Abstract
The interplay of transcription factors, histone modifiers, and DNA modification can alter chromatin structure that epigenetically controls gene transcription. During severe systemic inflammatory (SSI), the generation of facultative heterochromatin from euchromatin reversibly silences transcription of a set of acute proinflammatory genes. This gene-specific silencing is a salient feature of the endotoxin tolerant phenotype that is found in blood leukocytes of SSI patients and in a human THP-1 cell model of SSI. We previously reported that de novo induction of the NF-κB transcription factor RelB by endotoxin activation is necessary and sufficient for silencing transcription of acute proinflammatory genes in the endotoxin tolerant SSI phenotype. Here, we examined how RelB silences gene expression and found that RelB induces facultative heterochromatin formation by directly interacting with the histone H3 lysine 9 methyltransferase G9a. We found that heterochromatin protein 1 (HP1) and G9a formed a complex at the interleukin-1β promoter that is dependent on the Rel homology domain (RHD) of RelB. RelB knockdown disassociated the complex and reversed transcription silencing. We also observed that whereas RelB chromatin binding was independent of G9a, RelB transcriptional silencing required G9a accumulation at the silenced promoter. Binding between RelB and G9a was confirmed by glutathione S-transferase pulldown in vitro and coimmunoprecipitation in vivo. These data provide novel insight into how RelB is required to initiate silencing in the phenotype associated with severe systemic inflammation in humans, a disease with major morbidity and mortality.
Inflammation is an evolutionarily conserved stereotypic stress response primarily orchestrated by temporal alterations in gene expression, with important contributions from complement, coagulation, and neurogenic processes (1, 2). The genetic information encoded to generate inflammation regulates distinct functional sets of genes, including pro- and anti-inflammatory modifiers, directors of cell death, and mediators of cell respiration and metabolism (3). The initiating stage of virtually all inflammation depends on sensory receptors coupled to intracellular signals that activate the immunity master regulator NF-κB to generate p65 and p50 transactivating heterodimers at euchromatin promoters of a set of early response proinflammatory genes. When spread throughout the circulation, this early stage may precipitate the extreme stress response of severe systemic inflammation (SSI).4 Later stages of inflammation reprogramming often require protein synthesis and induce expression of distinct sets of genes with anti-inflammatory, survival, and energy regulation (4, 5). Recent data in humans from our laboratory and in animals from others highlight the role of epigenetics in regulating gene expression in the SSI phenotype (6–11). These epigenetic events provide specificity and plasticity among distinct sets of genes and depend on varied chromatin structure and modifications rather than distinct signaling pathways.
Chromatin exists in structural forms that provide access of DNA to transcription factors in responsive euchromatin and mask access to the transcriptional apparatus in compacted heterochromatin (12). Facultative heterochromatin defines chromatin that can switch between open euchromatin and compacted heterochromatin (13). The mechanisms that control compaction and de-compaction of facultative heterochromatin to influence phenotypes through epigenetic control of gene expression are poorly defined but involve the cooperative interplay of transcription factors and modifiers of nucleosomal histones and DNA (12, 14).
Regulation of chromatin structure through covalent and non-covalent histone modifications is critical for epigenetically regulating gene expression across the genome (15). Histone lysine methylation participates in various chromatin-associating functions, including transcriptional regulation, heterochromatin formation, DNA repair, and recombination (16, 17). G9a is a major mammalian methyltransferase responsible for mono- and dimethylation of histone H3K9 at euchromatin regions (18). Dimethylated H3K9 contributes to binding of heterochromatin protein 1 (HP1) and transition of accessible (open) euchromatin to compacted (closed) and transcriptionally silenced facultative heterochromatin (13). We and others reported that epigenetic alterations generate gene-specific control of SSI by Toll-like receptor 4 (TLR4) induces chromatin modifications in blood leukocytes and lung dendritic cells (6–11). These chromatin modifications are associated with reversibly silencing one class of genes, which includes proinflammatory mediators, and sustaining or accentuating transcription activation of other classes of genes, including anti-inflammatory and anti-microbial mediators (3). The epigenetic transcription silencing of acute proinflammatory genes requires G9a-dependent dimethylation of histone H3 lysine 9, recruitment of HP-1, CpG methylation of DNA, and assembly of H1 linker histone and high mobility group Box 1 (HMGB1) (7–9).
Information is limited on the nature of the molecular bridge between the transcription factors and epigenetic determinants. We discovered that de novo induction of NF-κB transcription factor RelB after TLR4 stimulation is necessary and sufficient for silencing transcription of TNFα and IL-1β in the SSI phenotype (8, 19). We also found that RelB can function in the same cell type as a dual transcription regulator in the SSI phenotype to deactivate transcription of acute proinflammatory genes while activating transcription of the NF-κB regulator IκBα (20). RelB also participates in constitutive silencing of inflammatory genes in fibroblasts by a process that supports regional methylation of CpG DNA (21), and when normally silenced fibroblasts are rendered RelB−/−, they become responsive to LPS (22).
In this study, we examined how RelB couples to epigenetically silence expression of acute proinflammatory genes and found that RelB initiates facultative heterochromatin formation by interacting with the histone H3 lysine methyltransferase G9a, which then mediates heterochromatin formation.
