Abstract
Activation-induced cytidine deaminase (AID) and APOBEC3G catalyze deamination of cytosine to uracil on single-stranded DNA, thereby setting in motion a regulated hypermutagenic process essential for human well-being. However, if regulation fails, havoc ensues. AID plays a central role in the synthesis of high affinity antibodies, and APOBEC3G inactivates human immunodeficiency virus-1. This minireview highlights biochemical and structural properties of AID and APOBEC3G, showing how studies using the purified enzymes provide valuable insight into the considerably more complex biology governing antibody generation and human immunodeficiency virus inactivation.
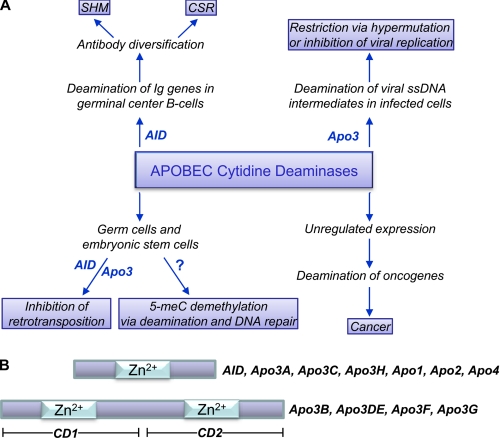
The APOBEC2 family of polynucleotide cytosine deaminases includes a group of 11 proteins with a common purpose: mutagenesis for the better. The founding member, Apo1 (APOBEC1), acts in a regulated manner to deaminate C to U on apoB mRNA to alter protein expression (1). Other family members have activity on ssDNA; in particular, AID is used to initiate affinity maturation of antibodies, and the Apo3 members (Apo3A–Apo3H) act in innate immunity to restrict retrotransposons and retroviruses (Fig. 1A) (1, 2). The ability of APOBEC enzymes to deaminate 5-MeC suggests a role in epigenetic regulation (Fig. 1A) (3). However, APOBEC enzymes acting on an inappropriate substrate can also have catastrophic consequences (Fig. 1A) (1, 2). Two family members, Apo2 and Apo4, still have no known activity or function (1). The Apo3 family in humans (Apo3A–Apo3H) was likely formed by gene duplications. Four of these deaminases (Apo3B, Apo3DE, Apo3F, and Apo3G) each have two homologous zinc-coordinating domains (CD1 and CD2), of which only CD2 is catalytically active (Fig. 1B) (1). Here, we discuss recent biological, biochemical, and structural data in an attempt to review current ideas about how mutations initiated by AID and Apo3G are responsible for immunological diversity and retroviral restriction, respectively.
FIGURE 1.
Overview of APOBEC cytidine deaminase functions and polypeptide domains. A, AID deaminations initiate antibody diversification pathways for SHM and CSR. Apo3 deamination restricts viruses with an ssDNA intermediate and may also physically inhibit the reverse transcription process. In germ cells and embryonic stem cells, APOBEC enzymes may restrict retrotransposons through RNA binding or deamination of reverse transcripts. APOBEC enzymes may also deaminate 5-MeC → T, which can initiate demethylation via DNA repair of T·G mismatches. Unregulated deaminations can initiate cell transformation, leading to cancer. B, the APOBEC family in humans is composed of seven members having a single Zn2+-coordinating deaminase domain per polypeptide chain and four others each having two homologous Zn2+-coordinating domains (CD1 and CD2) per chain, with only CD2 having deaminase activity. The N- and C-terminal domains are termed CD1 and CD2, respectively.
Importance of AID in Affinity Maturation
AID is necessary for adaptive immunity. It is induced in germinal center B-cells following low affinity antibody-antigen recognition (4) and shuttles between the cytoplasm and nucleus, with phosphorylation likely needed for entry into the nucleus (2). Once in the nucleus, AID initiates SHM and CSR by catalyzing C → U during transcription of IgV and S regions (Fig. 1A) (2). AID deaminates ssDNA in vivo and in vitro preferentially at C sites in 5′-WRC hot spot motifs (W = A or T; R = A or G) (2, 5). In vitro transcription studies using bacteriophage T7 RNA polymerase show favored C deaminations occurring in WRC sequences on the non-transcribed strand (2, 6), even when nucleosomes are present (7). AID also catalyzes 5-MeC → T by deamination but at a reduced rate compared with C → U (8, 9). Notably, AID “upmutants” with a higher activity give rise to increased antibody diversification (10).
