates that present the GRGDS and HHLGGAKQAGDV peptides—either alone or in combination—to compare the activities of the ligands in an adhesion assay of αIIbβ3-transfected CHO K1 cells. These biochemical findings are largely consistent with the recent structural data and support a common site for binding of the two peptide motifs. Indeed, we find that both ligands mediate cell adhesion and spreading through a common binding site on αIIbβ3. However, our results suggest that efficient cell adhesion does not require the presence of the lysine residue in the peptide. Rather, the peptide AGD retains the property of the full length γ peptide in mediating cell adhesion and spreading. Further, we compared cell adhesion to a peptide array to show that the platelet receptor recognizes peptides of the form XGD where X is either a basic residue (R or K) or one of the hydrophobic residues (G, A, V, L or I). Together with the recent structural report, these adhesion experiments reveal the promiscuity of the platelet receptor and identify the sequence determinants that are important for active interaction with the platelet receptor.
RESULTS
Experimental Design
We used model substrates that mimic the extracellular matrix to compare the roles played by the GRGDS and HHLGGAKQAGDV sequences in Fbg for their ability to mediate the attachment and spreading of cells in an αIIbβ3 integrin-dependent manner. The model substrates were self-assembled monolayers that presented the peptide ligands against a tri(ethylene glycol)-terminated background (Mrksich, 2009). The use of monolayers for studies of cell adhesion are notable because they allow the identities, densities and orientations of the peptides to be controlled and at the same time are effective at preventing the non-specific adsorption of protein and, therefore, remodeling of the matrix. The latter benefit owes to the terminal oligo(ethylene glycol) groups on the monolayer. Several previous reports have established the suitability of these model substrates for mechanistic studies of cell adhesion (Kato et al., 2004; Feng et al., 2004; Houseman et al, 2001; Houseman et al, 2003; Murphy et al., 2004).
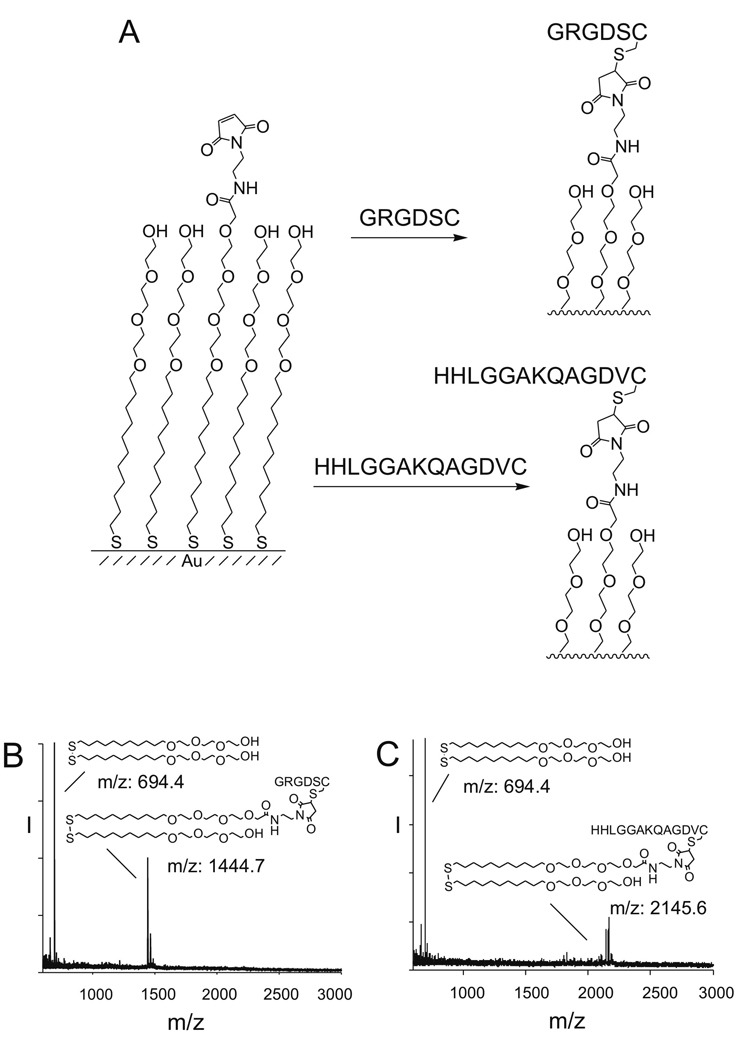
We first prepared monolayers that presented a mixture of maleimide groups and tri(ethylene glycol) groups with the former present at a density of 0.5% (relative to total alkanethiolate). We treated the monolayers with cysteine-terminated peptides to immobilize the ligands (Figure 1A). In each case we used mass spectrometry to verify immobilization of the peptides in high yield (Su et al., 2002). For example, a mass spectrum of monolayer to which the peptide GRGDSC was immobilized showed the expected peaks for the sodium adducts of symmetric glycol-substituted disulfide (m/z=694.4) and the mixed disulfide containing one glycol group and one GRGDSC terminated alkanethiol (m/z: 1444.7) (Figure 1B). Similarly, a mass spectrum of a monolayer to which the peptide HHLGGAKQAGDVC had been immobilized displayed peaks for the sodium adduct of the symmetrical glycol disulfide (m/z: 694.4) and the mixed disulfide composed of one glycol group and one HHLGGAKQAGDVC terminated alkanethiol (m/z: 2145.6) (Figure 1C). A SAMDI mass spectrum of the monolayer composed of both peptides showed peaks corresponding to each peptide-terminated disulfide and to the ethylene-glycol-terminated disulfide. The peak corresponding to the GRGDSC disulfide was larger than that for the HHLGGAKQAGDVC disulfide, likely due to a higher ionization efficiency of the former. We estimate that the relative density of the RGD peptide is between 0.35 and 0.65 in the monolayer.
Figure 1.
Model substrates used in this work. (A) Self-assembled monolayer presenting a maleimide group among tri(ethylene glycol) background. The maleimide group allows for the covalent immobilization of Cys-terminated peptides through a Michael addition. The glycol groups prevent nonspecific cell adhesion. (B) SAMDI characterization of a 0.5% GRGDSC SAM. Tri(ethylene glycol) disulfide and GRGDSC-terminated mixed disulfide were detected. All were sodium salts. (C) SAMDI characterization of a 0.5% HHLGGAKQAGDVC SAM. Tri(ethylene glycol) disulfide and HHLGGAKQAGDVC-terminated mixed disulfide were detected. All were sodium salts.
