Abstract
In mammals, a family of five acyl-CoA synthetases (ACSLs), each the product of a separate gene, activates long chain fatty acids to form acyl-CoAs. Because the ACSL isoforms have overlapping preferences for fatty acid chain length and saturation and are expressed in many of the same tissues, the individual function of each isoform has remained uncertain. Thus, we constructed a mouse model with a liver-specific knock-out of ACSL1, a major ACSL isoform in liver. Eliminating ACSL1 in liver resulted in a 50% decrease in total hepatic ACSL activity and a 25–35% decrease in long chain acyl-CoA content. Although the content of triacylglycerol was unchanged in Acsl1L−/− liver after mice were fed either low or high fat diets, in isolated primary hepatocytes the absence of ACSL1 diminished the incorporation of [14C]oleate into triacylglycerol. Further, small but consistent increases were observed in the percentage of 16:0 in phosphatidylcholine and phosphatidylethanolamine and of 18:1 in phosphatidylethanolamine and lysophosphatidylcholine, whereas concomitant decreases were seen in 18:0 in phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, and lysophosphatidylcholine. In addition, decreases in long chain acylcarnitine content and diminished production of acid-soluble metabolites from [14C]oleate suggested that hepatic ACSL1 is important for mitochondrial β-oxidation of long chain fatty acids. Because the Acsl1L−/− mice were not protected from developing either high fat diet-induced hepatic steatosis or insulin resistance, our study suggests that lowering the content of hepatic acyl-CoA without a concomitant decrease in triacylglycerol and other lipid intermediates is insufficient to protect against hepatic insulin resistance.
Acyl-CoA synthetase (ACSL)3 activates long chain fatty acid (FA) to acyl-CoA, thereby enhancing vectorial FA transport across the plasma membrane (1) and providing substrates for most downstream pathways that metabolize FA. ACSL1 is one of five ACSL isoforms, each encoded by a separate gene. Its mRNA expression is highest in adipose tissue, liver, and heart (2); and because Acsl1 mRNA and total ACSL1 activity increase 160-fold (3) and 100-fold (4), respectively, in differentiating 3T3-L1 adipocytes, ACSL1 has been thought to be important in activating FA destined for triacylglycerol (TAG) synthesis. In support of this idea, overexpressing ACSL1 in mouse heart increases cardiac myocyte TAG accumulation 12-fold and induces apoptotic pathways, cardiac hypertrophy, left ventricular dysfunction, and heart failure (5). However, Acsl1 mRNA expression is up-regulated in liver and adipose tissue by activators of peroxisome proliferator-activated factor α (PPARα) (6, 7) via a PPAR response element in the promoter region of Acsl1 (8), suggesting a possible function related to the β-oxidation of fatty acids. Moreover, overexpression of ACSL1 in rat primary hepatocytes increases oleate incorporation into diacylglycerol (DAG) but does not increase TAG mass (9). Thus, the exact role of ACSL1 in providing acyl-CoA for lipogenesis versus β-oxidation has remained uncertain.
It has been suggested that lipid intermediates, including FAs, long chain acyl-CoAs, DAG, ceramide, and phosphatidic acid, rather than TAG accumulation per se, might underlie the development of insulin resistance (10–13). For example, long chain acyl-CoAs directly affect glucose metabolism by allosterically inhibiting glycogen synthase, pyruvate dehydrogenase, glucose-6-phosphatase, and glucokinase (14, 15); and FAs or acyl-CoAs may also be ligands for hepatocyte nuclear factor 4α, a transcription factor that regulates aspects of lipoprotein and glucose metabolism (16). Further, DAG activates protein kinase C isoforms that can phosphorylate insulin receptor substrates on serine and threonine residues and thereby impair insulin signaling (10). However, alterations of acyl-CoAs do not always correlate with changes in insulin sensitivity (17). Although acyl-CoA content in muscle has been associated with diminished insulin sensitivity (18), in liver the relationship between acyl-CoAs and insulin sensitivity is less clear. For example, when lipoprotein lipase is overexpressed and FA flux into the liver from lipoprotein particles is increased, hepatic insulin resistance is associated with an increased hepatic content of TAG and acyl-CoA (19). However, when DAG acyltransferase-2 is overexpressed in liver, the mice retain normal hepatic and whole body insulin sensitivity despite hepatic steatosis and an elevated acyl-CoA content (20). Similarly, the absence of glycerol-3-phosphate acyltransferase 1 (Gpat1−/−) led to protection against hepatic insulin resistance despite a 64% increase in hepatic acyl-CoA content, arguing against the importance of long chain acyl-CoAs in promoting insulin resistance (17). Although a higher content of acyl-CoAs might contribute to insulin resistance in some models, it is not known whether protection against insulin resistance might be achieved by decreasing hepatic acyl-CoA content.
To understand the function of ACSL1 in liver, we constructed a mouse model with a liver-specific knock-out of ACSL1 (Acsl1L−/−). Eliminating ACSL1 in liver resulted in a 50% decrease in total hepatic ACSL activity and a 25–35% decrease in long chain acyl-CoA content. This model has allowed us to determine whether ACSL1 deficiency protects liver from hepatic steatosis and alters the incorporation of FAs into specific glycerolipids and to learn whether a decrease in long chain acyl-CoA content is sufficient to protect the liver from high fat diet-induced insulin resistance.
EXPERIMENTAL PROCEDURES
Generation of Acsl1L−/− Mice
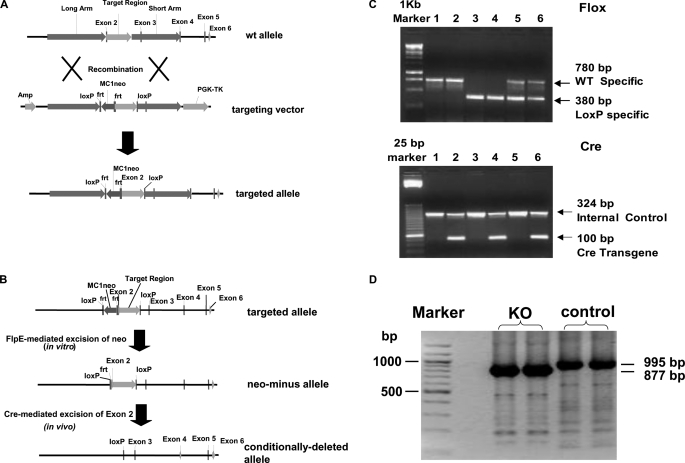
Liver-specific ACSL1 knock-out mice (Acsl1L−/−) were created by LoxP-Cre strategy (21). The gene-targeting vector was designed to produce a floxed Acsl1 exon 2 (Fig. 1A) and was constructed in a standard plasmid backbone containing neomycin phosphotransferase (neo) and thymidine kinase cassettes for positive and negative selection, respectively. The 5′ arm of homology was 4.5 kb in length and was derived from intron 1, whereas the 3′ arm was 3.8 kb in length and derived from a portion of intron 2, exon 3, and a portion of intron 3. Between the arms, a floxed exon 2 was cloned. The targeting vector was electroporated into E14Tg2A (E14) embryonic stem cells, and the cells were grown in medium supplemented with G418 and gancyclovir. Targeted cells were identified by PCR across both the 3′ and 5′ arms of homology using primers specific to neo and primers flanking the arms of homology. Neo was then excised from the targeted allele via transient expression of flpE recombinase (Fig. 1B). Targeted, neo-cells were microinjected into blastocysts derived from mouse strain C57BL/6 to produce transmitting chimeras. Twenty-one high quality chimeras were produced that transmitted the targeted allele. Duplex PCR was performed to distinguish wild-type and Flox alleles with wild-type specific primers (forward, 5′-AGCAAGCCACATGAAGGCATGTGTG-3′ and reverse, 5′-AAGTGGGGGACATAGGTGCCACT-3′) and LoxP-specific primers (forward, 5′-TAGAAAGTATAGGAACTTCGGCGCG-3′ and reverse, 5′-GCCCCTATATCACTTTTGGCGACA-3′). Mice heterozygous for the targeted allele (Acsl1Flox/+) were identified and back-crossed six times to C57BL/6 mice. The established Acsl1Flox/+ mice were then crossed with rat albumin promoter-Cre transgenic mice (Alb-CreTg/0), which express Cre recombinase exclusively in postpartum liver (22). The resulting double heterozygous mice carrying one floxed allele and Alb-Cre recombinase (Acsl1Flox/+-Alb-CreTg/0, i.e. ACSL1 heterozygote) were interbred to yield Acsl1L−/− mice (Acsl1Flox/Flox-Alb-CreTg/0), as well as five different groups of littermate controls (Fig. 1C). The presence or absence of the Alb-Cre transgene was determined by duplex PCR using Cre-specific primers (forward, 5′-GCGGTCTGGCAGTAAAAACTATC-3′ and reverse, 5′-GTGAAACAGCATTGCTGTCACTT-3′) and IL-2 as internal control for amplification (forward, 5′-CATGGCCACAGAATTGAAAGATCT-3′ and reverse, 5′-GTAGGTGGAAATTCTAGCATCATCC-3′) (Fig. 1C). To confirm the knock-out of exon 2 in Acsl1L−/− mice, total RNA was extracted (Qiagen RNeasy Mini kit) from Acsl1L−/− mice and control mice, and cDNA was synthesized by reverse transcription (Applied Biosystems). PCR was conducted with forward primer 5′-GCGGAGGAGAATTCTGCATAGAGAA-3′ and reverse primer 5′-ATATCAGCACATCATCTGTGGAAG-3′. PCR amplification of the region between coding exon 1 and exon 10 yielded a band of ∼1 kb in control mice and a smaller band of ∼900 bp in Acsl1L−/− mice (Fig. 1D). DNA sequencing (University of North Carolina Genome Analysis Facility) of the purified bands (QIAquick gel extraction kit) showed that in Acsl1L−/− mice, exon 1 was followed immediately by exon 3, proving that the shorter PCR product in Acsl1L−/− mice was due to the missing exon 2 (118 bp). The absence of exon 2 causes a frameshift and introduces a stop codon (TAG) immediately after exon 1, resulting a truncated peptide of 70 amino acids in the Acsl1L−/− mice compared with a 699-amino acid protein in the control mice (sequence not shown).
FIGURE 1.
Targeted disruption of the mouse ACSL1 gene in liver. A, ACSL1 conditional gene targeting. The targeting construct was designed to delete exon 2 of the ACSL1 gene when crossed to Cre-expressing animals. With excision of exon 2, splicing from exon 1 to downstream exons results in reading frame shifts and immediate translational stops. The positive selection marker (neo) was flanked by frt sites. Both neo and the target region were flanked by loxP sites. The vector also contained the thymidine kinase (TK) gene, allowing for positive-negative selection. B, ACSL1 FlpE/Cre-mediated excisions. Targeted cells were transiently transfected with a FlpE expression vector and screened for the loss of neo by PCR and by cell death when G418 was present in the medium. Targeted neo− cells were microinjected into blastocysts to produce transmitting chimeras. Cre recombinase eliminates exon 2. C, PCR amplification of loxP sites and Alb-Cre recombinase. Double heterozygous mice carrying one floxed allele and Alb-Cre recombinase (Acsl1Flox/+-Alb-CreTg/0, i.e. ACSL1 heterozygote) were interbred to obtain Acsl1L−/− mice along with five different groups of littermates. Duplex PCR yielded a 780 bp band for wild type allele and a 380 bp band for LoxP allele (upper panel). Another duplex PCR yielded a 324 bp band for internal control and a 100 bp band for Alb-Cre recombinase (lower panel). Lanes 4 show bands from an Acsl1L−/− mouse (Acsl1Flox/Flox-Alb-CreTg/0). D, PCR amplification of the region between exon 1 and exon 10. cDNA was synthesized from total RNA that was extracted from control and Acsl1L−/− mice. PCR with primers that bind exon 1 and exon 10 yielded a shorter band in Acsl1L−/− mice. Sequencing of the PCR products confirms that the sizes were 996 bp and 878 bp for control and Acsl1L−/− mice, respectively, and exon 2 (118 bp) was missing in Acsl1L−/− mice.
Animal Experiments
Male mice (age 12–17 weeks) were fed a standard diet (Prolab Isopro® RMH 3000, LabDiet) and killed after either a 4-h or a 24-h fast unless otherwise indicated. Plasma was collected from the retroorbital sinus. Liver, heart, and adipose tissues were snap frozen in liquid nitrogen and stored at −80 °C. For high fat diet studies, 8-week-old Acsl1L−/− and control (Acsl1Flox/Flox) mice were fed a high fat Western diet (45% calories from fat (mostly lard), 35% from carbohydrate (sucrose and corn starch) Research Diets D12451) or an isocaloric standard control diet (10% calories from fat (lard and soybean oil), 70% from carbohydrate (sucrose and corn starch) Research Diets D12450B) for 14–18 weeks. Mice were weighed weekly. Body composition was determined by magnetic resonance imaging. At ∼25 weeks, mice were killed, and tissues were snap frozen in liquid nitrogen and analyzed for enzyme activity, quantitative real time-PCR, Western blotting, and lipid content.
Primary Hepatocyte Culture and Oleate Labeling
Primary hepatocytes were obtained from fed 12-week-old male Acsl1Flox/Flox and Acsl1L−/− mice, by in situ liver perfusion using a modification of the collagenase method (23). Briefly, the livers were first perfused with a Ca2+-free perfusion buffer (10 mm HEPES, 132 mm NaCl, 20 mm dextrose, 6.7 mm KCl, 1 mm adenosine, 1.5 mm NaOH, 0.1 mm EGTA, 0.02 unit/ml insulin) and then with Ca2+-containing collagenase buffer (0.3 mg/ml; type CLS I, 230 units/ml; Worthington, Lakewood, NJ). Hepatocytes were collected after low speed centrifugation and seeded (1.0 × 106 cells/60-mm dishes) onto collagen-coated plates as described previously (9). The next day, hepatocytes were labeled with 2 ml of medium containing 0.5 μCi of [1-14C]oleate (final concentration, 500 μm oleate) for 3 h and then collected for cellular lipid extraction (24) and medium acid-soluble metabolites (ASMs) as a measure of incomplete FA oxidation (9). Lipid extracts were separated by thin layer chromatography in hexane:ethyl ether:acetic acid (80:20:1, v/v). The 14C-labeled lipids were detected and quantified with a Bioscan 200 imaging system.
Glucose and Insulin Tolerance Tests
Oral glucose tolerance tests (OGTTs) and insulin tolerance tests (ITTs) were performed in 5-month-old mice (after 3 months of high fat or control diet feeding). Mice were fasted for 4 h after the start of the light cycle and gavaged with glucose (2.5 g/kg) for OGTTs or injected intraperitoneally with insulin (0.75 unit/kg) for ITTs. Blood glucose was determined before and 15, 30, 60, 90, and 120 min after the glucose or insulin load with a glucometer (LifeScan, Inc.) (25).
Euglycemic-Hyperinsulinemic Glucose Clamp Experiments
In 11–13-week-old mice that had been fed a high fat (safflower oil) diet for the preceding 3 weeks, clamp studies were performed as described previously (17) with a prime-continuous [3-3H]glucose infusion 10-μCi bolus, 0.1 μCi/min) to determine rates of whole body glucose turnover followed by a primed-continuous insulin infusion (2.5 milliunits/kg/min; Humulin, Eli Lilly, Indianapolis, IN) to raise insulin levels within a physiologic range. A single 2-deoxy-d-[1-14C]glucose injection was administered at 75 min.
