Abstract
The acid-sensing ion channels (ASICs) open in response to extracellular acidic pH, and individual subunits display differential sensitivity to protons and calcium. ASIC1a acts as a high affinity proton sensor, whereas ASIC2a requires substantially greater proton concentrations to activate. Using chimeras composed of ASIC1a and ASIC2a, we determined that two regions of the extracellular domain (residues 87–197 and 323–431) specify the high affinity proton response of ASIC1a. These two regions appear to undergo intersubunit interactions within the multimeric channel to specify proton sensitivity. Single amino acid mutations revealed that amino acids around Asp357 play a prominent role in determining the pH dose response of ASIC1a. Within the same region, mutation F352L abolished PcTx1 modulation of ASIC1a. Surprisingly, we determined that another area of the extracellular domain was required for calcium-dependent regulation of ASIC1a activation, and this region functioned independently of high affinity proton sensing. These results indicate that specific regions play overlapping roles in pH-dependent gating and PcTx1-dependent modulation of ASIC1a activity, whereas a distinct region determines the calcium dependence of ASIC1a activation.
The acid-sensing ion channels (ASICs)3 are proton-gated ion channels expressed in neurons throughout the central and peripheral nervous system (1–3). ASICs are activated by extracellular acidosis, and protons act as ligands triggering channel opening (4). Disruption of the accn2 gene (which encodes ASIC1) dramatically reduces proton-gated currents in central neurons and alters a variety of behaviors, including fear, learning, and memory (5, 6). ASIC1 also contributes to neuronal damage and death during the prolonged acidosis accompanying cerebral ischemia (7). Specifically, mice with disruptions in the accn2 gene display 60% smaller lesion size compared with normal mice in models of stroke (8). Application of PcTx1, a venom peptide that prevents ASIC1a activation, is similarly neuroprotective, even when administered hours after injury (8, 9). Thus, ASIC1a represents a novel pharmacological target for the prevention of neuronal death following stroke.
Mammals have four genes encoding ASICs (accn1 to -4) that encode at least six different ASIC subunits (1–3, 10). Like all members of the DEG/ENaC family, individual ASIC subunits have two transmembrane regions separated by a large cysteine-rich extracellular region. Three ASIC subunits associate to form homomeric or heteromeric channels with distinct biophysical characteristics (11–14). Specifically, ASIC1a homomeric channels activate at pH values much closer to neutral pH compared with ASIC2a homomeric channels. The high affinity proton sensitivity of ASIC1a plays a prominent role in acidosis-induced neuronal death, and modulators that alter the pH dose response of ASIC1a affect neuronal sensitivity to prolonged acidosis (8, 9, 15). For example, the neuroprotective venom peptide PcTx1 increases the proton sensitivity of the ASIC1a channel, allowing the channel to desensitize at neutral pH and become unresponsive to subsequent acidic shifts in pH (16, 17). The large extracellular region of ASIC1a is thought to be the site of proton/modulator interaction and governs the characteristics of channel gating (10, 11, 18). However, the exact molecular mechanisms defining ASIC1a activation and the protein domains that are responsible for the apparent proton sensitivity of ASIC1a remain unclear. Here, we used chimeras containing specific regions from both ASIC1a and ASIC2a to identify the specific protein regions that confer high affinity proton sensing, PcTx1 sensitivity, and calcium modulation to ASIC1a.
EXPERIMENTAL PROCEDURES
Construction of Chimeras, Site-directed Mutagenesis, and Expression in Xenopus Oocytes
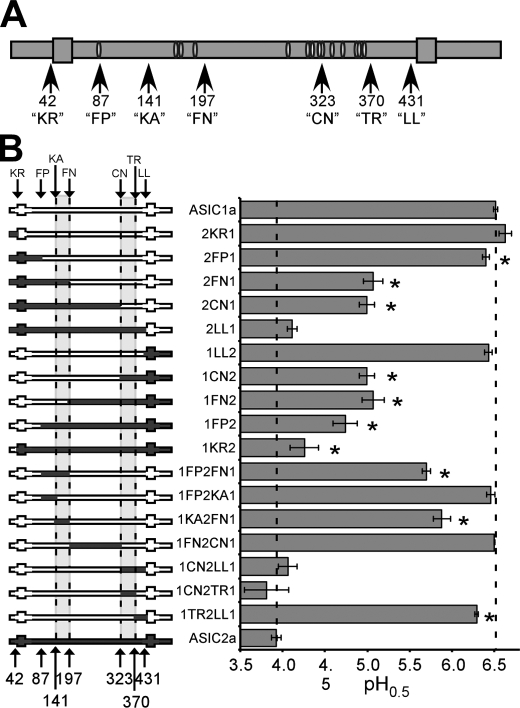
Human ASIC1a- and ASIC2a-encoding cDNAs in the pMT3 mammalian expression vector have been described previously (19, 20). These sequences correspond to GenBankTM accession number NM_001095 for human ASIC1a and NM_001094 for human ASIC2a. Chimeras were generated using overlap extension PCR (21). The specific locations of chosen chimeric break points in human ASIC1a or ASIC2a are shown in Fig. 1A and are as follows: “KR” is immediately preceding transmembrane region 1 (TM1) at K42 (1a) or R42 (2a); “FP” is at the conserved extracellular FPAVT sequence at F87 (1a) or F86 (2a); “KA” is at the conserved KANF sequence at K141(1a) or K140(2a); “FN” is at the conserved FNSG sequence at F197 (1a) or F196 (2a); “CN” is at the conserved CNCR sequence at C323 (1a) or C320 (2a); “TR” is at the third conserved TRY sequence at T370(1a) or T367(2a); and “LL” is just before transmembrane region two (TM2) at L431 (1a) and L428 (2a). When reported throughout, the numbering refers to the locations of corresponding residues in ASIC1a. Chimeras were cloned into pMT3 using unique restriction sites engineered into the PCR primers. Site-directed mutagenesis was employed on human ASIC1a using the QuikChange® kit from Stratagene (Cedar Creek, TX). All inserts were fully sequenced prior to electrophysiological analysis (Plant-Microbe Genomics Facility, Ohio State University, Columbus, OH). Qiagen Midi or Maxi prep kits (Valencia, CA) were used to prepare plasmid DNA for oocyte injection.
