Abstract
Paneth cells are a secretory epithelial lineage that release dense core granules rich in host defense peptides and proteins from the base of small intestinal crypts. Enteric α-defensins, termed cryptdins (Crps) in mice, are highly abundant in Paneth cell secretions and inherently resistant to proteolysis. Accordingly, we tested the hypothesis that enteric α-defensins of Paneth cell origin persist in a functional state in the mouse large bowel lumen. To test this idea, putative Crps purified from mouse distal colonic lumen were characterized biochemically and assayed in vitro for bactericidal peptide activities. The peptides comigrated with cryptdin control peptides in acid-urea-PAGE and SDS-PAGE, providing identification as putative Crps. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry experiments showed that the molecular masses of the putative α-defensins matched those of the six most abundant known Crps, as well as N-terminally truncated forms of each, and that the peptides contain six Cys residues, consistent with identities as α-defensins. N-terminal sequencing definitively revealed peptides with N termini corresponding to full-length, (des-Leu)-truncated, and (des-Leu-Arg)-truncated N termini of Crps 1–4 and 6. Crps from mouse large bowel lumen were bactericidal in the low micromolar range. Thus, Paneth cell α-defensins secreted into the small intestinal lumen persist as intact and functional forms throughout the intestinal tract, suggesting that the peptides may mediate enteric innate immunity in the colonic lumen, far from their upstream point of secretion in small intestinal crypts.
Antimicrobial peptides (AMPs)2 are released by epithelial cells onto mucosal surfaces as effectors of innate immunity (1–5). In mammals, most AMPs derive from two major families, the cathelicidins and defensins (6). The defensins comprise the α-, β-, and θ-defensin subfamilies, which are defined by the presence of six cysteine residues paired in characteristic tridisulfide arrays (7). α-Defensins are highly abundant in two primary cell lineages: phagocytic leukocytes, primarily neutrophils, of myeloid origin and Paneth cells, which are secretory epithelial cells located at the base of the crypts of Lieberkühn in the small intestine (8–10). Neutrophil α-defensins are stored in azurophilic granules and contribute to non-oxidative microbial cell killing in phagolysosomes (11, 12), except in mice whose neutrophils lack defensins (13). In the small bowel, α-defensins and other host defense proteins (14–18) are released apically as components of Paneth cell secretory granules in response to cholinergic stimulation and after exposure to bacterial antigens (19). Therefore, the release of Paneth cell products into the crypt lumen is inferred to protect mitotically active crypt cells from colonization by potential pathogens and confer protection against enteric infection (7, 20, 21).
Under normal, homeostatic conditions, Paneth cells are not found outside the small bowel, although they may appear ectopically in response to local inflammation throughout the gastrointestinal tract (22, 23). Paneth cell numbers increase progressively throughout the small intestine, occurring at highest numbers in the distal ileum (24). Mouse Paneth cells express numerous α-defensin isoforms, termed cryptdins (Crps) (25), that have broad spectrum antimicrobial activities (6, 26). Collectively, α-defensins constitute approximately seventy percent of the bactericidal peptide activity in mouse Paneth cell secretions (19), selectively killing bacteria by membrane-disruptive mechanisms (27–30). The role of Paneth cell α-defensins in gastrointestinal mucosal immunity is evident from studies of mice transgenic for human enteric α-defensin-5, HD-5, which are immune to infection by orally administered Salmonella enterica sv. typhimurium (S. typhimurium) (31).
The biosynthesis of mature, bactericidal α-defensins from their inactive precursors requires activation by lineage-specific proteolytic convertases. In mouse Paneth cells, inactive ∼8.4-kDa Crp precursors are processed intracellularly into microbicidal ∼4-kDa Crps by specific cleavage events mediated by matrix metalloproteinase-7 (MMP-7) (32, 33). MMP-7 null mice exhibit increased susceptibility to systemic S. typhimurium infection and decreased clearance of orally administered non-invasive Escherichia coli (19, 32). Although the α-defensin proregions are sensitive to proteolysis, the mature, disulfide-stabilized peptides resist digestion by their converting enzymes in vitro, whether the convertase is MMP-7 (32), trypsin (34), or neutrophil serine proteinases (35). Because α-defensins resist proteolysis in vitro, we hypothesized that Paneth cell α-defensins resist degradation and remain in a functional state in the large bowel, a complex, hostile environment containing varied proteases of both host and microbial origin.
Here, we report on the isolation and characterization of a population of enteric α-defensins from the mouse colonic lumen. Full-length and N-terminally truncated Paneth cell α-defensins were identified and are abundant in the distal large bowel lumen.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant and Synthetic Peptides
Native Crp4, disulfide-null (6C/A)-Crp4, proCrp4, (M19L)-Crp1, (M19L)-Crp6, and rhesus macaque β-defensin-4 (rhBD-4) were produced by recombinant methods and purified to homogeneity as described previously (27, 36). Recombinant peptides were expressed in E. coli as N-terminal His6-tagged fusion proteins from the EcoRI and SalI sites of the pET-28a expression vector (Novagen, Inc., Madison, WI) and subsequently purified as described (29, 36, 37). Peptide homogeneity was assessed by analytical reversed-phase HPLC, acid-urea polyacrylamide gel electrophoresis (AU-PAGE) (40), and peptide masses were confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, Voyager DE, PE Biosystems, Foster City, CA) (39).
Synthetic human β-defensins 1 and 3 (hBD-1 and hBD-3) were generously provided by Drs. Michael Selsted and Gÿorgÿ Ösapay (Dept. of Pathology and Laboratory Medicine, University of California, Irvine). Synthetic Crp2 and Crp3 peptides were a gift from Dr. Wuyuan Lu (Institute of Human Virology, University of Maryland School of Medicine).
Animals
All procedures on mice were performed with approval and in compliance with the policies of the Institutional Animal Care and Use Committee of the University of California, Irvine. Outbred Swiss Webster mice were 45-day-old males from Charles River Breeding Laboratories, Inc. (Wilmington, MA). Mice were housed under 12-h cycles of light and dark and had free access to standard rat chow and water.