Liver Insulin Signaling
Male mice (∼5 months old) were fasted overnight (16 h) and anesthetized with Avertin. Insulin (1 unit/kg of body weight) or phosphate-buffered saline was injected via the portal vein. Two min later, the left liver lobe was snap frozen in liquid nitrogen. Liver tissue was pulverized and homogenized on ice in a HEPES buffer (pH 7.4) containing 1% Nonidet P-40, 100 mm NaCl, 2% glycerol, 5 mm NaF, 1 mm EDTA, and proteinase inhibitor mixture and phosphatase inhibitor cocktails I and II (Sigma). Homogenates were centrifuged at 16,000 × g for 30 min at 4 °C, and supernatants were used for Western blotting with antibody against phosphorylated AKT.
Western Blotting
ACSL1 expression was determined in liver, gonadal adipose tissue, and heart using a polyclonal peptide antibody (a gift from Cell Signaling, 4047). Homogenates from liver and heart or total particulate from gonadal adipose tissue was separated by electrophoresis on an 10% polyacrylamide gel. Phosphorylation of AKT (Ser-473) was determined in the liver supernatant after injection of insulin through the portal vein. Anti-phospho-AKT (Ser-473) and anti-AKT were from Cell Signaling. Adobe Photoshop software was used for densitometry analysis.
ACSL, GPAT, and Acylglycerol-3-phosphate Acyltransferase (AGPAT) Activities
Initial rates of total ACSL activity in liver homogenates were measured with 2–4 μg of liver homogenate at 37 °C in the presence of 175 mm Tris (pH 7.4), 8 mm MgCl2, 5 mm dithiothreitol, 10 mm ATP, 250 μm CoA, 50 μm [1-14C]palmitic acid in 500 μm Triton X-100, and 10 μm EDTA in a total volume of 200 μl (9). GPAT specific activity was assayed with 20–40 μg of liver homogenate at room temperature in a 200-μl reaction mixture containing 75 mm Tris-HCl (pH 7.5), 4 mm MgCl2, 1 mg/ml bovine serum albumin (essentially FA-free), 1 mm dithiothreitol, 8 mm NaF, 800 μm [3H]glycerol 3-phosphate, and 80 μm palmitoyl-CoA (26). AGPAT activity was determined by measuring the conversion of [3H]lysophosphatidic acid (LPA) to [3H]phosphatidic acid in a 200-μl reaction mixture containing 100 mm Tris-HCl (pH 7.4), 10 μm oleoyl-LPA, 50 μm oleoyl-CoA, 0.25 μCi of [3H]oleoyl-LPA, and 1 mg/ml bovine serum albumin (essentially FA-free) (27). The reaction was started by adding 0.2–0.8 μg of liver total particulate followed by incubation for 6 min at 37 °C.
Liver and Plasma Metabolites
Lipids were extracted by a modified Folch method (28, 29), and liver TAG (Stanbio) and total cholesterol (Wako) mass were determined enzymatically. To determine hepatic lipid metabolite profiles, liver (100 mg) was homogenized with appropriate internal standards in specific organic reagents (17). After separation, purification, and elution, lipid metabolite extracts were separated by high performance liquid chromatography, and individual and total lipid species were analyzed by liquid chromatography/tandem mass spectrometry (17). Liver glycogen was extracted and digested with amyloglucosidase for 30 min at 37 °C (30). Glucose concentration was determined using the Glucose Autokit CII assay (Wako). For acylcarnitine species, male mice fed a standard diet were fasted 48 h before livers were collected. Specimens of powdered liver were homogenized in deionized water and extracted, and acylcarnitine species were measured by direct injection electrospray tandem mass spectrometry (31). For plasma chemistries and lipids, after Avertin anesthesia, blood was obtained from the tail vein or the retroorbital venous plexus. Plasma glucose, TAG, total cholesterol, free FA, β-hydroxybutyrate (Wako, Stanbio) and insulin (Roche Applied Science) were determined with commercially available kits (32).
RNA Extraction and Quantitative Real Time PCR Assay
Total liver RNA was isolated using the RNeasy Plus Mini kit (Qiagen), and 20 μg of total RNA was reverse transcribed to cDNA using the high capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR was performed with the iCycler Thermal Cycler instrument (Bio-Rad) in 25 μl using SYBR Green fluorescein reagent mix and 10 nm fluorescein as the calibration dye (Applied Biosystems). Each reaction contained cDNA derived from 20 ng of total RNA. Oligonucleotide primers were designed using the Primer-BLAST program (National Center for Biotechnology Information). Sequences of primers are listed in supplemental Table 1. The fold change in expression of the target genes was normalized to the endogenous control (β-actin) and was calculated relative to the control group using the 2−ΔΔCT method (33).
Statistical Analysis
Values are expressed as means ± S.E. Statistical comparisons between control and ACSL1 knock-out mice were determined using an unpaired, two-tailed Student's t test. Statistically significant effects of diet and genotype were identified using a two-way analysis of variance (ANOVA) with interaction. A p value < 0.05 was considered significant unless otherwise indicated.
RESULTS
Generation of Liver-specific ACSL1 Knock-out Mice (Acsl1L−/−)
The generation of targeting vector, embryonic stem cells containing floxed Acsl1 allele, and chimeras was described under “Experimental Procedures.” Acsl1L−/− mice (Acsl1Flox/Flox-Alb-CreTg/0) were produced by interbreeding double heterozygotes (Fig. 1C). For the experiments described, Acsl1L−/− were crossed with Acsl1Flox/Flox littermates lacking the Alb-Cre transgene (Acsl1Flox/Flox-Alb-Cre0/0) to yield Acsl1L−/− mice and Acsl1Flox/Flox mice. These two genotypes were born at the expected frequency (1:1), indicating that the combination of the floxed allele and the Cre transgene did not affect reproduction. Acsl1Flox/Flox mice were used as controls because pilot studies showed that neither the loxP sites nor the presence of albumin-Cre altered ACSL1 expression or phenotype (data not shown).
Liver-specific Deletion of ACSL1 (Acsl1L−/−)
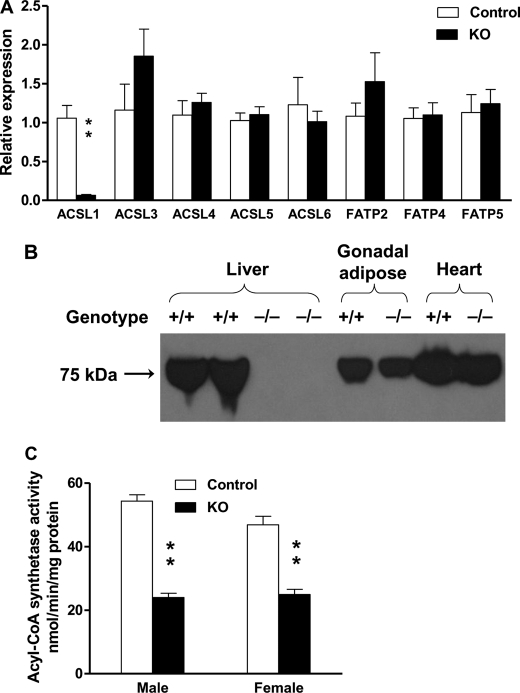
Acsl1 mRNA was virtually absent in Acsl1L−/− liver, confirming the ACSL1 deletion (Fig. 2A). No difference was observed for the mRNA from the other two major ACSL isoforms in liver, Acsl4 and Acsl5. The increase in the mRNA amount of the less abundant ACSL isoform, Acsl3, was not significant. The mRNA abundance for fatty acid transport proteins FATP2, 4, and 5, which have very long chain acyl-CoA synthetase activity, also remained unchanged in Acsl1L−/− liver.