FIGURE 1.
Two regions of the extracellular domain determine the pH dose response of ASIC1a and 2a. A, schematic of an ASIC subunit with the chimeric break points indicated (arrows). Large gray boxes represent transmembrane regions, and white ovals signify conserved cysteine residues. The number of the first amino acid within the break point motif of human ASIC1a is indicated (see “Experimental Procedures”). B, calculated pH0.5 of ASIC1a/2a chimeras. On the left are schematics of the chimeras. Areas in dark gray are ASIC2a sequence, and areas in white are ASIC1a sequence. Dotted lines, the locations of important break points. On the right is the graph of the pH0.5 for each chimera. Each value represents an n of 3–10 cells. Statistical significance was assessed using an unpaired t test. *, statistical significance with p < 0.05 compared with ASIC1a and ASIC2a. Error bars, S.E.
Unfertilized oocytes were harvested from wild-type oocyte positive Xenopus laevis females from Xenopus I (Dexter, MI) using standard procedures (22). ASIC expression plasmids at a 50–100 ng/μl concentration were injected into oocyte nuclei (located beneath the animal pole) using a PV820 Pneumatic Picopump (World Precision Instruments, Sarasota, FL). For co-expression studies, plasmids were injected at a 1:1 ratio (50 ng/μl of each construct). Oocytes were allowed to incubate at 18 °C for 18–72 h before experiments were performed.
Electophysiology
Macroscopic currents were recorded using the two-electrode voltage clamp technique at a holding potential of −60 mV. Electrodes (0.5–2 megaohms) were filled with 3 m KCl. Current was recorded using a Warner oocyte OC-725 clamp (Warner Instruments, Hamden, CT) and either a Powerlab 4SP digitizer with CHART software (ADInstruments, Colorado Springs, CO) or an Axon Digidata 1200 digitizer and pCLAMP-8 software (Molecular Devices, Sunnyvale, CA). The standard bath solution contained 116 mm NaCl, 2 mm KCl, 5 mm Hepes, 5 mm MES, 2 mm CaCl2, 1 mm MgCl2 and was pH-adjusted with 1 n HCl or 1 n NaOH. The pH of the standard bath solution was 7.4 unless otherwise indicated. Oocytes were placed in a bath chamber (500 μl), and acid applications were made using pump-driven perfusion directed toward the oocyte with a bath solution exchange rate of 2–3 ml/s (5 ml total). The cells were maintained in pH 7.4 for at least 2 min between acid applications to allow the cells to recover from desensitization. To minimize current run down, the peak current amplitude of evoked current from acidic test pH applications was normalized to the average of the peak current amplitudes evoked from saturating acidic pH (pH 3.0 or 5.0) applied before and after the test pH. PcTx1-containing venom from the tarantula Psalmopoeus cambridgei was purchased from SpiderPharm (Yarnell, AZ) and diluted to 1:3,333–1:5,000 for experiments.
The pH0.5 was calculated by fitting the data from individual pH dose-response experiments within each oocyte using the equation, I/IpHmax = 1/(1 + (EC50/[H+])n) = 1/(1 + 10n(pH − pH0.5)), where n is the Hill coefficient. EC50 and pH0.5 are the proton concentration and pH yielding half of the saturating peak current amplitude (IpHmax), respectively. The pHmax was determined as a saturating test pH for all channels within the experimental group. This was either pH 3.0 or pH 5.0. The Hill coefficient was 4.4 ± 0.46 (n = 18) for ASIC1a and 1.3 ± 0.05 ASIC2a (n = 13). Generally, the Hill coefficient correlated with the pH0.5 of mutant channels, with those showing smaller Hill coefficients also displaying a lower pH0.5. However, the variability in the calculated Hill coefficient of ASIC1a limited significance substantially. To quantify desensitization, IGOR (WaveMetrics, Portland, OR) was used to fit the decay phase of acid-activated currents to a single exponential equation, I = k0 + k1·e−t/τ. The τ of desensitization (τd) was calculated in seconds from the fitted curve. Two-tailed paired or unpaired t tests were done to analyze statistical significance as appropriate. A p value of <0.05 was considered significant.
RESULTS
Two Regions of the Extracellular Domain Specify the pH Sensitivity of ASIC1a
ASIC1a is activated by pH 6.7 and shows a pH of half-maximal activation (pH0.5) of ∼6.5. This pH0.5 corresponds to an EC50 for protons of 316 nm. ASIC2a requires a pH below 6.0 for activation, displays a pH0.5 of ∼3.9, and has an apparent EC50 for protons of 126 μm. Thus, ASIC1a requires an almost 400-fold lower proton concentration for half-maximal activation compared with ASIC2a. To identify the protein regions responsible for this difference in apparent proton affinity, chimeras were constructed with different segments of ASIC1a and ASIC2a sequence (Fig. 1A).
All chimeras produced transient proton-gated currents when expressed in Xenopus oocytes. A pH dose response was assessed for each construct, and the pH0.5 (the pH of half-maximal activation) was calculated. Substitution of several regions of ASIC1a with ASIC2a sequence affected the pH0.5. In particular, exchanging the region N-terminal to the “FP” site altered the pH0.5 slightly (see chimeras 2FP1, 1KR2, and 1FP2) (Fig. 1B). However, two other protein regions showed a more prominent role in defining the apparent proton sensitivity of ASIC1a. First, substitution of the region between amino acids 87 and 197 (1FP2FN1) of ASIC1a with ASIC2a sequence resulted in a dramatic reduction in the pH0.5 (Fig. 1B). Second, substitution in the region between amino acids 323 and 431 (1CN2LL1) of ASIC1a substantially diminished the proton dose response. Alternatively, exchanging the N-terminal region before amino acid 87 (2KR1), the C-terminal region after amino acid 431 (1LL2, 2LL1), or the middle region between amino acids 197 and 323 of ASIC1a with ASIC2a sequence (1FN2CN1) did little to alter the proton sensitivity.