Several “things” can happen following AID-catalyzed C deamination. The resulting U opposite G upon normal DNA replication leads to C → T transitions. On the other hand, U can be removed by UNG, and the resulting abasic site, when “copied” by an error-prone DNA polymerase that can insert T or C opposite the lesion, causes C → A and C → G transversions. Alternatively, U can undergo MMR or BER, which, in the presence of error-prone polymerases, can yield various transitions and transversions (2).
During SHM, replication and erroneous repair of U in IgV regions generate mutations at ∼10−3 to 10−4/base pair/cell division, which is roughly one million times higher than normal somatic mutation frequencies (2). In contrast to SHM, the presence of U in S regions provides sites for the initiation of dsDNA breaks required for CSR (11). CSR occurs by specific DNA deletions between S regions, enabling the VDJ segment of active IgM genes to be transferred to a downstream constant gene, thereby producing isotype IgG, IgA, or IgE instead of IgM (11). Without functional AID to initiate these processes, humans and mice develop HIGM-2 syndrome, which is exemplified by the absence of IgG, IgA, and IgE isotypes, caused by a loss of CSR, typically accompanied by a reduction in SHM, thereby creating a high susceptibility to autoimmunity and infection (4, 12).
Mechanisms That Target AID to IgV and S Regions Are Obscure
How AID is targeted selectively to IgV and S regions while avoiding other portions of the genome is not understood either at a global level, to explain why some genes are deaminated while others not, or at a local level, to address the distribution of C deaminations within a gene. Although active transcription of IgV and S regions provides ssDNA as a substrate for AID, transcription, while necessary, is not itself sufficient to account for AID targeting.
Data from cultured cell and mouse model studies have identified proteins and regulatory elements involved in AID targeting. SHM in B-cells is observed primarily in actively transcribed IgV and S regions of Ig genes, but also, albeit to a lesser degree, in non-Ig genes such as bcl6, cd79, cd83, and fas (2, 13). In a recent study examining the extent of genome deamination by AID, analysis of C → T mutations in ∼80 transcribed genes in MMR- and BER-deficient (ung−/−msh2−/−) mouse germinal center B-cells showed that about half the genes were deaminated above spontaneous levels (13). Those that were mutated at C sites exhibited 10–100-fold reduced mutation frequencies compared with IgV regions (13). Perhaps, as suggested by this study, AID targeting is relatively widespread, but C deaminations occurring in non-Ig genes are repaired in an error-free manner. This mechanism would maintain the genetic integrity of most non-Ig genes (13).
It is not known what causes the mutational gradient observed in the IgV region, in which mutations begin ∼150 nucleotides downstream from the IgV gene promoter and then decrease gradually over ∼1.5–2 kb farther downstream (2). Chromatin modifications, including histone H3 and H4 hyperacetylation and histone H2B phosphorylation, might be involved in regulating the density of mutations along the IgV region (2). Another unresolved issue is how AID can deaminate both the non-transcribed and transcribed strands in vivo (2).
Despite the current vagaries, it is well established that the targeting of AID to transcribed DNA involves protein cofactors and cis- and trans-acting DNA elements. Ig promoters and enhancers, the matrix attachment region, E-box motifs (2), and a large DNA region located to the 3′-side of the Ig constant region (14) may all contribute to AID targeting. However, the IgV region base sequence per se is not needed and can be replaced by other DNA sequences (2). The proteins that have been implicated in AID targeting include RNA pol II, eukaryotic single-stranded binding protein (RPA), and the β-catenin-like factor CTNNBL-1 (2, 15, 16). The RPA interaction is reported to require AID phosphorylation at Ser38 (2). The CTNNBL-1 interaction requires AID residues 39–43, but not phosphorylation (15).