FIGURE 2.
Acsl1L−/− liver contains little Acsl1 mRNA, no ACSL1 protein, and decreased ACSL activity. A, relative gene expression of Acsl and Fatp in Acsl1L−/− liver (n = 6 for each group). Total RNA was isolated from liver, and 20 μg was synthesized to cDNA for SYBR Green quantitative PCR. Target gene expression was normalized to the endogenous control β-actin and expressed as 2−ΔΔCT relative to the control. B, ACSL1 protein expression in the liver, heart, and adipose tissue of Acsl1L−/− mice. Extracts from 100 μg of liver, gonadal white adipose tissue, or heart were subjected to SDS-PAGE (10% gel). The proteins were transferred to a polyvinylidene difluoride membrane and probed with anti-ACSL1 peptide polyclonal antibody to detect endogenous ACSL1. This representative result was repeated four times. C, ACSL specific activity in liver. Homogenates from control and Acsl1L−/− liver were assayed in the presence of 50 μm [1-14C]palmitic acid. Data show means ± S.E. from 5 to 7 mice/group. **, p < 0.0001 versus control mice (Student's t test).
Using a rabbit peptide antibody against human ACSL1, we detected a ∼75 kDa band in liver from control liver, but not in liver from Acsl1L−/− mice, whereas the band was present in gonadal adipose tissue and heart from both control and knock-out mice, indicating that the deletion of ACSL1 was specific to liver (Fig. 2B). Consistent with the quantitative real time-PCR and Western blotting results, total ACSL activity in liver homogenates was 56 and 47% lower than in control male and female mice, respectively (Fig. 2C). This decrease in ACSL activity indicates that ACSL1 contributes approximately 50% of total ACSL activity in liver and that other ACSL isoforms did not compensate for the loss. The remaining ACSL activity probably represents activity from other ACSL and FATP isoforms present in liver.
At 12–14 weeks, Acsl1L−/− mice showed no significant difference from control mice for body weight, organ weights, adiposity, or plasma metabolites (supplemental Table 2). Because it has been reported that ACSL1 is up-regulated by PPARα, a transcription factor that increases FA β-oxidation (6, 7), we compared Acsl1L−/− and control mice after a 24-h fast. After a 24-h fast, both knock-out and control mice lost approximately 3 g of body weight and had similar changes in blood glucose, insulin, TAG, total cholesterol, ketone bodies, and hepatic glycogen (supplemental Table 3). No difference was observed when the 12–14-week-old mice were subjected to OGTTs and ITTs (data not shown), suggesting that ACSL1 deficiency in liver did not affect glucose metabolism.
Acsl1L−/− Liver Contained Less Acyl-CoA
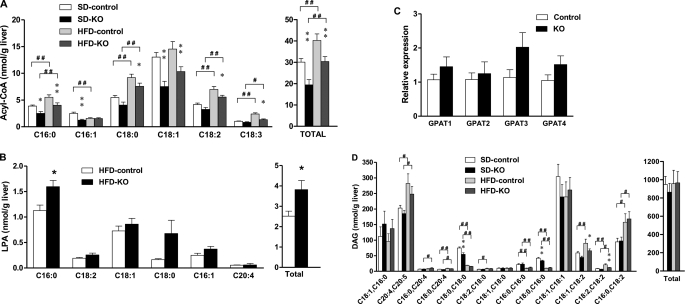
ACSL isoforms provide acyl-CoAs, not only for lipid metabolism, but also for lipid intermediates that are important for cell signaling. Previous studies have suggested that acyl-CoAs and DAG might affect insulin sensitivity in skeletal muscle and liver (10). To examine the effect of a deficiency of ACSL1 in liver, the amounts and composition of the FA-derived lipid intermediates, acyl-CoAs, LPA, and DAG were measured in mice fed matched low and high fat diets. The high fat diet contained 45% calories from predominantly saturated fat. As might be expected with a 56% decrease in hepatic ACSL activity in male Acsl1L−/− mice, both the amount and major species of acyl-CoA were affected. In both genotypes, the high fat diet increased the total and all major acyl-CoAs except C16:1-CoA and C18:1-CoA (Fig. 3A). When fed either the control or the high fat diet, the content of acyl-CoA in knock-out mice liver was 35 and 25% lower, respectively, than in liver from control mice (Table 1). The major acyl-CoA in liver, C18:1-CoA, was 42 and 29% lower in knock-out mice fed the control diet or the high fat diet, respectively (Fig. 3A). The decreases in multiple acyl-CoA species are consistent with the broad substrate range of ACSL1 (34).
FIGURE 3.
Acsl1L−/− liver contained less acyl-CoA than control liver. Control and Acsl1L−/− mice were fed a 45% fat diet (HFD, Research Diets D12451) or a matched 10% fat control diet (SD, Research Diets D12450B) from 8 to 23 weeks of age. Liver samples were snap frozen after the mice were fasted for 4 h. Lipid intermediates were extracted and measured by tandem mass spectrometry. A, total long chain acyl-CoA and acyl-CoA species. B, total long chain LPA and LPA species. C, relative expression of Gpat isoform mRNA in Acsl1L−/− liver (n = 6 for each group). Total RNA was isolated from liver, and 20 μg was synthesized to cDNA for SYBR Green quantitative PCR. Target gene expression was normalized to the endogenous control β-actin and expressed as 2−ΔΔCT relative to the control. D, total DAG and DAG species. Data show means ± S.E. from 10 mice/group. B, *, p < 0.05 versus control mice (Student's t test). A and C, *, p < 0.05; **, p < 0.01 versus control mice fed the same diet (two-way ANOVA); #, p < 0.05; ##, p < 0.01 versus 10% fat control diet in the same genotype (two-way ANOVA).
TABLE 1.
Liver lipid metabolites from Acsl1L−/− and Acsl1Flox/Flox mice fed a high fat or a control diet for 14–18 weeks
Samples were obtained after a 4-h fast. Data represent means ± S.E. The number of animals in each group is indicated in parentheses. KO, knock-out.
| Total liver lipid metabolites | 10% fat diet (control) |
45% fat diet (high fat) |
||
|---|---|---|---|---|
| Control (n = 10) | KO (n = 10) | Control (n = 10) | KO (n = 10) | |
| Acyl-CoA (nmol/g liver) | 30.1 ± 0.5 | 19.4 ± 0.4a | 40.2 ± 0.9b | 30.4 ± 0.6a,b |
| Lysophosphatidic acid (nmol/g liver) | 2.69 + 0.14 | 2.19 ± 0.32 | 2.52 ± 0.24 | 3.82 ± 0.44c |
| Diacylglycerol (nmol/g liver) | 949 ± 88 | 863 ± 98 | 957 ± 147 | 966 ± 123 |
a p < 0.01 compared with control mice fed the same diet (two-way ANOVA).
b p < 0.01 compared with 10% fat control diet in the same genotype (two-way ANOVA).
c p < 0.05 compared with control mice (Student's t test).
Despite the lower content of acyl-CoA, liver from Acsl1L−/− mice fed the high fat diet contained 52% more total LPA and 42% more C16:0-LPA than control liver (Fig. 3B). The 314 and 49% higher C18:0-LPA and C16:1-LPA were also close to statistical significance (p = 0.06, and p = 0.08, respectively). We measured the mRNA abundance and enzyme activity of GPAT, the enzyme that uses glycerol 3-phosphate and acyl-CoA to form LPA. For the four known GPAT isoforms, none of the changes in mRNA abundance was significant (including Gpat3 (p = 0.09) (Fig. 3C). Similarly, the 11% increase in total GPAT activity was not statistically significant (p = 0.06, 1.96 ± 0.07 nmol/min/mg of protein in control versus 2.17 ± 0.07 nmol/min/mg of protein in knock-out liver). Because the increase in LPA might also have been caused by a decrease in its metabolism, we measured the activity of AGPAT, which acylates LPA to form phosphatidic acid. No significant difference was found between control and knock-out mice (data not shown). Thus, the increase in LPA in Acsl1L−/− liver from high fat diet-fed mice remains unexplained. Total hepatic LPA content in mice fed the standard diet was similar in the two genotypes (Table 1).