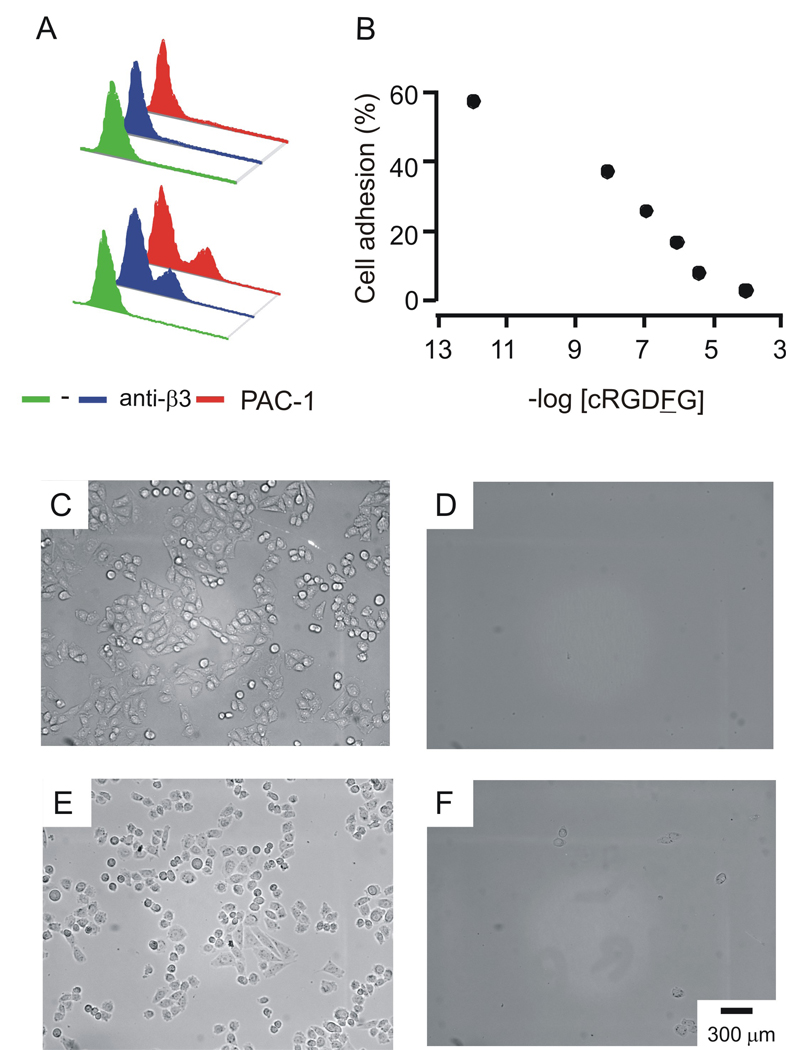
We used CHO K1 cells that were transfected with the αIIbβ3 receptor, as described previously (Salsmann et al., 2005). Cells were transfected with pcDNA3.1 vectors that coded for αIIb and β3 using FuGENE6 and analyzed by FACS after labeling with the PAC-1 (anti-αIIbβ3) and H-96 (anti-β3) antibodies. Approximately 30% of the cells expressed the receptor (Figure 2, A). Because the CHO K1 cells also express αv and β5 integrins on their surfaces, we performed adhesion experiments in the presence of the cyclic peptide cRGDFG, where F is a D amino acid. This peptide binds αvβ3 with an affinity that is approximately 1,400-fold greater than that for the αIIbβ3 integrin (Haubner et al., 1996). We verified that this peptide, at a concentration of 300 µM, could efficiently block the attachment of CHO K1 cells to a monolayer presenting the GRGDSC peptide (Figure 2, D), but did not interfere with adhesion of αIIbβ3 CHO K1 cells to the same monolayer (Figure 2, E). Treatment of transfected cells with both the cyclic peptide inhibitor and the PAC-1 antibody, however, prevented cell adhesion to the monolayer (Figure 2, F). These results establish that the cRGDFG inhibitor can be used to block αv and α5 integrins on CHO cells and therefore allow studies of αIIbβ3-mediated adhesion of cells. All experiments that follow, unless stated otherwise, were performed in the presence of 300 µM cRGDFG.
Figure 2.
αIIbβ3 CHO K1 cell line. (A) CHO K1 cells do not express αIIbβ3 (top). Transfected αIIbβ3 CHO K1 cells express αIIbβ3 at the cell surface, as detected by anti-β3 and PAC-1 antibody (bottom). (B) cRGDFG, a cyclic peptide that specifically binds to α5 and αV integrins, inhibits CHO K1 cell adhesion to 0.5% GRGDSC SAMs. (C) CHO K1 cells adhere to 0.5% GRGDSC SAMs in the absence of inhibitors, but (D) fail to adhere to the same substrate in the presence of [cRGDFG]=300 µM. (E) αIIbβ3 CHO K1 cells adhere to 0.5% GRGDSC SAMs in the presence of [cRGDFG]=300 µM, (F) this interaction is nulled by the presence of PAC-1.
Adhesion of αIIbβ3 CHO K1 Cells to GRGDSC and HHLGGAKQAGDVC
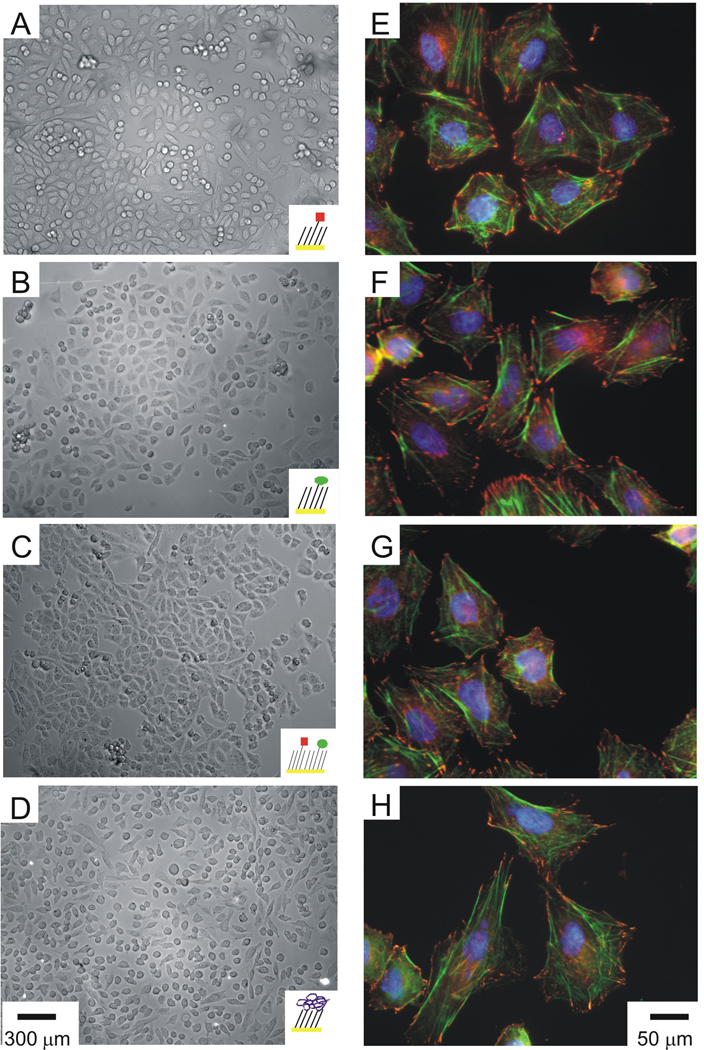
We compared the adhesion and spreading of αIIbβ3 CHO K1 cells to monolayers presenting GRGDSC, HHLGGAKQAGDVC and a mixture of both peptides. Cells were allowed to attach to the monolayers in serum-free F-12 K medium for 2 hours after which the media was exchanged with 10% FBS F-12K medium to facilitate cell spreading. We found that the cells adhered to monolayers presenting either the GRGDSC or the HHLGGAKQAGDVC peptides with comparable efficiency, in both cases assuming a flattened morphology with no refringent peripheral rim (Figure 3, A–C). The populations of cells on both substrates displayed a similar morphology, suggesting that both peptides have sufficient affinity for the receptor to support adhesion and spreading. The mixed monolayer supported cell adhesion in a similar manner as those that presented either peptide alone, suggesting that GRGDSC and HHLGGAKQAGDVC do not have differential roles in alphaIIb beta3-mediated cell adhesion and spreading. As a positive control, we adsorbed Fbg to a hydrophobic monolayer and observed a similar morphology for adherent cells (Figure 3, D). Several additional control experiments showed that the adhesion of cells on the model substrates was specific, including a lack of cell attachment to monolayers that present only glycol groups or unreacted maleimide groups (data not shown), and a lack of cell attachment to monolayers that presented the scrambled peptides GGRDGSC or GHHLGGADQAGKVC (data not shown).