Mutations in AID, such as S38A, which abolishes interactions with RPA (2), and HIGM-2 S43P, which fails to interact with CTNNBL-1 (15), significantly reduce SHM and CSR. However, the S38A and S43P mutants exhibit wild-type AID-specific activity (15, 17), but with altered deamination specificity (17). In vitro, the S43P mutant of AID deaminates the most commonly occurring S region motifs, 5′-GGC and 5′-AGC, with greater preference than wild-type AID (17). Aberrant deaminations combined with decreased targeting might disrupt the site-specific recombination process, providing a basis for the absence of CSR (17).
The current understanding of global and local AID targeting is derived primarily from genetic studies and limited biochemical assays involving prokaryotic transcription model systems (T7, T3, or Escherichia coli RNA polymerases). The prokaryotic transcription systems cannot be used to address specific interactions of AID with human or mouse Ig elements, transcription machinery, and potential recruiting cofactors. This point underscores the urgent need for studies with a human RNA pol II transcription system.
Error-prone Processing of AID-generated G·U Mispairs
Similar to AID targeting, there is a genetic road map for addressing the biochemical mechanisms of SHM, in which MMR and BER play a central role. Despite the availability of biochemical model systems that capture the essence of the standard error-free MMR and BER pathways in humans, it will be a formidable challenge to accommodate the specialized enzymes, such as pol η, and control signals, e.g. monoubiquitination of the sliding clamp proliferating cell nuclear antigen, required to model error-prone repair. It seems fair to say that the biochemical modeling process has barely begun. Here is a synopsis of what is known about the IgV region mutations generated by the processing of U.
The SHM mutation spectra in the IgV regions contain roughly equal mutation frequencies at C/G and A/T sites (2). Mutations at C/G sites occur by copying U or by copying an abasic site generated by the removal of U by UNG. Mutations at A/T sites occur when the elimination of G·U mispairs by MMR and BER creates a gapped DNA substrate surrounding the deamination site where error-prone DNA polymerases can act (2). pol η acting primarily at 5′-WA motifs is responsible for making A/T mutations (2), but pol ζ, pol κ, pol θ, and Rev1 can also generate mutations in the absence of pol η (2).
Genetic studies have identified many of the proteins required for error-prone processing of G·U mispairs during SHM; however, the biochemical mechanisms are another matter. Recent data suggest that mutations at A/T sites occur within 30–50 nucleotides of G·U sites (18, 19), yet typical MMR-generated gaps can often be hundreds of nucleotides long. Monoubiquitination of the sliding clamp proliferating cell nuclear antigen at Lys164 is needed for pol η to generate A/T mutations during error-prone gap-filling synthesis (20), but the mechanism remains unresolved. There may also be competition between MMR and BER to gain access to a G·U mispair, thereby initiating a long or short repair gap, respectively, but this is also uncertain (20, 21).
AID-induced mutations, needed for antibody maturation, are typically deleterious when introduced into non-Ig genes. AID-catalyzed deamination generates breaks in myc that allow translocations to occur with Ig genes in Burkitt lymphoma (22) and other proto-oncogenes in B-cell lymphomas (23). AID up-regulation in non-B-cells from viral infections (23) or hormonal changes (24) consistently correlates with cell transformation (Fig. 1A). With recent evidence that AID can deaminate genes outside the Ig locus (13), it is incumbent on B-cells to have properly segregated error-free and error-prone BER and MMR or suffer the inevitable cancerous consequences. This raises a broad scope of targeting questions involving the regulation of DNA repair pathways.
An approach using crude cell lysates to couple a G·U MMR assay with a genetic reversion assay may offer a biochemical entry point to address the processing of AID-generated G·U mismatches (25). Error-prone MMR was observed in lysates from activated B-cells and Ramos B-cells, but not from non-B HeLa cells. Mutations at A/T sites were 50% in tonsil cells and 25% in Ramos cells, which agrees with SHM distributions in vivo (2), compared with <5% in HeLa cells, which makes mutations almost exclusively at C/G sites (25). The future challenge is to use this type of assay to reconstitute error-prone MMR by identifying proteins from B-cell lysates that can switch MMR fidelity from high to low, with the aim of purifying the B-cell-specific proteins responsible for error-prone MMR.