The decrease in total acyl-CoA content did not affect the hepatic content of DAG in Acsl1L−/− liver, which was similar for both genotypes, when mice were fed either the control diet or the high fat diet (Fig. 3D and Table 1). The Acsl1L−/− DAG composition was similar to that of control mice, except for 27 and 21% less C18:0/C18:0-DAG and C18:0/C16:0-DAG, respectively, when mice were fed the control diet, and 27 and 44% less C18:1/C18:2-DAG and C18:2/C18:2-DAG, respectively, when mice were fed the high fat diet (Fig. 3D).
Incorporation of Oleate into Hepatocyte TAG and Long Chain FA Oxidation Were Impaired with ACSL1 Deficiency
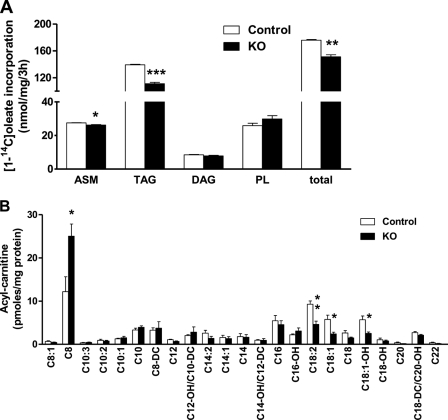
Based on substrate preferences, tissue location, subcellular location, and different sensitivity to inhibitors, we had hypothesized previously that each ACSL isoform provides acyl-CoAs destined for specific downstream pathways (35). Despite the normal liver content of TAG, when hepatocytes from control and Acsl1L−/− mice were incubated with [14C]oleate, the incorporation of the fatty acid into TAG was 20% lower in the knock-out cells (Fig. 4A). Labeled medium ASM, the product of incomplete FA oxidation, was 4.5% lower in the knock-out cells. In addition, [14C]oleate uptake into isolated Acsl1L−/− hepatocytes during a 2-min uptake experiment was similar to that of controls (data not shown). Further, Acsl1L−/− and control mice had similar plasma β-hydroxybutyrate concentrations after they were fed, fasted for 24 h, or fed with the high fat diet (supplemental Tables 1–3). However, a comprehensive analysis of the liver acylcarnitine species by tandem mass spectrometry showed that Acsl1L−/− liver contained 50%, 58%, and 55% less C18:2-carnitine, C18:1-carnitine, and C18:0-OH, respectively, and twice as much C8:0-carnitine (Fig. 4B). Long chain acylcarnitines are mitochondrial intermediates that are derived directly from their corresponding acyl-CoAs and thus reflect the flux of FA through mitochondrial FA oxidation (31). The decreases in long chain acylcarnitines were consistent with the lower incorporation of label from [14C]oleate into ASM and suggested that long chain FA oxidation in liver is impaired by ACSL1 deficiency. The increase in C8:0-carnitine, in contrast, suggested that peroxisomal FA oxidation had increased, and the mRNA for acyl-CoA oxidase 1, a key enzyme in peroxisomal oxidation, was 91% higher in Acsl1L−/− liver.
FIGURE 4.
ACSL1 deficiency decreased [1-14C]oleate incorporation into cell TAG and medium ASM, and liver contained fewer long chain acylcarnitines. A, primary hepatocytes were isolated from 12-week-old control and Acsl1L−/− mice after collagenase perfusion. Cells (1 × 106 cells/60-mm dish) were incubated with 500 μm [1-14C]oleate for 3 h and harvested, and medium was collected for ASMs, for the products of incomplete FA oxidation. [1-14C]Oleate incorporation into neutral lipid species was determined as described under “Experimental Procedures.” Data are reported as means ± S.E. from a representative experiment performed in triplicate dishes. *, p < 0.05; **, p < 0.01; ***, p < 0.001. B, male mice fed a chow diet (Prolab Isopro® RMH 3000) were fasted for 48 h before livers were collected. Acylcarnitine species were analyzed by tandem mass spectrometry. DC, dicarboxylated acylcarnitines; OH, hydroxylated acylcarnitines. Data show means ± S.E. for four animals/group. *, p < 0.05 for Acsl1L−/− mice versus control mice.
Phospholipid Composition Was Altered in Acsl1L−/− Liver
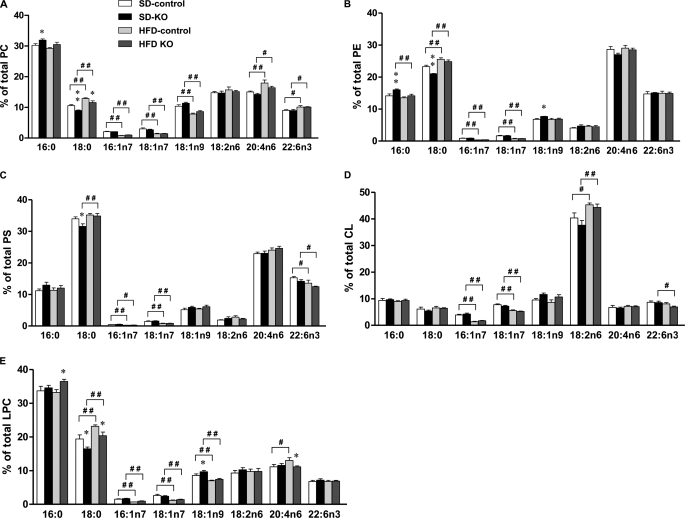
Because in vitro assays with purified ACSL1 showed activation of a broad range of saturated and unsaturated fatty acids with little apparent difference in FA preference (36, 37), the lipid profile in Acsl1L−/− liver was examined to determine whether a preference would be expressed in vivo. When fed the high fat diet, TAG increased in both genotypes, and Acsl1L−/− liver contained more DAG, FA, and lysophosphatidylcholine (Fig. 5). In Acsl1L−/− liver, small but significant differences were observed in the distribution of specific FAs in several phospholipid species. Acsl1L−/− liver PC and PE contained a higher percentage of C16:0, and PC, PE, and PS contained a lower percentage of C18:0 compared with control mice (Fig. 6, A–C). Compared with phospholipids in control liver, Acsl1L−/− liver LPC contained more C16:0 and C18:1 and less C18:0 and C20:4 (Fig. 6E). Few differences were observed in the genotypes for cardiolipin FA composition (Fig. 6D). These lower percentages of C18:0 in Acsl1L−/− liver phospholipids were also present after the mice were fed a high fat diet for 3 months, suggesting that ACSL1 makes a specific contribution of 18:0-CoAs to phospholipid composition either during de novo synthesis or during FA recycling.
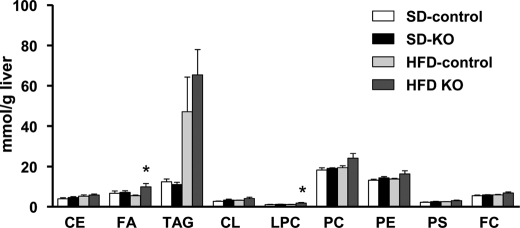
FIGURE 5.
Changes in neutral and phospholipids in livers from Acsl1L−/− and control mice fed control and high fat diets. Lipid classes from livers of mice fed a high fat diet (HFD) or a control diet (SD) for 14 weeks were extracted and analyzed (Lipomics Technologies, Inc). Data show means ± S.E. from five mice/group. *, p < 0.05 versus 10% fat control diet in the same genotype (two-way ANOVA). CL, cardiolipin; FC, free cholesterol.
FIGURE 6.