EXPERIMENTAL PROCEDURES
Cell Culture Model of SSI
THP-1 cells obtained from American Type Culture Collection were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10 units/ml penicillin G, 10 μg/ml streptomycin, 2 mm l-glutamine, and 10% fetal bovine serum (HyClone) at 37 °C and 5% CO2 in a humidified incubator. LPS-mediated tolerance that mimics the SSI phenotype in THP-1 cells was previously described (23). Briefly, LPS tolerance is generated by an initial stimulation with LPS (0111:B4; 1 μg/ml) for 16 h followed by re-stimulation with 1.0 μg/ml LPS for 3 h. This LPS acts exclusively through TLR4 receptor as determined in cells lacking TLR4. High concentrations of LPS are used to optimize the tolerant phenotype, although changes occur with doses as low as 10–100 ng/ml. Tolerance occurs within 3 h and sustains for at least 96 h (3). Normal and LPS tolerant THP-1 cells (1 × 106 cells/sample) were washed once with RPMI 1640, re-suspended in fetal bovine serum supplemented RPMI 1640 medium at 1 × 106 cells/ml, and stimulated with LPS 1 μg/ml for 3 h. Low passage number and log-phase cells were used for all experiments.
Chromatin Immunoprecipitation (ChIP) Assay
To assess p65, p50, RelB, G9a, HP1, and H3K9me2 binding to the IL-1β promoter in LPS tolerant and normal cells, ChIP assays (Upstate Biotechnology) were performed according to the manufacturer's instructions with the following modifications. Cells (5 × 106 cells/sample) were fixed by adding formaldehyde (from a 37% formaldehyde, 10% methanol stock (Calbiochem)) into the medium for a final formaldehyde concentration of 1% and incubated at room temperature for 10 min with gentle shaking. The chromatin was disrupted by sonication using a Diagenode Bioruptor (UCD-200TM-EX, Tosho Denk1 Co., Ltd). High power sonification (30 s on and 30 s off for 23 min) at this setting generated DNA fragments of ∼0.5–1.5 kilobases. Each sample was divided into two parts, providing an “input” sample that was not incubated with antibodies. The other portion was incubated overnight with antibodies specific for p65 (SC-372), p50 (SC-7178), RelB (SC-226), HP1 (SC-10215), and IgG (SC-2027) for the negative control (Santa Cruz Biotechnology, Santa Cruz, CA) and G9a (07–551) (Upstate Biotechnology). Purified DNA was re-suspended in 10 μl of distilled H2O.
Co-immunoprecipitation (IP) ChIP
The method of co-IP ChIP was performed according to a previous report (24). In brief, cells were fixed and chromatin-sheared as above. Immunoprecipitation was conducted with IgG, anti-RelB, or anti-G9a and 30 μl of protein A/G-agarose beads (50% slurry, Santa Cruz, SC-2003) with rotation overnight at 4 °C. Immunocomplexes were eluted by incubation with 10 mm dithiothreitol at 37 °C for 30 min and diluted 1:50 in IP dilution buffer. Elutes were then re-immunoprecipitated with second antibodies. Purified DNA was resuspended in 10 μl of distilled H2O.
Real-time PCR
The chromatin-immunoprecipitated DNA for each treatment was analyzed quantitatively for the amplification of the IL-1β promoter using specific primers (forward, 5′-cgtgggaaaatccagtattttaatg-3′, and reverse, 5′-caaatgtatcaccatgcaaatatgc-3′) and probe (5′-6-carboxyfluorescein-acatcaactgcacaacgattgtcaggaaaa-6-carboxytetramethylrhodamine-3′) (Applied Biosystems) as we used before. The real-time PCR reaction (total 25 μl) contained 3 μl of DNA, 12.5 μl of 2×TaqMan Universal Master Mix, 300 nm concentrations of each primer, and 100 nm dNTPs. The real-time PCR procedure was 2 min at 50 °C and 10 min at 95 °C followed by 40 cycles with 15 s at 95 °C and 1 min at 60 °C using the ABI Prism 7000 Sequence Detection System (Applied Biosystems). Data were normalized to the input DNA and are presented as -fold change relative to DNA from untreated cells.
To measure IL-1β mRNA, total RNA was isolated at various times after LPS stimulation using RNA STAT-60, according to the manufacturer's protocol (Tel-Test, Friendswood, TX). One microgram of RNA was reverse-transcribed to cDNA in a 20-μl volume containing 5 mm MgCl2, 1 mm dNTP, 2.5 μm oligo(dT), and 2.5 units/μl murine leukemia virus reverse transcriptase (Applied Biosystems). The PCR was performed using 4 μl of cDNA, and glyceraldehyde-3-phosphate dehydrogenase-pre-designed TaqMan primer/probe kits (Applied Biosystems) under the conditions described above. Data were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA and are presented as -fold change relative to mRNA from untreated cells.
Standard PCR
A standard PCR reaction (total 25 μl) containing 2 μl of ChIP DNA, 1 μm concentrations of each primer (as above), 2 mm MgCl2, 0.2 m dNTPs, and 0.03 units/μl AmpliTaq Gold DNA polymerase (Applied Biosystems) was performed to confirm the results of real-time PCR. The procedure was 1 cycle at 94 °C for 5 min, 35 cycles at 55 °C 30 s, and 72 °C for 30 s each plus a final cycle at 72 °C for 10 min. The PCR products were visualized on 1.5% agarose gels, and images were captured using Quantity One Imager (Bio-Rad).
Western Blot
Total, nuclear, and cytoplasmic protein was assayed by Western blot. The methods for protein extraction are the same as we used before (19). RelB, G9a, HP1, p65, p50, and HA antibodies (Santa Cruz) were used to visualize and quantify protein levels using ImageQuant software (GE Healthcare).