Figure 3.
αIIbβ3 CHO cell adhesion and cytoskeletal structure on model substrates. Optical micrographs of αIIbβ3 CHO cells incubated with [cRGDFG]=300 µM that adhered and spread on 0.5% SAMs presenting GRGDSC (A), HHLGGAKQAGDVC (B) and a mixed monolayer comprised of both peptides (C). As a control, αIIbβ3 CHO cells adhered and spread on a dodecanethiol monolayer with adsorbed Fbg (D). Scale bars are 300 µm. Immunofluorescence staining of αIIbβ3 CHO cells incubated with [cRGDFG]=300 µM that adhered and spread on 0.5% SAMs presenting GRGDSC (E), HHLGGAKQAGDVC (F), a mixed monolayer comprised of both peptides (G) and the Fbg control (H). Focal adhesions were visualized with an anti-vinculin antibody (red), actin stress fibers were visualized with AF 488 phalloidin (green), and nuclei were visualized with Hoescht (blue). Scale bars are 50 µm.
Cytoskeletal Structure of Cells Adherent to Model Substrates
We compared the assembly of focal adhesion and actin stress fibers of αIIbβ3 CHO K1 cells adhered to monolayers presenting GRGDSC, HHLGGAKQAGDVC, a mixture of both peptides, and adsorbed Fbg (Figure 3, E, F, G and H, respectively). We allowed αIIbβ3 CHO K1 cells to attach and spread on the monolayers for four hours, and then fixed the cells and probed with anti-vinculin IgG to visualize focal adhesions and with phalloidin-AF 488 to visualize actin stress fibers. Nuclei were observed with DAPI. The cells had well-developed cytoskeletal structures and mature focal adhesions along the perimeter. Cells that were allowed to adhere and spread on the three monolayers presenting the peptide ligands showed comparable cytoskeletal structures, demonstrating that the αIIbβ3-mediated adhesion to either of the peptide ligands is sufficient to support a well-formed cytoskeleton.
AGD is the Minimal Binding Motif for αIIbβ3
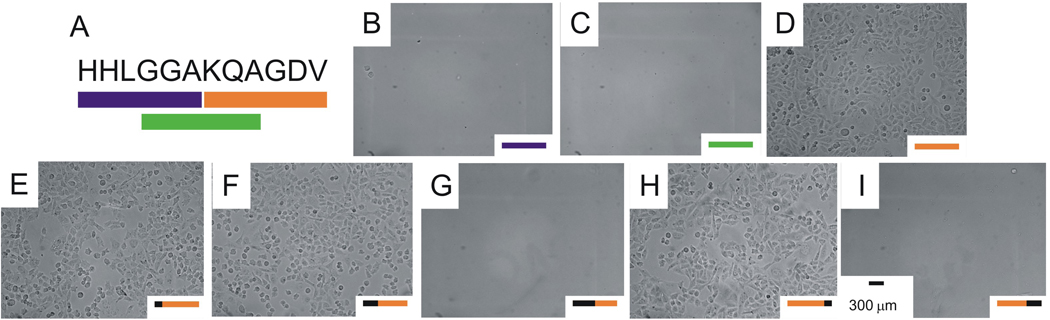
We next compared the adhesion of cells to monolayers presenting several fragments taken from the HHLGGAKQAGDV sequence in order to define the minimal binding motif. A comparison of cell adhesion to monolayers presenting the three peptides HHLGGAC, KQAGDVC and GGAKQAC—corresponding to the first, last and middle six residues in the sequence—revealed that only the second sequence supported cell adhesion (Figure 4, A–D). Further studies with monolayers presenting N and C terminal truncations of this peptide revealed that the AGD tripeptide supports efficient adhesion of cells (Figure 4, E–I). This result was surprising because the peptide has an obvious relevance to the well-characterized RGD motif, yet most integrins do not tolerate an Arg to Ala mutation in the ligand and because previous work has identified sequences that require the lysine residue.
Figure 4.
(A) Truncation of HHLGGAKQAGDV ligand to find minimal binding sequence. Optical micrographs of αIIbβ3 CHO K1 cells incubated with [cRGDFG]=300 µM that adhered and spread on 0.5% KQAGDVC (D) but failed to adhere to 0.5% HHLGGAC (B) and 0.5% GGAKQAC (C) SAMs. When KQAGDVC was truncated from the N terminus, αIIbβ3 CHO K1 cells adhered to 0.5% QAGDVC (E) and 0.5% AGDVC (F) SAMs, but failed to adhere to 0.5% GDVC SAMs (G). When KQAGDVC was truncated from the C terminus, αIIbβ3 CHO K1 cells adhered to 0.5% KQAGDC (H) SAMs, but failed to adhere to 0.5% KQAGC SAMs (I). Scale bars are 300 µm.
GRGDS and AGDV are Competitive Ligands for αIIbβ3
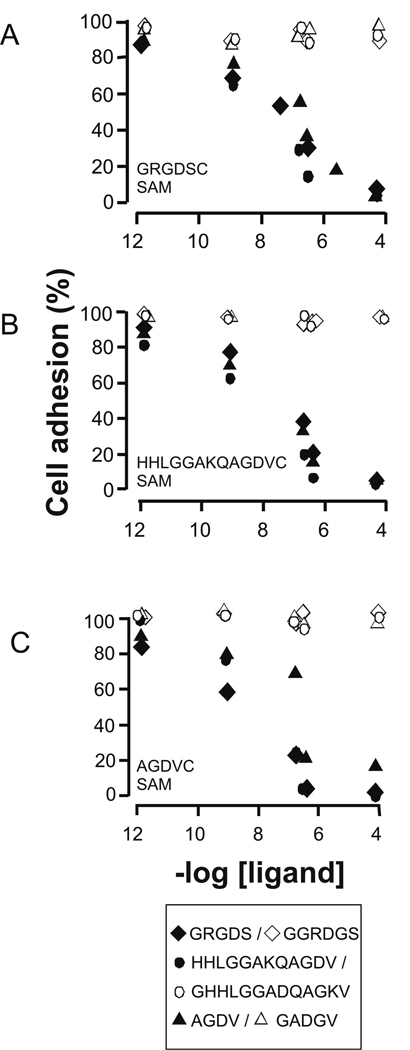
To ask whether the two peptides interact with the same site on αIIbβ3, we determined whether the peptides GRGDS, HHLGGAKQAGDV, and AGDV, in soluble form, could block the attachment of cells to the monolayers presenting these ligands. In these experiments we first incubated identical suspensions of cells (100,000/mL) in serum-free F-12K medium supplemented with cRGDFG (300 µM) and with the soluble peptides in concentrations ranging from 1 pM to 100 µM and then applied the mixtures to monolayers presenting either the GRGDSC, HHLGGAKQAGDVC, or AGDVC peptide (Figure 5). Cells were allowed to attach for 2 hours, after which the media was exchanged to 10% FBS F-12K medium (supplemented with cRGDFG) and cultured for an additional 2 hours. The slides were then rinsed and nuclei were stained with DAPI to facilitate counting of the adherent cells. We first found that each peptide could block the attachment of cells to monolayers presenting that peptide in a concentration-dependent manner. This experiment again verifies that the cell attachment is mediated by the immobilized peptide alone and not non-specific interactions between other cell-surface molecules and the monolayer. We also found, however, that each peptide could block cell attachment to monolayers presenting any of the three peptides, demonstrating that the ligands are in fact competitive in binding αIIbβ3. The soluble peptide HHLGGAKQAGDV, for example, blocked cell attachment to a monolayer presenting GRGDS, as did the soluble AGDV peptide. Control experiments confirmed that the use of scrambled peptides GGRDGS, GHHLGGADQAGKV and GADGV as soluble inhibitors had no effect on cell adhesion. Again, these results show that AGDV and GRGDS bind to the αIIbβ3 integrin competitively and suggest a common site on the receptor.
