Abstract
The TRPV4 (transient receptor potential vanilloid 4) ion channel, a member of the vanilloid subfamily of the transient receptor potential channels, is activated by membrane stretch, by non-noxious warm temperatures, and by a range of chemical activators. In the present study we examined the role of phosphorylation in modulating the activation of TRPV4. We expressed TRPV4 in HEK293 cells and activated the channel by cell swelling in a hypotonic solution. TRPV4 channel activation and serine phosphorylation were enhanced by exposure to the protein kinase C (PKC) activator phorbol 12-myristate 13-acetate or by application of bradykinin, which activates PKC via a G-protein-coupled mechanism. The enhancement was inhibited by the PKC inhibitors staurosporine, bisindolylmaleimide I, and rottlerin or by mutation of the serine/threonine residues Ser162, Thr175, and Ser189. The adenylate cyclase activator forskolin also enhanced activation of TRPV4, and the enhancement was antagonized by the selective cyclic AMP-dependent protein kinase (PKA) inhibitor H89 or by mutation of serine residue Ser824. Sensitization of TRPV4 by both PKC and PKA depended on the scaffolding protein AKAP79, because channel activation and phosphorylation were enhanced by co-transfection of AKAP79 and were antagonized by removal of AKAP79 using small interfering RNA. We conclude that the serine/threonine kinases PKC and PKA enhance activation of the TRPV4 ion channel by phosphorylation at specific sites and that phosphorylation depends on assembly of PKC and PKA by AKAP79 into a signaling complex with TRPV4.
TRPV4 was cloned from kidney, hypothalamus, and auditory epithelium and was given a number of names: OTRPC4 (Osm-9-like TRP channel 4) (1), VR-OAC (2), TRP12 (3), and VRL-2 (vanilloid receptor-like channel 2) (4). The gene for human TRPV4 is located on chromosome 12q23-q24.1 and has 15 exons, which code for a full-length protein with 871 amino acids. TRPV4 is a member of the transient receptor potential vanilloid subfamily of TRP2 channels, and like other members of this subfamily, it is a polymodal receptor activated by a wide variety of stimuli. TRPV4 is strongly expressed in kidney and is activated by hypotonicity, which has led to the suggestion that TRPV4 is an osmosensor important in regulating body fluid levels (2, 5–9). However, TRPV4 is also activated by innocuous heat with a threshold of >27 °C (6, 10, 11), by the phorbol ester 4α-phorbol 12,13-didecanoate (12, 13), by low pH (14), by endocannabinoids and arachidonic acid metabolites (15, 16), by the active compound, bisandrographolide A, of Andrographis paniculata, a Chinese herbal plant (17), and by nitric oxide (18). TRPV4 is expressed in a broad range of tissues, including lung, spleen, kidney, testis, fat, brain, cochlea, skin, smooth muscle, liver, and vascular endothelium (1–3); in the lamina terminalis of the mouse brain; in neurons of the arched vascular organ of the lamina terminalis; and in the median preoptic area, the optic chiasm, neurons of the subfornical organ, the ventral hippocampal commissure, anterior hypothalamic structures, and ependymal cells of the choroid plexus in the lateral ventricles, and dorsal root ganglia neurons (1–3). The broad spectrum of activators and the wide distribution of TRPV4 suggest that the functions of TRPV4 extend beyond osmosensation.
TRPV4 has been proposed to play a role in the mechanical hyperalgesia that is generated by the concerted action of inflammatory mediators present in inflamed tissues (19). After tissue injury, inflammatory mediators such as bradykinin, prostaglandin E2, 5-hydroxytryptamine, and histamine directly sensitize primary afferent neurons, resulting in hyperalgesia (reviewed in Ref. 20). Important intracellular signaling molecules contributing to inflammatory hyperalgesia include protein kinase C (PKC) (21, 22) and cyclic AMP-dependent protein kinase (PKA) (23). For example, the activation of the Gq-coupled B1 and B2 receptors by bradykinin leads to the release of a range of potential intracellular messengers, with a substantial body of evidence favoring the idea that the temperature threshold of TRPV1 is lowered by PKCϵ-mediated phosphorylation (21, 22, 24, 25). PKA, like PKC, is a critical intracellular signaling molecule mediating inflammatory hyperalgesia (26). In sensory neurons prostaglandin E2 activates both the EP1 receptor, which is Gq-coupled and therefore activates PKC, and the EP4 receptor, which is Gs-coupled and therefore activates PKA. Cyclic AMP analogues, the adenylate cyclase activator forskolin (FSK) or phosphodiesterase inhibitors enhance the mechanical and thermal hyperalgesic effects of prostaglandin E2 (27–29). Thus PKC and PKA have vital roles to play in the process of inflammatory hyperalgesia.
The speed and specificity of the action of kinases is in many cases enhanced by binding to scaffolding proteins, which preassemble the kinases into signaling complexes with their target substrates. The AKAP (a kinase-anchoring protein) family of scaffolding proteins was originally named for their ability to target PKA to appropriate substrates but are now known to assemble a wide range of kinases and phosphatases into signaling complexes with appropriate targets (30). A number of ion channels are subject to modulation by AKAPs, including glutamate receptors, calcium channels, and the M-type potassium channels (31–34). The heat-activated ion channel TRPV1, a member of the same subfamily as TRPV4, has recently been shown to be assembled into a signaling complex with PKA, PKC, and PP2B by AKAP79, and the sensitization of TRPV1 by PKC and PKA is critically reliant on binding to AKAP79 (35). The present study shows that PKC and PKA activation can sensitize TRPV4 to mechanical stimuli, identifies the relevant phosphorylation sites, and shows that the scaffolding protein AKAP79 plays a critical role in sensitization of TRPV4.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293 cells (ATCC) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, 50 μg/ml streptomycin, and 20 mm l-glutamine. The cells were transiently transfected using PolyFect reagent (Qiagen) in accordance with the manufacturer's instructions with some modification as follows. Briefly, 2 μg of DNA was diluted in a total volume of 100 μl of serum-free medium and mixed with 10 μl of PolyFect transfection reagent, followed by 10 min of room temperature incubation to allow DNA complex formation; 0.6 ml of growth medium was added to the transfection complexes; and DNA solution was transferred to the cells and was incubated for 24 h.
