Abstract
Interferon-α (IFNα) has shown promise in the treatment of various cancers. However, the development of IFN resistance is a significant drawback. Using conditions that mimic in vivo selection of IFN-resistant cells, the RST2 IFN-resistant cell line was isolated from the highly IFN-sensitive Daudi human Burkitt lymphoma cell line. The RST2 cell line was resistant to the antiviral, antiproliferative, and gene-induction actions of IFNα. Although STAT2 mRNA was present, STAT2 protein expression was deficient in RST2 cells. A variant STAT2 mRNA, which resulted from alternative splicing within the intron between exon 19 and 20, was expressed in several human cell lines but at relatively high levels in RST2 cells. Most importantly, the RST2 line showed an intrinsic resistance to apoptosis induced by a number of chemotherapeutic agents (camptothecin, staurosporine, and doxorubicin). Expression of STAT2 in RST2 cells not only rescued their sensitivity to the biological activities of IFNs but also restored sensitivity to apoptosis induced by these chemotherapeutic agents. The intrinsic resistance of the RST2 cells to IFN as well as chemotherapeutic agents adds a new dimension to our knowledge of the role of STAT2 as it relates to not only biological actions of IFN but also resistance to chemotherapy-induced apoptosis.
IFN2α/β regulates a number of cellular responses, such as proliferation, differentiation, and development (1). Although IFN triggers the death of some tumor cells by inducing proapoptotic proteins (tumor necrosis factor-related apoptosis-inducing ligand, PKR, etc.), IFN also promotes cell survival through a nuclear factor κB-dependent pathway (2–4). IFN is used to treat various human malignancies (chronic myeloid leukemia, non-Hodgkin lymphomas, Kaposi sarcoma, hairy cell leukemia, multiple myeloma, and malignant melanoma), viral infections, as well as various other diseases (5, 6). However, only a fraction of patients are responsive to IFN therapy, and many patients eventually develop resistance after chronic IFN exposure. The underlying mechanism for IFN resistance is still unclear, but it is reasonable to suggest that genetic variation and selection during prolonged IFN exposure may reflect IFN signaling defects.
IFN binds to its cell surface receptor resulting in the activation of JAK1 and TYK2 nonreceptor protein-tyrosine kinases, which phosphorylate STAT proteins (7). Phosphorylated STAT1 and STAT2 in a complex with IRF9 bind to a conserved IFN stimulus-response element (ISRE) present in the promoters of hundreds of IFN-stimulated genes (ISGs) inducing their expression. Mutant cell lines with defined signaling defects have made significant contributions in elucidating the IFN-activated JAK/STAT signal transduction pathway (7, 8). Such IFN-resistant mutants were isolated after multiple rounds of chemical mutagenesis and selection of IFN-resistant mutants. However, this procedure does not mimic in vivo what happens to patients who are subjected to long term IFN treatment. Moreover, these IFN-resistant cells have undergone multiple mutational events, because complementation of a single defect rescues aspects of IFN signaling but not sensitivity to all of the biological actions of IFN (8, 9). As an alternative approach, our laboratory has resorted to long term IFN treatment of cells to isolate naturally arising IFN-resistant mutants, which more closely resemble what occurs in vivo (10, 11). Using this strategy, we previously identified a STAT3-defective IFN-resistant cell line (11).
In this study, a mutant cell line (RST2) that was highly resistant to the antiviral, antiproliferative, and gene-inducing actions of IFN was isolated from the highly IFN-sensitive Daudi cell line by growth in the continuous presence of IFN. Sequencing of STAT2 mRNA identified an alternative splice site between exon 19 and 20 that is expressed in RST2 cells, causing translation termination at the beginning of the Src homology 2 domain of STAT2. Expression of STAT2 in RST2 cells rescued sensitivity to the antiviral, antiproliferative, and gene-inducing actions of IFN. Furthermore, although RST2 cells are intrinsically resistant to the induction of apoptosis by a variety of chemotherapeutic agents, reconstitution of STAT2 restored sensitivity to chemotherapy-induced apoptosis.
MATERIALS AND METHODS
Biological Reagents and Cell Culture
Recombinant human IFNα (IFNCon1) was generously provided by Dr. L. Blatt of InterMune (Brisbane, CA). Antibodies against the following proteins were used: STAT1, phospho-STAT1, STAT2, IRF9, and actin (Santa Cruz Biotechnology, Santa Cruz, CA); STAT3 (BD Biosciences); phospho-STAT2 and phospho-STAT3 (Upstate, Charlottesville, VA); and poly(ADP-ribose) polymerase (PARP) (Cell Signaling, Danvers, MA). Daudi and RST2 cells were maintained at ∼5 × 105 cells/ml in RPMI 1640 medium containing 10% defined bovine calf serum (Hyclone Laboratories, Logan, UT). Camptothecin, doxorubicin, and staurosporine were from Sigma.
Antiproliferative and Antiviral Assays
For antiproliferative assays, cells (1 × 105 cells/ml) were treated with the indicated IFNα concentrations. Cell numbers were determined after 72 h of IFNα treatment on a Beckman-Coulter counter (12). For antiviral activity, cells (1 × 105 cells/ml) were treated with the indicated IFNα concentrations for 24 h, followed by infection with vesicular stomatitis virus (VSV) for 1.5 h at 0.1 plaque-forming unit per cell. The virus yield in the medium was assayed by plaque formation at 24 h post-infection on Vero cells (3).