Figure 5.
Inhibition of αIIbβ3 CHO K1 cells on model substrates. Inhibition of αIIbβ3 CHO K1 cells incubated with [cRGDFG]=300 µM to monolayers presenting 0.5% GRGDSC (A), 0.5% HHLGGAKQAGDVC (B) and 0.5% AGDVC (C). Suspended cells were treated with either soluble GRGDS ( ◆), soluble HHLGGAKQAGDV (  ), or soluble AGDV (▲ ) at concentrations from 1 pM to 100 µM and then allowed to adhere to the model substrates. Cell adhesion was quantized as normalized units relative to adhesion in the absence of soluble peptides. The scrambled peptides GGRDGS ( ◇), GHHLGGADQAGKV (
), or soluble AGDV (▲ ) at concentrations from 1 pM to 100 µM and then allowed to adhere to the model substrates. Cell adhesion was quantized as normalized units relative to adhesion in the absence of soluble peptides. The scrambled peptides GGRDGS ( ◇), GHHLGGADQAGKV ( ) and GADGV (△) were used to examine selective inhibition. The standard error for each data point is within the range of 3–6%, which is smaller than the feature size for the data points.
) and GADGV (△) were used to examine selective inhibition. The standard error for each data point is within the range of 3–6%, which is smaller than the feature size for the data points.
Evaluating αIIbβ3 Specificity with a Peptide Array
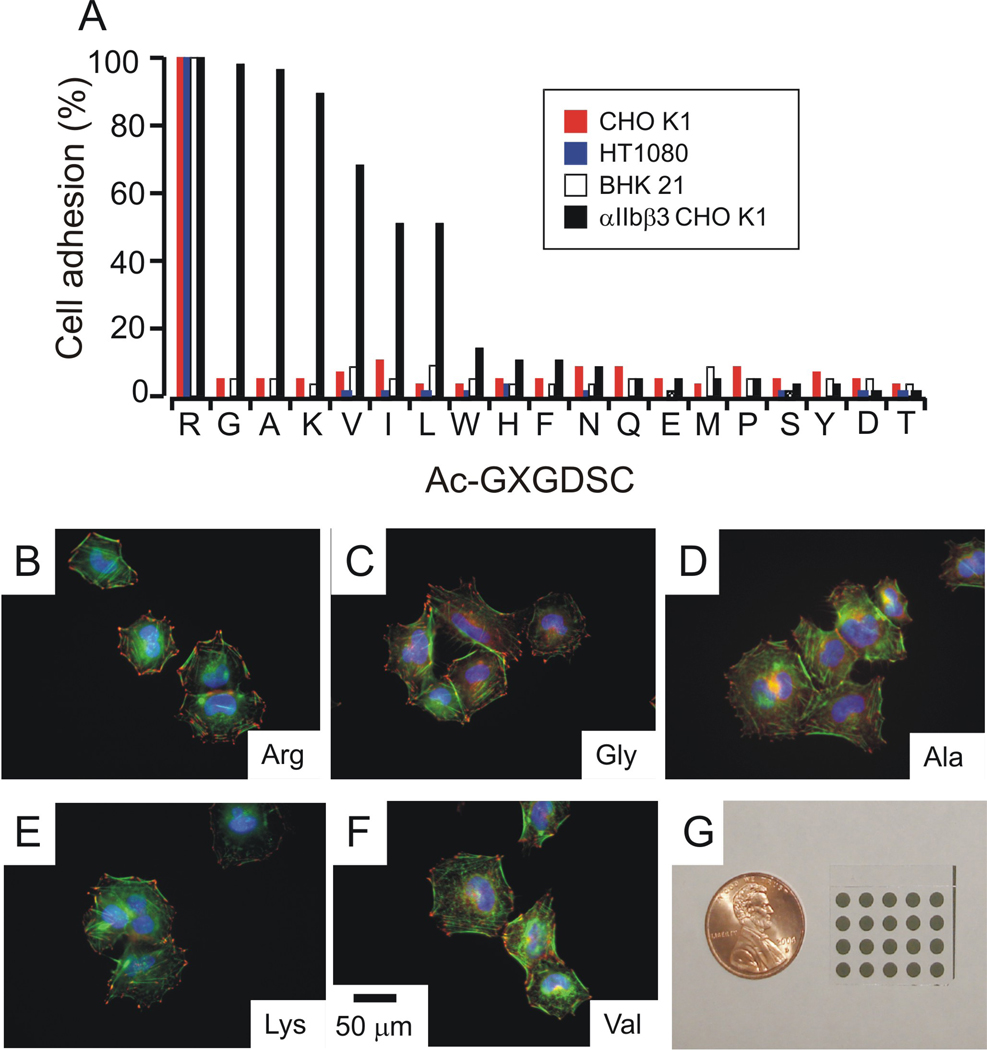
The αIIbβ3 integrin, like other integrins, tolerates amino acid substitutions on either side of the RGD sequence, but there have been no suggestions that the receptors tolerate substitution of the arginine residue with a hydrophobic residue. The report of the structure of the peptide-receptor complex, for example, showed that the lysine residue within the γ peptide substitutes for the arginine residue, but did not investigate a peptide lacking the lysine residue and does not provide information on the relative contribution of this interaction to the binding affinity. To further probe the promiscuity of the αIIbβ3 integrin for RGD peptide ligands, we used an array to assess the adhesion of cells to peptides having the arginine residue substituted with each amino acid other than cysteine. The library of Ac-GXGDSC peptides was synthesized and immobilized in an array with each peptide occupying a 1 mm diameter region of the monolayer (Figure 6, I). A suspension of αIIbβ3 CHO K1 cells was applied to the entire array and cells were allowed to attach to regions presenting the various peptides. The substrates were then rinsed and stained with DAPI, and the cells were counted to determine the relative efficiency of cell attachment to peptides in the library. We found that the Arg residue could be substituted with the other basic residue, Lys, or with hydrophobic residues (Figure 6, A). Substitution of the arginine residue with either glycine or alanine gave ligands that were comparable to the parent sequence and substitution with lysine and valine were only slightly less active. Substitution with leucine and isoleucine were also active, though at a lower level. Each of the other substitutions gave peptides that failed to support cell adhesion. αIIbβ3 CHO K1 cells also failed to adhere to 0.5% GGRDGSC, demonstrating the requirement for the GD scaffold for cell adhesion. For the five most active peptides from this array—those having Arg, Gly, Ala, Lys or Val at the first position—the αIIbβ3 CHO K1 cells had well-developed actin structures with clear vinculin-containing focal adhesions around the cell perimeter (Figure 6).
Figure 6.
Cell adhesion mediated by Ac-GXGDSC. (A) αIIbβ3 CHO K1 cells, CHO K1 cells, BHK 21 cells and HT 1080 cells were allowed to adhere to arrays of 0.5% GXGDSC . Cell adhesion was quantized as normalized units relative to adhesion in the 0.5% Ac-GRGDSC spot. Cells did not adhere to the spot presenting 0.5% GGRDGSC. Error bars are omitted for clarity. Immunostaining of peptides that mediated αIIbβ3 CHO K1 cell adhesion on the array. (B) 0.5% Ac-GRGDSC, (C) 0.5% Ac-GGGDSC, (D) 0.5% Ac-GAGDSC, (E) 0.5% Ac-GKGDSC, and (F) 0.5% Ac-GVGDSC. Focal adhesions were visualized with an anti-vinculin antibody (red), actin stress fibers were visualized with AF 488 phalloidin (green), and nuclei were visualized with Hoescht (blue).Scale bars are 50 µm. (G) 5X4 SAM array used for this work.