Anti-HIV-1 Cytosine Deaminase Activity of Apo3G
Along with other Apo3 deaminases, Apo3G can confer innate immunity in a variety of cells against virtually any retrovirus or virus that replicates using an ssDNA intermediate, e.g. HIV-1 or hepatitis B (Fig. 1A) (1). After encapsidation into a budding HIV virion through an RNA and/or Gag nucleocapsid interaction, Apo3G acts in the cytoplasm of T-cells to inactivate HIV during reverse transcription of the viral RNA (1). Apo3G preferentially deaminates 5′-YCC motifs (with the deamination site underlined) on the reverse-transcribed cDNA after the viral RNA template is degraded (1). Deamination requires ssDNA; therefore, synthesis of the second strand of DNA protects against further deamination but also creates large numbers of G → A mutations that effectively kill the virus. Alternatively, upon entry of the provirus to the nucleus, viral inactivation might occur by strand breakage at abasic sites caused by the removal of U.
Questions have arisen concerning whether Apo3G has a deamination-independent mode to inactivate HIV-1 because viral restriction has been reported using catalytically inactive Apo3G mutants (1). The latter data were generated in cell lines in which Apo3G is overexpressed. However, if less Apo3G is incorporated into the virion, as occurs in normal cell lines, then it appears that the sole means to restrict HIV is via deamination (26).
However, Apo3G does not get a free ride, for if it did, there would be no AIDS pandemic to deal with. To gain entry into the virion, Apo3G must first avoid proteasomal degradation mediated by the HIV-encoded Vif, which binds to Apo3G in its CD1 domain (Fig. 1B) and facilitates its polyubiquitination (1). A potential anti-HIV therapeutic strategy might be to screen for small molecules or peptides that interfere with Vif binding to Apo3G (1).
Structures of Apo2 and Apo3G CD2
High resolution structures have been determined for human Apo2 (27) and for the catalytic CD2 domain of Apo3G (Fig. 2) (28–30). A feature of APOBEC proteins required for catalytic activity is the presence of a Zn2+-coordinating motif (His-X-Glu-X23–28-Pro-Cys-X2–4-Cys) and bound water molecule. The closely positioned water molecule can be activated by Glu to become a zinc hydroxide for nucleophilic attack in the hydrolytic deamination reaction (31).
FIGURE 2.
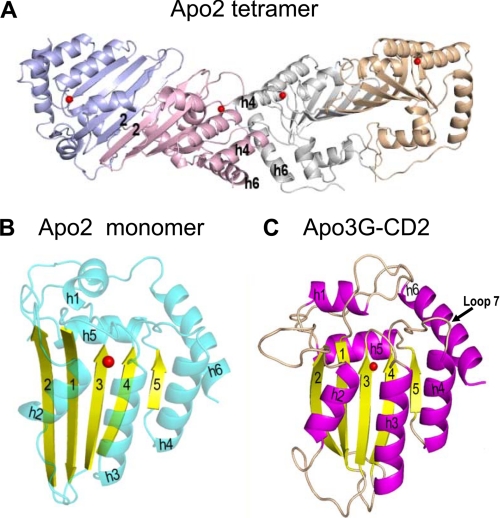
Comparison of Apo2 and Apo3G CD2 structures found by x-ray crystallography. Apo2 in crystal form appears as an elongated tetramer of identical polypeptide chains (Protein Data Bank code 2NYT) (A). The Apo2 polypeptide structure (B) shows α-helices (blue) and β-strands (yellow) arranged in much the same way as in the Apo3G CD2 structure (Protein Data Bank code 3E1U) (C), with its α-helices (red) and β-strands (yellow). In C, loop 7 is identified as the likely determinant of deamination motif specificity in Apo3G. The bound zinc ion is represented by a red dot. The figure was adapted from Ref. 28.
The x-ray crystal structure of Apo2, the first APOBEC structure to be determined (27), indicates a rod-shaped tetramer (Fig. 2A). Apo2 dimerizes through β-2 strand contacts and tetramerizes in a head-to-head orientation (Fig. 2A). The presence of α-helices h4 and h6 in Apo2 (Fig. 2A) prevents formation of the kind of square-shaped tetramer observed for free cytidine deaminases (27). Although human Apo2 has no known activity, it has been used successfully as a guide to understand how mutations in patients with HIGM-2 syndrome inactivate AID (27) and to model Apo3G residues important for Vif binding (32).