Plasmid and DNA Construction
Potential TRPV4 PKC and PKA phosphorylation sites were sought by using GPS (36), PredPhospho (37), NetPhosK (38), and ScanSite (39). Potential phosphorylation sites were targeted when they were identified with high probability by one or more of these packages.
The siRNA against AKAP79 was constructed in a plasmid co-expressing green fluorescent protein to facilitate identification of successfully transfected cells, as previously described (35). We received the following kind gifts of plasmids: hTRPV4 tagged with V5 from Dr. David Cohen, AKAP79 from Dr. John Scott, and hB2 receptor from Dr. Tanya O'Neill.
Point mutations were introduced into TRPV4 by using QuikChange (Stratagene) site-directed mutagenesis in accordance with the manufacturer's instructions. The S189A/T175A double mutant was constructed by combination of the S189A site mutant together with the T175A mutant. The S162A/S189A/T175A triple mutant was constructed by combination of the S162A site mutant together with the double mutant S189A/T175A. All of the constructs were verified by DNA sequencing (Department of Biochemistry, University of Cambridge). The cDNAs were subcloned into the pcDNA3 vector (Invitrogen) for amplification and transfection.
Imaging of Intracellular Calcium and Data Analysis
Calcium imaging was performed as described previously (47). Briefly, HEK293 cells were transiently transfected with 2 μg of TRPV4 cDNA (WT or mutant), or co-transfected with 0.67 μg of WT TRPV4 cDNA + 1.33 μg of AKAP79 or siRNA AKAP79 for 24 h. The cells were plated onto 13-mm polylysine-treated coverslips and grown for 1 day, loaded with 10 μm fluo-4 AM (Molecular Probes) for 40 min, and then mounted into a chamber that was continuously perfused with an isotonic solution (100 mm NaCl, 2 mm CaCl2, 4 mm KCl, 1 mm MgCl2, 10 mm HEPES, 10 mm glucose, 100 mm mannitol, pH 7.4, osmolarity 320 ± 5 mosm). [Ca2+]i was monitored with an inverted confocal microscope (Bio-Rad). Cell swelling was induced by omitting mannitol from this solution (230 ± 5 mosm). Fluo-4 AM was excited at 488 nm, and images of the fluorescence, F, were captured every 3 s. At the end of the experiment, the maximal fluorescence (Fmax) was obtained by application of the calcium ionophore ionomycin (10 μm). The Ca2+-dependent fluorescence changes were calibrated as described (40). The data are expressed as F/Fmax. The inhibitors were applied for 30 min before treatment with e.g. PMA, FSK, or bradykinin. Significance was tested using one-way analysis of variance with Bonferroni's post hoc test (SPSS for Windows). The significance levels in figures are given as: ns, not significant, >5%; *, <5%; **, <1%; ***, <0.1%. All of the errors shown as bars in the figures or quoted in the text are ± S.E. All of the experiments were performed at room temperature (20–22 °C).
Immunoprecipitation and Immunoblotting
The following antibodies were used: anti-phosphoserine antibody (PSR-45;l Sigma) and anti-V5 (Invitrogen). Following treatment as specified in figure legends, transfected cells were solubilized in lysis buffer (20 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA, 2 mm EGTA, 1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 50 mm NaF, 10% protease inhibitors (Roche Applied Science), and 1× phosphatase inhibitor (Sigma)), the cell lysates were centrifuged at 12,000 rpm for 10 min, and cleared supernatant was mixed with 1 μl mouse anti-V5 to precipitate TRPV4 and 30 μl of protein A-agarose (Santa Cruz) and was incubated for 3 h. Immunocomplexes were collected by centrifugation, and the immunoprecipitates were washed three times with lysis buffer followed by boiling for 5 min in SDS-PAGE sample buffer. Agarose beads were removed by centrifugation prior to loading the samples onto 7.5% polyacrylamide gels. The proteins were transferred from the resolved SDS-PAGE gels to Hybond-P membrane. The blots were blocked in blocking buffer (phosphate-buffered saline containing 0.1% Tween 20 and 1.5% gelatin) at room temperature for 1 h and then incubated with diluted primary antibody (PSR-45 1:1000, anti-V5: 1:10,000 in phosphate-buffered saline) at 4 °C for 3 h. The blots were then washed in the washing buffer (phosphate-buffered saline with 0.1% Tween 20) prior to the addition of horseradish peroxidase-conjugated sheep anti-mouse (for PSR-45 and V5) for 1 h at room temperature. The blots were washed repeatedly and developed using ECL chemiluminescent reagent (Amersham Biosciences) before exposure to x-ray film. All of the bands were quantified with National Institutes of Health Image 1.62 software. Because the expression levels of TRPV4 were somewhat variable (see lower blots in Figs. 2A and 5A), the phosphoserine band densities were expressed relative to the relevant TRPV4 band densities. All of the blots shown in the figures are typical of at least three similar results.