Preparation of Cell Lysates and Nuclear Proteins
Cells were washed twice with ice-cold phosphate-buffered saline, and lysates were prepared by incubation in lysis buffer (30 min, 4 °C) containing 50 mm Tris (pH 7.4), 150 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 15% glycerol, 1 mm NaF, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml soybean trypsin inhibitor, 5 μg/ml leupeptin, and 1.75 μg/ml benzamidine. Whole cell lysates were precleared by centrifugation (12,000 × g, 15 min). Nuclear proteins were prepared using CelLytic NuCLEAR extraction kit (Sigma). In brief, nuclei were isolated by lysing cells on ice for 20 min in lysis buffer containing 10 mm HEPES (pH 7.9), 1.5 mm MgCl2, 15 mm KCl, 1 mm dithiothreitol, 0.6% Nonidet P-40, 1 mm NaF, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml soybean trypsin inhibitor, 5 μg/ml leupeptin, and 1.75 μg/ml benzamidine; and centrifuged (10,000 × g, 30 s). Nuclei were washed with lysis buffer, and nuclear proteins were extracted with buffer containing 20 mm HEPES (pH 7.9), 1.5 mm MgCl2, 420 mm NaCl, 0.2 mm EDTA, 1 mm dithiothreitol, and 25% glycerol with 1 mm NaF, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml soybean trypsin inhibitor, 5 μg/ml leupeptin, and 1.75 μg/ml benzamidine (13). Protein was determined by the Bradford method (Bio-Rad).
Immunoblot Analysis
Total cell lysates (25 μg) were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore), and immunoblotted with the indicated antibodies, followed by anti-mouse or -rabbit IgG coupled with horseradish peroxidase (Santa Cruz Biotechnology). Blots were visualized by enhanced chemiluminescence (Pierce).
DNA Binding Activity Assays
Nuclear extracts (10 μg) were incubated with 32P-labeled probes for the highly conserved ISRE in the promoter region of ISG15 (5′-GATCCATGCCTCGGGAAAGGGAAACCGAAACTGAAGCC-3′) or the high affinity c-sis inducible element (SIE) in the c-fos gene (5′-AGCTTCATTTCCCGTAATCCCTAAAGCT-3′), respectively, and subjected to electrophoretic mobility shift assay (EMSA) (11). To define the presence of specific STAT proteins in DNA-protein complexes, nuclear extracts were preincubated with a 1:50 dilution of anti-STAT antibodies at 25 °C for 20 min before EMSA. Bands were quantified by phosphorimage autoradiography.
Quantitative Real Time PCR (qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen), and qPCR was performed on an iCyclerIQ (Bio-Rad) using iScript One-Step RT-PCR kit with SYBR Green (Bio-Rad). Reaction parameters were as follows: cDNA synthesis at 50 °C for 20 min, transcriptase inactivation at 95 °C for 5 min, PCR cycling at 95 °C for 10 s, and 60 °C for 30 s for 40 cycles. The following primers were used for qPCR: β-actin, 5′-AGAAGGAGATCACTGCCCTG-3′ (forward) and 5′-CACATCTGCTGGAAGGTGGA-3′ (reverse); GBP1, 5′-AGGAGTTCCTTCAAAGATGTGGA-3′ (forward) and 5′-TTCTGAACAAAGAGACGATAGCC-3′ (reverse); ISG15, 5′-TCCTGGTGAGGAATAACAAGGG-3′ (forward) and 5′-GTCAGCCAGAACAGGTCGTC-3′ (reverse); IFIT1, 5′-TCAGGTCAAGGATAGTCTGGAG-3′ (forward) and 5′-AGGTTGTGTATTCCCACACTGTA-3′ (reverse); STAT2, 5′-TCGGGCAGAACTGTAGGACT-3′ (forward) and 5′-ACCTCCTTCGTGTACGGTTG-3′ (reverse); STAT2 splicing variant, 5′-TCGGGCAGAACTGTAGGACT-3′ (forward) and 5′-ACCTTCCGAGGGTTTGAAGT-3′ (reverse); and total STAT2, 5′-CACCAGCTTTACTCGCACAG-3′ (forward) and 5′-TGGAAGAATAGCATGGTAGCCT-3′ (reverse).
RNA Preparation and Microarray Analysis
Total cellular RNA was extracted with TRIzol reagent (Invitrogen) (14) and submitted to Genome Explorations Inc. (Memphis, TN) for labeling and hybridization to human U133A version 2.0 GeneChips (Affymetrix Inc.). Expression values were determined using Affymetrix Microarray Suite 5.0 software. All data analysis was performed using GeneSpring software 7.0 (Silicon Genetics, Inc.), and expression values for each gene were normalized as described previously (14). The average fold-change in gene expression from three independent sets of GeneChip data for Daudi and RST2 cells was subjected to nonparametric t testing.
Apoptosis Assay
Cells were treated with the indicated drug for 24 h. The induction of apoptosis was monitored by immunoblotting whole cell extracts for the caspase-dependent PARP cleavage or DNA fragmentation using the cell death detection ELISAPLUS (Roche Applied Science).
Plasmid Constructs and Transfection Conditions
To determine the role of STAT2, RST2 cells were transfected with empty vector pEGFP-N1 (Clontech) or green fluorescent protein-tagged STAT2 expression vector pEGFP-STAT2 (generously provided by Dr. Nancy Reich, University of Stony Brook, NY) (15). Transfection was accomplished by electroporation (capacitance 300 microfarads, 250 V) with 50 μg of salmon sperm DNA and 20 μg of plasmid DNA for each sample (4). Three rounds of cell sorting resulted in the isolation of stable green fluorescent protein-positive cells.