To further define the important residues within these regions, chimeras containing only part of the regions 87–197 and 323–431 were generated. Exchanging only region 141–197 of ASIC1a with ASIC2a sequence (chimera 1KA2FN1) recapitulated the proton dose response of chimera 1FP2FN1 in which the entire 87–197 region was exchanged (Fig. 1B). However, exchanging the region between 87 and 141 (1FP2KA1) showed a pH0.5 indistinguishable from ASIC1a. Furthermore, exchanging just the N-terminal area of amino acids 323–370 (1CN2TR1) resulted in a chimera with a pH dose response indistinguishable from the chimera in which the entire region was exchanged (Fig. 1B). Exchanging amino acids 370–431 (1TR2LL1) also reduced the apparent proton sensitivity slightly. Thus, amino acids within regions 141–197 and 323–370 appear to play a dominant role in specifying the pH dose response of ASIC1a, with residues 323–370 being the most important.
Specific Amino Acids between 323 and 370 Determine the Apparent Proton Affinity of ASIC1a
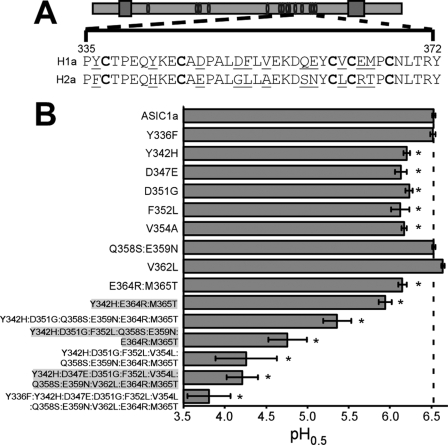
Exchanging the region between amino acids 323 and 370 of ASIC1a with ASIC2a sequence decreased the pH0.5 from 6.5 to 3.8, a more than 400-fold decrease in apparent proton sensitivity. There are 11 amino acids within this region that differ between ASIC1a and ASIC2a. To determine which amino acids are important for proton sensitivity, individual amino acids in ASIC1a were converted to the corresponding amino acids in ASIC2a (Fig. 2A). Y342H, D347E, D351G, F352L, V354A, and the double mutant E364R/M365T all displayed smaller relative currents in response to pH 6.5, and their pH0.5 was substantially smaller compared with ASIC1a (Fig. 2B). However, the lowest pH0.5 elicited from these single mutants was 6.19, and none of these single mutations decreased pH sensitivity to the level of 1CN2TR1 (pH0.5 = 3.8), suggesting that changes in multiple amino acids are responsible for the effect of exchanging all amino acids between 323 and 370 with ASIC2a sequence. To determine whether combining mutations could decrease the pH0.5 further, subunits with multiple mutations were constructed and analyzed (Fig. 2B). Only when all 11 substitutions were made was the pH0.5 of 1CN2TR1 recapitulated. These results probably indicate that all of these residues contribute to high affinity proton sensing of ASIC1a.
FIGURE 2.
Conversion of amino acids within region 323–370 of ASIC1a to the corresponding residues in ASIC2a eliminates high affinity proton sensing. A, schematic of an ASIC subunit with conserved cysteines indicated as white ovals. Below is the sequence of ASIC1a and ASIC2a within the region indicated by dashed lines. Amino acids 323–335 are identical between ASIC1a and ASIC2a and are not shown. Residues in boldface type are conserved cysteines. Residues that are different between ASIC1a and ASIC2a are underlined. B, calculated pH0.5 of the indicated mutants (n = 3–10). Note that the sequence of the mutant with a substitution of all 11 residues is identical to chimera 1CN2TR1. Statistical significance was assessed using an unpaired t test. *, p < 0.05 compared with human ASIC1a. Error bars, S.E.
Asp357 and Surrounding Residues Play a Critical Role in Proton-dependent Gating
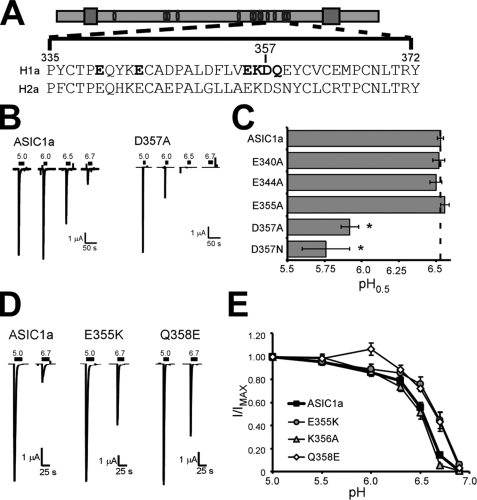
Amino acids with negatively charged side chains have been hypothesized to play a role in determining proton binding (23, 24). We reasoned that because both ASIC1a and ASIC2a are activated by protons (albeit at different concentrations), conserved acidic residues might be necessary for proton-dependent gating. Therefore, we tested the effect of mutation of acidic residues within region 323–370 that are conserved between ASIC1a and ASIC2a (Fig. 3A). Surprisingly, ASIC1a mutants with alterations in acidic residues Glu340, Glu344, and Glu355 displayed a normal proton dose response. However, D357A displayed a decreased response to pH 6.5 solutions (Fig. 3B). Quantification of the pH0.5 revealed that D357A showed a relatively large decrease in pH0.5 (5.92 ± 0.06 for D357A (n = 7, p = 8.5 × 10−14 compared with ASIC1a) (Fig. 3C). Similar results were obtained with D357N, suggesting that the charge of the aspartic acid side chain is important.
FIGURE 3.
Asp357 and surrounding amino acids are important for proton sensitivity of ASIC1a. A, schematic of an ASIC subunit with the indicated sequence of ASIC1a and ASIC2a indicated below. Amino acids in boldface type were mutated. B, representative traces of ASIC1a and D357A in response to pH 5.0, 6.0, 6.5, and 6.7 application. C, calculated pH0.5 values of ASIC1a channels with mutations in acidic residues (n = 6–20). Statistical significance was assessed using an unpaired t test. *, p < 0.05 compared with ASIC1a. D, representative traces of the pH 5.0 and pH 6.7 response of ASIC1a, E355K, and Q358E. E, pH dose-response of channels with mutations around Asp357. I/IMAX is the peak amplitude of current evoked by the test pH (indicated on the x axis) divided by the peak current amplitude of the pH 5.0 response (n = 5–18).