Preparation and Purification of Enteric α-Defensins
Peptides were isolated using procedures described previously except where noted (8). Segments of ileum and distal large bowel, consisting of the colon and rectum only, were excised from mice immediately following euthanasia by halothane inhalation followed by cervical dislocation. Protein extracts were prepared from “complete” tissue, consisting of tissue plus luminal contents. Alternatively, extracts were generated from tissue and luminal contents processed separately. To remove the luminal contents, intestinal segments were opened by longitudinal incisions, and the luminal contents were washed away from the segments by dipping the organ into ice-cold water. Samples were homogenized in 100 ml of ice-cold 60% acetonitrile plus 1% trifluoroacetic acid and incubated at 4 °C overnight prior to clarification by centrifugation and lyophilization. Lyophilized samples were resuspended in 5 ml of 5% acetic acid and chromatographed on a 10 × 60-cm Bio-Gel P-60 column (Bio-Rad), and 350 drop fractions were collected. Fractions containing α-defensins were identified by the presence of rapidly migrating peptides in AU-PAGE stained with Coomassie Brilliant Blue, a characteristic of α-defensins in this system (8, 40). The fraction of total extracted protein in α-defensin containing fractions was determined by Bradford Assay (Bio-Rad Protein Assay).
Samples of combined α-defensins were purified further by cation-exchange chromatography before analysis by SDS-PAGE, in bactericidal peptide assays, and by peptide sequencing. Lyophilized α-defensin pools were resuspended in 100 mm ammonium formate loading buffer (pH 6.2) and applied to a 3-ml CM-Sepharose column equilibrated with the same buffer and washed with 10 ml of loading buffer. Resin-bound peptides were eluted using 10-ml volumes of 0.2 m, 1.25 m, and 2.0 m ammonium acetate (pH 5.2) followed by a final wash of 2.0 m ammonium acetate. α-Defensins eluted from the resin with 1.25 m ammonium acetate. Fractions were lyophilized, resuspended in 5 ml of 5% acetic acid, dialyzed against 5 liters of 5% acetic acid, and lyophilized again.
Peptide Analysis by Mass Spectrometry
Samples were separated by C18 reversed-phase HPLC that was developed with an aqueous 15–35% gradient of acetonitrile using 0.1% trifluoroacetic acid as the ion-pairing agent (41) delivered in 90 min at 1 ml/min. Samples of HPLC fractions were mixed with an equal volume of 10 mg/ml α-cyano-4-hydroxycinnamic acid in 60% acetonitrile containing 0.1% trifluoroacetic acid and left to dry completely prior to analysis by MALDI-TOF MS (Voyager-DE, PE Biosystems) in the Mass Spectroscopy Facility of the Dept. of Chemistry at the University of California, Irvine.
HPLC fractions with masses consistent with known or predicted mouse α-defensins were subjected to performic oxidation to detect the modification of cysteine and methionine residues. Performic acid reagent was prepared by mixing 1 part hydrogen peroxide with 19 parts 97% formic acid and incubating on ice for 1 h. Lyophilized samples were dissolved in this reagent and incubated at ambient temperature for 30 min, dried in vacuo, washed once with 50 μl of ice-cold H2O, and twice washed with 20 μl of H2O. Samples of oxidation reactions were mixed 1:1 with 10 mg/ml α-cyano-4-hydroxycinnamic acid in 60% acetonitrile containing 0.1% trifluoroacetic acid prior to analysis by MALDI-TOF MS as before. Upon oxidation, cystine and cysteine are converted to cysteic acid by addition of three oxygen atoms per cysteine; methionine residues are converted to methionine sulfone by the addition of two oxygen atoms to each methionine sulfur atom. Consequently modification of peptides or proteins containing 6 cysteine residues results in a predicted increase in mass of 288 atomic mass units. Performic acid oxidation of Crps containing 6 Cys and 1 or 2 Met residues would increase peptide mass by 320 or 352 atomic mass units, respectively.
SDS-PAGE
SDS-PAGE was performed using 10–20% Tris-Tricine precast ready-gels (Bio-Rad) with samples that were diluted 1:2 with SDS-PAGE sample buffer (Bio-Rad) prepared according to manufacturer's instructions with 1% β-mercaptoethanol and boiled for 10 min. 5 μl of Kaleidoscope (Bio-Rad) prestained molecular weight standards were run on each gel.
Peptide Extraction from AU-PAGE Gels and Peptide Sequencing
α-Defensin preparations from mouse complete ileum and complete colon were run in AU-PAGE and electroblotted to 0.1 μm polyvinylidene difluoride membrane (Millipore Immobilon PSQ) using 5% acetic acid as the transfer buffer. Individual peptide bands were excised from Coomassie-stained membranes prior to N-terminal sequencing. Eleven rounds of Edman degradation were performed on an ABI 494-HT Procise Edman Sequencer by the Molecular Structure Facility at the University of California, Davis.
Bactericidal Peptide Assays
E. coli ML35, Listeria monocytogenes 10403S, and Salmonella enterica sv. typhimurium ΔphoP were target organisms for assays testing bactericidal activity of α-defensin preparations as described previously (42). Bacteria growing exponentially in Trypticase soy broth (TSB) at 37 °C were collected by centrifugation, washed, and resuspended in 10 mm PIPES (pH 7.4) supplemented with 0.01 vol. of (1% v/v) TSB (PIPES-TSB) (28, 37). Bacteria (5 × 106/ml) were exposed to varied peptide concentrations for 1 h at 37 °C in 50 μl of PIPES-TSB. Samples were diluted 1:100 with 10 mm PIPES (pH 7.4) and plated on Trypticase soy agar plates using an Autoplate 4000 (Spiral Biotech, Inc., Bethesda, MD). Surviving bacteria were quantified as colony-forming units per milliliter (CFUs/ml) after 10–18 h of incubation.
Defensin Sensitivity to Trypsin and Chymotrypsin Proteolysis
Recombinant and synthetic α-defensins and β-defensins were digested with trypsin (Sigma, T-8253) or α-chymotrypsin (Sigma, C-9135) as described below and analyzed for susceptibility to proteolysis by AU-PAGE and MALDI-TOF MS as above. Samples (5 μg) were incubated with each proteinase at 37 °C for 2 h at a substrate to enzyme molar ratio of 50:1 in 50 mm ammonium bicarbonate (pH 8.0). Samples consisting of 85% of each digest were analyzed by AU-PAGE, and the remainder of each digest was subjected to MALDI-TOF MS.