RESULTS
Characterization of IFN-resistant RST2 Cells
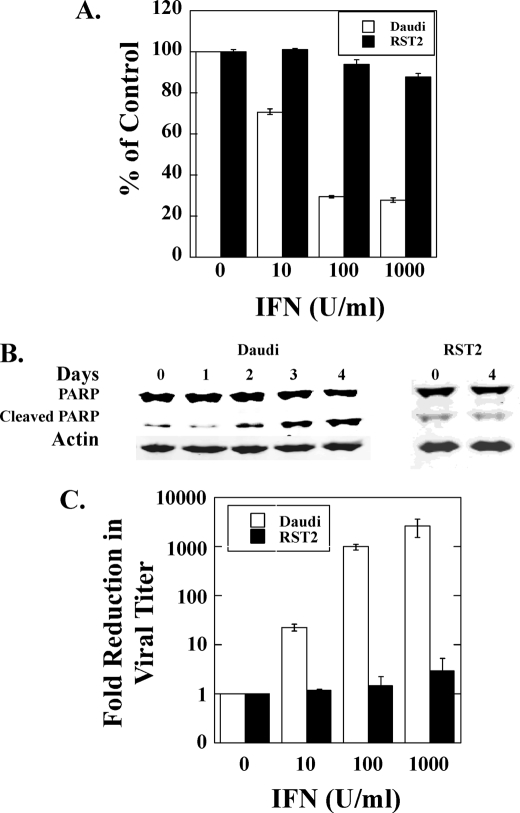
To understand the mechanisms that underlie the clinical resistance of patients after prolonged IFN exposure, we employed the highly IFN-sensitive Daudi human lymphoblastoid cell line. As shown in Fig. 1, Daudi cell proliferation was inhibited by ∼70% at an IFN concentration of 100 IU/ml, and cell death was detected in IFN-treated cells by day 2 as determined by the appearance of the p85 product of cleaved PARP. To isolate IFN-resistant cells, Daudi cells were maintained in the continuous presence of IFN at a concentration of 100 IU/ml. During the first 2 weeks of IFN treatment, massive cell death was observed, and only a small fraction of IFN-treated Daudi cells survived. The surviving cells, maintained in the continuous presence of IFN (100 IU/ml) for another month, grew at the same rate as untreated parental Daudi cells. The cell culture was then treated sequentially for 1 month with IFN at 1000 IU/ml and an additional month at 5000 IU/ml, during which IFN-induced inhibition of cell proliferation and apoptosis were not detected. After culture in the absence of IFN for several months, the proliferation of these cells (denoted as RST2 cells) was unaffected by IFN at 1000 IU/ml, and no IFN-induced apoptosis was observed (Fig. 1, A and B).
FIGURE 1.
IFN induced antiproliferative, apoptotic, and antiviral activities in Daudi and RST2 cells. A, Daudi and RST2 cells were treated with the indicated concentrations of IFNα. At day 3, cells were counted in a Coulter counter. B, Daudi and RST2 cells were treated with 100 IU/ml IFNα, and apoptosis was assayed for cleaved PARP by immunoblotting. C, Daudi and RST2 cells were treated overnight with the indicated concentrations of IFNα. The cells were then infected with VSV at 0.1 plaque-forming unit/cell, and the virus yield in the medium was determined by plaque formation.
IFN inhibits the replication of a wide range of DNA and RNA viruses. However, cells resistant to the antiproliferative activity of IFN can retain sensitivity to the antiviral activity of IFN (16). We next examined the sensitivity of RST2 cells to the antiviral activity of IFN against VSV infection. As shown in Fig. 1C, treatment with as little as 10 IU/ml of IFN reduced VSV production by more than 10-fold, and 1000 IU/ml reduced viral titer by >3 logs in Daudi cells. In contrast, treatment with 1000 IU/ml IFN did not reduce the VSV production in RST2 cells. Taken together these results demonstrate that RST2 cells are resistant to IFN-induced antiproliferative, antiviral, and apoptotic activities.
Identification of a JAK/STAT Signaling Defect in RST2 Cells
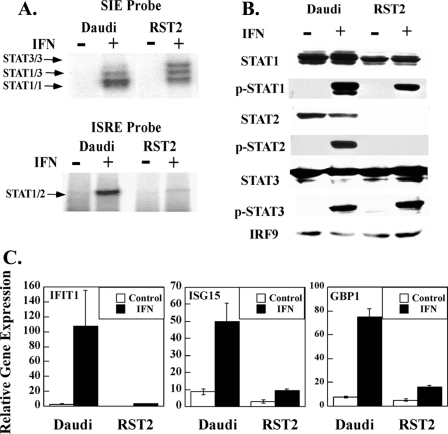
RST2 cells are highly IFN-resistant, suggesting that they may have a defective IFN-signaling pathway. IFN signals through the activation of the IFN-stimulated gene factor 3 (ISGF3) complex, composed of STAT1, STAT2, and IRF9 (17), as well as STAT1 and STAT3 dimers that bind to ISG promoter elements (11). As shown in Fig. 2A using the SIE probe, IFN induces distinctive formation of STAT1, STAT3, and STAT1/STAT3 dimer complexes in both Daudi and RST2 cells. The relative level of STAT1 dimers in Daudi cells is higher than in RST2 cells. In contrast, as shown in Fig. 2A using the ISRE probe there was barely detectable activation of ISGF3 in RST2 cells, although there was clear activation in Daudi cells, suggesting a defective ISGF3-signaling pathway in RST2 cells. To further characterize this defect, cell lysates were immunoblotted with STAT and IRF9 antibodies. Fig. 2B shows that STAT3 and IRF9 were expressed at similar levels in Daudi and RST2 cells. Although RST2 cells express somewhat lower STAT1 levels, STAT2 was undetectable in these cells as compared with Daudi cells. Moreover, IFN induced the tyrosine phosphorylation of STAT1 and STAT3 in Daudi and RST2 cells at levels reflecting their protein expression. These results localize the defect in RST2 cells at the level of the STAT2 protein.
FIGURE 2.
RST2 cells have a defect in ISGF3 activation by IFN. A, nuclear proteins were prepared from Daudi and RST2 cells treated with IFNα (1000 IU/ml) for 30 min and then subjected to electrophoretic mobility shift assays with a 32P-labeled SIE or ISRE probe. STAT complexes were identified by supershifts with anti-STAT1, -STAT2, or -STAT3 (data not shown). B, total cell lysates were prepared from Daudi and RST2 cells treated with IFNα (1000 IU/ml) for 15 min and immunoblotted with indicated antibodies. C, qPCR was performed on cDNA prepared from Daudi and RST2 cells treated with IFNα for 5 h. Gene expression was normalized to actin expression in each sample and presented as mean ± S.D. (n = 3).