The D357A mutation showed the largest change in pH dose response of any single mutation examined in these studies. We reasoned that the region surrounding Asp357 might play a prominent role in defining the pH sensitivity of ASIC1a. Since our previous results indicated that E355A and Q358S did not alter pH sensitivity (Figs. 2B and 3C), we tested mutations that more significantly affected the amino acids. Surprisingly, mutants E355K and Q358E showed increased current in response to pH 6.7 application (Fig. 3D). Further quantification revealed that the proton dose response of these mutant channels was shifted such that the pH0.5 was greater than ASIC1a (Fig. 3E). The pH0.5 for E355K was 6.64 ± 0.02 (n = 8, p = 0.003 compared with ASIC1a), and the pH0.5 of Q358E was 6.66 ± 0.03 (n = 7, p = 0.00032 compared with ASIC1a). We also assessed the impact of K356A mutation and determined that pH sensitivity was slightly reduced, although the data narrowly reached statistical significance. (pH0.5 of K356A = 6.48 ± 0.02, n = 8, p = 0.049 compared with ASIC1a) (Fig. 3E). These results indicate that the area around Asp357 plays an important role in the pH dose response of ASIC1a.
Two Regions Are Sufficient for High Affinity Proton Sensing
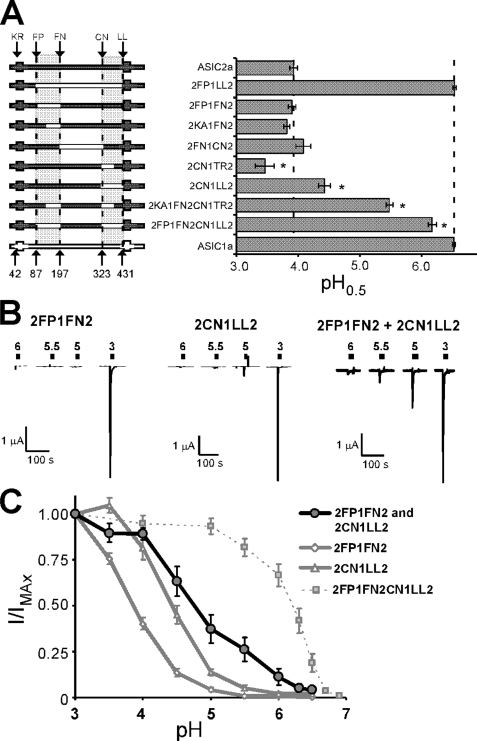
ASIC1a sequences between amino acids 87–197 and 323–431 are required for high affinity proton sensing. To determine whether either region is sufficient, we analyzed ASIC2a chimeras with ASIC1a sequence within these regions (Fig. 4A). The presence of ASIC1a sequence in the region between 87 and 197 alone did not increase the apparent proton sensitivity of ASIC2a. When the region of ASIC2a between 323 and 431 was exchanged with ASIC1a sequence, the pH sensitivity was increased (pH0.5 of 2CN1LL2 = 4.43 ± 0.10, n = 6, p = 0.0003 compared with ASIC2a). Replacing regions 141–197 and 323–370 (2KA1FN2CN1TR2) increased the pH0.5, but only to 5.48 ± 0.05, n = 3. However, when both regions 87–197 and 323–431 of ASIC2a were exchanged with ASIC1a sequence (2FP1FN2CN1LL2), the chimera displayed a dramatic increase in proton sensitivity (pH0.5 of 2FP1FN2CN1LL2 = 6.17 ± 0.07, n = 7, p = 1 × 10−6 compared with ASIC2a) (Fig. 4A). Although this chimera was still less sensitive than ASIC1a (p = 0.0005), these results suggest that ASIC1a sequence in the region between amino acid 87 and 197 as well as the region between 323 and 431 is sufficient to confer high affinity proton sensing and a pH0.5 higher than 6.0.
FIGURE 4.
Two regions of ASIC1a encompassing amino acids 87–197 and 323–431 are sufficient for high affinity proton sensing and undergo intersubunit interaction. A, pH0.5 values of ASIC2a chimeras with ASIC1a sequence. A schematic of chimeras tested is shown on the left (n = 5–8). *, p < 0.05 compared with human ASIC1a. B, representative traces of proton-gated currents in oocytes expressing 2FP1FN2, 2CN1LL2, or both 2FP1FN2 and 2CN1LL2. Scale bars, 1 μA (y axis) and 100 s (x axis). C, pH dose response of proton-gated current in oocytes injected with both 2FP1FN2 and 2CN1LL2 (dark line) compared with those injected with only 2FP1FN2, 2CN1LL2, or 2FP1FN2CN1LL2 (n = 3–10). Error bars, S.E.
Our data indicate that two regions of ASIC1a must be present for a pH0.5 greater than 6.0. These results suggest that these regions may interact to determine the pH sensitivity of ASIC1a. Functional ASIC channels are formed by association of three ASIC subunits. Thus, the interaction between these regions could be within the same subunit (intrasubunit), or one region of a subunit could interact with the other region from another subunit (intersubunit). To determine whether intersubunit interactions between these two regions affect pH sensitivity of ASICs, we co-expressed chimeras 2FP1FN2 and 2CN1LL2 and measured proton-gated current (Fig. 4, B and C). Oocytes expressing both 2FP1FN2 and 2CN1LL2 displayed appreciable proton-activated current with pH 5.5 (Fig. 4B). However, very little proton-activated current was observed in response to pH 5.5 in oocytes expressing either chimera alone. The pH dose response of oocytes expressing both chimera was also shifted toward more neutral pH values (Fig. 4C). Although the pH sensitivity of proton-gated currents when 2FP1FN2 and 2CN1LL2 were co-expressed was not as high as for 2FP1FN2CN1LL2, this increase in proton sensitivity suggests that regions 87–197 and 323–431 undergo intersubunit interactions, at least in part, to specify the proton dose response.