RNA Interference
Pools of control (SC-37007), RelB-specific (SC-36402), G9a-specific (SC-43777), and HP1-specific (SC-37737) small interfering RNAs (siRNAs; Santa Cruz) were transfected by electroporation with 8 μl (0.5 μm) of siRNA in 100 μl of transfection medium (Nucleofector; Amaxa, Gaithersburg, MD) as previously described (19). Each pool included three siRNA sequences that specifically target different sequence of the target gene to ensure specific knockdown. To check for off-target effects by siRNA, our previous experiments indicated that the expression of other inflammatory proteins, like IκBα and p65, is not affected by the introduction of these siRNA species into tolerant cells. In addition, Western blot data confirm that RelB or G9a knockdown by siRNA did not affect the expression of the reciprocal gene. Thus, no off-target effects have been identified. Immediately after transfection, cells were left unstimulated or treated with 1 μg/ml LPS to induce tolerance. After 48 h, cells were harvested, washed, and stimulated for 3 h with 1 μg/ml LPS. Cells were harvested, and RNA was isolated and used to determine the IL-1β mRNA expression levels.
Immunoprecipitation
In vivo binding of G9a and RelB was assayed by IP. Cells (∼30 × 106) were harvested in cold phosphate-buffered saline (PBS) and washed with PBS. Then the cell pellet was suspended in EBS lysis buffer (50 mm Tris-HCl, 120 mm NaCl, 0.5% Nonidet P-40, and protease inhibitor) and incubated on ice for 30 min. After centrifugation of cell lysates (15 min, 14,000 × g, 4 °C), samples of supernatant termed “cell extract” were used for IP. Samples of total extract (1 mg of protein) plus 4 μg of G9a antibody (ab40542-100), RelB antibody (ab33907-100) (Abcam Biotechnology) or normal rabbit IgG (SC-2027) (Santa Cruz), and 40 μl of protein A/G beads (50% slurry, Santa Cruz, SC-2003) were incubated at 4 °C overnight with agitation. Beads were washed 6 times with NETN buffer (20 mm Tris-HCl, 1 mm EDTA, 900 mm NaCl, 0.5% Nonidet P-40) then washed once with NETN buffer with 100 mm NaCl before elution (95 °C, 5 min) of bound proteins in 100 μl of gel-loading buffer. Samples of eluted proteins were assayed Western blot and incubated with RelB or G9a antibody followed by appropriate horseradish peroxidase-conjugated secondary antibodies and Super Signal Chemiluminescent substrate (Pierce). Procedures used for IP and Western blotting were identical in all experiments with quantities adjusted for scale of each. All observations were replicated at least three times.
Plasmid Construction
It has been shown that the C-terminal part of the Rel homology domain (between amino acids 264 and 379) is essential for effective RelB dimerization (25). RelB mutant (truncated RelB with amino acid 264–379 deleted) was generated by GenScript Corp. DNA fragments encoding N-terminal G9a (amino acids 1–500) and C-terminal G9a (amino acids 501–1000) were generated with the Expand High Fidelity PCR System (Roche Applied Science) from human G9a pEGFP-C3 plasmid (kindly provided by Dr Yoichi Shinkai, Institute for Virus Research, Kyoto University). The primers used for amplifying the N-terminal G9a domain were 5′- aacagggatccagcatgagtgatgatgtccactcactgg-3′ (forward) and 5′-ctgtggaattcctcagttggctccagcctgcagcagcac-3′ (reverse). That for C-terminal G9a was 5′-aacagggatccagcatgataaatgcagtggacaaacagc-3′ (forward). Amplified DNA was cloned into the pGEX-4T-1 vector (GE Healthcare) and generated recombinant glutathione S-transferase (GST) fusion proteins. The constructs were verified by automatic DNA sequencing, and their protein expression was confirmed by Western blotting.
GST Pulldown Assay
GST, GST N/C-terminal G9a plasmids were transformed into RosettaTM(DE3)pLysS Competent Cells from Novagen, and the protein expression was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside up to a concentration of 1 mm at 37 °C for 2.5 h. To generate [35S]Met-labeled proteins, RelB template generated with Expand High Fidelity PCR System (Roche Applied Science) from pCDNA3-RelB (gift from Dr. G. Bren) was added to the TNT Quick Coupled Transcription/Translation System (Promega). The primers used for producing RelB template were 5′-ggatcctaatacgactcactataggaacagccaccatgcttcggtctgggccagcctct-3′ (forward) and 5′-ctacgtggcttcaggccccggggataggaggcc-3′ (reverse). GST pulldown was performed using ProFound pulldown GST protein-protein interaction kit (Pierce). After pulldown, proteins were eluted out and analyzed by Western blot.
Statistical Analysis
Statistical analysis and graphic presentations were performed using Microsoft Excel XP. Results shown are the average of three experiments. Significance was taken at p < 0.05 (Student's t test).
RESULTS
RelB, G9a, and HP1 Form a Complex at the Silenced IL-1β Promoter
To test whether G9a and HP1 form a complex with RelB at the IL-1β promoter during transcription silencing, we employed the co-IP and ChIP assay that uses a sequel of primary and secondary antibodies to assess potential in vivo cooperation among promoter regulators. Using anti-RelB as the primary antibody and G9a and HP1 as secondary antibodies, we first demonstrated that a complex of the three proteins forms at the promoter of silenced cells (Fig. 1). Input DNA and IgG were used as controls. A similar paradigm occurred when we used G9a as the primary antibody and RelB and HP1 as secondary antibodies. Under these conditions, p65 did not accumulate at the promoter (not shown). Although these data suggest that a cooperative complex of the transcription factor and histone modifiers form at the silenced IL-1β promoter, they do not establish the functional relevance of this relationship nor do they establish whether the binding interactions of the three proteins are direct or indirect.