Recent x-ray and NMR studies of the catalytic Apo3G CD2 domain revealed that there is much similarity to the core secondary structure of Apo2, namely a five-stranded β-sheet core flanked by six α-helices (Fig. 2, B and C) (28–30). However, differences are observed in the active center loops, which are likely involved in ssDNA binding (Fig. 2, B and C) (28, 31).
The determinants of deamination specificity in the APOBEC family have been studied with AID, Apo3G, and also Apo3F. Mutants of Apo3G (28) and AID (33) designed from structural inferences and of Apo3F designed from amino acid homology comparisons with Apo3C (34) identified an 8–9-residue region for determining deamination specificity. This region is located at the position corresponding to loop 7 in Apo3G-(313–320) (Fig. 2C), AID-(113–123), and Apo3F-(305–313) (31). A wholesale change of mutational specificity can result by swapping all of these residues between APOBEC enzymes (33). From the perspective of the DNA, nucleoside analog substitutions in trinucleotide deamination motifs and nearby bases indicate that recognition of the 5′-CCC motif by Apo3G is determined by contacts with pyrimidine ring positions 3 and 4 one to two nucleotides 5′ of the target C (35).
AID and Apo3G Subunit Composition
Apo2 crystallized in the form of a tetramer, whereas in solution, dimers and tetramers were identified by size exclusion chromatography (27). Apo3G CD2 crystallized as a monomer (28) and is also a monomer according to molecular exclusion chromatography (36). Using low resolution small angle x-ray scattering, full-length Apo3G appears to dimerize in an elongated shape roughly similar to the Apo2 tetramer (37). Atomic force microscopy data show that Apo3G is present in four forms (monomer, dimer, trimer, tetramer), where the relative amount of each form depends on the presence or absence of ssDNA substrate and on the concentration of salt (38).
The main message conveyed by the data is that a combination of Apo3G/ssDNA/salt conditions may determine the form(s) of the enzyme in vitro and perhaps in vivo. Chemical cross-linking indicates that Tyr124 and Trp127 are involved in RNA-facilitated dimerization at the N-terminal interface (39), suggesting that interface mutant W127A is excluded from HIV virions because of an inability to bind RNA (40). However, co-immunoprecipitation and in-cell quenched Förster resonance energy transfer observations suggest that dimerization may occur primarily at the C-terminal interface, using residues 209–336, and may not be influenced by the presence of ssDNA or RNA (41). Because larger oligomeric states have also been observed by atomic force microscopy, e.g. tetramers, perhaps oligomerization may occur in various orientations (38), head-to-head, tail-to-tail, and maybe even head-to-tail.
The biochemical data strongly indicate that Apo3G oligomer formation in solution is a dynamic process. The same conclusions likely pertain to all of the APOBEC proteins, including AID. Of course, the central questions are what forms are present in vivo, and if there is more than a single dominant form, what are the roles for each?
AID and Apo3G DNA-scanning Mechanisms
AID and Apo3G catalyze multiple C deaminations on ssDNA (5, 38). Enzymes that scan DNA are typically designed to search for the proverbial “needle-in-a-haystack,” looking for a rare target such as an aberrant DNA base or a restriction site. In contrast, AID (5) and Apo3G3 encounter numerous deamination motifs but stochastically deaminate cytosines, creating small clusters of Us separated by stretches of non-deaminated DNA. When using AID in a model T7 RNA polymerase transcription system, stochastic processive deaminations are present (6, 7). In the case of Apo3G, deaminations from virions containing one or a few molecules were also in local clusters of the sequenced env region, consistent with processive deaminations (26). The diversity in the deamination spectra is in keeping with the need to generate numerous different types of mutations to neutralize the potentially ”staggering“ number of different antigen molecules, in the case of AID, and to incapacitate a rapidly evolving HIV, in the case of Apo3G. However, similar to dsDNA-scanning enzymes, such as UNG and EcoRV, AID and Apo3G scan DNA processively via combinations of sliding and jumping (or hopping) (6, 38).