We repeated this adhesion experiment with CHO-K1, BHK 21 and HT1080 cells and found that only Ac-GRGDSC promoted significant cell adhesion, which is consistent with the known specificity of the α5 and αv integrin receptors. These cells also failed to adhere to 0.5% GGRDGSC. Hence, these experiments verify that the peptide arrays provide reliable information on the activity of peptides for mediating cell adhesion and therefore reveal that the αIIbβ3 integrin is less specific in its recognition of peptide ligands and prefers ligands of the form XGD where X is either a basic or a hydrophobic residue.
DISCUSSION
The peptides GRGDSC and AGDVC mediate αIIbβ3 CHO K1 cell adhesion
The most significant result from this work is the finding that the platelet receptor can recognize the γ carboxy-terminal sequence by primarily interacting with the AGD motif. Further, AGD and RGD bind competitively to αIIbβ3 and have a comparable ability to mediate the adhesion of CHO cells engineered with the receptor. Indeed, we found that αIIbβ3 CHO K1 cells adhered and spread efficiently on model substrates presenting either the GRGDSC or HHLGGAKQAGDVC peptides and were similar to cells that were adherent on Fbg-coated substrates. The well-developed cytoskeletal structure in cells—including the distributed focal adhesions and clear actin stress filaments—suggests that each of the peptide ligands has sufficient affinity for the platelet receptor to mediate biologically relevant adhesion. Further, control experiments establish that the adhesion of cells is mediated primarily by the immobilized ligand, since monolayers presenting no peptide or scrambled forms of the peptide failed to support cell adhesion. Inhibition experiments reveal that the two peptides bind competitively to the receptor, since either peptide in soluble form showed a dose-dependent inhibition of the attachment of cells to monolayers presenting either peptide. This result, as discussed below, provides biochemical data that is consistent with a recent x-ray structure which makes a strong argument that the RGD and γ peptides bind to a common site on αIIbβ3. Taken together, these results show that GRGDS and HHLGGAKQAGDV bind to a common site in αIIbβ3 and can independently mediate efficient cell adhesion, spreading and cytoskeletal organization.
Structural data of αIIbβ3 bound to Fbg peptides is consistent with cell adhesion data
The recent report by Springer and coworkers described crystal structures of the complexes formed between αIIbβ3 and either the HHLGGAKQAGDV or HHLGGAKQRGDV peptides (Springer et al., 2008). The structures revealed that the binding site for the γ peptide overlaps that for the RGD peptide. Therefore, this structural data provides strong evidence that the peptides share a common binding site and as stated above appears to settle this open question by ruling out separate, and potentially synergistic, binding sites for the two peptides. Our experimental finding that the peptides can competitively block the adhesion of cells is consistent with this report and together, the two studies provide structural and biochemical evidence that the peptides bind a common site in αIIbβ3.
A second question that has been debated and still remains open concerns the minimal peptide motif within the γ peptide that is sufficient for high affinity binding to the receptor. We approached this question by comparing cell attachment to monolayers presenting different fragments of the γ peptide and we found that the AGD peptide alone efficiently mediates cell adhesion and spreading. Further, inhibition experiments revealed that both the RGD and AGD peptides, at similar concentrations, prevented cell attachment to a monolayer presenting the RGD adhesion ligand, suggesting that the two peptides have comparable affinity for the αIIbβ3 integrin. This result was surprising, as the Arg to Ala substitution involves dramatic changes in charge and size of the residue side chain. Other reports that are inconsistent with our finding include the failure of an AGDV peptide to inhibit Fbg binding to activated platelets (Plow et al., 1985). This study used soluble peptides to compete for binding of radiolabeled Fbg to platelets. But because platelets have other cell surface receptors that bind to RGD—including αvβ3 and α5β1 (Kunicki, 1989)—these integrins can mediate Fbg binding even in the presence of platelet receptor antagonists (Humphries et al., 2006). RGD peptides, by contrast, are efficient inhibitors of Fbg binding to platelets because they block multiple integrins. AGD peptides, on the other hand, can only block Fbg binding to integrin αIIbβ3. We also suggest that valency of the fibrinogen protein—which is a dimer having both RGD and AGD motifs—is important for efficient aggregation of platelets, and that fibrinogen mutants that delete either of these functions can have an impaired ability to mediate aggregation.
Other published results are consistent, but not affirmative, of our finding that AGD is the minimal binding sequence in HHLGGAKQAGDV. Mice homozygous for a modified γ sequence that lacked the QAGDV sequence displayed highly defective platelet aggregation. The Fbg variant that included this mutation also failed to bind to αIIbβ3 in solid phase assays. However, we are unaware of a study that has tested explicitly the ability of the AGD tripeptide to block the interaction of the platelet γ chain with the receptor.
The x-ray structures of the γ-αIIbβ3 complex led to the interpretation that the minimal binding motif is KQAGD. A comparison of the complexes formed on binding of either the KQAGD peptide or the RGD peptide reveal that in both cases the Asp side chain interacts with a Mg2+ metal-ion dependent adhesion site (MIDAS) and the Gly residue resides in a cleft between the α and β subunits. Hence, αIIbβ3 has a strict requirement for the glycine and aspartate residues in the ligand. In the complex formed on binding RGD, the side chain of Arg lies along a cleft and positions the guanidinium group for hydrogen bonding to αIIb residue Asp 224 and to an αIIb backbone carbonyl. For the KQAGD peptide, the methyl side chain of alanine is directed towards the guanidinium binding pocket but cannot reach the pocket. Instead, the peptide ligand interacts with an extended binding site on the integrin and has a turn in the backbone that leads to insertion of the lysine side chain into the arginine binding pocket, so that the ammonium group participates in the same hydrogen bonding pattern as does the guanidinium group of the RGD peptide. This structure suggests a requirement for a basic residue in the peptide ligand, but allows the basic residue to be separated from the Gly-Asp diad provided the peptide can adopt a conformation that positions the lysine side chain in the guanidinium binding pocket.
The crystal structure does not provide information on the energetic contribution made by the lysine residue within KQAGD to the binding affinity of the peptide for the receptor. To address this point, we compared cell adhesion to monolayers presenting Ac-GAKQAGDVC, Ac-GASQAGDVC, GRGDSC and AGDVC and found that all ligands supported cell adhesion without significant differences (data not shown). Hence, the lysine appears to not provide a strong contribution to the binding energy, at least not for the densities of peptide used in our experiments. We are not aware of any biochemical reports that address the importance of K406 in Fbg for binding to αIIbβ3. A report by Hantgan and coworkers addressed the binding of αIIbβ3 with the disintegrin echistatin and used sedimentation velocity and fluorescence anisotropy assays to suggest that the Arg residue in RGD accounted for only 20% of the binding energy between the peptide ligand and αIIbβ3—that is, an echistatin analog that substituted AGD for RGD retained 80% of the activity for binding to αIIbβ3 (Hantgan et al., 2006). While the crystal structure reveals a direct contact between the integrin and Lys 406, we do not know whether the apparently favorable hydrogen bonding interaction formed by K406 in HHLGGAKQAGDV with αIIbβ3 could be compensated by an energetic penalty for the turn in the backbone to position the lysine side chain.