Because STAT2 plays a critical role in regulating ISG expression, we examined the effect of IFN on several ISGs that are highly induced in Daudi cells (18). As shown in Fig. 2C, whereas IFIT1, ISG15, and GBP1 were well induced by IFN treatment in Daudi cells (50–120-fold), these ISGs were poorly induced in RST2 cells (<3-fold).
Identification of a STAT2 mRNA Splicing Variant in RST2 Cells
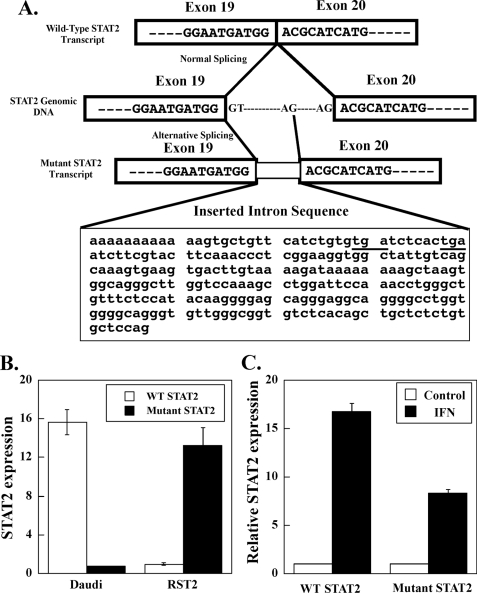
Although STAT2 protein was not detected in RST2 cells, we found similar expression levels of STAT2 mRNA in Daudi and RST2 cells by microarray and qPCR analysis. We therefore examined whether there were mutations in the STAT2 gene that could affect STAT2 protein expression. The human STAT2 gene is located on human chromosome 12 and contains 23 exons. To test for STAT2 mutations, we carried out PCR using probes corresponding to nucleotides 1–770, 770–1448, and 1448–2556 of the STAT2 open reading frame. An extra ∼200-bp insertion was found in STAT2 mRNA isolated from RST2 cells between bases 1448 and 2556. Sequencing the open reading frame of STAT2 cDNA identified a 247-bp sequence inserted in the mRNA (Fig. 3A), resulting in the early termination of STAT2 protein at the beginning of the Src homology 2 domain. A search of the inserted 247-bp sequence shows that this sequence originates from the intron between exon 19 and 20. As depicted in Fig. 3A, the mutant STAT2 mRNA apparently results from the use of an alternative splice site. Sequencing the genomic DNA from RST2 cells between exon 19 and 20 showed that the DNA sequence was identical in DNA isolated from Daudi and RST2 cells, indicating that a genomic mutation was not responsible for the presence of the splice variant in RST2 cells. As shown in Fig. 3B, qPCR analysis of wild-type and mutant STAT2 transcripts showed that >90% of the STAT2 transcripts in Daudi cells were wild-type STAT2, although in RST2 cells ∼90% were mutant STAT2 transcripts. This splice variant of STAT2 was present in other human cells (Daudi, MCF7, and HUH7) as determined by qPCR, albeit at lower levels relative to RST2 cells (data not shown). Both wild-type STAT2 mRNA and the splicing variant were up-regulated by IFN treatment of Daudi cells (Fig. 3C).
FIGURE 3.
Alternative splice sites identified in the STAT2 gene of RST2 cells. A, schematic depiction of normal and alternative splicing in STAT2 transcripts. The inserted intron sequence is shown in the box. Stop codons are underlined. B, qPCR was performed on cDNA prepared from Daudi cells and RST2 cells. Expression of wild-type (WT) and mutant STAT2 transcripts was normalized to actin expression and presented as mean ± S.D. (n = 3). C, qPCR was performed on cDNA prepared from Daudi cells treated with IFNα for 5 h. The IFN-induced levels of wild-type and mutant STAT2 transcripts were presented relative to untreated cells as mean ± S.D. (n = 3).
IFN-resistant RST2 Cells Are Resistant to Several Chemotherapeutic Agents
To gain insight into other alterations in RST2 cells, we performed genome-wide expression analysis using Affymetrix DNA microarray technology. RNA was isolated from Daudi or RST2 cells and processed for hybridization to human U133A version 2.0 GeneChips, which probe 14,500 human genes. We compared the gene expression levels between Daudi cells and RST2 cells. Statistical analysis using Wilcoxin 2 sample rank test revealed 168 annotated genes whose expression levels were significantly (p < 0.05) different in the RST2 cells as compared with Daudi cells. The expression of 78 genes was down-regulated from 2- to 39-fold, and the expression of 24 genes was up-regulated from 2- to 6-fold (supplemental Table). Consistent with our qPCR data, the level of the STAT2 transcript was similar in RST2 and Daudi cells.
To further characterize the biological role of STAT2, we analyzed the differentially regulated genes in RST2 cells by using the Gene Ontology (GO) classification. As shown in Table 1, a number of genes involved in apoptosis was down-regulated in RST2 cells as compared with Daudi cells, such as PKR (EIF2AK2), STAT1, PML, MCL1, as well as genes that play a role in sterol/steroid metabolism (p < 0.004) and fatty acid β-oxidation (p < 0.01). Steroid metabolism is implicated in multidrug resistance especially against various chemotherapeutic agents (19, 20).
TABLE 1.