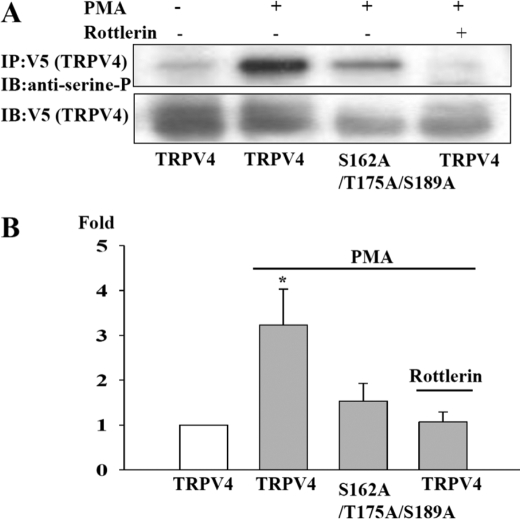
FIGURE 2.
TRPV4 is phosphorylated in response to activation of PKC. A, the upper blot shows phosphoserine detected as described under “Experimental Procedures” following 10 min of exposure to PMA (1 μm) or rottlerin (10 μm). The lower blot shows TRPV4 expression levels in the same experiment detected by stripping and reprobing using anti-V5 antibody. B, collected results from three experiments. Phosphoserine band densities (upper blot in A) expressed relative to TRPV1 expression levels (lower blot). The ordinate shows phosphoserine normalized to level in absence of PMA. *, p ≤ 0.05 compared with bar 1. IP, immunoprecipitation; IB, immunoblotting.
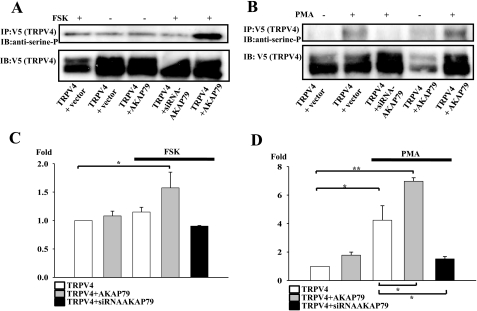
FIGURE 5.
AKAP79 enhances TRPV4 phosphorylation levels following activation of PKA and PKC. A, upper panel shows serine phosphorylation levels of TRPV4 following activation of PKA (treatment with 100 μm FSK for 40 min; first, fourth, and fifth lanes). The lower panel shows same membrane reprobed with anti-V5 antibody to control for variations in TRPV1 expression. The phosphoserine level was strongly enhanced following exposure to FSK in cells co-expressing TRPV4 and AKAP79 (fifth lane). B, similar experiment in which PKC was activated with PMA (1 μm, 20 min; second, third, and fifth lanes). C, summary of experiments with PKA activation, similar to that shown in A. Phosphoserine band density normalized to TRPV1 band density (lower panel in A) and expressed relative to level in absence of FSK and AKAP (bar 1). The bars show the means ± S.E. (n = 3). The significance level was p < 0.05 (*) compared with the first bar. D, summary of experiments with PKC activation, similar to that shown in B. The significance levels were p < 0.05 (*) and p < 0.01 (**) (n = 3) compared with the first bar (comparisons shown above bars) and compared with the third bar (comparisons shown below bars). IP, immunoprecipitation; IB, immunoblotting.
RESULTS
TRPV4 Sensitization by PKC Activation
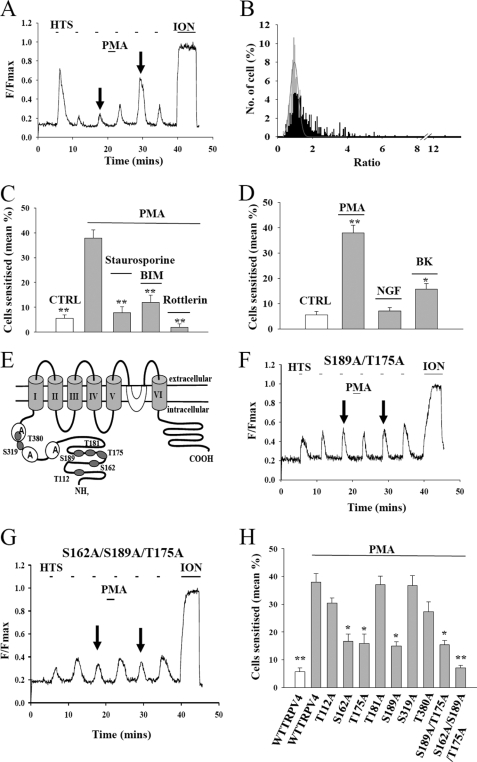
We used HEK293 cells transfected with plasmids containing cDNAs coding for TRPV4 to investigate the sensitization of TRPV4. Hypotonic solutions were used to activate TRPV4 (1–3). The calcium influx through the TRPV4 channel, whether activated by heat or by hypotonic solutions, is sensitized by inflammatory mediators in a similar manner (6, 19). Transfected cells were loaded with the intracellular calcium indicator fluo-4 and imaged in a confocal microscope. The increase in calcium-dependent fluorescence (ΔF) caused by applying a hypotonic solution was used as an index of the activation of TRPV4 (Fig. 1A). There was no increase in calcium-dependent fluorescence either in untransfected cells or in cells transfected with an empty vector. Activation of PKC by application of PMA, a specific PKC activator, increased the ΔF observed in response to application of hypotonic solution (Fig. 1A). TRPV4 has been reported to be directly activated by phorbol esters at high concentrations (12, 13), so to eliminate this direct effect as a possible explanation for sensitization, we removed PMA before applying the first test pulse of hypotonic solution (Fig. 1A). We calculated the ratio between the values of ΔF when sensitization had reached a peak to that immediately before exposure to PMA (Fig. 1A, arrows). Without PMA, the distribution of these ratios was well fitted by a Gaussian function (Fig. 1B). As an index of the sensitization of TRPV4 following exposure to PMA, we calculated the percentage of cells in which the ratio exceeded the 95% confidence limit of the control distribution (Fig. 1B). Sensitization was markedly inhibited by the broad spectrum kinase inhibitor staurosporine, by the PKC-specific inhibitor bisindolylmaleimide I, and by the inhibitor rottlerin (Fig. 1C). We note that rottlerin had been proposed to be a specific inhibitor of PKCδ but is now thought to be less specific or even to act by other routes (41). In summary, these experiments suggest that phosphorylation by PKC plays an important role in the sensitization of TRPV4.