FIGURE 1.
RelB, G9a, and HP1 form a complex at the silenced IL-1β promoter. A, a diagrammatic representation of the IL-1β proximal promoter region analyzed by ChIP. The NF-κB site and the location of the primers used in PCR are shown. B, chromatin from normal or silenced cells stimulated for 3 h with 1.0 μg/ml LPS was cross-linked and immunoprecipitated with RelB antibody (1°). To demonstrate that G9a and HP1 form a complex with RelB at the silenced IL-1β promoter, RelB-specific ChIP complexes were further immunoprecipitated with anti-G9a or HP1 antibodies (2°) as described under “Experimental Procedures.” Input to the RelB ChIP and IgG controls is also shown. These results are representative of three independent observations. N, normal; S, silenced.
G9a and HP1 Binding to the IL-1β Promoter Is Dependent on RelB
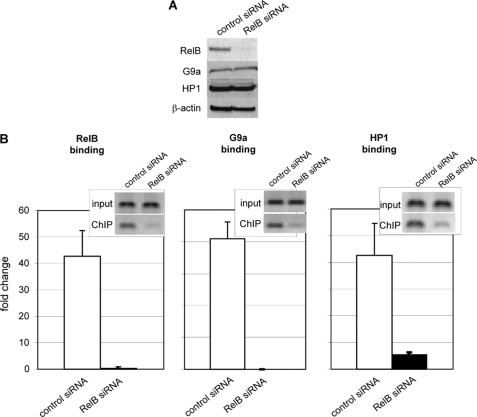
To assess whether RelB might function as an initiating step in assembling facultative heterochromatin during transcription repression, we used RelB siRNA to determine the effect of removing RelB on the promoter binding of G9a and HP1. Immunoblotting confirmed the decreased expression of RelB protein after transfection with RelB siRNA without demonstrable changing the levels of G9a and HP1 proteins (Fig. 2A). After siRNA RelB knockdown, ChIP assays showed reduced enrichment of RelB, G9a, and HP1 at the IL-1β promoter compared with the results using control siRNA (Fig. 2B). The small insets show results using standard PCR, and the real-time PCR data are plotted as bar graphs. Three independent experiments were performed, and the observed changes were statistically significant. The disruption of the complex of transcription and chromatin mediators also correlated with reduced generation of the H3K9 dimethylation silencing mark and increased binding of p65 (not shown). Together, these results supported that the accumulation of G9a and HP1 at the IL-1β promoter is dependent on the presence of RelB and that together these chromatin modifiers may support a RelB-dependent shift in the histone code from activating to silencing through histone H3K9 dimethylation.
FIGURE 2.
G9a and HP1 binding to the IL-1β promoter is dependent on RelB. A, nuclear extracts from silenced cells transfected with control siRNA or RelB siRNA were analyzed by immunoblot as described under “Experimental Procedures” and show decreased expression of RelB. No detectable change in G9a or HP1 protein levels was observed. β-Actin gel loading control is also shown. Results shown are representative of three independent experiments. B, RelB knockdown in silenced cells significantly (p ≤ 0.05) decreases RelB binding to the IL-1β promoter. Silenced cells were transfected with control siRNA (open bars) or RelB siRNA (closed bars). RelB (left), G9a (middle), or HP1 (right) ChIP DNA was analyzed for the presence of IL-1β promoter sequences by standard PCR (inset) or real-time PCR (bar graph) as described under “Experimental Procedures.” ChIP analysis also shows that RelB knockdown significantly decreased G9a and HP1 at the IL-1β promoter. Results shown are the mean ± S.E. from three independent real-time PCR ChIP experiments as described under “Experimental Procedures.”.
IL-1β Promoter Silencing and RelB-dependent G9a Binding to the IL-1β Promoter Require the RelB Rel Homology Domain (RHD)
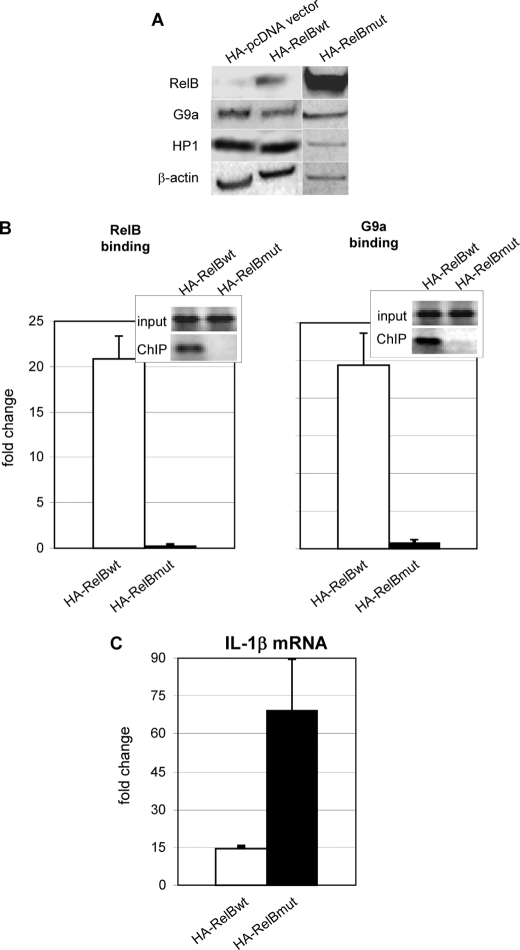
We previously reported that transfection of wild type RelB represses transcription of IL-1β in LPS-responsive THP-1 cells (19). This approach was feasible as little RelB is expressed in LPS-responsive/normal cells, whereas de novo induction of RelB occurs in transcriptionally silenced THP-1 cells. To test whether we could induce interactions of RelB and G9a in vivo by increasing levels of RelB, we transfected a wild type (HA-RelB) plasmid or a plasmid containing a mutant of the RHD region of RelB to overexpress RelB in normal THP-1 cells. RelB protein expression increased in nuclear extracts from cells transfected with RelB wild type or RelB mutant (Fig. 3A); there was no effect of transfection on levels of G9a or HP1 proteins. Fig. 3B shows that transfection of RelB wild type, but not RelB RHD mutant, induced binding of both RelB and G9a to the IL-1β promoter. Fig. 3C depicts that reduced levels of IL-1β mRNA occurred after transfection of wild type RelB but not RelB RHD mutant. These data support that RelB initiates silencing by a process that requires the RHD domain and that nuclear levels of the protein may provide the rate-limiting step for silencing.