Genes related to apoptosis affected in RST2 cells relative to Daudi cells
Apoptotic genes were defined according to Gene Ontology classification.
| Ratio of RST2 to Daudi | |
|---|---|
| Genes down-regulated in RST2 cells | |
| CSTA | −4.8 |
| DYRK2 | −2.0 |
| EIF2AK2 (PKR) | −3.5 |
| HDAC2 | −2.0 |
| IFI16 | −2.1 |
| IFI6 | −7.1 |
| ITGA4 | −2.5 |
| MCL1 | −2.3 |
| NFKBIA | −2.0 |
| NMI | −2.7 |
| OAS1 | −3.1 |
| OAS2 | −4.4 |
| PML | −16.5 |
| RRAS2 | −2.0 |
| STAT1 | −4.2 |
| TNFSF10 | −2.2 |
| Genes up-regulated in RST2 cells | |
| REL (c-Rel) | 2.9 |
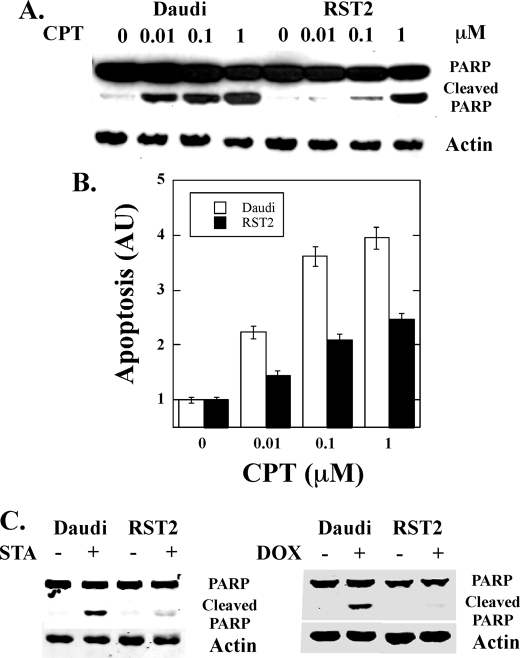
Because the data in Fig. 1 demonstrated that RST2 cells were resistant to IFN-induced apoptosis, we next examined the sensitivity to apoptosis induced by chemotherapeutic agents. Daudi and RST2 cells were treated with the indicated concentrations of camptothecin (CPT), and apoptosis was monitored by immunoblotting for the 89-kDa caspase-3 cleavage product of PARP or DNA fragmentation by enzyme-linked immunosorbent assay. As shown in Fig. 4, A and B, both PARP cleavage and DNA fragmentation were detected in Daudi cells treated with 0.01 μm CPT, reaching a maximal PARP cleavage at 1 μm CPT. In contrast, almost no PARP cleavage and DNA fragmentation were detected in RST2 cells at 0.1 μm CPT, and the levels of PARP cleavage and DNA fragmentation at 1 μm CPT in RST2 cells were similar to that induced at 0.01 μm CPT in Daudi cells. These results suggest that RST2 cells were ∼100-fold less sensitive to CPT-induced apoptosis than Daudi cells. Moreover, as shown in Fig. 4C, RST2 cells are more resistant to the induction of apoptosis by staurosporine and doxorubicin as compared with Daudi cells.
FIGURE 4.
Resistance of RST2 cells to chemotherapeutic drugs induced cell death. A and B, Daudi and RST2 cells were treated with the indicated concentrations of CPT for 24 h, and apoptosis was assayed for PARP cleavage by immunoblotting (A) or DNA fragmentation by enzyme-linked immunosorbent assay (B). AU, absorbance units. C, Daudi and RST2 cells were treated with 1 μm staurosporine (STA) and 1 μm doxorubicin (Dox) for 24 h, and apoptosis was assayed for PARP cleavage by immunoblotting.
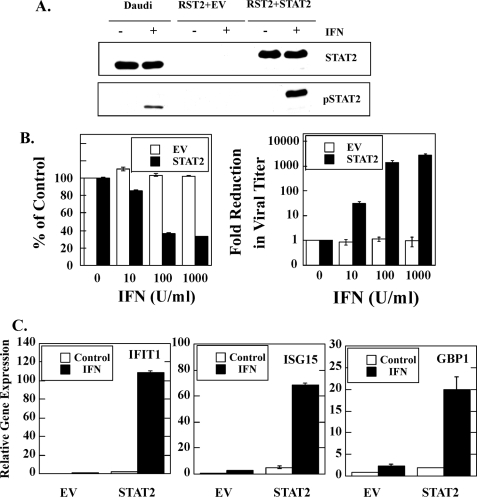
STAT2 Expression in RST2 Cells Rescues Sensitivity to IFN-induced Biological Activity and Sensitizes Cells to Chemotherapy-induced Cell Death
Our results identified STAT2 as a major defect in the RST2 cells; however, other defects in the IFN-signaling pathway could be responsible for the altered phenotype of RST2 cells. We then determined whether STAT2 expression in RST2 cells restored sensitivity to the biological activities of IFN. Fig. 5A shows that stable STAT2 expression in RST2 cells (RST2+STAT2) rescues IFN-induced STAT2 tyrosine phosphorylation, as detected by blotting with anti-phospho-STAT2 antibody. IFN treatment does not induce the rapid tyrosine phosphorylation of STAT2 in RST2 cells transfected with empty vector (RST2+EV) but does in the parental Daudi cell line as expected. Thus, transfection with wild-type STAT2 restored “normal” STAT2 expression in RST2 cells and IFN-induced tyrosine phosphorylation of STAT2. To determine whether STAT2 rescues sensitivity to the antiproliferative action of IFN, RST2 cells stably expressing wild-type STAT2 or empty vector were treated with IFN, and antiproliferative assays were performed. RST2 cells transfected with empty vector were highly resistant to the antiproliferative action of IFN with little inhibition observed at IFN concentrations as high as 1,000 IU/ml (Fig. 5B). In contrast, IFN treatment inhibited the proliferation of STAT2-expressing RST2 cells at concentrations as low as 10 IU/ml, and near maximal inhibition of proliferation was observed at 100 IU/ml. Moreover, STAT2 expression in RST2 cells also rescued IFN-induced antiviral activity. Although IFN treatment did not reduce the viral titer in RST2 cells transfected with empty vector, IFN treatment of RST2 cells expressing wild-type STAT2 resulted in a 3 log reduction in viral titer at 100 IU/ml (Fig. 5B).
FIGURE 5.