FIGURE 1.
TRPV4 is sensitized by activation of PKC. A, representative trace from a single HEK293 cell transfected with TRPV4 shows increase in [Ca2+]i, measured from the [Ca2+]i-dependent fluorescence of fluo-4 in response to successive applications of hypotonic solution (HTS, 230 ± 5 mosm, pulsed onto the cells for 42 s every 5 min). Untransfected cells showed no increase in [Ca2+]i on application of HTS. Activation of the TRPV4 channel is sensitized by application of PMA (1 μm). The arrows show responses whose ratio was used to measure sensitization (see B). Calcium-dependent fluorescence calibrated at end of experiment by application of the calcium ionophore ionomycin (ION, 10 μm). B, sample histogram of ratios obtained as in A under control (CTRL) conditions (gray) and with application of PMA (black). Sensitization calculated as percentage of ratios exceeding upper 5% confidence level of control distribution (Gaussian best fit shown with red line). C, sensitization of TRPV4 induced by 1 μm PMA was blocked by the broad spectrum kinase inhibitor staurosporine (200 nm), by the PKC-specific inhibitor bisindolylmaleimide I (BIM; 1 μm), and by the inhibitor rottlerin (10 μm). All of the inhibitors were applied for 10 min in advance and also throughout the experiment. The concentration of bisindolylmaleimide I used, 1 μm, is substantially above the K½ for inhibition of PKC in isolated enzyme preparations (7.9 nm) (46), as is often necessary in intact cell preparations, but note that it is below the K½ for other isolated enzymes, for example myosin light chain kinase (∼2 μm) or PKA (insensitive) (46). D, sensitization of TRPV4 by PMA (1 μm, 2 min), nerve growth factor (NGF; 100 ng/ml, 8 min, cells co-transfected with TrkA receptors), and bradykinin (BK; 1 μm, 2 min, cells co-transfected with B2 receptors). E, positions of TRPV4 serine and threonine residues identified by prediction software (see “Experimental Procedures”) as likely PKC phosphorylation sites, shown in the context of a putative transmembrane topology model. F and G, sensitization induced by PMA (1 μm) was abolished by a S189A/T175A mutation (F) and by a S162A/S189A/T175A mutation (G). H, summary of sensitization of WT and phospho-site mutant TRPV4 in experiments such as those shown in A, F, and G. All of the bars show the means ± S.E. The bars are (from left, with 1 μm PMA apart from the first bar): WT TRPV4 (nexp = 6; ncell = 499); WT TRPV4 (nexp = 6; ncell = 594); T112A TRPV4 (nexp = 4; ncell = 278); S162A TRPV4 (nexp = 5; ncell = 240); T175A TRPV4 (nexp = 4; ncell = 329); T181A TRPV4 (nexp = 4; ncell = 235); S189A TRPV4 (nexp = 6; ncell = 211); S319A TRPV4 (nexp = 4; ncell = 199); T380A TRPV4 (nexp = 4; ncell = 152); S189A/T175A TRPV4 (nexp = 5; ncell = 166); S162A/T175A/S189A TRPV4 (nexp = 8; ncell = 380). The significance levels relative to WT TRPV4 plus PMA were p ≤ 0.05 (*) an p ≤ 0.005 (**).
Bradykinin causes sensitization of TRPV1 by activating PKCϵ (22), which in turn potentiates activation of TRPV1 by phosphorylating serine residues 502 and 800 (residue numbering for rat TRPV1) (24). Nerve growth factor causes sensitization of TRPV1 by activating the tyrosine kinase Src, which phosphorylates human TRPV1 at tyrosine 200 (40). We therefore investigated whether bradykinin and nerve growth factor can sensitize TRPV4 by co-transfecting the appropriate receptors with TRPV4 (see “Experimental Procedures”). There was no significant increase in the percentage of sensitized cell following exposure to nerve growth factor. Bradykinin, however, gave a significant enhancement (Fig. 1D), showing that activation of PKC by a physiologically important pro-inflammatory mediator can potentiate gating of TRPV4. No enhancement was observed in the absence of transfected B2 receptors (not shown).
TRPV4 Serine and Threonine Residues Involved in Sensitization by PKC
Using in silico prediction methods, we identified seven serine or threonine residues in the TRPV4 N-terminal cytoplasmic domain as candidate substrates for PKC-dependent phosphorylation (Fig. 1E). Although these residues could potentially be phosphorylated by PKC, not all may in practice be accessible to the kinase; so to identify the functionally important sites, these serine or threonine residues were individually replaced with alanine, and the resulting mutant proteins were investigated for enhancement following PKC activation as above. Sensitization of the majority of the single mutants was not significantly different from that observed for wild type TRPV4, but three of the candidate PKC phosphorylation sites, Ser162, Thr175, and Ser189, showed a significantly lower enhancement when compared with WT TRPV4 (Fig. 1H). The double mutant S189A/T175A (Fig. 1F) did not totally abolish PMA-induced sensitization, but mutating all three sites simultaneously completely abolished functional sensitization following PMA application with no significant effect on the activation of TRPV4 by hypotonic solution (Fig. 1, G and H).