RESULTS
Apparent Paneth Cell α-Defensins in Distal Colonic Lumen
Paneth cell α-defensins were tentatively identified in protein extracts from mouse distal colonic lumen following protein separation by gel-permeation chromatography. Because mouse enteric α-defensins are highly mobile in AU-PAGE, Bio-Gel P-60 (P-60) fractions containing apparent α-defensins were subjected to AU-PAGE analysis to identify fractions containing rapidly migrating peptides of low molecular weight (40).
As expected (8), peptides with mobilities characteristic of α-defensins were detected in P-60 separations of complete (tissue plus luminal contents) mouse ileum (boxed region, Fig. 1A). In AU-PAGE, proteins resolve on the basis of charge-to-size ratio rather than as a more direct function of molecular weight alone. In mice, production of α-defensin mRNAs and peptides occurs exclusively in Paneth cells in the small intestine (8–10, 31, 43, 44). In contrast, α-defensin production in the colon has not been detected at any level (8–10, 31, 43, 44). Nevertheless, when protein extracts of complete large intestine were separated as shown for the complete ileum sample, highly mobile peptides of low molecular weight were readily apparent in AU-PAGE (boxed region, Fig. 1B). In replicate experiments, putative α-defensin-containing fractions from complete ileum and complete large bowel were combined, as were the remaining fractions containing all other extracted proteins. Less total protein was consistently extracted from complete large intestine compared with the complete ileum; however, the measurements also showed that the combined α-defensin-containing fractions represented approximately an equivalent ∼4% of all protein extracted from both sources. Additionally, putative α-defensins were visualized when similar experiments were performed for samples of complete rectum, the most distal portion of the large bowel (data not shown).
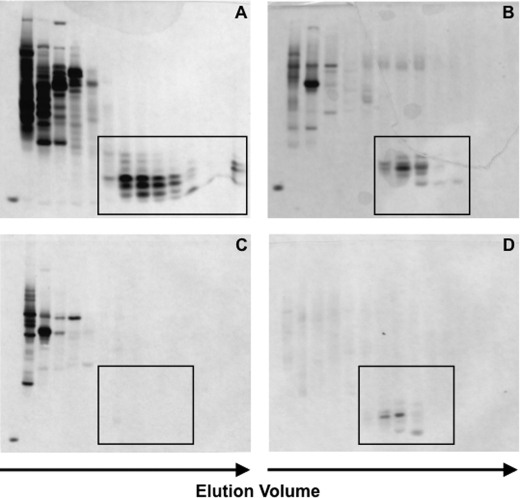
FIGURE 1.
Isolation of putative α-defensins from mouse colon. Proteins extracted from complete ileum (organ plus luminal contents) of 12 mice (A), complete colon (organ plus luminal contents) of 12 mice (B), colonic tissue of 9 mice (C), and colonic luminal contents of 9 mice (D) were separated by P-60 gel-permeation chromatography (“Experimental Procedures”). Beginning 10 fractions after elution of the void volume, every third fraction was analyzed in AU-PAGE, and gels were stained with Coomassie Blue. The arrows indicate increasing elution time from the P-60 columns. The boxed region denotes either α-defensins (A) or the anticipated position of α-defensin elution from P-60 gel-permeation chromatography columns and rapid migration in AU-PAGE (B–D). Note that no apparent α-defensin peptides were evident in the colonic tissue extract (C). Recombinant Crp4 was loaded in the far left lane in panels A–C.
Putative α-defensins among proteins extracted from complete mouse large bowel derive from colonic lumen. Proteins extracted separately from large bowel tissue and luminal contents were analyzed by gel-permeation chromatography and AU-PAGE. Consistent with the absence of both Paneth cells and α-defensin mRNAs in mouse colon (10), no apparent α-defensin peptides were detected in extracts of colonic tissue (Fig. 1C). In contrast, putative α-defensins were both evident and abundant among proteins extracted from the luminal contents of the distal colon (boxed region, Fig. 1D). Thus, the finding of putative α-defensins in the colonic lumen is consonant with previous studies showing that α-defensins are not produced in the colonic tissue and must derive from another source. Given that α-defensins have been isolated and characterized from both the mouse and human small intestinal lumen (34, 41) and Paneth cells are the only cells that express α-defensin genes in mouse small bowel, Paneth cell secretions released into small intestinal crypts are the most likely source of these peptides.
Structural Integrity of Colonic α-Defensins
To test the possibility that colonic luminal α-defensins may contain cryptic intrastrand scissions, preparations of combined ileal α-defensins and of putative colonic α-defensins were analyzed by SDS-PAGE (Fig. 2). Although the α-defensin canonical disulfide array (Fig. 3) confers resistance to proteolysis in vitro (35, 36), we considered that the complex milieu of host and microbial-derived proteases in the colonic lumen could cleave within the α-defensin polypeptide backbone, but such intrastrand scissions might not be evident in peptides stabilized by disulfide bonds. The presence of intrastrand scissions would result in the peptides disintegrating into multiple fragments upon reduction and their disappearance from the gel. However, under reducing conditions, putative α-defensins from the colon showed no evidence of cryptic scissions, co-migrating at 4 kDa with native, intact ileal α-defensins and with Crp3 and Crp4 control peptides (Fig. 2). This finding shows that the apparent α-defensins in colon are intact and resistant to intrastrand cleavage events in the highly proteolytic environment of the colonic lumen.
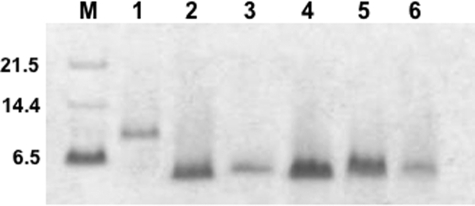
FIGURE 2.
Colonic α-defensins are structurally intact. α-Defensin preparations from ileum tissue, complete ileum, and complete colon were analyzed by SDS-PAGE under reducing conditions (“Experimental Procedures”) along with pro-Crp4, Crp4, and Crp3 control peptides. Pro-Crp4 has a molecular mass of 8221 Da, and mature, processed α-defensins have molecular masses of ∼3700–4500 Da. 5% of the total defensin preparation generated from protein extracts from four mice separated by gel-permeation chromatography and CM-Sepharose was electrophoresed in the lanes as described. Lanes: M, molecular weight markers; 1, proCrp4; 2, Crp4; 3, Crp3; 4–6, α-defensin preparations from ileal tissue, complete ileum, and complete colon, respectively. Because of their similar sizes, reduced α-defensins migrate with indistinguishable mobilities in SDS-PAGE, as observed. The combined colonic α-defensins migrate as a single band with the same apparent molecular weights as control α-defensins, indicating that intact α-defensins were isolated from the colon.