FIGURE 3.
IL-1β promoter silencing and RelB-dependent G9a binding to the IL-1β promoter requires the RHD domain of RelB. A, nuclear extracts from THP-1 cells transfected with HA-pcDNA, HA-RelBwt, or HA-RelBmut expression vectors were analyzed by immunoblotting as described under “Experimental Procedures.” RelB expression was increased in nuclear extracts from cells transfected with RelBwt or RelBmut. No detectable difference in G9a or HP1 protein levels was observed. Results shown are representative of three independent experiments. B, THP-1 cells were transfected with HA-RelBwt (open bars) or HA-RelBmut (closed bars). RelB (left) or G9a (right) chromatin-immunoprecipitated DNA was analyzed for the presence of IL-1β promoter sequences by standard PCR (inset) or real-time PCR (bar graph) as described under “Experimental Procedures.” The lack of RHD in the RelBmut significantly (p ≤ 0.05) decreased RelB as well as G9a binding to the IL-1β promoter. Results shown are the mean ± S.E. from three independent real-time PCR experiments as described under “Experimental Procedures.” C, real time measurement of IL-1β mRNA shows that promoter silencing requires the RHD domain of RelB (compare HA-RelBwt (open bars) with HA-RelBmut (closed bars)). Results shown are the mean ± S.E. from three independent measurements of IL-1β mRNA. wt, wild type.
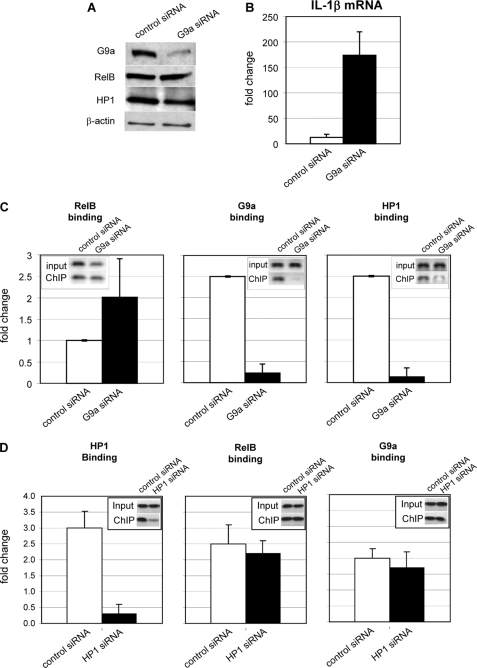
RelB-dependent Silencing Is Mediated by G9a
Because RelB is both necessary and sufficient for transcription silencing (19) and because the data in this study thus far supported a RelB-dependent proximal process, we determined whether the transcription silencing activity of RelB is mediated by a link to G9a. To test this we used G9a-specific siRNA to deplete G9a protein, a process that did not substantially alter RelB or HP1 protein levels as assessed by immunoblotting (Fig. 4A). We then assessed whether depletion of G9a by siRNA would reverse silencing, as determined by mRNA analysis with control siRNA versus G9a siRNA. Fig. 4B shows that responsivity of the promoter to LPS stimulation of IL-1β mRNA is restored in the G9a knockdown. To determine the effect of the G9a knockdown on occupancy of the promoter by RelB and HP1 in the presence of reductions in G9a and reversal of transcription silencing, we used ChIP analysis. The results in Fig. 4C support that RelB remains bound to the IL-1β promoter (although at lower levels) after G9a knockdown, whereas HP1 and G9a are removed. This indicates that RelB silencing activity likely requires G9a-dependent modifications of chromatin.
FIGURE 4.
RelB-dependent gene silencing is mediated by G9a. A, nuclear extracts from silenced cells transfected with control siRNA or G9a siRNA were analyzed by immunoblotting as described under “Experimental Procedures” and show decreased expression of G9a. No detectable change in RelB or HP1 protein levels was observed. Results shown are representative of three independent experiments. B, real-time PCR measurement of IL-1β mRNA shows that promoter silencing is mediated by G9a (compare control siRNA (open bars) with G9a siRNA (closed bars)). Results shown are the mean ± S.E. from three independent real-time PCR measurements of IL-1β mRNA as described under “Experimental Procedures.” C, silenced cells were transfected with control siRNA (open bars) or G9a siRNA (closed bars). RelB (left), G9a (middle), or HP1 (right) chromatin-immunoprecipitated DNA was analyzed by standard PCR (inset) or real-time PCR (bar graph) for the presence of IL-1β promoter sequences as described under “Experimental Procedures.” G9a knockdown in silenced cells did not affect (p ≥ 0.05) RelB binding to the IL-1β promoter. ChIP analysis also shows that G9a knockdown significantly (p ≤ 0.05) decreased HP1 binding at the IL-1β promoter. Results shown are the mean ± S.E. from three independent experiments. D, silenced cells were transfected with control siRNA (open bars) or HP1 siRNA (closed bars). Chromatin was immunoprecipitated with HP1, RelB, or G9a antibody, and chromatin-immunoprecipitated DNA was analyzed was analyzed by standard PCR (inset) or real-time PCR as described in C. HP1 knockdown did not affect (p ≥ 0.05) RelB or G9a binding to the IL-1β promoter. Results shown are the mean ± S.E. from three independent experiments.