TRPV4 Sensitization by PKC Is Associated with Increased Serine Phosphorylation
We quantified total serine phosphorylation of TRPV4 with an anti-phosphoserine antibody. TRPV4 was observed to be partially phosphorylated in the basal state, and following exposure to PMA the normalized band density increased by ∼2-fold (Fig. 2). Mutating the three phosphorylation sites identified above (S162A/T175A/S189A) inhibited the increased level of phosphorylation following exposure to PMA, as did exposure to the inhibitor rottlerin (Fig. 2).
TRPV4 Sensitization by PKA
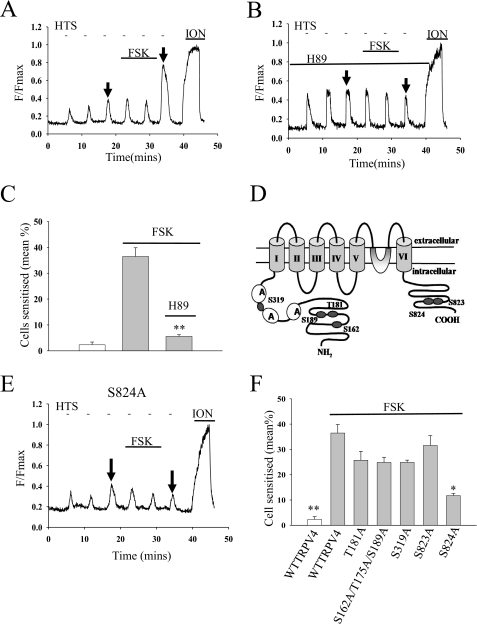
Application of the adenylate cyclase activator FSK increased ΔF in a subpopulation of TRPV4-transfected cells (Fig. 3, A and C). The specific PKA inhibitor H89 (5 μm) abolished the sensitization of TRPV4 by FSK (Fig. 3, B and C). Using in silico methods, we identified six serine/threonine residues in the TRPV4 N- and C-terminal domains as candidate substrates for PKA-dependent phosphorylation (Fig. 3D). These residues were individually replaced with alanine, and the resulting mutant proteins were subjected to functional analysis as above. Most mutants were found to exhibit sensitization in response to PKA activation similar to WT TRPV4, but mutation of one C-terminal residue, Ser824, largely inhibited the sensitization caused by FSK (Fig. 3, E and F).
FIGURE 3.
TRPV4 is sensitized by activation of PKA. A, representative trace shows effect of PKA activator FSK (100 μm) on [Ca2+]i increase in response to hypotonic solution (HTS; details as in Fig. 1). B, similar experiment in the presence of PKA blocker H89 (5 μm, applied 30 min prior to the start). C, summary of effect of FSK and H89. **, p < 0.01 relative to second bar. D, positions of TRPV4 serine and threonine residues identified by prediction software as likely PKA phosphorylation sites. E, representative trace of effect of FSK (100 μm) on S824A mutant. F, summary of effect of PKA activation on TRPV4 phosphosite mutants. Bars are (from left, with 100 μm FSK apart from the first bar): WT TRPV4 (nexp = 6; ncell = 499); WT TRPV4 (nexp = 5; ncell = 376); T181A TRPV4 (nexp = 3; ncell = 268); S162A/T175A/S189A TRPV4 (nexp = 3; ncell = 435); S319A TRPV4 (nexp = 3; ncell = 295); S823A TRPV4 (nexp = 3; ncell = 118); and S824A TRPV4 (nexp = 7; ncell = 403). The significance levels with respect to the second bar are p < 0.05 (*). ION, ionomycin.
AKAP79 Binds to and Enhances Sensitization of TRPV4 by PKA and PKC
AKAP79 binds to several members of the thermo-TRP family of ion channels, most strongly to TRPV1 and TRPV4 (35). The binding to TRPV4 suggests that AKAP79 may play a role in PKA and PKC-mediated sensitization of TRPV4, as it does in the case of TRPV1 (35). We therefore investigated whether AKAP79 mediates PKA- and PKC-dependent sensitization of TRPV4.
Co-expression of TRPV4 and AKAP79 enhances sensitization by FSK (Fig. 4, A and E) and by PMA (Fig. 4, C and F), showing that augmenting the endogenous levels of AKAP79 enhances the sensitization of TRPV4 by both PKA and PKC. We next down-regulated endogenous expression of AKAP79 by the use of siRNA against AKAP79, which was co-expressed with green fluorescent protein in the same vector to allow identification of transfected cells. In HEK293 cells transfected with siRNA against AKAP79, the expression of AKAP79 is reduced to 10% or less (35).
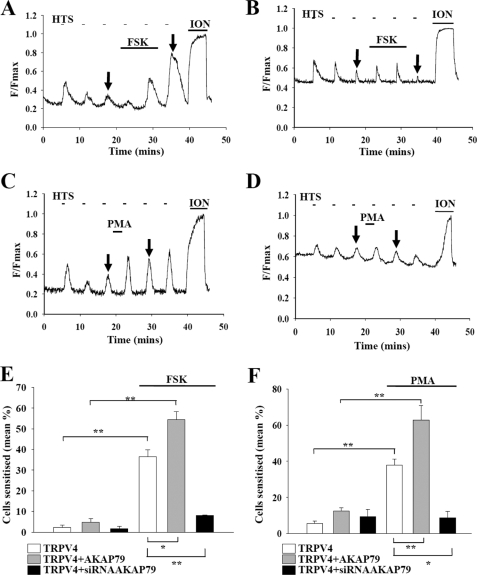
FIGURE 4.