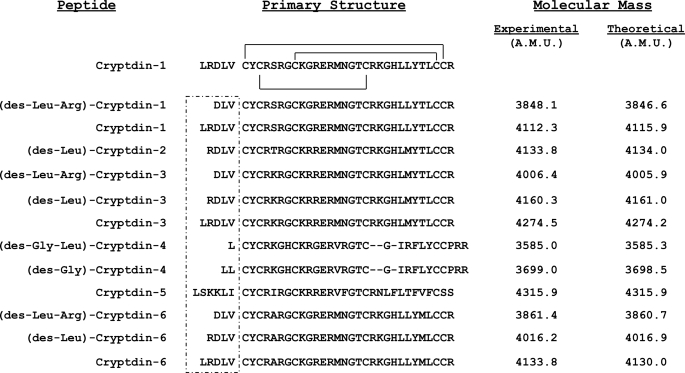
FIGURE 3.
Primary structures of colonic α-defensins deduced by MALDI-TOF MS. Masses of colonic α-defensins were determined by MALDI-TOF MS (“Experimental Procedures”). Experimental masses were compared with the theoretical masses deduced from previously characterized mouse Paneth cell α-defensin peptide and mRNA sequences and used to deduce peptide primary structures and identities. All masses are given in atomic mass units (A.M.U.). Dash characters were introduced into the alignment of certain colonic α-defensin primary structures to maintain cysteine spacing. The canonical α-defensin disulfide array is depicted for Crp1 above the aligned colonic α-defensins. Residues proximal to the first cysteine are boxed to highlight the variable N termini of colonic α-defensins. A form of each Crp was identified in the colon.
Full-length and Truncated Forms of Paneth Cell α-Defensins in Large Bowel Lumen
Putative α-defensins from colonic lumen consist of full-length Paneth cell α-defensins as well as Crp variants with truncated N termini. Preparations of nominal colonic luminal α-defensins were purified further using C18 reversed-phase HPLC, and molecular masses of the separated peptides were determined using MALDI-TOF MS. Experimental peptide masses corresponded to those of abundant Crp peptides, allowing the identities and primary structures of α-defensins isolated from the colonic lumen to be inferred (Fig. 3). For example, colonic peptides with the same molecular masses as full-length Crps 1, 3, 4, 5, and 6 were detected readily (Fig. 3). Additional MS data corresponded to variants of Crps 1, 2, 3, 4, and 6 lacking one or two amino acids from the peptide N terminus (Fig. 3). These findings are consistent with detection of full-length and N-terminally truncated α-defensins from luminal rinses of mouse ileum (41).
Chemical modification of putative α-defensins from the colon provided further evidence that they were Paneth cell α-defensins (supplemental Table S1). For example, performic acid oxidation preferentially modifies the sulfur atoms of Cys and Met residues in polypeptide chains, increasing the atomic mass of each Cys by 48 atomic mass units by addition of 3 oxygen atoms and each Met by 32 atomic mass units by addition of 2 oxygen atoms. When coupled with MALDI-TOF MS, the method provides further evidence that a molecule is a defensin in that all defensins contain six Cys residues. Specifically, a peptide mass shift of +288 atomic mass units would correspond to oxidation of a peptide that contains 6 Cys, a mass increase of 320 atomic mass units would correspond to 6 Cys and 1 Met, and an increase of 352 atomic mass units is predicted for peptides containing 6 Cys and 2 Met residues. When peptides tentatively identified as full-length or truncated Crps 1–6 were oxidized, their modified masses were consistent with their deduced identities (supplemental Table S1). For example, performic acid treatment of deduced Crp1-derived peptides increased peptide atomic masses by 320 atomic mass units, corresponding to the oxidation of 6 Cys plus 1 Met. In addition, these apparent colonic Crps were recognized by an anti-Crp1 antibody in Western blot analyses (data not shown), providing further evidence of their identities as α-defensins. These collective findings provided strong evidence that the peptides are α-defensins.
Nine putative colonic luminal α-defensins were identified definitively by N-terminal sequence analysis. Apparent α-defensins from mouse complete colon were separated in AU-PAGE (Fig. 4A), electroblotted to a polyvinylidene difluoride membrane, and stained with Coomassie Blue. Individual peptide bands were excised from the membrane (see “Experimental Procedures”). Because the N termini at the majority of mouse Paneth cell α-defensins are identical LRDLVC sequences with the first variable residue in Crps 1–3 and 6 occurring at position ten, 11 cycles of automated Edman sequence analysis were performed on each peptide band. These N-terminal sequence analyses disclosed the presence of one or two predominant sequences at the N terminus of each peptide band (Fig. 4B). Empty cycles, characteristic of unmodified cysteines, were interpreted as Cys residues at those positions. Results show that the sequenced peptides correspond to full-length, des-Leu, or des-Leu-Arg variants of Crps 1–3 or 6 as well as full-length Crp4 (Fig. 4B). These findings are compelling evidence that Paneth cell α-defensins and their N-terminal truncated forms persist intact in the lumen of the colon.
FIGURE 4.
N-terminal sequencing confirms the identities of colonic α-defensins. A, contains the following samples electrophoresed in a non-reducing AU-PAGE gel: Crp3 (lane 1), Crp4 (lane 2), a preparation of α-defensins from complete ileum (lane 3), a preparation of α-defensins from complete colon (lane 4). Individual bands from the complete colon α-defensin preparation in lane 4 are labeled with lowercase letters a–e. Peptide bands were transferred to polyvinylidene difluoride membrane as preparation for 11 cycles of N-terminal sequencing (“Experimental Procedures”) with the resulting deduced peptide identities and primary structures shown in B. Underlined sequence represents amino acid residues determined by sequencing. Because residue position 10 is the first variable amino acid among full-length Crps 1–3 and 6, it is boxed to highlight the identities of individual peptides.