FA composition of phospholipids from Acsl1L−/− and control livers. PC (A), PE (B), PS (C), CL (D), and LPC (E) from livers of mice fed a high fat diet (HFD) or a control diet (SD) for 14 weeks were extracted and analyzed (Lipomics Technologies, Inc.). Data show means ± S.E. from five mice/group. *, p < 0.05; **, p < 0.01 versus control mice fed the same diet (two-way ANOVA). #, p < 0.05; ##, p < 0.01 versus 10% fat control diet in the same genotype (two-way ANOVA). CL, cardiolipin.
Unlike the changes in phospholipids, the hepatic content of neutral lipids, including CE and TAG, did not change significantly in Acsl1L−/− liver (Fig. 7). High fat diet feeding decreased the percentages of C16:1 and C18:1n7 in TAG in both control and knock-out mice, but no difference between genotype was observed. Acsl1L−/− liver contained a lower percentage of C20:4 and C22:6 in TAG and tended to have lower percentage of C18:2 TAG (p = 0.05) when fed the high fat diet. These data indicate that neither a 50% decrease in total ACS activity nor a specific deficiency of ACSL1 alters the predominant TAG species.
FIGURE 7.
FA composition of neutral lipids from Acsl1L−/− and control livers. TAG (A) and CE (B) from livers of mice fed a high fat diet (HFD) or a control diet (SD) for 14 weeks were extracted and analyzed (Lipomics Technologies, Inc). Data show means ± S.E. from five mice/group. *, p < 0.05 versus control mice fed the same diet (two-way ANOVA). #, p < 0.05; ##, p < 0.01 versus 10% fat control diet in the same genotype (two-way ANOVA).
ACSL1 Deficiency Did Not Protect Mice from Developing Insulin Resistance
Because the Acsl1L−/− liver contained 25–35% less acyl-CoA than control mice, we questioned whether Acsl1L−/− mice would be protected from developing hepatic and whole-body insulin resistance. OGTTs and ITTs were performed in mice fed a high fat diet for 3 months. Compared with mice fed the control diet, mice fed the high fat diet had higher peak blood glucose levels and a larger area under the curve during the OGTTs, and but no differences between genotypes were observed in either the OGTTs or the ITTs (supplemental Fig. 1, A and B, respectively).
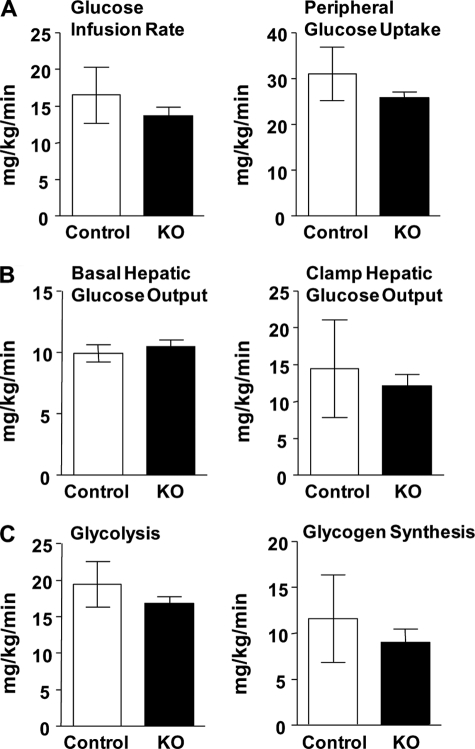
To examine the possibility that long term feeding might cause a compensatory adaptation, we examined the tissue and whole body insulin sensitivity using euglycemic-hyperinsulinemic clamp experiments in young mice (11–13 weeks) after they were fed a high fat (safflower oil) diet for 3 weeks. Hepatic ACSL1 deficiency had no effect on hepatic and whole body insulin sensitivity, hepatic glucose production, or peripheral glucose utilization (Fig. 8). Thus, a hepatic ACSL1 deficiency did not protect the mice from diet-induced whole body or hepatic insulin resistance. Further, direct insulin injection into the portal vein increased Akt Ser-473 phosphorylation similarly in control and knock-out mice (supplemental Fig. 2), consistent with the interpretation that the 25–35% lower hepatic acyl-CoA content observed in the knock-out liver (Table 1) did not enhance hepatic insulin signaling.
FIGURE 8.
Euglycemic-hyperinsulinemic clamp study. Control (KO) and Acsl1L−/− mice fed a high fat diet for 3 weeks were studied after 16 h of food deprivation. With an infusion of 2.5 milliunits of insulin/kg/min, plasma glucose was maintained by an intravenous variable 20% glucose infusion, and rates were determined for glucose infusion rate and peripheral glucose uptake (A), basal and clamp hepatic glucose output (B), and glycolysis and glycogen synthesis (C). Results are expressed as means ± S.E. (n = 10 for each genotype).
DISCUSSION
Long chain ACS isoforms activate FAs of between 10 and 22 carbons and provide acyl-CoAs for lipid synthesis, lipid oxidation, and lipid signaling. Since the discovery of the first ACSL isoform, ACSL1, in 1990 (36), four additional mammalian ACSL isoforms have been identified, each with several splice variants (38). Major progress has been made in our knowledge of ACSL primary structure, enzymatic kinetics, tissue distribution, subcellular location, and nutritional regulation. However, because no specific inhibitors for individual ACSL isoforms have been identified, the exact role of each ACSL in lipid metabolism and lipid signaling remained unknown. We hypothesized previously that specific ACSL isoforms provide acyl-CoAs for particular metabolic pathways and that individual ACSL isoforms might have tissue-specific functions. Supporting evidence includes in vitro overexpression studies of ACSL1 and ACSL5 (9, 39). Overexpressing ACSL1 in primary hepatocytes increases the incorporation of oleate into DAG and phospholipid and decreases incorporation into CE but has no effect on TAG. In contrast, overexpressing ACSL5 in hepatoma cells partitions exogenous FA toward TAG synthesis and storage but not toward phospholipid or CE synthesis. However, gain-of-function studies are fraught with possible ambiguities. The overexpressed protein may be mislocated, or the overexpressed enzymatic product may overwhelm the capacity of downstream metabolic pathways. Thus, we constructed a mouse deficient in ACSL1 in liver to elucidate the role of ACSL1 in hepatic metabolism.
Using the LoxP-Cre strategy, we knocked out the Acsl1L−/− gene specifically in liver, demonstrated by the absence of the exon 2 sequence, the >94% knockdown of Acsl1L−/− mRNA by quantitative real time-PCR, and the absence of ACSL1 protein. The specific activity of ACSL in liver was 50% lower than in controls, confirming that ACSL1 is a major hepatic ACSL isoform. Changes in mRNA from the other major ACSL and FATP isoforms were not significant, and the loss of ACSL specific activity suggested that a complementary increase of other proteins with ACSL activity had not occurred. Overall, the lack of ACSL1 and 50% of ACSL activity in liver did not have a major effect on mouse phenotype, viability, adiposity, or growth. Liver and adipose tissues were of normal weight, and the plasma concentrations of FA, TAG, total cholesterol, and fasting ketone bodies did not change.
The ability of ACSL to convert lipophilic FAs into water-soluble acyl-CoAs that are trapped within cells stimulated the concept that proteins with ACSL activity might facilitate cellular FA uptake (1). In fact, ACSL1 was identified in an unbiased search designed to clone proteins that increase the uptake of long chain FAs into cells (40). However, when overexpressed 3.7-fold in rat primary hepatocytes, ACSL1 did not increase total metabolized FA (9), either because ACSL1 does not enhance FA uptake in hepatocytes or because this gain-of-function model could not further increase an already high basal rate of FA uptake. In the present study, the 25–35% lower total acyl-CoA and major acyl-CoA species in the Acsl1L−/− liver could be due solely to the decrease in ACSL activity or to the combined effect of decreased ACSL activity and decreased ACSL1-mediated FA uptake. Arguing against the latter possibility is the observation that plasma nonesterified FA concentrations were similar in control and Acsl1L−/− mice under all conditions tested and the finding that FA uptake was similar in control and Acsl1L−/− hepatocytes. Further, cellular FA content was unchanged or increased in Acsl1L−/− liver.