Effects of AKAP79 on sensitization of TRPV4 by PKA and PKC activation. A, sensitization of TRPV4 with co-transfected AKAP79 following activation of PKA with FSK. Same protocol as in Fig. 3A. B, similar experiment on cell co-expressing TRPV4 and siRNA against AKAP79. Note the elevated base line caused by the presence of green fluorescent protein, indicating successful transfection of the cell by the siRNA-expressing plasmid. C, cell co-expressing TRPV4 and AKAP79, showing sensitization following activation of PKC by PMA. D, similar experiment on cell co-expressing TRPV4 and siRNA against AKAP79. E, percentage of cells sensitized following activation of PKA in experiments similar to those shown in A and B, compared with cells not transfected with AKAP79 or siRNA against AKAP79. The bars are (from left, with 100 μm FSK applied in last three bars): TRPV4 (nexp = 6; ncell = 594); TRPV4 + AKAP79 (nexp = 4; ncell = 222); TRPV4 + siRNA (nexp = 3; ncell = 239); TRPV4 + FSK (nexp = 5; ncell = 376); TRPV4 + AKAP79 + FSK (nexp = 3; ncell = 262); and TRPV4 + siRNA + FSK (nexp = 3; ncell = 206). All of the bars indicate the means ± S.E. The significance levels are p < 0.05 (*) and p < 0.01 (**) compared with controls (comparisons shown above bars) and compared with the fourth bar (comparisons shown below bars). F, percentage of cells sensitized following activation of PKC in experiments similar to those shown in C and D, compared with cells not transfected with AKAP79 or siRNA against AKAP79. The bars are (from left, with 1 μm PMA applied in the last three bars): TRPV4 (nexp = 6; ncell = 594); TRPV4 + AKAP79 (nexp = 4; ncell = 222); TRPV4 + siRNA (nexp = 3; ncell = 239); TRPV4 + PMA (nexp = 6; ncell = 499); TRPV4 + AKAP79 + PMA (nexp = 3; ncell = 170); and TRPV4 + siRNA + PMA (nexp = 3; ncell = 235). All of the bars indicate the means ± S.E. The significance levels are p < 0.05 (*) and p < 0.01 (**) compared with control (comparisons shown above bars) and compared with the fourth bar (comparisons shown below bars). ION, ionomycin; HTS, hypotonic solution.
In calcium imaging experiments the background fluorescence caused by the transfected green fluorescent protein can be seen as an enhanced base line (Fig. 4, B and D). Fig. 4B shows that down-regulation of AKAP79 almost completely inhibits sensitization by FSK (results summarized in Fig. 4E). Fig. 4D shows that a similar abolition of the sensitization caused by PMA is also observed when AKAP79 is down-regulated (results summarized in Fig. 4F). Thus the presence of AKAP79 is essential for the functional sensitization of TRPV4 by both PKA and PKC.
AKAP79 Enhances Phosphorylation of TRPV4 by PKA and PKC
Activation of PKA with FSK caused only a marginal enhancement of TRPV4 serine phosphorylation (Fig. 5A), probably because phosphorylation at the single Ser824 site identified above does not have a powerful effect in raising overall phosphorylation above the base-line level caused by background phosphorylation of other sites. Transfection of AKAP79 or down-regulation of AKAP79 using siRNA also had only small effects on TRPV4 phosphorylation levels (Fig. 5, A and C). However, the combined effects of transfection of AKAP79 together with activation of PKA with FSK did cause a significantly enhanced level of serine phosphorylation (Fig. 5, A and C).
Activation of PKC with PMA, on the other hand, caused a significant increase in serine phosphorylation (Fig. 5, B and D), as might be expected in view of the experiments in Figs. 1 and 2, which show that more than one site is phosphorylated by PKC. Co-transfection of AKAP79 had no significant effect on basal levels of serine phosphorylation (Fig. 5D, second bar) but did significantly enhance the increase following PKC activation (Fig. 5, B, final lane, and D, fourth bar). Down-regulation of AKAP79 expression by transfection with siRNA abolished the enhancement caused by PKC activation (Fig. 5, B and D, last bar). These results reinforce the functional studies above in showing that AKAP79 is involved in mediating phosphorylation of TRPV4 by both PKA and PKC.
DISCUSSION
The TRPV4 ion channel shows substantial homology to TRPV1, the first member of the vanilloid subclass of TRP ion channels to be cloned. Like TRPV1, TRPV4 is activated by a wide range of stimuli, which among the possible physiological activators include membrane stretch caused by cell swelling or mechanical stress, warm temperatures above ∼27 °C, low pH, nitric oxide, and a variety of intracellular lipid messengers. The physiological role of TRPV4 and which of these possible activating stimuli is physiologically relevant remain unknown. TRPV4 is highly expressed in renal nephron and in hypothalamus, and through its osmosensory properties may play a role in regulating body fluids (2, 42), but is also expressed in bladder epithelium, where it may play a role in bladder voiding (9, 43) and in sensory neurons, where roles both in detection of strong mechanical stimuli (5) and sensation of warm temperatures (10) have been proposed.
Tissue damage and inflammation cause the release of a range of pro-inflammatory mediators, with bradykinin and prostaglandins prominent among them. These mediators activate of intracellular signaling pathways and downstream kinases, among which PKA and PKC are known to be physiologically important (20). Both PKA and PKC enhance activation of TRPV1 (44). In the present paper we examined whether TRPV4 is modulated in a similar way to TRPV1 by phosphorylation by PKA and PKC. We used membrane stretch as a convenient activator of TRPV4, and we monitored activation of TRPV4 from the calcium influx when the channel opens. We carried out our experiments in a HEK293 cell expression system. We note that in the case of the related TRPV1 ion channel, studies in our lab and elsewhere have found that expression systems provide a highly reliable guide to the behavior of the ion channel in its native environment (35, 40, 45).