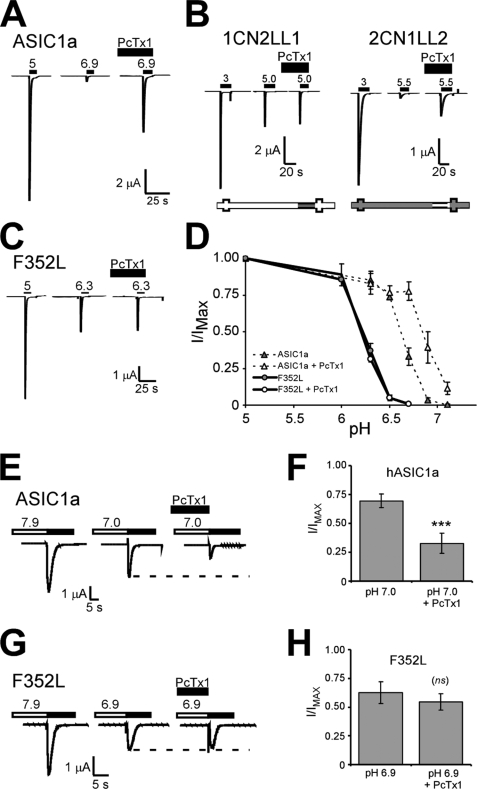
F352L Eliminates PcTx1 Sensitivity of ASIC1a
In addition to increased proton sensitivity, ASIC1a is also sensitive to the venom peptide PcTx1, whereas ASIC2a is not (17). PcTx1 limits ASIC1a activity by increasing the apparent proton sensitivity of ASIC1a and promoting steady-state desensitization at pH 7.4 (16–18, 25). If steady-state desensitization is not induced, then PcTx1 potentiates ASIC1a activation by shifting the pH0.5 of activation to more neutral pH values (16, 25). Under such conditions, ASIC1a currents evoked by non-saturating pH values are substantially enhanced in the presence of PcTx1 (Fig. 5A). We asked whether the same domains that determine high affinity proton sensing also confer sensitivity to PcTx1. ASIC1a chimeras containing ASIC2a in the region between amino acids 323 and 431 (1CN2LL1) were not affected by PcTx1-containing venom (Fig. 5B). Conversely, ASIC2a chimeras containing ASIC1a sequence in this same region (2CN1LL2) were enhanced by PcTx1-containing venom (Fig. 5B). These results suggest that ASIC1a sequence in the region between amino acids 323 and 431 is both necessary and sufficient for PcTx1 enhancement of the pH dose response and are in agreement with recently published observations (26). To determine the specific amino acids in this region involved in the PcTx1 sensitivity of ASIC1a, we analyzed channels with mutations converting amino acids in this region of ASIC1a to amino acids present in ASIC2a. For most mutants, the effect of PcTx1 was retained (data not shown). However, ASIC1a channels with mutation F352L were unaffected by PcTx1-containing venom (the pH 6.3 response was 36.9 ± 5.0% maximal current without PcTx1 and 31.2 ± 1.8% in the presence of PcTx1, n = 5, p = 0.22) (Fig. 5C). A thorough pH dose-response analysis in the presence of PcTx1 revealed that F352L was unaffected by PcTx1 (Fig. 5D). The pH0.5 of F352L was 6.23 ± 0.01 (n = 4) in the absence of PcTx1 and was 6.21 ± 0.04 (n = 4, p = 0.72) in the presence of PcTx1. Under the same conditions, the pH dose response of ASIC1a was substantially shifted toward more neutral pH values (pH0.5 of ASIC1a in the absence of PcTx1 was 6.59 ± 0.04 (n = 5), and in the presence of PcTx1, it was 6.81 ± 0.06 (n = 3), p = 0.02). These results indicate that the F352L mutation eliminated PcTx1-induced potentiation of the pH dose response of ASIC1a.
FIGURE 5.
PcTx1 modulation is eliminated by the F352L mutation. A–C, representative traces of the PcTx1 effect on the response to submaximal proton concentrations of human ASIC1a (A), chimeras 1CN2LL1 and 2CN1LL2 (B), and point mutation F352L (C). For human ASIC1a and F352L, the basal pH was maintained at pH 7.9 to eliminate PcTx1 action on steady-state desensitization (see below). PcTx1-containing venom (1:3,000–1:5,000) was present in the extracellular bath solution for 2 min prior to the application of saturating acidic pH solutions. The activating pH values chosen produced maximal (left) and submaximal (right) responses from the individual channels. Similar results were attained when pH 5 was used to activate 2CN1LL2, and pH 5.5 was used to activate 1CN2LL1 (not shown). D, pH dose response of human ASIC1a and F352L in the presence and absence of PcTx1 (1:5,000). Basal pH was maintained at pH 7.9 before application of acidic pH values. Peak current amplitude was normalized to that evoked from pH 5 in each cell (n = 5–9). E and F, representative traces of PcTx1 modulation of steady-state desensitization of human ASIC1a (E) and F352L (F). Basal pH was maintained at 7.9 and dropped to the indicated conditioning pH (either 7.0 for ASIC1a or 6.9 for F352L) for 2 min, and pH 5.0 was used to activate proton-gated currents (dark bar above trace). PcTx1 was applied for 2 min at pH 7.9 as well as during the conditioning and activating pH. Quantification of the effect of PcTx1 on steady-state desensitization of human ASIC1a and F352L is shown in F and H, respectively. The peak current amplitude of pH 5.0-evoked currents following exposure to the conditioning test pH (either 7.0 or 6.9) was normalized to peak current amplitude following exposure to pH 7.9 solutions for 2 min (n = 5–6). PcTx1 was applied as above. Statistical significance was assessed using a paired t test. ***, p < 0.0005. Error bars, S.E.
PcTx1 modulation of steady-state desensitization was also lost in F352L. Steady-state desensitization is induced by incubating cells in a slightly acidic pH (conditioning pH) for 2 min prior to pH 5.0 application (Fig. 5, E and F). When PcTx1 was present, steady-state desensitization of human ASIC1a was enhanced, and subsequent activation of the channels produced a substantially smaller current (Fig. 5, E and F). However, steady-state desensitization of F352L was not significantly different in the presence of PcTx1 (Fig. 5, G and H). Thus, the F352L mutation prevents PcTx1 modulation of the pH-dependence of both activation and steady-state desensitization.