Expression of STAT2 in RST2 cells rescued the IFN-induced biological activities. A, cells were treated with 1000 IU/ml IFNα for 15 min, and 50 μg of the total cell lysate was separated on 7.5% SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and immunoblotted with anti-STAT2 or anti-phospho-STAT2 antibodies. Experiments were performed in parallel with IFN-sensitive Daudi cells. B, antiproliferative and antiviral activities induced by IFN in STAT2-expressing RST2 cells were assayed as described in Fig. 1. C, IFN induction of IFIT1, ISG15, and GBP1 gene expression in RST2 cells reconstituted with STAT2. qPCR was performed on cDNA prepared from IFN-treated cells (1000 IU/ml for 5 h). Gene expression was normalized to β-actin and presented as mean ± S.D. (n = 3).
In addition, STAT2 expression in RST2 cells also rescued ISG expression. RST+EV and RST2+STAT2 cells were treated with IFN; total RNA was extracted, and the expression of IFIT1, ISG15, and GBP1 was determined. As shown in Fig. 5C, IFN induced the expression of these ISGs in RTS2 cells expressing STAT2 but not in RST2 cells transfected with the empty vector. Moreover, the IFN-induced expression levels in RST2+STAT2 cells are comparable with what is observed in parental Daudi cells (Fig. 2C). Thus, STAT2 expression in RST2 cells rescues sensitivity to antiproliferative, antiviral, apoptotic, and gene-inducing actions of IFN, demonstrating that the defective IFN response in RST2 cells resides in STAT2.
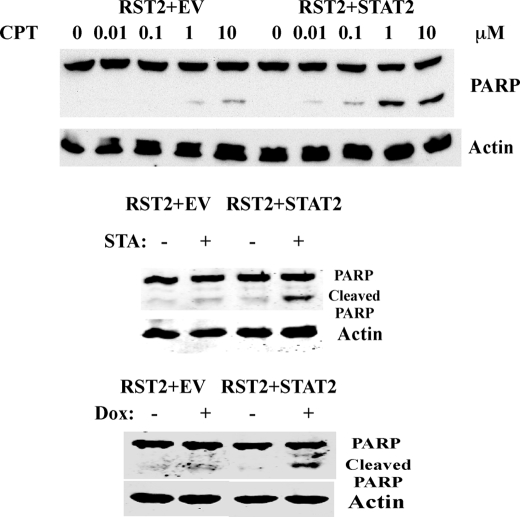
Finally, we examined the effect of STAT2 expression on the sensitivity of the RST2 cells to various chemotherapy-induced cell death. RST2 cells transfected with empty vector and RST2 cells expressing wild-type STAT2 were treated with camptothecin, staurosporine, or doxorubicin, and apoptosis was monitored by immunoblotting for the 89-kDa caspase-3 cleavage product of PARP. As shown in Fig. 6, expression of STAT2 in RST2 cells sensitized the cells to camptothecin-induced cell death. PARP cleavage was detected in RST2 cells expressing STAT2 at 0.01 μm CPT, reaching a maximal PARP cleavage at 1 μm CPT. In contrast, PARP cleavage was first detected in RST2 cells transfected with empty vector at 0.1 μm CPT, and the level of PARP cleavage at 10 μm CPT in RST2+EV cells was similar to that induced at 0.1 μm CPT in RST2+STAT2 cells. Thus, STAT2-expressing RST2 cells were ∼100-fold more sensitive to CPT-induced apoptosis when compared with EV-expressing cells. In addition, as shown in Fig. 6 RST2 cells expressing STAT2 are more sensitive to the induction of apoptosis by staurosporine and doxorubicin as compared with RST2 cells transfected with empty vector.
FIGURE 6.
Expression of STAT2 in RST2 cells restored the sensitivity to chemotherapeutic drugs. Cells were treated with indicated concentrations of CPT, 1 μm staurosporine (STA), and 1 μm doxorubicin (Dox) for 24 h, and total cell lysates were prepared, and apoptosis was assayed by immunoblotting for PARP cleavage.
DISCUSSION
IFNα is used to treat various cancers as well as viral infections. However, the development of resistance to IFN has been a significant drawback. In this study, we report the isolation of a STAT2-deficient IFN-resistant cell line by maintaining cells in the continuous presence of IFN. Our results suggest that the STAT2 protein could function not only as the classically defined IFN-activated transcription factor that regulates proliferation and antiviral functions of the cells but may also play a role in other cell functions such as apoptosis. The finding of an intrinsic drug resistance of these IFN-resistant cells adds a new dimension to our knowledge of the role of STAT2 in signaling pathways.
STAT2 was previously shown to be required for the IFN-induced STAT1 activation (21). In STAT2 null U6A cells, STAT1 is not activated by IFN but can be activated upon STAT2 reconstitution. In contrast we found that although STAT2 protein was not detected in RST2 cells, STAT1 was activated by IFN. The discrepancy may due to cell type-specific differences in the cells studied. U6A cells were derived from a fibrosarcoma, while our RST2 cells were isolated from a B-cell lymphoma. Similarly, STAT1 is activated in macrophages but not in fibroblasts isolated from STAT2 knock-out mice (22). These results indicate that the IFN-signaling pathway is cell type-dependent.
Different splicing products have been reported for various STAT proteins (23, 24). These splicing isoforms are usually truncated at the C terminus of the proteins and function differently from their normal isoforms. For example, the STAT1β isoform functions as a dominant negative for IFNα signaling (25). However, we were unable to detect the presence of a STAT2 isoform. Previous studies identified a STAT2 splicing variant mRNA that uses an alternative splicing site present in the intron between exon 20 and 21 (26). In contrast, we have identified an mRNA transcript resulting from alternative splicing in the intron between exon 19 and 20, which can be detected at low levels in B-lymphoblastoid, hepatoma, and breast cancer cells. In addition, we were unable to detect the STAT2 protein in RST2 cells using a number of monoclonal and polyclonal antibodies directed against various epitopes located throughout the STAT2 protein, suggesting that the STAT2 protein in RST2 cells is rapidly degraded or not translated.