Activation of PKC by PMA, a potent and specific activator, substantially enhanced both the gating and the phosphorylation of TRPV4. The physiological pro-inflammatory mediator bradykinin, which activates Gq and hence PKC, also potentiated activation of TRPV4. The effect of PMA was inhibited by the broad spectrum kinase inhibitor staurosporine, by the specific PKC inhibitor bisindolylmaleimide I, and by the inhibitor rottlerin, which has been proposed to be a specific inhibitor of PKCδ (but see Ref. 41). These results show that PKC sensitizes TRPV4, as it does in the case of TRPV1. We identified three phosphorylation sites close together in the N-terminal domain, Ser162, Thr175, and Ser189, mutation of each of which partially inhibited the enhancement caused by PKC. Mutation of other candidate residues was without effect. Mutation of all three sites together completely abolished the effect of PKC activation without a significant effect on gating of TRPV4. Similar experiments on TRPV1 have also identified candidate PKC phosphorylation sites, but in different parts of the molecule: Ser502 located in the cytoplasmic domain linking putative transmembrane domains S1 and S2, close to the binding site for capsaicin, and Ser801 located in the C-terminal domain (24, 25).
Activation of PKA by application of the adenylate cyclase activator FSK also enhanced gating of TRPV4. The specific PKA inhibitor H89 abolished the effect of FSK, confirming that PKA is involved. A number of candidate phosphorylation sites was investigated, and among these sites a substantial inhibition of the enhancement, although not a complete abolition, was observed following mutation of a single site, Ser824, in the C-terminal domain.
Finally, by using a combination of co-expression and knockdown approaches, AKAP79 was shown to play a vital role in tethering PKA and PKC to TRPV4 to modulate its gating. Functional studies using calcium imaging showed that AKAP79 overexpression enhanced sensitization of TRPV4 by FSK (Fig. 4, A and E) and PMA (Fig. 4, B and F), whereas down-regulation of AKAP79 using siRNA inhibited sensitization. These functional studies were supported by studies of the effect of AKAP79 overexpression or down-regulation on the phosphorylation of TRPV4, in which it was shown that overexpression of AKAP79 enhanced the phosphorylation induced by PMA (Fig. 5, C and D), whereas knockdown with siRNA against AKAP79 decreased the effect of PKC activation on phosphorylation (Fig. 5, C and D).
In summary, the gating of the TRPV4 ion channel by cell swelling is modulated by phosphorylation by the serine/threonine kinases PKA and PKC in a manner reminiscent of TRPV1, although at sites located in quite different places on the protein. As is the case with TRPV1, AKAP79 orchestrates the action of PKC and PKA by tethering these kinases to TRPV4 so as to enhance the function and phosphorylation of the targeted ion channel. Manipulating this signaling integrator could be a promising target for the development of novel analgesics.
Footnotes
- TRP
- transient receptor potential
- PMA
- phorbol 12-myristate 13-acetate
- PKC
- protein kinase C
- FSK
- forskolin
- PKA
- cyclic AMP-dependent protein kinase
- siRNA
- small interfering RNA
- WT
- wild type.
REFERENCES
- 1.Strotmann R., Harteneck C., Nunnenmacher K., Schultz G., Plant T. D. (2000) Nat. Cell Biol. 2, 695–702 [DOI] [PubMed] [Google Scholar]
- 2.Liedtke W., Choe Y., Martí-Renom M. A., Bell A. M., Denis C. S., Sali A., Hudspeth A. J., Friedman J. M., Heller S. (2000) Cell 103, 525–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wissenbach U., Bödding M., Freichel M., Flockerzi V. (2000) FEBS Lett. 485, 127–134 [DOI] [PubMed] [Google Scholar]
- 4.Delany N. S., Hurle M., Facer P., Alnadaf T., Plumpton C., Kinghorn I., See C. G., Costigan M., Anand P., Woolf C. J., Crowther D., Sanseau P., Tate S. N. (2001) Physiol. Genomics 4, 165–174 [DOI] [PubMed] [Google Scholar]
- 5.Alessandri-Haber N., Yeh J. J., Boyd A. E., Parada C. A., Chen X., Reichling D. B., Levine J. D. (2003) Neuron 39, 497–511 [DOI] [PubMed] [Google Scholar]
- 6.Gao X., Wu L., O'Neil R. G. (2003) J. Biol. Chem. 278, 27129–27137 [DOI] [PubMed] [Google Scholar]
- 7.Nilius B., Prenen J., Wissenbach U., Bödding M., Droogmans G. (2001) Pflügers Arch. 443, 227–233 [DOI] [PubMed] [Google Scholar]
- 8.Vriens J., Watanabe H., Janssens A., Droogmans G., Voets T., Nilius B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birder L., Kullmann F. A., Lee H., Barrick S., de Groat W., Kanai A., Caterina M. (2007) J. Pharmacol. Exp. Ther. 323, 227–235 [DOI] [PubMed] [Google Scholar]
- 10.Güler A. D., Lee H., Iida T., Shimizu I., Tominaga M., Caterina M. (2002) J. Neurosci. 22, 6408–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe H., Vriens J., Suh S. H., Benham C. D., Droogmans G., Nilius B. (2002) J. Biol. Chem. 277, 47044–47051 [DOI] [PubMed] [Google Scholar]
- 12.Watanabe H., Davis J. B., Smart D., Jerman J. C., Smith G. D., Hayes P., Vriens J., Cairns W., Wissenbach U., Prenen J., Flockerzi V., Droogmans G., Benham C. D., Nilius B. (2002) J. Biol. Chem. 277, 13569–13577 [DOI] [PubMed] [Google Scholar]