Calcium Dependence Is Determined by Amino Acids 197– 323
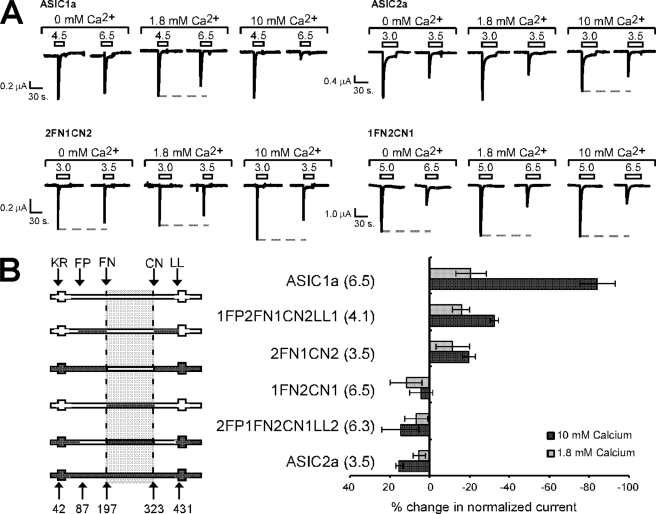
The pH dose response of ASIC1a is also markedly affected by extracellular calcium, with increasing concentrations of calcium increasing the concentration of protons required for channel activation (4, 27–29). In fact, it has been proposed that calcium tonically inhibits ASIC1a activation and protons activate ASICs by facilitating calcium unbinding (23, 24, 30). This effect can be observed by the decrease in the relative peak current amplitude evoked by pH 6.5 in the presence of either 1.8 or 10 mm calcium (Fig. 6A). To determine whether calcium affects ASIC1a and ASIC2a similarly, we tested the calcium dependence of ASIC2a activation. Surprisingly, we find that calcium enhanced proton sensitivity of ASIC2a, as indicated by the relative increase in pH 3.5-evoked currents (Fig. 6A). Quantification of the percentage change in normalized currents reveals a negative change for ASIC1a and a positive change for ASIC2a with increasing calcium (Fig. 6B). We used our ASIC1a/2a chimeras to identify the regions that are responsible for the difference in calcium dependence between ASIC1a and ASIC2a. We found that exchanging amino acids 197–323 of ASIC1a with ASIC2a sequence (1FN2CN1) eliminated the ASIC1a-like response to calcium (Fig. 6, A and B). Similarly, replacing the residues of ASIC2a between amino acids 197–323 with ASIC1a sequence (2FN1CN2) was sufficient to allow calcium inhibition of proton sensitivity (as indicated as a negative change in percentage of normalized current in Fig. 6B), although the channel had a pH0.5 of 4.1. Exchanging the regions we previously identified as playing a prominent role in the dose response to protons (FP-FN and CN-LL) did not determine the response to calcium. Thus, the region between amino acids 197 and 323 is responsible for the differential calcium sensitivity of ASIC1a and ASIC2a, whereas the region responsible for the pH0.5 differences lies in regions 87–197 and 323–431.
FIGURE 6.
The calcium dependence of ASIC1a activation does not segregate with regions controlling the pH dose response. A, representative traces of the effect of calcium on ASIC1a, ASIC2a, and the indicated chimeras. Oocytes were incubated in either nominal calcium (0 mm calcium added), 1.8 mm calcium, or 10 mm calcium. Proton-gated currents were activated with a pH to induce a maximal response (left) and a submaximal response (right). The gray dotted line indicates the maximal response in the specific calcium concentration. Note that pH 6.5-evoked current is reduced compared with the maximal response as calcium concentration increases for ASIC1a, and such a response is not observed with ASIC2a. B, quantification of the calcium response. A schematic of chimeras tested is shown on the left, and the change in normalized current evoked from the submaximal pH application in either 1.8 or 10 mm calcium compared with nominal calcium is shown on the right. The activating pH values used were those indicated in parentheses. Because different pH values were used to activate current for different channels, the trend (whether positive or negative changes with increased calcium) and not the absolute values was considered for comparison.
DISCUSSION
The apparent proton sensitivity of ASIC1a determines channel activation and the extent of depolarization and calcium influx induced by that activation. Most modulators of ASIC1a function, including zinc, redox reagents, lactate, and PcTx1, act by altering the apparent proton sensitivity of the channel (16, 18, 25, 31–34). These compounds also impact ASIC1a-induced neuronal death (8, 9, 15, 31). We find that two protein regions, located between amino acids 87 and 197 and between amino acids 323–431 of the extracellular domain are both necessary and sufficient for high affinity proton sensing of ASIC1a.
The published crystal structure of desensitized chicken ASIC1a indicates that the channel is a trimer with individual subunits making multiple intra- and intersubunit contacts (11). Each subunit has a “wrist domain,” which connects the extracellular domain to transmembrane regions as well as a “β-ball,” “finger,” “palm,” and “thumb” domain. The exact role of each individual domain is not known, and there is still much debate about the mechanism of ASIC activation by protons (11). Specifically, where the proton-binding site(s) is located (if specific sites exist), how protons trigger channel activation, how modulators alter the apparent proton sensitivity of the channel, and the exact role of calcium in proton-dependent ASIC gating remain unknown.
Currently, there are two different theories on the location of the proton binding sites of ASIC1a. Experiments using chimeras composed of proton-insensitive and proton-sensitive ASICs suggest that specific residues within the wrist domain of ASICs are required for proton-dependent activation (35–37). Channels that are unresponsive to protons are produced when these residues are mutated, and because some of these residues can be titrated by protons, it has been suggested that they represent the proton-binding site(s). The involvement of these residues in proton-mediated activation has been confirmed by other studies looking at the role of conserved acidic residues (38). Whether these residues are required for the allosteric changes induced by proton binding or if they act as protonation sites has yet to be experimentally determined. Our experiments did not identify these regions as playing a large role in specifying high affinity proton sensing of ASIC1a.