In the reverse experiment we knocked down HP1 by siRNA and measured RelB and G9a binding to the promoter. The results (Fig. 4D) showed no decrease both in RelB and G9a binding after HP1 depletion, suggesting that their assembly at the IL-1β promoter is proximal to HP1 binding.
RelB and G9a Co-immunoprecipitate in Vivo in Silenced Cells and Directly Interact in Vitro
To test whether RelB and G9a proteins might directly interact, we used a series of co- immunoprecipitations. In vivo assessments included co-immunoprecipitations of native proteins in silenced cells and of normal cells transfected with the wild type HA-RelB. RelB and G9a were used as both primary and secondary antibodies in the in vivo assays. In vitro interactions of RelB and G9a were analyzed using GST pulldown assays. Both the N and C termini of G9a were linked to GST as fusion proteins. Fig. 5, A and B, show that RelB and G9a co-immunoprecipitate with primary or secondary antibodies after transfection of HA RelB and in silenced cells in the native state. This supports that RelB and G9a couple in vivo either directly or indirectly. Fig. 5B shows that in vitro translated RelB directly interacts with GST-linked G9a in vitro and that this interaction predominates at the N terminus (1–500 amino acids) of G9a. GST vector and resin alone controls were negative. The co-immunoprecipitation experiments were repeated three times under each condition. Together these data support that RelB can initiate silencing by directly binding to G9a, which mediates formation of facultative heterochromatin, as indicated by the presence of HP1 (13).
FIGURE 5.
RelB and G9a co-immunoprecipitate in silenced cells and in vitro. A, silenced cells were transfected with empty vector or HA-RelBwt construct as described under “Experimental Procedures.” Whole cell extracts were prepared and incubated with anti-G9a, anti-HA RelB, or control IgG antibodies. Immunocomplexes were isolated, and co-immunoprecipitated proteins were analyzed by immunoblotting. Primary (1°) antibodies were used for isolation and purification of immune complexes; secondary (2°) antibodies were used for visualization of immunocomplex by immunoblot and show that HA-RelBwt forms a protein complex with G9a. These results are representative of three independent experiments. B, GST-G9a bacterially expressed fusion proteins were tested for direct protein-protein interaction with in vitro expressed RelB as described under “Experimental Procedures.” GST-G9a (N-terminal) fusion protein forms a stable complex with RelB, whereas GST-G9a (C-terminal) fusion protein does not show interaction with RelB above background (GST vector). Resin alone and RelB input to the GST pulldown assay (TNT/RelB) are also shown. These results are representative of three independent experiments.
DISCUSSION
This is the first report on a mechanism responsible for the link between an NF-κB family member and the epigenetic events that condense chromatin to heterochromatin via a link to a lysine methyltransferase and heterochromatin protein 1. These data provide novel insight into how RelB initiates silencing in the phenotype associated with severe systemic inflammation in humans, a disease with major morbidity and mortality. Silencing is dependent on the de novo induction of RelB by TLR4 activation. The data support a direct interaction of RelB with the methyltransferase G9a, which through dimethylation of histone H3 at lysine 9 (H3K9) generates a stable binding site for HP1 chromatin adapter protein and supports a switch in the histone code from active to silenced transcription (26). We further show that whereas RelB is a key initiator of this silencing cascade, G9a is a central mediator in chromatin compaction. This interpretation is based on results showing that RelB binds the promoter in the absence of G9a (G9a knockdown), which occurs at a time that transcription silencing is abrogated. Under these conditions we also show that HP1 does not bind, supporting that facultative heterochromatin chromatin reverses to euchromatin. In contrast, RelB knockdown results in removal of both G9a and HP1 from the promoter as well as reversal of both dimethylation of H3K9 and transcription silencing. Transfection of RelB in LPS-responsive cells that do not express RelB induces accumulation of RelB, G9a, and HP1 at the IL-1β promoter and silences transcription. Our in vivo studies further support that the RelB RHD is required for G9a-mediated gene silencing. Additional in vitro experiments show that the N-terminal domain of G9a participates in this interaction by directly interacting with RelB.
Although published data indicate that transcription factors and their interplay with cognate DNA are linked to gene silencing by both histone and DNA methylation responses, there are limited data about how these molecular bridges form. However, precedent for G9a or histones and DNA interactions with transcription factors or co-factors exist. The growth factor-independent 1 (Gfi1) transcriptional regulatory oncoprotein couples to G9a to silence p21 gene transcription (27). CCAAT displacement protein cut homolog also recruits G9a to repress transcription of p21 (28). In the innate immune response of cells to viral infection that is recognized by TLR, PRDI-BF1 DNA-binding protein initiates post-induction silencing of interferon-β by direct interaction with G9a, which thereafter mediates chromatin modifications (29). This scenario is somewhat analogous to what we report herein for RelB induction in response to TLR4 stimulation. Methylation and deamination of CpGs generate p53 binding sites on a genomic scale and thereby may control silencing through altering accessibility of transcription factors (30). Msx1 transcription factor and the negative regulator of muscle differentiation core enhancer region interact with linker histone protein H1 to inhibit transcription and myogenesis (31).