Colonic Luminal α-Defensins Retain Bactericidal Activity
Colonic α-defensins were bactericidal against all bacterial species assayed, showing that they exist in a functional state (Fig. 5). Colonic α-defensin activities were compared with those of 1) corresponding pooled ileal α-defensins, 2) a mixture of control Crp peptides, and 3) Crp4. The colonic α-defensins were bactericidal in the low micromolar range, as were corresponding ileal α-defensins. Native α-defensins from both sources were most active against Listeria monocytogenes, with the colonic α-defensins exhibiting slightly greater activity than the ileal peptides at lower peptide concentrations (Fig. 5A). The colonic and ileal α-defensin samples had similar bactericidal peptide activities against E. coli ML35 (Fig. 5C) and S. enterica sv. typhimurium ΔphoP (Fig. 5E). Generally, peptide mixtures were less active than individual control peptides (Fig. 5 and supplemental Fig. S1). Results shown are representative of several experiments, and the reproducibility of these assays was demonstrated by performing determinations in triplicate at selected peptide concentrations (supplemental Fig. S2). The bactericidal activity of individual Crps within the Crp mix has been confirmed (supplemental Fig. S3). Thus, the in vitro bactericidal peptide assays show that the colonic α-defensins are microbicidal, killing ∼99% of all bacterial species tested (Fig. 5, A, C, and E).
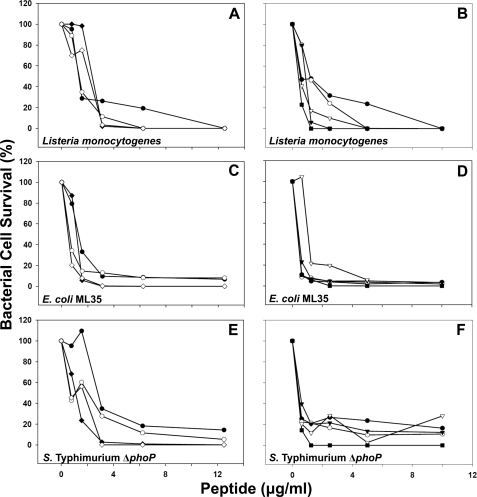
FIGURE 5.
α-Defensins isolated from the complete colon and colonic lumen retain bactericidal activity. Bactericidal peptide assays were performed against L. monocytogenes (A and B), E. coli ML35 (C and D) and S. enterica sv. typhimurium ΔphoP (E and F), surviving bacteria were counted, and data were expressed as percent of no peptide control. A, C, and E symbols: ♦, Crp4; ♢, Crp Mix; ●, α-defensins from complete ileum; ○, α-defensins from complete colon. B, D, and F symbols: ■, Crp3; ●, α-defensins from complete ileum; ○, α-defensins from complete colon; ▼, α-defensins from ileal tissue; ▽, α-defensins from colonic lumen. α-Defensin preparations from the complete colon and colonic luminal contents were bactericidal. Data are representative of multiple experiments.
To confirm that the microbicidal activities of α-defensins prepared from complete colon are derived from luminal α-defensins, bactericidal peptide activities of α-defensins purified specifically from the colonic lumen alone were determined. Colonic luminal α-defensins were compared with α-defensin preparations from complete colon, complete ileum, ileal tissue, and synthetic Crp3. As anticipated, colonic luminal α-defensins killed all bacterial species tested (Fig. 5, B, D, and F). Against L. monocytogenes, all peptide samples, including Crp3, had equivalent activities at 10 μg/ml (Fig. 5B). We conclude, therefore, that colonic luminal α-defensins are bactericidal in vitro, thereby retaining potential functional roles in innate mucosal immunity in the large bowel.
Differential Susceptibility of α- and β-Defensins to Proteolysis
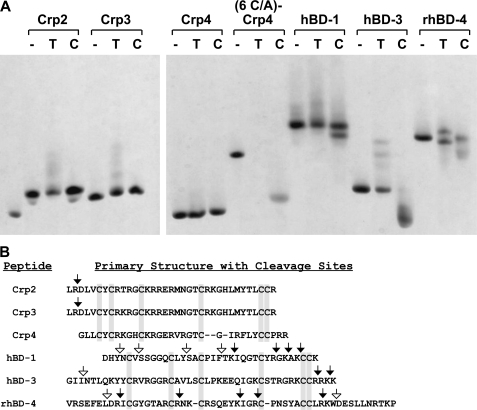
Exposure of representative α- and β-defensins to trypsin and chymotrypsin in in vitro digests showed that α-defensins were less readily proteolyzed than β-defensins (Fig. 6). Although α-defensins are not produced in the colon, certain β-defensin genes are expressed in colonocytes (45–50). Accordingly, in response to finding Paneth cell α-defensins in the colonic lumen, we assessed the relative susceptibility of certain α-defensins and β-defensins to in vitro proteolysis by trypsin and chymotrypsin. Following incubation with these proteases, samples of digests were assayed for evidence of proteolysis by AU-PAGE and MALDI-TOF MS analyses. By these indices, Crp2, Crp3, and Crp4, were more resistant to proteolysis than hBD-1, hBD-3, and rhBD-4, the prototype β-defensins tested in these assays. Native Crp4 was completely resistant to both proteases, and Crp2 and Crp3 were insensitive to chymotrypsin but were altered by trypsin as shown by their modified mobilities in AU-PAGE (Fig. 6A). MALDI-TOF MS analysis of trypsin digested Crps showed that full-length forms of Crps 2–4 remained abundant, evidence of their inherent proteolytic stability. N-terminal des-Leu-Arg variants of Crps 2 and 3 also were detected in trypsin but not chymotrypsin digests (Fig. 6B), consistent with the enzymatic specificities of the two proteinases. As anticipated, disulfide null (6C/A)-Crp4 was degraded by both enzymes (Fig. 6).
FIGURE 6.
Differential susceptibility of α-defensins and β-defensins to proteolysis by trypsin and chymotrypsin. Representative mouse α-defensins, including Crp2, Crp3, and Crp4, and β-defensins, hBD-1, hBD-3, and rhBD-4, were incubated without enzyme (−), with trypsin (T), or with chymotrypsin (C). (6 C/A)-Crp4, a peptide lacking the disulfide bonds common to all α-defensins, was included as a susceptible control peptide. Samples representing 85% of each digest were analyzed by AU-PAGE (A), and the remainder of each digest was subjected to MALDI-TOF MS to determine the masses of enzymatic cleavage products larger than 2 kDa. In B, the primary structures of the peptides are aligned with the canonical cysteine positions boxed in gray. Trypsin cleavage sites deduced from the MALDI-TOF MS analysis are shown with filled arrows, and chymotrypsin cleavage sites are shown with open arrows.