In any event, our results are clear in demonstrating that the AGD peptide is sufficient, and comparable to RGD, for binding the integrin and that a basic residue is not necessary for the activity of HHLGGAKQAGDV. Because of the obvious similarity of the AGD and RGD motifs, we used a peptide array to compare the adhesion of cells to peptides having the XGD sequence. We observed that three cell lines that lacked αIIbβ3 demonstrated adhesion only on the RGD peptide, consistent with the extensive literature on integrin specificity (Ginsberg et al, 1990; Li et al., 2003). The αIIbβ3 CHO K1 cells, by contrast, showed efficient adhesion to peptides having a basic residue in the first position (Arg and Lys) but also to those having small hydrophobic residues in this position (including Gly, Ala and Val, and to a lesser extent, Leu and Ile). Further, cells on this latter set of peptides were well spread, had distributed focal adhesions and clear stress fibers and appeared indistinguishable from cells on RGD. The GD scaffold was necessary for adhesion, as demonstrated by the lack of adhesion of all cells used to 0.5% GGRDGSC. Previous reports have discussed the importance of the GD scaffold in integrin binding. For the integrin αIIbβ3, Plow and coworkers found that conservative mutations Gly to Ala and Asp to Glu in GRGDSP peptides were >100-fold less potent in inhibiting platelet aggregation. (Plow et al., 1985). Lazarus and coworkers made variants of the snake venom kistrin where each residue was substituted with Ala. They found that D51A variant was >200-fold less potent in platelet aggregation inhibition than wild-type kistrin. The D51E mutant was >100-fold less potent in platelet aggregation inhibition (Dennis et al., 1993). Hence, this result reveals the expanded binding specificity of αIIbβ3 and is consistent with the general understanding that this receptor is promiscuous relative to other integrins.
It is significant that the atypical integrin ligands we identified above are present in the C terminal γ chain of several species. Lamprey and xenopus γ chains contain an RGD sequence, while human, dog and mouse contain an AGD sequence. Rat and chicken γ chains contain a VGD sequence, while zebrafish and carp contain IGD and LGD triads, respectively (Springer et al., 2008). This pattern is consistent with the evolution of Fbg and αIIbβ3 to introduce residues in the first position that abrogate binding of these peptides to the other integrin receptors that mediate adhesion in tissue and the endothelial cells lining the luminal vessel.
Our results are further significant in that they suggest a novel class of αIIbβ3 antagonists that may have increased specificity for the platelet receptor. Several cyclic peptides found in snake venoms have been characterized as potent anti-thrombotic agents and operate by using either a RGD or KGD motif to bind the integrin (Scarborough et al., 1993; Oshiwaka et al., 1999). The cyclic peptide eptifibatide—which has a homoarginine residue in the first position—was derived from barbourin (Scarborough et al. 1991) and is now used in clinical settings to prevent platelet aggregation. Eptifibatide has gained widespread use, as it overcomes the immunogenicity and clearance time problems faced by abciximab, a Fab fragment used to inhibit platelet aggregation. Although eptifibatide does not cause immune response and follows renal body clearance (Scarborough, 1999), one of its secondary effects is prolonged bleeding in patients (Curran et al, 2005). Eptifibatide also binds the αIIbβ3 integrin on endothelial cells with lower affinity, and it has been suggested that the peptide binds this integrin at therapeutic concentrations, though the consequences of this interaction are not yet well characterized (Lele et al., 2001). In this respect, inhibitors that display a greater specificity for αIIbβ3 relative to other integrins may be valuable. Our results suggest that linear or cyclic peptides having the AGD motif, or other hydrophobic residues in the first position, may serve as highly selective inhibitors of platelet aggregation. Current efforts in our laboratory are not investigating the design of novel anti-aggregation molecules using the XGD scaffold, where X is a hydrophobic residue.
Self-assembled monolayers as models of the extracellular matrix
This work is also significant because it demonstrates the utility of self-assembled monolayers as mimics of the extracellular matrix. Studies of cell adhesion, and of the ligands and receptors that mediate adhesion, remain challenging because of the difficulties in controlling the activities of ligands that are tethered to culture substrates. ‘Inert’ monolayers that present oligo(ethylene glycol) chains are effective platforms for the study of ligand-mediated cell adhesion. Further, the ability to chemically control the composition of the monolayer—including the density, site of attachment and nature of the tether of the ligand—together with the use of mass spectrometry to confirm the structure of the surface makes these ECM mimics broadly applicable to studies of the interactions of cells with ECM. We are particularly intrigued by the possibility of using arrays prepared from hundreds of peptides to identify novel adhesion ligands (Derda et al, 2007).
SIGNIFICANCE
The regulated adhesion of cells is important in homeostasis and disease. Studies of adhesion and the identification of the ligands and receptors that mediate adhesion often use protein-coated substrates and are complicated by the difficulty in controlling the activities, identities and densities of the ligands. The development of model substrates—and of self assembled monolayers of alkanethiolates on gold in particular—now provides routes to prepare structurally defined surfaces for controlling the ligand-receptor interactions that mediate adhesion. The present work has applied the monolayer substrates to identify the peptide ligands in Fbg that interact with the platelet integrin. We find that αIIbβ3 CHO K1 cells attach, spread and assemble cytoskeleton on monolayers presenting both the RGD and the HHLGGAKQAGDVC ligands. Further, inhibition experiments reveal that these two ligands bind competitively to the receptor, in agreement with a recent structural study. Of greater significance, this work compared cell adhesion to a peptide array and revealed that the platelet receptor recognizes peptides of the form XGD, where X can either be a basic or hydrophobic group. This relaxed specificity for the receptor is unprecedented and suggests the design of antagonists that better discriminate in binding to the platelet receptor relative to other integrins, and that may lead to a new class of anti-thrombotic agents.
EXPERIMENTAL
Antibodies and Reagents
All reagents were used as received. All amino acids and peptide synthesis reagents were purchased from Anaspec, with the exception of Fmoc-Asp(WangLL)-ODmab resin, obtained from EMD Chemicals. Anti-integrin antibody PAC-1 (anti-αIIbβ3) was obtained from BD Biosciences. Other anti-integrin antibodies, M-148 (anti-αIIb) and H-96 (anti-β3) were purchased from Santa Cruz Biotechnology. Monoclonal anti-vinculin antibody was obtained from Sigma. Texas Red goat anti-mouse IgG, AF 488 goat anti-mouse IgG, AF 488 goat anti-rabbit IgG and AF 488 phalloidin were obtained from Invitrogen. 4´,6-Diamidino-2-phenylindole (DAPI) was obtained from Molecular Probes. Human Fbg was obtained from Sigma. FuGENE transfection reagent was purchased from Roche. F-12K cell culture medium was purchased from ATCC. All other cell culture media and reagents were obtained from Gibco, with the exception of geneticin and zeocin, which were from Invitrogen.
Peptide Synthesis
Linear peptides were synthesized manually following standard Fmoc peptide synthesis protocols using Fmoc-Rink amide MBHA resin. The cyclic peptide cRGDFG, where F is a D amino acid, was synthesized using standard Fmoc peptide synthesis protocol using Fmoc-Asp(WangLL)-ODmab resin. The Dmab group was cleaved using 2% hydrazine in DMF for 20 minutes, and the peptide was cyclized by treatment with 3 equivalents of HOBt and DIC for 4 hours. All peptides were purified by reverse phase HPLC using a C18 column (Waters) and characterized with Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS).