Analysis of the crystal structure of chicken ASIC1a has highlighted a second region, termed the “acid pocket,” as the proton-binding site (11, 39). This highly acidic region contains several pairs of carboxyl-carboxylate interactions between the side chains of several amino acids. Specifically, contacts between Asp238 and Asp350 (Asp351 in human ASIC1a), Glu239 and Asp346 (Asp347 in human ASIC1a), Glu220 and Asp408, and Glu80 and Glu417 define the four pairs (11). It has been suggested that protonation of these residues confers pH sensing. Although mutations to some of these residues reduce apparent proton affinity (11), mutation to others does not affect apparent proton sensitivity, and individual mutations in this region do not eliminate proton-dependent gating (as mutations in the wrist domain do) (38). This could be due to redundancy between the residue pairs. Our data highlight this region and these residues as playing a role in high affinity proton sensing of ASIC1a versus ASIC2a. Specifically, our studies found that two of these residues, Asp347 and Asp351 in ASIC1a, were involved in specifying the difference in proton sensitivity between ASIC1a and ASIC2a. Thus, our data support a role for the acid pocket in determining apparent proton sensitivity. However, we also find that residues along the thumb and palm domains are also responsible for the low affinity proton sensing of ASIC2a. Further, we find that residues 141–197 of ASIC1a, located throughout the finger, palm, and β-Ball regions, are also important for proton sensing. Except for Arg191, residues in this region have not been previously highlighted as playing a direct role in the acidic pocket of chicken ASIC1a (11). Thus, it appears that the entire region is important for defining the sensitivity of proton sensing.
Our data suggest that intersubunit interactions between 87 and 197 and between 323 and 431 are important for specifying the pH dose response. In the crystal structure of desensitized ASIC1a, these regions make several intersubunit contacts, one near the base of the thumb domain and one near the tip of the thumb domain (11). The importance of Asp357 in acid sensing supports the idea that these intersubunit contacts are important. Mutation of this residue decreases proton sensitivity more profoundly than any single amino acid mutation we tested. In the crystal structure of chicken ASIC1a, this residue is located near Lys355 (Lys356 in human ASIC1a) and Asn357 (Gln358 in human ASIC1a), two residues that form strong intersubunit contacts in the crystal structure (11). Interestingly, these residues make contact around Glu178 and Arg176 of the adjacent subunit, residues that fall within the second region (residues 87–197) that we find necessary and sufficient for high affinity proton sensing. Further support for the importance of these regions comes from previous data indicating that Asp351, Gln358, and Asn359 are required in Lamprey ASIC1a to form active ASIC1a channels, suggesting that these residues are critical for proton-dependent gating (37). We find that mutations Q358E and E355K increase the apparent proton sensitivity of ASIC1a. It is interesting to speculate that substitution of a glutamine with glutamic acid could add additional negative changes to aid in proton binding, and substitution of glutamic acid with a lysine could mimic proton binding. However, whether these regions are required for allosteric conformational changes that accompany gating or are directly involved in setting the pKa of the proton-binding site is unknown. Yet, these results indicate that this intersubunit contact between the thumb and palm domain is very important for proton-dependent gating.
The PcTx1 venom peptide enhances the overall proton sensitivity of ASIC1a, suggesting that PcTx1 may directly or indirectly modulate proton sensing. We find that residue Phe352 plays a prominent role in PcTx1 sensitivity. Specifically, the F352L substitution in ASIC1a eliminates PcTx1 action. Several studies have used computer modeling of PcTx1 docking to ASIC1a to determine the PcTx1 binding site and the specific residues that probably make contact with PcTx1 (26, 40). Asp351, a residue experimentally shown to reduce PcTx1 affinity, is thought to directly interact (18, 26, 40). However, a role for Phe352 has not been noted. In the crystal structure, the phenylalanine side chain points inward from the presumed PcTx1 docking site (11). Although our studies show that Phe352 is required for PcTx1 action, further studies will be required to determine whether this residue is involved in binding of PcTx1 or transduction of PcTx1 binding to changes in channel activity. Interestingly, PcTx1 action also requires intersubunit interactions with regions that overlap with amino acids 87–197 and 323–370 (18). The fact that the apparent proton sensitivity of ASIC1a and PcTx1 sensitivity are determined by overlapping regions suggests a possible molecular explanation for PcTx1-based modulation of the pH dose response.
Calcium is another modulator of ASIC1a proton sensitivity. Moreover, calcium has several effects on ASIC1a activity. First, calcium at high concentration can block the channel, an effect that requires residues Glu425 and Asp432 of rat ASIC1a (23). In addition, the proton dependence of ASIC activation is inversely dependent on calcium concentration (24, 28). This has led to speculation that the proton-binding site and this second calcium binding site are the same and that protons activate the channel by displacing calcium (24, 27, 30, 41). Here, we report that the proton-gated ASIC2a does not show calcium dependence of activation, like that observed in ASIC1a and ASIC3. Although the high concentration of protons required for activation could mask any possible calcium dependence for ASIC2a gating, our studies of ASIC1a and ASIC2a chimeras indicate that the calcium dependence of activation and apparent proton sensitivity do not segregate together. For example, a chimera with a pH0.5 of 4.0 shows calcium sensitivity (2FN1CN2), and one with a pH0.5 of 6.5 does not (1FN2CN1). In fact, we found that the calcium dependence of ASIC1a activation involved a completely distinct region (between 197 and 323) compared with those involved in the differential pH sensitivity of ASIC1a and ASIC2a. This indicates that the sites important for calcium-dependent activation are distinct from the identified sites that specify apparent proton sensitivity and modulator action.
Acknowledgments
We thank Dr. Michael J. Welsh, John Wemmie, Tatiana Rokhlina, and Maggie Price of the University of Iowa for providing the human ASIC1a and ASIC2a clones used in these studies. Ellen Yang, Kirsten Loomis, Tom McCartney, and Anita Pryor provided basic support for these studies. We also thank K. Mykytyn for thoughtful editing of the manuscript.
This work was supported by National Science Foundation Grant IBN-0416920.