The major findings in the liver-specific Acsl1L−/− mice were the changes in phospholipid FA composition and the decreases in TAG and ASM synthesis by isolated hepatocytes. The observed relative increases in C16:0 and C18:1 and decreases in C18:0 and C20:4 in several phospholipid species point out limitations in drawing conclusions about substrate preference based on in vitro assays. Thus, the reported FA preferences of purified ACSL1 (37) did not predict the changes in actual FA composition observed in liver phospholipids, a composition that likely depends on substrate availability, enzyme affinity, and lysophospholipid acyltransferases activities. Further, one would not have predicted that FA composition would change in only some glycerolipids, such as LPC, PC, and PE, but not in others, such as TAG, and that diet would have so little influence on FA composition.
With regard to TAG synthesis, the function of ACSL1 in liver has been unclear. On the one hand, hepatic Acsl1 mRNA is higher in obese and hypertriglyceridemic rats with fatty livers (41, 42), suggesting a link with TAG synthesis. However, PPARα ligands that stimulate genes required for FA β-oxidation strongly induce hepatic Acsl1 mRNA (6, 7) via a PPAR response element in the gene promoter (8). Moreover, ACSL1 is located both in liver endoplasmic reticulum (43) and in mitochondria (44), the organelles responsible for TAG synthesis and FA β-oxidation, respectively. Experimentally, overexpressed ACSL1 in NIH 3T3 fibroblasts, mouse heart, or mouse liver increased TAG synthesis (5, 45, 46), but adenovirus-mediated overexpression in rat primary hepatocytes did not affect oleate incorporation into TAG (9), and siRNA targeting of Acsl1 in human hepatoma Huh7 cells did not alter oleate incorporation into TAG (47). In the current study, the ACSL1 deficiency did not protect mice from diet-induced hepatic steatosis; however, in isolated hepatocytes, [14C]oleate incorporation into TAG was diminished by 20%. Although our findings suggest that ACSL1 is not critical for hepatic TAG synthesis, it is possible that Acsl1L−/− liver had adapted to the ACSL1 deficiency by altering the transcription of genes that are involved in the uptake and/or metabolism of lipids.
Despite the lack of difference in plasma β-hydroxybutyrate concentration between genotypes at baseline, after fasting for 24 h, or after feeding low or high fat diets, a defect in β-oxidation is supported by several observations. First, Acsl1 mRNA in liver is up-regulated by PPARα, a major transcription factor that increases the expression of genes involved in FA β-oxidation. Second, despite the low, but significant, 4.5% decrease in incorporation of [14C]oleate into ASM in isolated hepatocytes, the 50–58% decreases in liver C18-carnitines are consistent with a reduced flux of acyl-CoAs into the pathway of mitochondrial β-oxidation (31), and the 2-fold increase in C8:0-carnitine and the 91% increase in acyl-CoA oxidase-1 mRNA suggest a compensatory increase of FA oxidation in peroxisomes. Taken as a whole, our data suggest that both TAG synthesis and β-oxidation of long chain FA are impaired in Acsl1L−/− liver.
In the context of excess TAG stores in nonadipose tissues, increases in acyl-CoA, DAG, and ceramide have each been linked to insulin resistance (10–12). In Acsl1L−/− mice fed a high fat diet, hepatic total acyl-CoA content and major acyl-CoA species were 25% lower than in controls, and we wondered whether the mice would be protected from developing insulin resistance. The ACSL1 deficiency also caused an increase in hepatic LPA content and a decrease in several DAG species but did not change the total amount of DAG. Despite these alterations in lipid intermediates, Acsl1L−/− mice were normal in their glucose metabolism, in glucose and insulin tolerance, and in the ability of insulin to stimulate AKT phosphorylation. When challenged with a 45% fat diet for 15 weeks, Acsl1L−/− mice became as obese and insulin-resistant as control mice. Further, a euglycemic-hyperinsulinemic clamp study showed no significant differences between the genotypes in hepatic or whole body insulin sensitivity in young mice fed a high fat diet for 3 weeks. A previous study showed that an increase in hepatic acyl-CoA content does not result in insulin resistance (17); our current study suggests that a decrease in acyl-CoA content does not protect against insulin resistance when liver TAG content is elevated.
In summary, this is the first knock-out model designed to study the role of ACSL1 in liver and in whole body metabolism. Despite the absence of hepatic ACSL1, a 50% decrease in ACSL specific activity, and a 25–35% decrease in acyl-CoA content, no obvious changes were observed in liver morphology or appearance, organ weights, or plasma lipid concentrations, and Acsl1L−/− mice were not protected from diet-induced hepatic steatosis. Small, but consistent changes were present in the FA composition of the major phospholipids, and both TAG synthesis and β-oxidation were impaired.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants DK59935 (to R. A. C.), DK40936 (to G. I. S.), U24 DK59635 (to G. I. S.), P30 DK34987 (to G. I. S.), a grant from the American Diabetes Association (to D. M. M.), a postdoctoral fellowship from the American Heart Association Mid-Atlantic Region (to L. O. L.), Predoctoral Training Grant HL069768 in Integrative Vascular Biology (to J. M. E.) and P30 DK034987 and P30 DK056350.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and supplemental Tables 1–4.
- ACSL
- long chain acyl-CoA synthetase
- AGPAT
- acylglycerol-3-phosphate acyltransferase
- ANOVA
- analysis of variance
- ASM
- acid-soluble metabolite
- CE
- cholesteryl ester
- DAG
- diacylglycerol
- FA
- fatty acid
- FATP
- fatty acid transport protein
- GPAT
- glycerol-3-phosphate acyltransferase
- ITT
- insulin tolerance test
- LPA
- lysophosphatidic acid
- LPC
- lysophosphatidylcholine
- neo
- neomycin
- OGTT
- oral glucose tolerance test
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- PPAR
- peroxisome proliferator-activated factor
- TAG
- triacylglycerol.