Our data support that binding of RelB and G9a on the promoter depends on the RHD of Rel B and the N-terminal 500 amino acids of G9a. This is not surprising for RelB, as the RHD domain is required for its binding to partners such as NF-κB p100, which are required to stabilize structurally labile RelB (25, 32). The N-terminal region of G9a also provides a structural site for protein-to-protein interactions, perhaps through its polyglutamic acid domain or cytosine-rich domain (33, 34). However, the precise N-terminal site(s) required for binding transcription factors has not been identified. The G9a ankyrin repeats that bind to methylation sites on histones and the SET domains that are enzymatically active are both located in the C terminus. Also, the precise region(s) of RelB that interacts after RelB binds to the promoter by the RHD domain are not known.
G9a is a crucial multifunctional mediator of epigenetic silencing. A recent report links the G9a chromatin compaction process both to histone H3K9 dimethylation and to methylation of lysine 27 on linker histone H1 isoform 4 (H1.4) (35). Both methylation sites recruit HP1, which facilitates facultative heterochromatin formation, and the process supports locating and retaining H1.4 at gene promoters. Another novel observation in this study is that the coordinated accumulation of H1 and HP1 can be reversed by the H1.4 lysine 27 demethylase JMJD2/KDMA, an event that could contribute to a return to the state of euchromatin and competent transcription.
This paradigm of facultative heterochromatin formation dependent on both histone H3K9 and linker histone H1 methylation modifications controlled by G9a and requiring HP1 may be important in the SSI and endotoxin-tolerant THP-1 cell. Recently, we reported that removal of H1 isoforms (including H1.4 that is referred to as H1b in humans) and HMGB1 by siRNA removes RelB from the promoters of acute proinflammatory genes IL-1β and TNFα, ablates H3K9 dimethylation, removes HP1, and reverses transcription silencing in endotoxin-tolerant THP cells (9). This paradigm also appears relevant to humans with SSI, as in the same study we note that linker histone H1 and HMGB1 accumulate in vivo at the TNFα and IL-1β promoters, but not the IkBα promoter, in peripheral blood leukocytes (9). We also find that RelB is constitutively expressed, and G9a and HP1 bind to the TNFα promoter of peripheral blood SSI human leukocytes (8, 19).
We advocate that RelB induction generates a pivotal feed-back and feed-forward transcription regulator for both activation and repression of distinct sets of genes in a cell- and context-specific manner during inflammatory processes, such as SSI. An important question is how the NF-κB pathway might control both activation of proinflammatory and anti-inflammatory phenotypes. It is known that a first wave of p65 may induce immediate response genes including TNFα and IL-1β but also inducing transcription factors that control downstream reprogramming for expression of both activators and repressors, whose dominance changes over time. This coupling process may include a second wave of p65 activation by an alternative signaling pathway such as IκB kinase α and require protein synthesis. Our unpublished preliminary data support this possibility.
There is a another implication of the importance of the NF-κB-dependent master control system. Because there are many NF-κB cognate DNA sites across the genome, our findings predict an extensive inventory for the RelB potential influence on the genetic code that regulates inflammation. This RelB-dependent reprogramming may differ among more localized inflammatory diseases like rheumatoid arthritis and inflammatory bowel disease (3), an important concept to investigate. Also, the nature of the intracellular specificity that distinguishes between transcription repression and activation by RelB is unknown, but differences in nucleosome topography among the chromosomes and/or germ line-encoded DNA proximal promoter sequences in relation to CpG sites for methylation may contribute.
Based on the results of our previous reports and the findings described herein, we provide in Fig. 6 a model for epigenetic regulation of a set of acute proinflammatory genes during SSI. SSI is often initiated by TLR4-dependent processes that require NF-κB activation and binding of p65 with cooperating transactivators and co-factors to multiple gene promoters, including those of rapid-response acute proinflammatory mediators as well as downstream factors. Epigenomic reprogramming follows the early events. A critical component of the temporal sequence of gene expression and the epigenetic reprogramming of SSI is expression of the NF-κB factor RelB, which functions as a key initiator of the silencing cascade by its direct interaction with G9a. G9a acts as a key mediator in gene silencing through dimethylation of H3K9, which provides a site for recruitment of HP1. Other coordinated events that participate in chromatin compaction and are supported, at least in part by G9a and HP1, include CpG methylation and recruitment of linker histone H1 and HMGB1 at proximal promoters. The resulting state of silenced facultative heterochromatin may be sustained for days but can return to euchromatin under certain experimental conditions and in SSI survivors. Exploiting this epigenetic plasticity may provide opportunities for specific therapies for inflammatory diseases.
FIGURE 6.
Model for TLR-4-induced RelB-dependent formation of facultative heterochromatin and transcription silencing of acute proinflammatory genes in the SSI phenotype. This conceptual model is based on this study and components of our previously published work. SSI is initiated when euchromatin is activated by membrane sensors such as TLR4 that couple to NF-κB activation and subsequent binding of p65 and p50 heterodimers to the promoters of a set of acute proinflammatory genes. Subsequent induction of RelB leads to its recruitment and direct binding to G9a. G9a dimethylates H3K9 to which HP1 binds and links to Dnmt3 binding and DNA CpG methylation. Linker histone H1 and HMGB1 (not shown) are recruited and may facilitate remodeling of the nucleosome(s) that “locks” compacted chromatin and stabilizes the p50/RelB/G9a bond.