In contrast to Crp resistance to trypsin and chymotrypsin exposure, each of the three β-defensins tested were more extensively proteolyzed as judged by their modified mobilities in AU-PAGE gels (Fig. 6A). MALDI-TOF MS revealed that the β-defensins were proteolyzed N-terminal of the Cys1–Cys5 bond, and hBD-3 and rhesus BD-4 peptides also were cleaved C-terminal of the Cys3–Cys6 disulfide bond (Fig. 6B). No cleavage events were detectable in the disulfide-stabilized polypeptide chain of hBD-3. However, hBD-1 and rhBD-4 were susceptible to extensive intramolecular proteolysis within the disulfide bonds as deduced from peptide masses evident in mass spectra of their digests (Fig. 6B). Thus, under these in vitro conditions, α-defensins exhibited greater inherent resistance to proteolysis than β-defensins.
DISCUSSION
Microbicidal α-defensins occur at abundant levels in the lumen of the distal colon, even though the peptides are released exclusively by Paneth cells found only in the small intestine. The colonic α-defensin peptides are not synthesized by cells that reside in colonic tissue (8–10, 25, 43, 44) but accumulate in association with colonic luminal contents. Full-length Crps and Crps truncated N-terminally by one or two amino acid residues were identified, indicating involvement of an unknown aminopeptidase, and certain N-terminal truncations were replicated in vitro by peptide exposure to trypsin. The colonic α-defensins have overall bactericidal peptide activity equivalent to that of native α-defensin preparations extracted from ileal tissue. Accordingly, the removal of one or two N-terminal amino acids does not appear to modulate antibacterial activity or predispose the molecules to more extensive degradation. We speculate that persistence of Paneth cell α-defensins far from their sites of secretion to the most distal regions of the large intestine indicates that these molecules may also mediate mucosal immunity in the colon.
Functional, luminal α-defensins potentially operate in innate immunity alongside antimicrobial products made locally in the large bowel. Although α-defensins are not produced in the colon, colonic epithelial cells express AMPs of the β-defensin and cathelicidin peptide families. In mice, for example, β-defensin-1 (mBD-1) and the cathelicidin CRAMP are constitutively expressed in colon (51). Human colonocytes constitutively produce cathelicidin LL-37 (52, 53) and hBD-1 (47), and hBD-2, hBD-3, and hBD-4 are induced upon inflammation (47, 54). Both Gram-negative and Gram-positive bacteria produce proteases that inactivate linear AMPs, including LL-37 and CRAMP (55). Human β-defensins are susceptible to inactivating proteolysis by endogenous host proteases in airway epithelium (56), by Proteus mirabilis ZapA protease (57), and by Porphyromonas gingivalis culture supernatants, which also cleave HNP-1, a human neutrophil α-defensin (58). Although both α- and β-defensins have tridisulfide arrays and similar folded conformations, the Cys spacing and disulfide connectivities of the peptide families differ (4). Although very few peptides have been analyzed, the disulfide array appears not to be a determinant of microbicidal activity for either peptide family. On the other hand disruptions to disulfide pairing induces peptide susceptibility to proteolysis in vitro (36, 59, 60). In contrast to the sensitivity of the hBD-1 and rhBD-4 polypeptide backbones to trypsin cleavage, mouse α-defensins resisted proteolysis between Cys1 and Cys6 (Figs. 3, 4, and 6, and data not shown). Overall, the three representative β-defensins analyzed here were more susceptible than α-defensins to in vitro proteolysis with trypsin and chymotrypsin (Fig. 6). These findings suggest that β-defensins also would be more susceptible than α-defensins to degradation in the colonic lumen by varied proteinases of cellular or microbial origin. We speculate that, if this were the case, colonic AMPs, including β-defensins, may function locally at their sites of secretion from colonocytes, and α-defensins could contribute to a greater extent in the lumen.
Extracellular proteolysis of the luminal α-defensins may be mediated by varied proteases. The conversion of Paneth cell α-defensins from inactive precursors to bactericidal mature peptides differs in mice and humans. For example, human Paneth cells accumulate inactive pro-α-defensins that are converted to active forms after secretion (34), but mouse Paneth cell α-defensin precursors are processed intracellularly by MMP-7 as the activating convertase (32). Although intracellular pro-Crp processing is dependent on MMP-7, we have found that exposure of pro-Crp molecules to trypsin, cathepsin G, elastase, proteinase 3, and thrombin results in proregion proteolysis that generates native Crp peptides. The identification of des-Leu-Arg variants of several Crps in the colonic lumen is consistent with the possibility that a trypsin-like enzyme may modify Paneth cell α-defensins post-secretion. However, such variants have not been detected in protein extracts of wild-type or MMP-7-null mouse ileal tissue. On the other hand, isolation of full-length and des-Leu variant Crps from colon indicates that N-terminal truncation in the lumen is not complete or uniform and supports a potential role for multiple proteases in luminal processing of α-defensins.
Processed, microbicidal α-defensins in the large bowel milieu may participate both directly and indirectly as effectors of colonic innate immunity. For example, they may confer protection against acute infection by colonic pathogens or possibly influence the composition of the colonic microflora. Enteric α-defensins exhibit direct in vitro microbicidal activity against small intestinal pathogens, S. aureus, E. coli, L. monocytogenes, S. typhimurium, V. cholerae, and Giardia lamblia (25, 61–63). In mice, the appearance of Paneth cells during postnatal crypt development is crucial for the ability to clear in vivo infections by Shigella sp., an organism with a colonic tropism (64). Our findings provide rationale for assessing the sensitivity of Shigella sp. and additional colonic pathogens to direct killing by α-defensins.
Colonic luminal α-defensins may also influence composition of the microflora in both small intestine and large intestine (65–67). For example, quantitative and qualitative variation of the α-defensin population along the intestinal tract could contribute to differential influences of the peptides on the commensal flora, shaping the population. Achieving an understanding of the interactions between α-defensins and this microbial ecosystem is complicated, because most commensal microbes are unculturable obligate anaerobes, and the effects of defensins on these species are largely unexplored. Whether colonic α-defensins protect against pathogens directly or by influencing the microflora, we speculate that the abundance of Crps in the mouse colonic lumen suggests a function beyond the small intestine. The finding also suggests that, in enteric immunity, the site of effector molecule synthesis and secretion, e.g. Paneth cells, may not mean that function is restricted to that particular location. In conclusion, Paneth cell α-defensins resist proteolysis in vivo to persist in the distal colonic lumen, a finding that may redefine the environment in which Paneth cell α-defensins mediate innate immunity.