- 13.Xu F., Satoh E., Iijima T. (2003) Br. J. Pharmacol. 140, 413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki M., Mizuno A., Kodaira K., Imai M. (2003) J. Biol. Chem. 278, 22664–22668 [DOI] [PubMed] [Google Scholar]
- 15.Vriens J., Owsianik G., Fisslthaler B., Suzuki M., Janssens A., Voets T., Morisseau C., Hammock B. D., Fleming I., Busse R., Nilius B. (2005) Circ. Res. 97, 908–915 [DOI] [PubMed] [Google Scholar]
- 16.Watanabe H., Vriens J., Prenen J., Droogmans G., Voets T., Nilius B. (2003) Nature 424, 434–438 [DOI] [PubMed] [Google Scholar]
- 17.Smith P. L., Maloney K. N., Pothen R. G., Clardy J., Clapham D. E. (2006) J. Biol. Chem. 281, 29897–29904 [DOI] [PubMed] [Google Scholar]
- 18.Yoshida T., Inoue R., Morii T., Takahashi N., Yamamoto S., Hara Y., Tominaga M., Shimizu S., Sato Y., Mori Y. (2006) Nat. Chem. Biol. 2, 596–607 [DOI] [PubMed] [Google Scholar]
- 19.Alessandri-Haber N., Dina O. A., Joseph E. K., Reichling D., Levine J. D. (2006) J. Neurosci. 26, 3864–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J., Zhang X., McNaughton P. A. (2006) Semin. Cell Dev. Biol. 17, 638–645 [DOI] [PubMed] [Google Scholar]
- 21.Cesare P., McNaughton P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15435–15439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cesare P., Dekker L. V., Sardini A., Parker P. J., McNaughton P. A. (1999) Neuron 23, 617–624 [DOI] [PubMed] [Google Scholar]
- 23.Bhave G., Zhu W., Wang H., Brasier D. J., Oxford G. S., Gereau R. W., 4th (2002) Neuron 35, 721–731 [DOI] [PubMed] [Google Scholar]
- 24.Numazaki M., Tominaga T., Toyooka H., Tominaga M. (2002) J. Biol. Chem. 277, 13375–13378 [DOI] [PubMed] [Google Scholar]
- 25.Bhave G., Hu H. J., Glauner K. S., Zhu W., Wang H., Brasier D. J., Oxford G. S., Gereau R. W., 4th (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12480–12485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aley K. O., Levine J. D. (1999) J. Neurosci. 19, 2181–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cunha F. Q., Teixeira M. M., Ferreira S. H. (1999) Br. J. Pharmacol. 127, 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taiwo Y. O., Bjerknes L. K., Goetzl E. J., Levine J. D. (1989) Neuroscience 32, 577–580 [DOI] [PubMed] [Google Scholar]
- 29.Taiwo Y. O., Levine J. D. (1991) Neuroscience 44, 131–135 [DOI] [PubMed] [Google Scholar]
- 30.Smith F. D., Langeberg L. K., Scott J. D. (2006) Trends Biochem. Sci. 31, 316–323 [DOI] [PubMed] [Google Scholar]
- 31.Altier C., Dubel S. J., Barrère C., Jarvis S. E., Stotz S. C., Spaetgens R. L., Scott J. D., Cornet V., De Waard M., Zamponi G. W., Nargeot J., Bourinet E. (2002) J. Biol. Chem. 277, 33598–33603 [DOI] [PubMed] [Google Scholar]
- 32.Colledge M., Dean R. A., Scott G. K., Langeberg L. K., Huganir R. L., Scott J. D. (2000) Neuron 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 33.Hoshi N., Langeberg L. K., Scott J. D. (2005) Nat. Cell Biol. 7, 1066–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshi N., Zhang J. S., Omaki M., Takeuchi T., Yokoyama S., Wanaverbecq N., Langeberg L. K., Yoneda Y., Scott J. D., Brown D. A., Higashida H. (2003) Nat. Neurosci. 6, 564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X., Li L., McNaughton P. A. (2008) Neuron 59, 450–461 [DOI] [PubMed] [Google Scholar]
- 36.Zhou F. F., Xue Y., Chen G. L., Yao X. (2004) Biochem. Biophys. Res. Commun. 325, 1443–1448 [DOI] [PubMed] [Google Scholar]
- 37.Kim J. H., Lee J., Oh B., Kimm K., Koh I. (2004) Bioinformatics 20, 3179–3184 [DOI] [PubMed] [Google Scholar]
- 38.Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. (2004) Proteomics 4, 1633–1649 [DOI] [PubMed] [Google Scholar]
- 39.Yaffe M. B., Leparc G. G., Lai J., Obata T., Volinia S., Cantley L. C. (2001) Nat. Biotechnol. 19, 348–353 [DOI] [PubMed] [Google Scholar]
- 40.Zhang X., Huang J., McNaughton P. A. (2005) EMBO J. 24, 4211–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soltoff S. P. (2007) Trends Pharmacol. Sci. 28, 453–458 [DOI] [PubMed] [Google Scholar]
- 42.Liedtke W., Friedman J. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13698–13703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gevaert T., Vriens J., Segal A., Everaerts W., Roskams T., Talavera K., Owsianik G., Liedtke W., Daelemans D., Dewachter I., Van Leuven F., Voets T., De Ridder D., Nilius B. (2007) J. Clin. Invest. 117, 3453–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J., Zhang X., McNaughton P. A. (2006) Curr. Neuropharmacol. 4, 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vellani V., Mapplebeck S., Moriondo A., Davis J. B., McNaughton P. A. (2001) J. Physiol. 534, 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martiny-Baron G., Kazanietz M. G., Mischak H., Blumberg P. M., Kochs G., Hug H., Marmé D., Schächtele C. (1993) J. Biol. Chem. 268, 9194–9197 [PubMed] [Google Scholar]
- 47.Bonnington J. K., M. Naughton P. A. (2003) J. Physiol. 551, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]