Most importantly, we found that IFN-resistant RST2 cells were also resistant to the apoptotic action of various chemotherapeutic drugs. However, RST2 cells were as sensitive as parental Daudi cells to apoptosis induced by etoposide (data not shown), which differs from the other chemotherapeutic agents studied in that etoposide functions by binding topoisomerase II and forming a ternary complex with DNA (27, 28). Cancer-specific mutations in the components of the IFN-signaling pathway are believed to contribute to the failure of IFN as an effective antitumor agent (16, 29, 30). However, the relationship between IFN resistance and resistance to chemotherapeutic agents has not been well characterized. A previous study found that sensitivity to chemotherapeutic agents is coupled to IFN sensitivity, such that selection for resistance to one leads to resistance to the other (31). These results also have significant implications for IFN treatment of cancer patients, especially those who have developed IFN resistance during the course of treatment. The choice of alternative therapeutic agents in such patients should be carefully considered, because we found that RST2 cells are resistant to various chemotherapeutic agents.
Although STAT2 expression restored the sensitivity of RST2 cells to chemotherapeutic drugs, the underlying mechanism is presently unknown. However, as shown in Table 1 and supplemental Table several genes involved in the apoptotic pathway are down-regulated in RST2 cells. For example, the promyelocytic leukemia (PML) gene is the most down-regulated gene in RST2 cells. The PML gene encodes a tumor suppressor, and PML appears to be involved in multiple apoptotic pathways (32). Therefore, low PML expression in RST2 cells would be consistent with their resistance to apoptosis induced by various chemotherapeutic agents. Moreover, STAT1, which is down-regulated in RST2 cells, has been shown to mediate apoptosis in fibrosarcoma cells through regulation of the caspase pathway (33). However, the role of STAT2 in this pathway is unclear because STAT2 is necessary for STAT1 activation in fibrosarcoma cells. In addition, two important genes in the NF-κB pathway are affected in RST2 cells, i.e. Rel (c-Rel) is up-regulated and NFκB1A (IκBα) is down-regulated. The NF-κB pathway antagonizes the apoptotic actions of various chemotherapeutic agents (34–36). Both c-Rel up-regulation and IκBα down-regulation would be expected to promote NF-κB activity in RST2 cells, and hence make these cells relatively resistant to chemotherapeutic agents, which is what we observed.
In conclusion, our results expand upon the role that STAT2 plays in the classical IFN-signaling pathway, which leads to inhibition of cell proliferation and viral replication through ISG expression. We show that STAT2 also regulates a novel signaling pathway to regulate apoptosis by IFN and various chemotherapeutic agents. It is enticing to consider the possibility that this role of STAT2 is through regulation of gene expression in its well known role as a transcription factor. Consistent with this possibility, we observe the down-regulation of basal expression of a number of genes in RST2 cells that are known ISGs (PKR, STAT1, PML, and MCL1), as well as genes that play a role in sterol/steroid metabolism and fatty acid β-oxidation.
STAT2 is the only member of the STAT family that is exclusively used by a single cytokine, IFNα/β. It is of particular interest that we previously showed that STAT2 was “activated” as evidenced by its nuclear translocation by epidermal growth factor in intestinal epithelial cells (37). However, no STAT2 target genes were examined in that study. STAT proteins are usually considered to function upon their tyrosine phosphorylation and subsequent dimerization to regulate transcription. However, recent studies showed that STATs may play a broader role in gene regulation, as non-tyrosine-phosphorylated STAT proteins also appear to regulate gene transcription (38). In addition, STAT3 may act as an adapter for the activation of phosphatidylinositol 3-kinase (39) and nuclear factor κB (11). Moreover, a recent study indicates that STAT3 is also present in the mitochondria and functions in cellular respiration (40). Thus, in future studies it will be highly interesting to determine whether STAT2 functions in regulating the apoptotic response of cells via its role as a transcription factor and regulating the expression of apoptotic or anti-apoptotic genes, or is localized to another cellular compartment such as to the mitochondria and regulates apoptosis through other pathways.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA73753 (to L. M. P.). This work was also supported by funds from the Muirhead Chair Endowment at the University of Tennessee Health Science Center (to L. M. P.).

The on-line version of this article (available at http://www.jbc.org) contains a supplemental table.
- IFN
- interferon
- ISRE
- IFN-stimulus response element
- ISG
- IFN-stimulated gene
- SIE
- c-sis inducible element
- VSV
- vesicular stomatitis virus
- qPCR
- quantitative real time-PCR
- PARP
- poly (ADP-ribose) polymerase
- CPT
- camptothecin
- EV
- empty vector
- PML
- promyelocytic leukemia
- PKR
- protein kinase R.