Preparation of Monolayers
Monolayers were prepared as previously described (Houseman, et al., 2003). Glass coverslips were sonicated for 30 minutes in deionized ultrafiltered (DIUF) water and then 30 minutes in ethanol and then dried under a stream of nitrogen. Titanium (4 nm) and then gold (29 nm) were evaporated onto the coverslips using an electron beam evaporator (Boc Edwards) at a rate of 0.05–0.10 nm/s and at a pressure of 1 µTorr. The gold-coated coverslips were immersed in an ethanolic solution of maleimide-terminated disulfide (1%) and tri(ethylene glycol)-terminated disulfide (99%) overnight at room temperature. The total disulfide concentration was 1 mM. The substrates were washed with DIUF water, ethanol and then dried under a stream of nitrogen.
cDNA Constructs, Transfection and Cell Culture
Constructs for αIIb and β3 were a gift from Professor Joel Bennet. CHO K1, BHK 21 and HT1080 cells were purchased from ATCC. All CHO cells were cultured at 37 °C and 5% CO2 using F12K medium supplemented with 10% FBS and pen/strep. Other cell lines were cultured in the same conditions using medium suggested by ATCC. CHO K1 cells were transfected with αIIb and β3 cDNA with FuGENE following the supplier’s protocol. Transfected cells were selected with 1 mg/mL zeocin and 1 mg/mL geneticin.
Cell adhesion assay
Cysteine-terminated peptides were immobilized onto maleimide-presenting SAMs by immersing the chip in 1 mM peptide solution and incubating at 37 °C for 1 hour. Substrates were rinsed with DIUF water and ethanol and dried under a stream of nitrogen. αIIbβ3 CHO K1 cells were detached from culture plates with 1 mM EGTA/2 mM EDTA in PBS, rinsed with serum-free F-12K medium, centrifuged and resuspended in serum-free F-12K medium supplemented with cRGDFG (300 µM) and 1X penicillin/streptomycin. After 15-minute of incubation, cells (200,000/mL) were added to substrates in 24-well culture plates. After 2 hour incubation at 37 °C and 5% CO2, media was exchanged for F-12K medium supplemented with 10% FBS, 1X penicillin/streptomycin and cRGDFG (300 µM). After 2 hours, the substrates were washed gently with warm PBS, and adherent cells were immediately fixed with 4% formaldehyde for 30 minutes, visualized with a 20X objective of a Zeiss Axiovert 200 inverted microscope, and imaged. For cell adhesion assays mediated by Ac-GXGDSC, αIIbβ3 CHO K1 cells were incubated on a 5x4 circle (1 mm diameter) array for 1 hour with serum-free F-12K medium supplemented with cRGDFG (300 µM) and 1X penicillin/streptomycin, followed by media exchange to F-12K medium supplemented with 10% FBS, 1X penicillin/streptomycin and cRGDFG (300 µM). The substrates were washed with warm PBS, and adherent cells were immediately fixed and permebealized with 4% formaldehyde and 0.5% Triton-X for 30 minutes. Fixed cells were stained with DAPI, visualized and imaged with a 20X objective as described. The number of adherent cells per field of view was counted using Image J software. Cells were counted in at least three fields of view for each circle, and each experiment was repeated 3 times. The degree of adhesion is reported as a percentage of cells that adhere relative to control experiments with the peptide Ac-GRGDSC.
Immunostaining
αIIbβ3 CHO K1 cells were allowed to adhere and spread on substrates as described above. Substrates were washed gently with PBS, and fixed and permebealized as described above. Cells were stained with monoclonal anti-vinculin IgG (1:1000) to visualize focal adhesions, phalloidin (1:500) to visualize stress fibers, and DAPI (1:5000) to visualize the nucleus. Substrates were rinsed thoroughly with PBS and mounted on microscope slides with Aqua Poly/Mount (PolySciences Inc.). Substrates were visualized with a 100 X objective of a Zeiss Axiovert 200 inverted microscope, and imaged.
Cell adhesion inhibition assay
Suspensions of αIIbβ3 CHO K1 cells (100,000/mL) were incubated with cRGDFG (300 µM) in serum-free F-12K medium with 1 mg/mL BSA and 1X penicillin/streptomycin for 15 minutes at 37 °C and 5% CO2. These suspensions were then supplemented with soluble peptides at concentrations raging from 1 pM to 100 µM and further incubated for 15 minutes at 37 °C and 5% CO2. Cells were then added to monolayers and incubated at 37 °C and 5% CO2 for 2 hours, then media was exchanged for F-12K medium supplemented with 10% FBS and 1X penicillin/streptomycin. The substrates were further incubated for 2 hours. The substrates were washed gently with warm PBS, and adherent cells were immediately fixed and permebealized with 4% formaldehyde and 0.5% Triton-X for 30 minutes. Fixed cells were stained with DAPI, visualized and imaged with a 20X as described previously. The number of adherent cells per field of view was counted using Image J. Cells were counted in at least five adjacent fields for each substrate, and each experiment was repeated 3 times. The degree of inhibition is reported as a percentage of cells that adhere relative to control experiments in the absence of soluble peptide.
ACKNOWLEDGMENTS
We thank Professor Joel Bennett for supplying αIIb and β3 constructs. This work was supported by the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
REFERENCES
- Bennett J, Shattil S, Power J, Gartner T. Interaction of Fibrinogen With Its Platelet Receptor. Differential Effects of Alpha and Gamma Chain Fibrinogen Peptides of the Glycoprotein IIb-IIIa Complex. J. Biol. Chem. 1988;263:12948–12953. [PubMed] [Google Scholar]
- Bini A, Haidaris P, Kudryk B. In: Encyclopedic Reference of Vascular Biology and Pathology. Bikfalvi A, editor. Heidelberg, Germany: Springer; 2000. pp. 107–125. [Google Scholar]
- Curran MP, Keating GM. Eptifibatide: A Review of Its use in Patients with Acute Coronary Syndromes and/or Undergoing Percutaneous Coronary Intervention. Drugs. 2005;65:2009–2035. doi: 10.2165/00003495-200565140-00007. [DOI] [PubMed] [Google Scholar]
- Dennis MS, Carter P, Lazarus RA. Binding Interactions of Kistrin with Platelet Glycoprotein IIb-IIIa: Analysis by Site-Directed Mutagenesis. Proteins. 1993;15:312–321. doi: 10.1002/prot.340150308. [DOI] [PubMed] [Google Scholar]
- Derda R, Li L, Orner BP, Lewis RL, Thomson JA, Kiessling LL. Defined Substrates for Human Embryonic Stem Cell Growth Identified from Surface Arrays. ACS Chem. Biol. 2007;2:347–355. doi: 10.1021/cb700032u. [DOI] [PubMed] [Google Scholar]
- Doolittle R, Goldbaum D, Doolittle L. Designation of the Sequences Involved in the “Coiled-Coil” Interdomainal Connections in Fibrinogen: Construction of an Atomic Scale Study. J. Mol. Biol. 1978;120:311–325. doi: 10.1016/0022-2836(78)90070-0. [DOI] [PubMed] [Google Scholar]