Use of MMR- and BER-deficient mice (ung−/−msh2−/−) has provided a connection between in vivo mutagenesis and AID scanning on ssDNA in vitro. Sequencing of DNA clones containing an IgV gene (Jh4 intron) region revealed heterogeneous patterns of closely clustered deaminations separated by relatively long segments containing numerous non-deaminated C sites (42). A recent study quantified the targeting of purified AID acting on the non-transcribed or transcribed strand of the Jh4 intron DNA (43). The deaminations were compared on a site-by-site basis with mutations on the same DNA in mice defective both in MMR and BER, so mutations are caused solely by AID. A high correlation (r ∼ 0.7) between the biochemical and mutational data on the non-transcribed strand, but not on the transcribed strand (r ∼ 0.3), indicates that the inherent enzymatic properties of AID remain highly visible despite the large number of factors operating on the IgV region in vivo, e.g. AID-interacting proteins, transcription, nucleosome positioning, and many more. Of course, in the presence of DNA repair, AID-initiated mutations are strongly attenuated. It also seems likely that excessive deaminations might impose too high a mutational load in germinal center B-cells, so cells that have acquired nonproductive mutations may be swept from the immune system via apoptosis (44).
In contrast to AID, which shows no spatial bias, Apo3G deaminations favor the 5′-direction on ssDNA substrates in the presence of salt (45). Perhaps with two DNA-binding domains per polypeptide chain, in addition to sliding and jumping, Apo3G could remain bound while wrapping the DNA (38) or may engage in intersegmental transfer (46), either of which could impose a deamination bias (38). One point seems certain, that the bias requires the positively charged CD1 domain; by itself, the catalytic CD2 domain is neither directional nor processive irrespective of salt (28). Consistent with the biochemical data, a 5′ → 3′ gradient of G → A hypermutation in regions of the HIV genome has been observed in vivo (47) and may reflect a combination of the time that the cDNA remains single-stranded during reverse transcription (47) and local gradients from the intrinsic deamination polarity of Apo3G (38, 47).
Compendium of Loose Ends
AID and Apo3G are just 2 of 11 APOBEC family members, of which 9 have been shown to catalyze C → U on ssDNA (1). Like AID, these C deaminases, when acting in the wrong place at the wrong time, can create chaos in the cell. Compared with AID, less is known about the effects of Apo3 enzymes on cell transformation. Apo3 enzymes are overexpressed in many lymphoma tissues (1) and are able to deaminate human papillomavirus, which could influence the development of tumors (48). Given their widespread distribution and biological importance, it is essential to determine how each of the APOBEC proteins are regulated in vivo and to relate the biochemical and structural properties of these deaminases to their biological function. Part of the regulation appears to involve RNA. When recombinant AID is isolated from baculovirus-infected insect cells or E. coli, and Apo3G or Apo3F is isolated from T-cells, the enzymes are associated with RNA (8, 49, 50). AID is barely active except when first treated with RNase (8). For Apo3G, the RNA is enriched with that of the Alu transposon. Apo3G-binding Alu RNA appears to restrict transposition (1). Is restricting transposons the functional origin of APOBEC enzymes? Finally, here is a remarkable new observation to “chew on.” DNA demethylation arising by the combined action of AID or Apo2a/b and a DNA glycosylase (Mbd4) has been observed by microinjection of an in vitro methylated dsDNA fragment into a zebrafish embryonic cell (3). Is it possible that some of the APOBEC proteins may be involved in programmed demethylation during vertebrate development?
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R37GM21422 and ES013192. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
P. Pham, L. Chelico, and M. F. Goodman, unpublished data.
- APOBEC
- apolipoprotein B mRNA-editing complex
- C
- cytosine
- U
- uracil
- A
- adenine
- T
- thymine
- G
- guanine
- ssDNA
- single-stranded DNA
- AID
- activation-induced cytidine deaminase
- SHM
- somatic hypermutation
- CSR
- class switch recombination
- IgV
- immunoglobulin variable gene or region
- S
- switch region
- UNG
- uracil glycosylase
- MMR
- mismatch repair
- BER
- base excision repair
- dsDNA
- double-stranded DNA
- HIGM-2
- hyper-IgM type 2
- pol
- polymerase
- RPA
- replication protein A
- HIV
- human immunodeficiency virus
- Vif
- viral infectivity factor.