REFERENCES
- 1.Platanias L. C. (2005) Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 2.Du Z., Wei L., Murti A., Pfeffer S. R., Fan M., Yang C. H., Pfeffer L. M. (2007) J. Cell. Biochem. 102, 1087–1094 [DOI] [PubMed] [Google Scholar]
- 3.Yang C. H., Murti A., Pfeffer S. R., Basu L., Kim J. G., Pfeffer L. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13631–13636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang C. H., Murti A., Pfeffer S. R., Kim J. G., Donner D. B., Pfeffer L. M. (2001) J. Biol. Chem. 276, 13756–13761 [DOI] [PubMed] [Google Scholar]
- 5.Pfeffer L. M., Dinarello C. A., Herberman R. B., Williams B. R., Borden E. C., Bordens R., Walter M. R., Nagabhushan T. L., Trotta P. P., Pestka S. (1998) Cancer Res. 58, 2489–2499 [PubMed] [Google Scholar]
- 6.De Clercq E. (2004) J. Clin. Virol. 30, 115–133 [DOI] [PubMed] [Google Scholar]
- 7.Darnell J. E., Jr., Kerr I. M., Stark G. R. (1994) Science 264, 1415–1421 [DOI] [PubMed] [Google Scholar]
- 8.Stark G. R. (1997) Harvey Lect. 93, 1–16 [PubMed] [Google Scholar]
- 9.Brierley M. M., Fish E. N. (2005) J. Biol. Chem. 280, 13029–13036 [DOI] [PubMed] [Google Scholar]
- 10.Pfeffer L. M., Donner D. B. (1990) Cancer Res. 50, 2654–2657 [PubMed] [Google Scholar]
- 11.Yang C. H., Murti A., Pfeffer L. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5568–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenkraft B. L., Nanus D. M., Albino A. P., Pfeffer L. M. (1991) Cancer Res. 51, 5881–5887 [PubMed] [Google Scholar]
- 13.Yang C. H., Shi W., Basu L., Murti A., Constantinescu S. N., Blatt L., Croze E., Mullersman J. E., Pfeffer L. M. (1996) J. Biol. Chem. 271, 8057–8061 [DOI] [PubMed] [Google Scholar]
- 14.Wei L., Sandbulte M. R., Thomas P. G., Webby R. J., Homayouni R., Pfeffer L. M. (2006) J. Biol. Chem. 281, 11678–11684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banninger G., Reich N. C. (2004) J. Biol. Chem. 279, 39199–39206 [DOI] [PubMed] [Google Scholar]
- 16.Pfeffer L. M., Wang C., Constantinescu S. N., Croze E., Blatt L. M., Albino A. P., Nanus D. M. (1996) J. Urol. 156, 1867–1871 [PubMed] [Google Scholar]
- 17.Schindler C., Fu X. Y., Improta T., Aebersold R., Darnell J. E., Jr. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7836–7839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang C. H., Wei L., Pfeffer S. R., Du Z., Murti A., Valentine W. J., Zheng Y., Pfeffer L. M. (2007) J. Immunol. 178, 986–992 [DOI] [PubMed] [Google Scholar]
- 19.Zelcer N., Reid G., Wielinga P., Kuil A., van der Heijden I., Schuetz J. D., Borst P. (2003) Biochem. J. 371, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mack J. T., Townsend D. M., Beljanski V., Tew K. D. (2007) Curr. Drug Metab. 8, 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung S., Qureshi S. A., Kerr I. M., Darnell J. E., Jr., Stark G. R. (1995) Mol. Cell. Biol. 15, 1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park C., Li S., Cha E., Schindler C. (2000) Immunity 13, 795–804 [DOI] [PubMed] [Google Scholar]
- 23.Yan R., Qureshi S., Zhong Z., Wen Z., Darnell J. E., Jr. (1995) Nucleic Acids Res. 23, 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong Z., Wen Z., Darnell J. E., Jr. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4806–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caldenhoven E., van Dijk T. B., Solari R., Armstrong J., Raaijmakers J. A., Lammers J. W., Koenderman L., de Groot R. P. (1996) J. Biol. Chem. 271, 13221–13227 [DOI] [PubMed] [Google Scholar]
- 26.Sugiyama T., Nishio Y., Kishimoto T., Akira S. (1996) FEBS Lett. 381, 191–194 [DOI] [PubMed] [Google Scholar]
- 27.Kingma P. S., Burden D. A., Osheroff N. (1999) Biochemistry 38, 3457–3461 [DOI] [PubMed] [Google Scholar]
- 28.Binaschi M., Bigioni M., Cipollone A., Rossi C., Goso C., Maggi C. A., Capranico G., Animati F. (2001) Curr. Med. Chem. Anticancer Agents 1, 113–130 [DOI] [PubMed] [Google Scholar]
- 29.Xu B., Grandér D., Sangfelt O., Einhorn S. (1994) Blood 84, 1942–1949 [PubMed] [Google Scholar]
- 30.Wong L. H., Krauer K. G., Hatzinisiriou I., Estcourt M. J., Hersey P., Tam N. D., Edmondson S., Devenish R. J., Ralph S. J. (1997) J. Biol. Chem. 272, 28779–28785 [DOI] [PubMed] [Google Scholar]
- 31.Khodarev N. N., Minn A. J., Efimova E. V., Darga T. E., Labay E., Beckett M., Mauceri H. J., Roizman B., Weichselbaum R. R. (2007) Cancer Res. 67, 9214–9220 [DOI] [PubMed] [Google Scholar]
- 32.Wang Z. G., Ruggero D., Ronchetti S., Zhong S., Gaboli M., Rivi R., Pandolfi P. P. (1998) Nat. Genet. 20, 266–272 [DOI] [PubMed] [Google Scholar]
- 33.Kumar A., Commane M., Flickinger T. W., Horvath C. M., Stark G. R. (1997) Science 278, 1630–1632 [DOI] [PubMed] [Google Scholar]
- 34.Rayet B., Gélinas C. (1999) Oncogene 18, 6938–6947 [DOI] [PubMed] [Google Scholar]
- 35.Wang C. Y., Cusack J. C., Jr., Liu R., Baldwin A. S., Jr. (1999) Nat. Med. 5, 412–417 [DOI] [PubMed] [Google Scholar]
- 36.Wang C. Y., Mayo M. W., Baldwin A. S., Jr. (1996) Science 274, 784–787 [DOI] [PubMed] [Google Scholar]
- 37.Johnson L. R., McCormack S. A., Yang C. H., Pfeffer S. R., Pfeffer L. M. (1999) Am. J. Physiol. 276, C419–C425 [DOI] [PubMed] [Google Scholar]
- 38.Yang J., Chatterjee-Kishore M., Staugaitis S. M., Nguyen H., Schlessinger K., Levy D. E., Stark G. R. (2005) Cancer Res. 65, 939–947 [PubMed] [Google Scholar]
- 39.Pfeffer L. M., Mullersman J. E., Pfeffer S. R., Murti A., Shi W., Yang C. H. (1997) Science 276, 1418–1420 [DOI] [PubMed] [Google Scholar]
- 40.Wegrzyn J., Potla R., Chwae Y. J., Sepuri N. B., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., Moh A., Moghaddas S., Chen Q., Bobbili S., Cichy J., Dulak J., Baker D. P., Wolfman A., Stuehr D., Hassan M. O., Fu X. Y., Avadhani N., Drake J. I., Fawcett P., Lesnefsky E. J., Larner A. C. (2009) Science 323, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.