Supplementary Material
Acknowledgments
We appreciate gifts of Crp2 and Crp3 from Dr. Wuyuan Lu (Institute of Human Virology, University of Maryland School of Medicine) and hBD-1 and hBD-3 from Dr. Michael Selsted and Dr. Gÿorgÿ Ösapay (Dept. of Pathology and Laboratory Medicine, University of California, Irvine). Additionally, we gratefully acknowledge Dr. Michael T. Shanahan and R. Alan Llenado for their aid in preliminary studies and for useful discussions. We also thank Dr. Jack Presley at the University of California, Davis Molecular Structure Facility for assistance with Edman sequence analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants DK044632 and AI059346. This work was also supported by the Human Frontiers Science Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- AMP
- antimicrobial peptide
- Crp
- cryptdin
- MMP-7
- matrix metalloproteinase-7
- pro-Crp
- pro-cryptdin
- rhBD
- rhesus macaque β-defensin
- hBD
- human β-defensin
- HPLC
- high-performance liquid chromatography
- AU
- acid-urea
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- TSB
- trypticase soy broth
- CFU
- colony forming unit(s)
- MS
- mass spectrometry
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Lehrer R. I., Ganz T. (2002) Curr. Opin. Hematol. 9, 18–22 [DOI] [PubMed] [Google Scholar]
- 2.Lehrer R. I., Ganz T. (2002) Curr. Opin. Immunol. 14, 96–102 [DOI] [PubMed] [Google Scholar]
- 3.Schutte B. C., McCray P. B., Jr. (2002) Annu. Rev. Physiol. 64, 709–748 [DOI] [PubMed] [Google Scholar]
- 4.Selsted M. E., Ouellette A. J. (2005) Nat. Immunol. 6, 551–557 [DOI] [PubMed] [Google Scholar]
- 5.Mukherjee S., Vaishnava S., Hooper L. V. (2008) Cell Mol. Life Sci. 65, 3019–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zasloff M. (2002) Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 7.Ganz T. (2003) Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 8.Selsted M. E., Miller S. I., Henschen A. H., Ouellette A. J. (1992) J. Cell Biol. 118, 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patil A., Hughes A. L., Zhang G. (2004) Physiol. Genomics 20, 1–11 [DOI] [PubMed] [Google Scholar]
- 10.Ouellette A. J., Cordell B. (1988) Gastroenterology 94, 114–121 [DOI] [PubMed] [Google Scholar]
- 11.Ganz T., Selsted M. E., Szklarek D., Harwig S. S., Daher K., Bainton D. F., Lehrer R. I. (1985) J. Clin. Invest. 76, 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selsted M. E., Ouellette A. J. (1995) Trends Cell Biol. 5, 114–119 [DOI] [PubMed] [Google Scholar]
- 13.Eisenhauer P. B., Lehrer R. I. (1992) Infect. Immun. 60, 3446–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwig S. S., Tan L., Qu X. D., Cho Y., Eisenhauer P. B., Lehrer R. I. (1995) J. Clin. Invest. 95, 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu X. D., Lloyd K. C., Walsh J. H., Lehrer R. I. (1996) Infect. Immun. 64, 5161–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porter E. M., Bevins C. L., Ghosh D., Ganz T. (2002) Cell Mol. Life Sci. 59, 156–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cash H. L., Whitham C. V., Behrendt C. L., Hooper L. V. (2006) Science 313, 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peeters T., Vantrappen G. (1975) Gut 16, 553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayabe T., Satchell D. P., Wilson C. L., Parks W. C., Selsted M. E., Ouellette A. J. (2000) Nat. Immunol. 1, 113–118 [DOI] [PubMed] [Google Scholar]
- 20.Ganz T. (2004) C R Biol. 327, 539–549 [DOI] [PubMed] [Google Scholar]
- 21.Ouellette A. J. (2005) Springer Semin. Immunopathol. 27, 133–146 [DOI] [PubMed] [Google Scholar]
- 22.Sommers S. C. (1966) Gastroenterology 51, 841–850 [PubMed] [Google Scholar]
- 23.Boulton R. A., Usselmann B., Mohammed I., Jankowski J. (2003) World J. Surg. 27, 1014–1017 [DOI] [PubMed] [Google Scholar]
- 24.Trier J. S. (1966) Gastroenterology 51, 560–562 [PubMed] [Google Scholar]
- 25.Ouellette A. J., Hsieh M. M., Nosek M. T., Cano-Gauci D. F., Huttner K. M., Buick R. N., Selsted M. E. (1994) Infect. Immun. 62, 5040–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolls J. K., McCray P. B., Jr., Chan Y. R. (2008) Nat. Rev. Immunol. 8, 829–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satchell D. P., Sheynis T., Kolusheva S., Cummings J., Vanderlick T. K., Jelinek R., Selsted M. E., Ouellette A. J. (2003) Peptides 24, 1795–1805 [DOI] [PubMed] [Google Scholar]
- 28.Satchell D. P., Sheynis T., Shirafuji Y., Kolusheva S., Ouellette A. J., Jelinek R. (2003) J. Biol. Chem. 278, 13838–13846 [DOI] [PubMed] [Google Scholar]
- 29.Tanabe H., Qu X., Weeks C. S., Cummings J. E., Kolusheva S., Walsh K. B., Jelinek R., Vanderlick T. K., Selsted M. E., Ouellette A. J. (2004) J. Biol. Chem. 279, 11976–11983 [DOI] [PubMed] [Google Scholar]
- 30.Hadjicharalambous C., Sheynis T., Jelinek R., Shanahan M. T., Ouellette A. J., Gizeli E. (2008) Biochemistry 47, 12626–12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salzman N. H., Ghosh D., Huttner K. M., Paterson Y., Bevins C. L. (2003) Nature 422, 522–526 [DOI] [PubMed] [Google Scholar]
- 32.Wilson C. L., Ouellette A. J., Satchell D. P., Ayabe T., López-Boado Y. S., Stratman J. L., Hultgren S. J., Matrisian L. M., Parks W. C. (1999) Science 286, 113–117 [DOI] [PubMed] [Google Scholar]