- ASIC
- acid-sensing ion channel
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Wemmie J. A., Price M. P., Welsh M. J. (2006) Trends Neurosci. 29, 578–586 [DOI] [PubMed] [Google Scholar]
- 2.Voilley N. (2004) Curr. Drug Targets Inflamm. Allergy 3, 71–79 [DOI] [PubMed] [Google Scholar]
- 3.Krishtal O. (2003) Trends Neurosci. 26, 477–483 [DOI] [PubMed] [Google Scholar]
- 4.Waldmann R., Champigny G., Bassilana F., Heurteaux C., Lazdunski M. (1997) Nature 386, 173–177 [DOI] [PubMed] [Google Scholar]
- 5.Wemmie J. A., Askwith C. C., Lamani E., Cassell M. D., Freeman J. H., Jr., Welsh M. J. (2003) J. Neurosci. 23, 5496–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wemmie J. A., Chen J., Askwith C. C., Hruska-Hageman A. M., Price M. P., Nolan B. C., Yoder P. G., Lamani E., Hoshi T., Freeman J. H., Jr., Welsh M. J. (2002) Neuron 34, 463–477 [DOI] [PubMed] [Google Scholar]
- 7.Xiong Z. G., Chu X. P., Simon R. P. (2007) Front Biosci. 12, 1376–1386 [DOI] [PubMed] [Google Scholar]
- 8.Xiong Z. G., Zhu X. M., Chu X. P., Minami M., Hey J., Wei W. L., MacDonald J. F., Wemmie J. A., Price M. P., Welsh M. J., Simon R. P. (2004) Cell 118, 687–698 [DOI] [PubMed] [Google Scholar]
- 9.Pignataro G., Simon R. P., Xiong Z. G. (2007) Brain 130, 151–158 [DOI] [PubMed] [Google Scholar]
- 10.Kellenberger S., Schild L. (2002) Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 11.Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 12.Benson C. J., Xie J., Wemmie J. A., Price M. P., Henss J. M., Welsh M. J., Snyder P. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2338–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Askwith C. C., Wemmie J. A., Price M. P., Rokhlina T., Welsh M. J. (2004) J. Biol. Chem. 279, 18296–18305 [DOI] [PubMed] [Google Scholar]
- 14.Hesselager M., Timmermann D. B., Ahring P. K. (2004) J. Biol. Chem. 279, 11006–11015 [DOI] [PubMed] [Google Scholar]
- 15.Wang W. Z., Chu X. P., Li M. H., Seeds J., Simon R. P., Xiong Z. G. (2006) J. Biol. Chem. 281, 29369–29378 [DOI] [PubMed] [Google Scholar]
- 16.Chen X., Kalbacher H., Gründer S. (2006) J. Gen. Physiol. 127, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escoubas P., De Weille J. R., Lecoq A., Diochot S., Waldmann R., Champigny G., Moinier D., Ménez A., Lazdunski M. (2000) J. Biol. Chem. 275, 25116–25121 [DOI] [PubMed] [Google Scholar]
- 18.Salinas M., Rash L. D., Baron A., Lambeau G., Escoubas P., Lazdunski M. (2006) J. Physiol. 570, 339–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swick A. G., Janicot M., Cheneval-Kastelic T., McLenithan J. C., Lane M. D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1812–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Askwith C. C., Cheng C., Ikuma M., Benson C., Price M. P., Welsh M. J. (2000) Neuron 26, 133–141 [DOI] [PubMed] [Google Scholar]
- 21.Horton R. M., Cai Z. L., Ho S. N., Pease L. R. (1990) BioTechniques 8, 528–5352357375 [Google Scholar]
- 22.Adams C. M., Snyder P. M., Price M. P., Welsh M. J. (1998) J. Biol. Chem. 273, 30204–30207 [DOI] [PubMed] [Google Scholar]
- 23.Paukert M., Babini E., Pusch M., Gründer S. (2004) J. Gen. Physiol. 124, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Immke D. C., McCleskey E. W. (2003) Neuron 37, 75–84 [DOI] [PubMed] [Google Scholar]
- 25.Chen X., Kalbacher H., Gründer S. (2005) J. Gen. Physiol. 126, 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qadri Y. J., Berdiev B. K., Song Y., Lippton H. L., Fuller C. M., Benos D. J. (2009) J. Biol. Chem. 284, 17625–17633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berdiev B. K., Mapstone T. B., Markert J. M., Gillespie G. Y., Lockhart J., Fuller C. M., Benos D. J. (2001) J. Biol. Chem. 276, 38755–38761 [DOI] [PubMed] [Google Scholar]
- 28.de Weille J., Bassilana F. (2001) Brain Res. 900, 277–281 [DOI] [PubMed] [Google Scholar]
- 29.Babini E., Paukert M., Geisler H. S., Grunder S. (2002) J. Biol. Chem. 277, 41597–41603 [DOI] [PubMed] [Google Scholar]
- 30.Zhang P., Sigworth F. J., Canessa C. M. (2006) J. Gen. Physiol. 127, 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu X. P., Close N., Saugstad J. A., Xiong Z. G. (2006) J. Neurosci. 26, 5329–5339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu X. P., Wemmie J. A., Wang W. Z., Zhu X. M., Saugstad J. A., Price M. P., Simon R. P., Xiong Z. G. (2004) J. Neurosci. 24, 8678–8689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho J. H., Askwith C. C. (2008) J. Neurophysiol. 99, 426–441 [DOI] [PubMed] [Google Scholar]
- 34.Immke D. C., McCleskey E. W. (2001) Nat. Neurosci. 4, 869–870 [DOI] [PubMed] [Google Scholar]
- 35.Smith E. S., Zhang X., Cadiou H., McNaughton P. A. (2007) Neurosci. Lett. 426, 12–17 [DOI] [PubMed] [Google Scholar]
- 36.Chen X., Polleichtner G., Kadurin I., Gründer S. (2007) J. Biol. Chem. 282, 30406–30413 [DOI] [PubMed] [Google Scholar]
- 37.Coric T., Zheng D., Gerstein M., Canessa C. M. (2005) J. Physiol. 568, 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paukert M., Chen X., Polleichtner G., Schindelin H., Gründer S. (2008) J. Biol. Chem. 283, 572–581 [DOI] [PubMed] [Google Scholar]
- 39.Shaikh S. A., Tajkhorshid E. (2008) Biophys. J. 95, 5153–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pietra F. (2009) J. Chem. Inf. Model. 49, 972–977 [DOI] [PubMed] [Google Scholar]
- 41.Todorović N., Corić T., Zhang P., Canessa C. (2005) Ann. N.Y. Acad. Sci. 1048, 331–336 [DOI] [PubMed] [Google Scholar]