REFERENCES
- 1.Mashek D. G., Coleman R. A. (2006) Curr. Opin. Lipidol. 17, 274–278 [DOI] [PubMed] [Google Scholar]
- 2.Mashek D. G., Li L. O., Coleman R. A. (2006) J. Lipid Res. 47, 2004–2010 [DOI] [PubMed] [Google Scholar]
- 3.Marszalek J. R., Kitidis C., Dararutana A., Lodish H. F. (2004) J. Biol. Chem. 279, 23882–23891 [DOI] [PubMed] [Google Scholar]
- 4.Coleman R. A., Reed B. C., Mackall J. C., Student A. K., Lane M. D., Bell R. M. (1978) J. Biol. Chem. 253, 7256–7261 [PubMed] [Google Scholar]
- 5.Chiu H. C., Kovacs A., Ford D. A., Hsu F. F., Garcia R., Herrero P., Saffitz J. E., Schaffer J. E. (2001) J. Clin. Invest. 107, 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoonjans K., Staels B., Grimaldi P., Auwerx J. (1993) Eur. J. Biochem. 216, 615–622 [DOI] [PubMed] [Google Scholar]
- 7.Frederiksen K. S., Wulff E. M., Sauerberg P., Mogensen J. P., Jeppesen L., Fleckner J. (2004) J. Lipid Res. 45, 592–601 [DOI] [PubMed] [Google Scholar]
- 8.Schoonjans K., Watanabe M., Suzuki H., Mahfoudi A., Krey G., Wahli W., Grimaldi P., Staels B., Yamamoto T., Auwerx J. (1995) J. Biol. Chem. 270, 19269–19276 [DOI] [PubMed] [Google Scholar]
- 9.Li L. O., Mashek D. G., An J., Doughman S. D., Newgard C. B., Coleman R. A. (2006) J. Biol. Chem. 281, 37246–37255 [DOI] [PubMed] [Google Scholar]
- 10.Savage D. B., Petersen K. F., Shulman G. I. (2007) Physiol. Rev. 87, 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postic C., Girard J. (2008) J. Clin. Invest. 118, 829–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wymann M. P., Schneiter R. (2008) Nat. Rev. Mol. Cell Biol. 9, 162–176 [DOI] [PubMed] [Google Scholar]
- 13.Shulman G. I. (2000) J. Clin. Invest. 106, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faergeman N. J., Knudsen J. (1997) Biochem. J. 323, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson A. L., Cooney G. J. (2000) Diabetes 49, 1761–1765 [DOI] [PubMed] [Google Scholar]
- 16.Petrescu A. D., Hertz R., Bar-Tana J., Schroeder F., Kier A. B. (2002) J. Biol. Chem. 277, 23988–23999 [DOI] [PubMed] [Google Scholar]
- 17.Neschen S., Morino K., Hammond L. E., Zhang D., Liu Z. X., Romanelli A. J., Cline G. W., Pongratz R. L., Zhang X. M., Choi C. S., Coleman R. A., Shulman G. I. (2005) Cell Metab. 2, 55–65 [DOI] [PubMed] [Google Scholar]
- 18.Timmers S., Schrauwen P., de Vogel J. (2008) Physiol. Behav. 94, 242–251 [DOI] [PubMed] [Google Scholar]
- 19.Kim J. K., Fillmore J. J., Chen Y., Yu C., Moore I. K., Pypaert M., Lutz E. P., Kako Y., Velez-Carrasco W., Goldberg I. J., Breslow J. L., Shulman G. I. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7522–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monetti M., Levin M. C., Watt M. J., Sajan M. P., Marmor S., Hubbard B. K., Stevens R. D., Bain J. R., Newgard C. B., Farese R. V., Sr., Hevener A. L., Farese R. V., Jr. (2007) Cell Metab. 6, 69–78 [DOI] [PubMed] [Google Scholar]
- 21.Kos C. H. (2004) Nutr. Rev. 62, 243–246 [DOI] [PubMed] [Google Scholar]
- 22.Postic C., Magnuson M. A. (2000) Genesis 26, 149–150 [DOI] [PubMed] [Google Scholar]
- 23.Berry M. N., Friend D. S. (1969) J. Cell Biol. 43, 506–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 25.Buhl E. S., Jessen N., Pold R., Ledet T., Flyvbjerg A., Pedersen S. B., Pedersen O., Schmitz O., Lund S. (2002) Diabetes 51, 2199–2206 [DOI] [PubMed] [Google Scholar]
- 26.Coleman R. A., Haynes E. B. (1983) J. Biol. Chem. 258, 450–456 [PubMed] [Google Scholar]
- 27.Agarwal A. K., Barnes R. I., Garg A. (2006) Arch. Biochem. Biophys. 449, 64–76 [DOI] [PubMed] [Google Scholar]
- 28.Samuel V. T., Liu Z. X., Qu X., Elder B. D., Bilz S., Befroy D., Romanelli A. J., Shulman G. I. (2004) J. Biol. Chem. 279, 32345–32353 [DOI] [PubMed] [Google Scholar]
- 29.Folch J., Lees M., Sloane Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 30.Suzuki Y., Lanner C., Kim J. H., Vilardo P. G., Zhang H., Yang J., Cooper L. D., Steele M., Kennedy A., Bock C. B., Scrimgeour A., Lawrence J. C., Jr., DePaoli-Roach A. A. (2001) Mol. Cell Biol. 21, 2683–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An J., Muoio D. M., Shiota M., Fujimoto Y., Cline G. W., Shulman G. I., Koves T. R., Stevens R., Millington D., Newgard C. B. (2004) Nat. Med. 10, 268–274 [DOI] [PubMed] [Google Scholar]
- 32.Hammond L. E., Gallagher P. A., Wang S., Hiller S., Kluckman K. D., Posey-Marcos E. L., Maeda N., Coleman R. A. (2002) Mol. Cell Biol. 22, 8204–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 34.Iijima H., Fujino T., Minekura H., Suzuki H., Kang M. J., Yamamoto T. (1996) Eur. J. Biochem. 242, 186–190 [DOI] [PubMed] [Google Scholar]
- 35.Coleman R. A., Lewin T. M., Van Horn C. G., Gonzalez-Baró M. R. (2002) J. Nutr. 132, 2123–2126 [DOI] [PubMed] [Google Scholar]
- 36.Suzuki H., Kawarabayasi Y., Kondo J., Abe T., Nishikawa K., Kimura S., Hashimoto T., Yamamoto T. (1990) J. Biol. Chem. 265, 8681–8685 [PubMed] [Google Scholar]
- 37.Fujino T., Kang M.-J., Suzuki H., Iijima H., Yamamoto T. (1996) J. Biol. Chem. 271, 16748–16752 [DOI] [PubMed] [Google Scholar]
- 38.Mashek D. G., Bornfeldt K. E., Coleman R. A., Berger J., Bernlohr D. A., Black P., DiRusso C. C., Farber S. A., Guo W., Hashimoto N., Khodiyar V., Kuypers F. A., Maltais L. J., Nebert D. W., Renieri A., Schaffer J. E., Stahl A., Watkins P. A., Vasiliou V., Yamamoto T. T. (2004) J. Lipid Res. 45, 1958–1961 [DOI] [PubMed] [Google Scholar]
- 39.Mashek D. G., McKenzie M. A., Van Horn C. G., Coleman R. A. (2006) J. Biol. Chem. 281, 945–950 [DOI] [PubMed] [Google Scholar]
- 40.Schaffer J. E., Lodish H. F. (1994) Cell 79, 427–436 [DOI] [PubMed] [Google Scholar]
- 41.Shimomura I., Tokunaga K., Jiao S., Funahashi T., Keno Y., Kobatake T., Kotani K., Suzuki H., Yamamoto T., Tarui S., Matsuzawa Y. (1992) Biochim. Biophys. Acta 1124, 112–118 [DOI] [PubMed] [Google Scholar]
- 42.Kuriyama H., Yamashita S., Shimomura I., Funahashi T., Ishigami M., Aragane K., Miyaoka K., Nakamura T., Takemura K., Man Z., Toide K., Nakayama N., Fukuda Y., Lin M. C., Wetterau J. R., Matsuzawa Y. (1998) Hepatology 27, 557–562 [DOI] [PubMed] [Google Scholar]
- 43.Lewin T. M., Kim J. H., Granger D. A., Vance J. E., Coleman R. A. (2001) J. Biol. Chem. 276, 24674–24679 [DOI] [PubMed] [Google Scholar]
- 44.Distler A. M., Kerner J., Peterman S. M., Hoppel C. L. (2006) Anal. Biochem. 356, 18–29 [DOI] [PubMed] [Google Scholar]
- 45.Souza S. C., Muliro K. V., Liscum L., Lien P., Yamamoto M. T., Schaffer J. E., Dallal G. E., Wang X., Kraemer F. B., Obin M., Greenberg A. S. (2002) J. Biol. Chem. 277, 8267–8272 [DOI] [PubMed] [Google Scholar]
- 46.Parkes H. A., Preston E., Wilks D., Ballesteros M., Carpenter L., Wood L., Kraegen E. W., Furler S. M., Cooney G. J. (2006) Am. J. Physiol. Endocrinol. Metab. 291, E737–E744 [DOI] [PubMed] [Google Scholar]
- 47.Yao H., Ye J. (2008) J. Biol. Chem. 283, 849–854 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.