- 33.Ayabe T., Wulff H., Darmoul D., Cahalan M. D., Chandy K. G., Ouellette A. J. (2002) J. Biol. Chem. 277, 3793–3800 [DOI] [PubMed] [Google Scholar]
- 34.Ghosh D., Porter E., Shen B., Lee S. K., Wilk D., Drazba J., Yadav S. P., Crabb J. W., Ganz T., Bevins C. L. (2002) Nat. Immunol. 3, 583–590 [DOI] [PubMed] [Google Scholar]
- 35.Kamdar K., Maemoto A., Qu X., Young S. K., Ouellette A. J. (2008) J. Biol. Chem. 283, 32361–32368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maemoto A., Qu X., Rosengren K. J., Tanabe H., Henschen-Edman A., Craik D. J., Ouellette A. J. (2004) J. Biol. Chem. 279, 44188–44196 [DOI] [PubMed] [Google Scholar]
- 37.Shirafuji Y., Tanabe H., Satchell D. P., Henschen-Edman A., Wilson C. L., Ouellette A. J. (2003) J. Biol. Chem. 278, 7910–7919 [DOI] [PubMed] [Google Scholar]
- 38.Deleted in proof
- 39.Weeks C. S., Tanabe H., Cummings J. E., Crampton S. P., Sheynis T., Jelinek R., Vanderlick T. K., Cocco M. J., Ouellette A. J. (2006) J. Biol. Chem. 281, 28932–28942 [DOI] [PubMed] [Google Scholar]
- 40.Selsted M. E. (1993) Genet. Eng. 15, 131–147 [DOI] [PubMed] [Google Scholar]
- 41.Ouellette A. J., Satchell D. P., Hsieh M. M., Hagen S. J., Selsted M. E. (2000) J. Biol. Chem. 275, 33969–33973 [DOI] [PubMed] [Google Scholar]
- 42.Ayabe T., Satchell D. P., Pesendorfer P., Tanabe H., Wilson C. L., Hagen S. J., Ouellette A. J. (2002) J. Biol. Chem. 277, 5219–5228 [DOI] [PubMed] [Google Scholar]
- 43.Ouellette A. J., Greco R. M., James M., Frederick D., Naftilan J., Fallon J. T. (1989) J. Cell Biol. 108, 1687–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouellette A. J., Miller S. I., Henschen A. H., Selsted M. E. (1992) FEBS Lett. 304, 146–148 [DOI] [PubMed] [Google Scholar]
- 45.Rodenburg W., Keijer J., Kramer E., Roosing S., Vink C., Katan M. B., van der Meer R., Bovee-Oudenhoven I. M. (2007) BMC Microbiol. 7, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cunliffe R. N., Mahida Y. R. (2004) J. Leukoc Biol. 75, 49–58 [DOI] [PubMed] [Google Scholar]
- 47.O'Neil D. A., Porter E. M., Elewaut D., Anderson G. M., Eckmann L., Ganz T., Kagnoff M. F. (1999) J. Immunol. 163, 6718–6724 [PubMed] [Google Scholar]
- 48.Tarver A. P., Clark D. P., Diamond G., Russell J. P., Erdjument-Bromage H., Tempst P., Cohen K. S., Jones D. E., Sweeney R. W., Wines M., Hwang S., Bevins C. L. (1998) Infect. Immun. 66, 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ou G., Baranov V., Lundmark E., Hammarström S., Hammarström M. L. (2009) Scand. J. Immunol. 69, 150–161 [DOI] [PubMed] [Google Scholar]
- 50.Takahashi A., Wada A., Ogushi K., Maeda K., Kawahara T., Mawatari K., Kurazono H., Moss J., Hirayama T., Nakaya Y. (2001) FEBS Lett. 508, 484–488 [DOI] [PubMed] [Google Scholar]
- 51.Iimura M., Gallo R. L., Hase K., Miyamoto Y., Eckmann L., Kagnoff M. F. (2005) J. Immunol. 174, 4901–4907 [DOI] [PubMed] [Google Scholar]
- 52.Hase K., Eckmann L., Leopard J. D., Varki N., Kagnoff M. F. (2002) Infect. Immun. 70, 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schauber J., Svanholm C., Termén S., Iffland K., Menzel T., Scheppach W., Melcher R., Agerberth B., Lührs H., Gudmundsson G. H. (2003) Gut 52, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fahlgren A., Hammarstrom S., Danielsson A., Hammarstrom M. L. (2004) Clin. Exp. Immunol. 137, 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidtchen A., Frick I. M., Andersson E., Tapper H., Björck L. (2002) Mol. Microbiol. 46, 157–168 [DOI] [PubMed] [Google Scholar]
- 56.Taggart C. C., Greene C. M., Smith S. G., Levine R. L., McCray P. B., Jr., O'Neill S., McElvaney N. G. (2003) J. Immunol. 171, 931–937 [DOI] [PubMed] [Google Scholar]
- 57.Belas R., Manos J., Suvanasuthi R. (2004) Infect. Immun. 72, 5159–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carlisle M. D., Srikantha R. N., Brogden K. A. (2009) J. Innate Immun. 1, 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson C. L., Schmidt A. P., Pirilä E., Valore E. V., Ferri N., Sorsa T., Ganz T., Parks W. C. (2009) J. Biol. Chem. 284, 8301–8311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoover D. M., Wu Z., Tucker K., Lu W., Lubkowski J. (2003) Antimicrob. Agents Chemother. 47, 2804–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aley S. B., Zimmerman M., Hetsko M., Selsted M. E., Gillin F. D. (1994) Infect. Immun. 62, 5397–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisenhauer P. B., Harwig S. S., Lehrer R. I. (1992) Infect. Immun. 60, 3556–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harwig S. S., Eisenhauer P. B., Chen N. P., Lehrer R. I. (1995) Adv. Exp. Med. Biol. 371A, 251–255 [DOI] [PubMed] [Google Scholar]
- 64.Fernandez M. I., Regnault B., Mulet C., Tanguy M., Jay P., Sansonetti P. J., Pédron T. (2008) J. Immunol. 180, 4924–4930 [DOI] [PubMed] [Google Scholar]
- 65.Bik E. M., Eckburg P. B., Gill S. R., Nelson K. E., Purdom E. A., Francois F., Perez-Perez G., Blaser M. J., Relman D. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shanahan F. (2002) Best Pract. Res. Clin. Gastroenterol. 16, 915–931 [DOI] [PubMed] [Google Scholar]
- 67.Salzman N. H., Underwood M. A., Bevins C. L. (2007) Semin. Immunol. 19, 70–83 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.