- D’ Souza S, Ginsberg M, Burke T, Lam S, Plow E. Localization of an Arg-Gly-Asp Recognition Site Within an Integrin Adhesion Receptor. Science. 1988;242:91–93. doi: 10.1126/science.3262922. [DOI] [PubMed] [Google Scholar]
- D’ Souza S, Ginsberg M, Burke T, Plow E. The Ligand Binding Site of the Platelet Integrin Receptor GPIIb-IIIA is Proximal to the Second Calcium Binding Domain of Its α Subunit. J. Biol. Chem. 1990;265:3440–3446. [PubMed] [Google Scholar]
- Farrell D, Thiagarajan P, Chung D, Davie E. Role of Fibrinogen Alpha and Gamma Chain Sites in Platelet Aggregation. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10729–10732. doi: 10.1073/pnas.89.22.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Mrksich M. The Synergy Peptide PHSRN and the Adhesion Peptide RGD Mediate Cell Adhesion through a Common Mechanism. Biochemistry. 2004;43:15811–15821. doi: 10.1021/bi049174+. [DOI] [PubMed] [Google Scholar]
- Ginsberg MH, Loftus JC, Plow EF. Ligand Binding to Integrins: Common and Ligand-Specific Recognition Mechanisms. Cell Differ. Dev. 1990;32:203–213. doi: 10.1016/0922-3371(90)90033-s. [DOI] [PubMed] [Google Scholar]
- Hantgan RR, Stahle MC, Connor JH, Horita DA, Rocco M, McLane MA, Yakovlev S, Medved L. Integrin αIIbα3: Ligand Interactions are Linked to Binding-Site Remodeling. Protein Sci. 2006;15:1893–1906. doi: 10.1110/ps.052049506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubner R, Gratias R, Diefenbach B, Goodman S, Jonczyk A, Kessler H. Structural and Functional Aspects of RGD-Containing Cyclic Pentapeptides as Highly Potent and Selective Integrin αVβ3 Antagonists. J. Am. Chem. Soc. 1996;118:7461–7472. [Google Scholar]
- Hawiger J, Timmons S, Kloczewiak M, Strong D, Doolittle R. γ and α Chains of Human Fibrinogen Possess Sites Reactive With Human Platelet Receptors. Proc. Natl. Acad. Sci. U.S.A. 1982;79:2068–2071. doi: 10.1073/pnas.79.6.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmbäck K, Danton MJS, Suh TT, Daugherty CC, Degen JL. Impaired Platelet Aggregation and Sustained Bleeding in Mice Lacking the Fibrinogen Motif Bound by Integrin αIIbα3. Embo J. 1996;15:5760–5771. [PMC free article] [PubMed] [Google Scholar]
- Houseman BT, Gawalt ES, Mrksich M. Maleimide-Functionalized Self-Assembled Monolayers for the Preparation of Peptide and Carbohydrate Biochips. Langmuir. 2003;19:1522–1531. [Google Scholar]
- Hu D, White C, Panzer-Knodle S, Page J, Nicholson N, Smith J. A New Model of Dual Interacting Ligand Binding Sites on Integrin αIIbβ3. J. Biol. Chem. 1999;274:4633–4639. doi: 10.1074/jbc.274.8.4633. [DOI] [PubMed] [Google Scholar]
- Humphries JD, Byron A, Humphries MJ. Integrin Ligands at a a Glance. J. Cell. Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Mrksich M. Using Model Substrates To Study the Dependence of Focal Adhesion Formation on the Affinity of Integrin-Ligand Complexes. Biochemistry. 2004;43:2699–2707. doi: 10.1021/bi0352670. [DOI] [PubMed] [Google Scholar]
- Kloczewiak M, Timmons S, Lukas T, Hawiger J. Platelet Receptor Recognition Site on Human Fibrinogen. Synthesis and Structure-Function Relationship of Peptides Corresponding to the Carboxy-Terminal Segment of the Gamma Chain. Biochemistry. 1984;23:1767–1774. doi: 10.1021/bi00303a028. [DOI] [PubMed] [Google Scholar]
- Kunicki TJ. Platelet Membrane Glycoproteins and their Function: an Overview. Blut. 1989;59:30–34. doi: 10.1007/BF00320245. [DOI] [PubMed] [Google Scholar]
- Lam SC, Plow E, Smith M, Andriux A, Ryckwaert J, Margerie G, Ginsberg M. Evidence that Arginyl-Glycyl-Aspartate Peptides and Fibrinogen γ Chain Peptides Share a Common Binding Site on Platelets. J. Biol. Chem. 1987;262:947–950. [PubMed] [Google Scholar]
- Lele M, Sajid M, Wajih N, Stouffer GA. Eptifibatide and 7E3, but not Triptofiban, Inhibit αVβ3 Integrin-Mediated Binding of Smooth Muscle Cells to Thrombospondin and Prothrombin. Circulation. 2001;104:582–587. doi: 10.1161/hc3101.092199. [DOI] [PubMed] [Google Scholar]
- Li R, Hoess R, Bennett J, DeGrado W. Use of Phage Display to Probe the Evolution of Binding Specificity and Affinity in Integrins. Protein Engineering. 2003;16:65–72. doi: 10.1093/proeng/gzg002. [DOI] [PubMed] [Google Scholar]
- Mrksich M. Using Self-Assembled Monolayers to Mimic the Extracellular Matrix. Acta Biomaterialia. 2009;5:832–841. doi: 10.1016/j.actbio.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WL, Mercurius KO, Mrksich M. Substrates for Cell Adhesion Prepared via Active Site-directed Immobilization of a Protein Domain. Langmuir. 2004;20:1026–1030. doi: 10.1021/la035733m. [DOI] [PubMed] [Google Scholar]
- Oshiwaka K, Terada S. Ussuristatin 2, a Novel KGD-Bearing Disintegrin from Agkistrodon ussuriensis Venom. J. Biochem. 1999;125:31–35. doi: 10.1093/oxfordjournals.jbchem.a022264. [DOI] [PubMed] [Google Scholar]
- Plow EF, Pierscbacher MD, Rouslahti E, Marguerie GA, Ginsberg MH. The Effect of Arg-Gly-Asp Containing Peptides on Fibrinogen and von Willebrand Factor Binding to Platelets. Proc. Natl. Acad. Sci. U.S.A. 1985;82:8057–8061. doi: 10.1073/pnas.82.23.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsmann A, Schaffner-Reckinger E, Kabile F, Plançon S, Kieffer N. A New Functional Role of the Fibrinogen RGD Motif as the Molecular Switch That Selectively Triggers Integrin αIIbβ3-Dependent RhoA Activation During Cell Spreading. J. Biol. Chem. 2005;280:33610–33619. doi: 10.1074/jbc.M500146200. [DOI] [PubMed] [Google Scholar]
- Scarborough RM. Development of Eptifibatide. Am. Heart. J. 1999;138:1093–1104. doi: 10.1016/s0002-8703(99)70075-x. [DOI] [PubMed] [Google Scholar]
- Scarborough RM, Rose JW, Hsu MA, Phillips DR, Fried VA, Campbell AM, Nannizzi L, Cahro IF. Barbourin A. GPIIb-IIIa-Specific Integrin Antagonist from the Venom of Sistrurus m. barbouri. J. Biol. Chem. 1991;266:9359–9362. [PubMed] [Google Scholar]
- Scarborough RM, Naughton MN, Teng W, Rose JW, Phillips DR, Nannizzi L, Arfsten A, Campbell AM, Cahro IF. Design of Potent and Specific Integrin Antagonists. Peptide Antagonists with High Specificity for Glycoprotein IIB-IIIa. J. Biol. Chem. 1993;268:1066–1073. [PubMed] [Google Scholar]
- Springer T, Zhu J, Xiao T. Structural Basis for Distinctive Recognition of Fibrinogen γC Peptide by the Platelet Integrin αIIbβ3. J. Cell Biol. 2008;182:791–800. doi: 10.1083/jcb.200801146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Mrksich M. Using Mass Spectrometry to Characterize Self-Assembled Monolayers Presenting Peptides, Proteins and, Carbohydrates. Angew. Chem. Int. Ed. 2002;41:4715–4718. doi: 10.1002/anie.200290026. [DOI] [PubMed] [Google Scholar]
- Ugarova T, Budzynski A, Shattil S, Ruggeri Z, Gingsberg M, Plow E. Conformational Changes in Fibrinogen Elicited by Its Interaction With Platelet Membrane Glycoprotein GPIIb-IIIA. J. Cell. Biol. 1993;182:791–800. [PubMed] [Google Scholar]