REFERENCES
- 1.Chiu Y. L., Greene W. C. (2008) Annu. Rev. Immunol. 26, 317–353 [DOI] [PubMed] [Google Scholar]
- 2.Peled J. U., Kuang F. L., Iglesias-Ussel M. D., Roa S., Kalis S. L., Goodman M. F., Scharff M. D. (2008) Annu. Rev. Immunol. 26, 481–511 [DOI] [PubMed] [Google Scholar]
- 3.Rai K., Huggins I. J., James S. R., Karpf A. R., Jones D. A., Cairns B. R. (2008) Cell 135, 1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. (2000) Cell 102, 553–563 [DOI] [PubMed] [Google Scholar]
- 5.Pham P., Bransteitter R., Petruska J., Goodman M. F. (2003) Nature 424, 103–107 [DOI] [PubMed] [Google Scholar]
- 6.Bransteitter R., Pham P., Calabrese P., Goodman M. F. (2004) J. Biol. Chem. 279, 51612–51621 [DOI] [PubMed] [Google Scholar]
- 7.Shen H. M., Poirier M. G., Allen M. J., North J., Lal R., Widom J., Storb U. (2009) J. Exp. Med. 206, 1057–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bransteitter R., Pham P., Scharff M. D., Goodman M. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan H. D., Dean W., Coker H. A., Reik W., Petersen-Mahrt S. K. (2004) J. Biol. Chem. 279, 52353–52360 [DOI] [PubMed] [Google Scholar]
- 10.Wang M., Yang Z., Rada C., Neuberger M. S. (2009) Nat. Struct. Mol. Biol. 16, 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stavnezer J., Guikema J. E., Schrader C. E. (2008) Annu. Rev. Immunol. 26, 261–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., Tezcan I., Ersoy F., Kayserili H., Ugazio A. G., Brousse N., Muramatsu M., Notarangelo L. D., Kinoshita K., Honjo T., Fischer A., Durandy A. (2000) Cell 102, 565–575 [DOI] [PubMed] [Google Scholar]
- 13.Liu M., Duke J. L., Richter D. J., Vinuesa C. G., Goodnow C. C., Kleinstein S. H., Schatz D. G. (2008) Nature 451, 841–845 [DOI] [PubMed] [Google Scholar]
- 14.Blagodatski A., Batrak V., Schmidl S., Schoetz U., Caldwell R. B., Arakawa H., Buerstedde J. M. (2009) PLoS Genet. 5, e1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conticello S. G., Ganesh K., Xue K., Lu M., Rada C., Neuberger M. S. (2008) Mol. Cell 31, 474–484 [DOI] [PubMed] [Google Scholar]
- 16.Chaudhuri J., Khuong C., Alt F. W. (2004) Nature 430, 992–998 [DOI] [PubMed] [Google Scholar]
- 17.Pham P., Smolka M. B., Calabrese P., Landolph A., Zhang K., Zhou H., Goodman M. F. (2008) J. Biol. Chem. 283, 17428–17439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unniraman S., Schatz D. G. (2007) Science 317, 1227–1230 [DOI] [PubMed] [Google Scholar]
- 19.Frieder D., Larijani M., Collins C., Shulman M., Martin A. (2009) Mol. Cell. Biol. 10.1128/MCB.00647-00609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynaud C. A., Delbos F., Faili A., Guéranger Q., Aoufouchi S., Weill J. C. (2009) Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schanz S., Castor D., Fischer F., Jiricny J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5593–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbiani D. F., Bothmer A., Callen E., Reina-San-Martin B., Dorsett Y., Difilippantonio S., Bolland D. J., Chen H. T., Corcoran A. E., Nussenzweig A., Nussenzweig M. C. (2008) Cell 135, 1028–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okazaki I. M., Kotani A., Honjo T. (2007) Adv. Immunol. 94, 245–273 [DOI] [PubMed] [Google Scholar]
- 24.Pauklin S., Sernández I. V., Bachmann G., Ramiro A. R., Petersen-Mahrt S. K. (2009) J. Exp. Med. 206, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pham P., Zhang K., Goodman M. F. (2008) J. Biol. Chem. 283, 31754–31762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Browne E. P., Allers C., Landau N. R. (2009) Virology 387, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prochnow C., Bransteitter R., Klein M. G., Goodman M. F., Chen X. S. (2007) Nature 445, 447–451 [DOI] [PubMed] [Google Scholar]