Acknowledgments
We thank Jean Hu and Sue Cousart for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1AI-09169 and RO1AI-065791 (to C. E. M.).
- SSI
- severe systemic inflammation
- HP1
- heterochromatin protein 1
- IP
- immunoprecipitation
- ChIP
- chromatin immunoprecipitation
- GST
- glutathione S-transferase
- H3K9
- histone H3 lysine 9
- HMGB1
- high mobility group box protein 1
- IL-1β
- interleukin 1β
- LPS
- lipopolysaccharide endotoxin
- RHD
- Rel homology domain
- TLR4
- Toll-like receptor 4
- TNFα
- tumor necrosis factor α
- HA
- hemagglutinin
- siRNA
- small interfering RNA.
REFERENCES
- 1.Hotchkiss R. S., Karl I. E. (2003) N. Engl. J. Med. 348, 138–150 [DOI] [PubMed] [Google Scholar]
- 2.Riedemann N. C., Guo R. F., Ward P. A. (2003) J. Clin. Invest. 112, 460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCall C. E., Yoza B. K. (2007) Am. J. Respir. Crit. Care Med. 175, 763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobb J. P., Buchman T. G., Karl I. E., Hotchkiss R. S. (2000) Surg. Infect. (Larchmt) 1, 207–215 [DOI] [PubMed] [Google Scholar]
- 5.Ramirez-Carrozzi V. R., Nazarian A. A., Li C. C., Gore S. L., Sridharan R., Imbalzano A. N., Smale S. T. (2006) Genes Dev. 20, 282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan C., Li L., McCall C. E., Yoza B. K. (2005) J. Immunol. 175, 461–468 [DOI] [PubMed] [Google Scholar]
- 7.El Gazzar M., Yoza B. K., Hu J. Y., Cousart S. L., McCall C. E. (2007) J. Biol. Chem. 282, 26857–26864 [DOI] [PubMed] [Google Scholar]
- 8.El Gazzar M., Yoza B. K., Chen X., Hu J., Hawkins G. A., McCall C. E. (2008) J. Biol. Chem. 283, 32198–32208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Gazzar M., Yoza B. K., Chen X., Garcia B. A., Young N. L., McCall C. E. (2009) Mol. Cell. Biol. 29, 1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster S. L., Hargreaves D. C., Medzhitov R. (2007) Nature 447, 972–978 [DOI] [PubMed] [Google Scholar]
- 11.Wen H., Dou Y., Hogaboam C. M., Kunkel S. L. (2008) Blood 111, 1797–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 13.Trojer P., Reinberg D. (2007) Mol. Cell 28, 1–13 [DOI] [PubMed] [Google Scholar]
- 14.Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 15.Ko M., Sohn D. H., Chung H., Seong R. H. (2008) Mutat. Res. 647, 59–67 [DOI] [PubMed] [Google Scholar]
- 16.Kim J. K., Samaranayake M., Pradhan S. (2008) Cell. Mol. Life Sci. 66, 596–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akbarian S., Huang H. S. (2009) Biol. Psychiatry 65, 198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y. (2005) Genes Dev. 19, 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoza B. K., Hu J. Y., Cousart S. L., Forrest L. M., McCall C. E. (2006) J. Immunol. 177, 4080–4085 [DOI] [PubMed] [Google Scholar]
- 20.Chen X., Yoza B. K., El Gazzar M., Hu J. Y., Cousart S. L., McCall C. E. (2009) Clin. Vaccine Immunol. 16, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruys V., Thompson P., Beutler B. (1993) J. Exp. Med. 177, 1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia Y., Chen S., Wang Y., Mackman N., Ku G., Lo D., Feng L. (1999) Mol. Cell. Biol. 19, 7688–7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaRue K. E., McCall C. E. (1994) J. Exp. Med. 180, 2269–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Rauch T., Chen Z. X., Szabó P. E., Riggs A. D., Pfeifer G. P. (2006) J. Biol. Chem. 281, 19489–19500 [DOI] [PubMed] [Google Scholar]
- 25.Ryseck R. P., Novotny J., Bravo R. (1995) Mol. Cell. Biol. 15, 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eskeland R., Eberharter A., Imhof A. (2007) Mol. Cell. Biol. 27, 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan Z., Zarebski A., Montoya-Durango D., Grimes H. L., Horwitz M. (2005) Mol. Cell. Biol. 25, 10338–10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishio H., Walsh M. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11257–11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyory I., Wu J., Fejér G., Seto E., Wright K. L. (2004) Nat. Immunol. 5, 299–308 [DOI] [PubMed] [Google Scholar]
- 30.Zemojtel T., Kielbasa S. M., Arndt P. F., Chung H. R., Vingron M. (2009) Trends Genet. 25, 63–66 [DOI] [PubMed] [Google Scholar]
- 31.Lee H., Habas R., Abate-Shen C. (2004) Science 304, 1675–1678 [DOI] [PubMed] [Google Scholar]
- 32.Fusco A. J., Savinova O. V., Talwar R., Kearns J. D., Hoffmann A., Ghosh G. (2008) J. Biol. Chem. 283, 12324–12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milner C. M., Campbell R. D. (1993) Biochem. J. 290, 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Estève P. O., Patnaik D., Chin H. G., Benner J., Teitell M. A., Pradhan S. (2005) Nucleic. Acids Res. 33, 3211–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trojer P., Zhang J., Yonezawa M., Schmidt A., Zheng H., Jenuwein T., Reinberg D. (2009) J. Biol. Chem. 284, 8395–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]