- 28.Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., Chen X. S. (2008) Nature 456, 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Furukawa A., Nagata T., Matsugami A., Habu Y., Sugiyama R., Hayashi F., Kobayashi N., Yokoyama S., Takaku H., Katahira M. (2009) EMBO J. 28, 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harjes E., Gross P. J., Chen K. M., Lu Y., Shindo K., Nowarski R., Gross J. D., Kotler M., Harris R. S., Matsuo H. (2009) J. Mol. Biol. 389, 819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bransteitter R., Prochnow C., Chen X. S. (2009) Cell. Mol. Life Sci. DOI:10.1007/s00018-00009-00070-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K. L., Mangeat B., Ortiz M., Zoete V., Trono D., Telenti A., Michielin O. (2007) PLoS ONE 2, e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohli R. M., Abrams S. R., Gajula K. S., Maul R. W., Gearhart P. J., Stivers J. T. (2009) J. Biol. Chem. 284, 22898–22904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langlois M. A., Beale R. C., Conticello S. G., Neuberger M. S. (2005) Nucleic Acids Res. 33, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rausch J. W., Chelico L., Goodman M. F., Le Grice S. F. (2009) J. Biol. Chem. 284, 7047–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen K. M., Martemyanova N., Lu Y., Shindo K., Matsuo H., Harris R. S. (2007) FEBS Lett. 581, 4761–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wedekind J. E., Gillilan R., Janda A., Krucinska J., Salter J. D., Bennett R. P., Raina J., Smith H. C. (2006) J. Biol. Chem. 281, 38122–38126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chelico L., Sacho E. J., Erie D. A., Goodman M. F. (2008) J. Biol. Chem. 283, 13780–13791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huthoff H., Autore F., Gallois-Montbrun S., Fraternali F., Malim M. H. (2009) PLoS Pathogens. 5, e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huthoff H., Malim M. H. (2007) J. Virol. 81, 3807–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett R. P., Salter J. D., Liu X., Wedekind J. E., Smith H. C. (2008) J. Biol. Chem. 283, 33329–33336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rada C., Di Noia J. M., Neuberger M. S. (2004) Mol. Cell 16, 163–171 [DOI] [PubMed] [Google Scholar]
- 43.MacCarthy T., Kalis S. L., Roa S., Pham P., Goodman M. F., Scharff M. D., Bergman A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8629–8634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaheen A., Boulianne B., Parsa J. Y., Ramachandran S., Gommerman J. L., Martin A. (2009) Blood 114, 547–554 [DOI] [PubMed] [Google Scholar]
- 45.Chelico L., Pham P., Calabrese P., Goodman M. F. (2006) Nat. Struct. Mol. Biol. 13, 392–399 [DOI] [PubMed] [Google Scholar]
- 46.Nowarski R., Britan-Rosich E., Shiloach T., Kotler M. (2008) Nat. Struct. Mol. Biol. 15, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 47.Suspène R., Rusniok C., Vartanian J. P., Wain-Hobson S. (2006) Nucleic Acids Res. 34, 4677–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vartanian J. P., Guétard D., Henry M., Wain-Hobson S. (2008) Science 320, 230–233 [DOI] [PubMed] [Google Scholar]
- 49.Gallois-Montbrun S., Holmes R. K., Swanson C. M., Fernández-Ocaña M., Byers H. L., Ward M. A., Malim M. H. (2008) J. Virol. 82, 5636–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiu Y. L., Soros V. B., Kreisberg J. F., Stopak K., Yonemoto W., Greene W. C. (2005) Nature 435, 108–114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.