Abstract
Matrix metalloproteinases are maintained in an inactive state by a bond between the thiol of a conserved cysteine in the prodomain and a zinc atom in the catalytic domain. Once this bond is disrupted, MMPs become active proteinases and can act on a variety of extracellular protein substrates. In vivo, matrilysin (MMP7) activates pro-α-defensins (procryptdins), but in vitro, processing of these peptides is slow, with about 50% conversion in 8–12 h. Similarly, autolytic activation of promatrilysin in vitro can take up to 12–24 h for 50% conversion. These inefficient reactions suggest that natural cofactors enhance the activation and activity of matrilysin. We determined that highly sulfated glycosaminoglycans (GAG), such as heparin, chondroitin-4,6-sulfate (CS-E), and dermatan sulfate, markedly enhanced (>50-fold) the intermolecular autolytic activation of promatrilysin and the activity of fully active matrilysin to cleave specific physiologic substrates. In contrast, heparan sulfate and less sulfated forms of chondroitin sulfate did not augment matrilysin activation or activity. Chondroitin-2,6-sulfate (CS-D) also did not enhance matrilysin activity, suggesting that the presentation of sulfates is more important than the overall degree of sulfation. Surface plasmon resonance demonstrated that promatrilysin bound heparin (KD, 400 nm) and CS-E (KD, 630 nm). Active matrilysin bound heparin (KD, 150 nm) but less so to CS-E (KD, 60 μm). Neither form bound heparan sulfate. These observations demonstrate that sulfated GAGs regulate matrilysin activation and its activity against specific substrates.
Matrix metalloproteinases (MMPs)3 comprise a family of endopeptidases that act on a variety of extracellular proteins, such as chemokines, antimicrobial peptides, matrix components, and more, to effect numerous repair, immune, and disease processes (1–3). For many substrates, MMP cleavage results in gain-of-function processing, such as the activation of latent antimicrobial peptides (4, 5) and cytokines (1), or altered biologic activity, as with limited proteolysis of chemokines (6, 7) and shedding of cell surface proteins (8). Thus, the mechanisms controlling zymogen activation and proteinase activity against specific substrates would sit high in the hierarchy of events controlling many host response pathways. As for all proteinases, the activity of MMPs is regulated at four points: gene expression, compartmentalization (i.e. pericellular accumulation of enzyme), proenzyme (or zymogen) activation, and enzyme inactivation, and is further controlled by substrate availability, concentration, and affinity.
ProMMPs are kept in a catalytically inactive state by the interaction between the thiol of the conserved prodomain cysteine and the zinc ion of the catalytic site. To become active, the thiol-Zn2+ interaction, commonly called the “cysteine switch,” must be disrupted (9), which can be mediated by proteolysis of the prodomain, post-translational modification of the thiol, allosteric interactions with other macromolecules, or other possible mechanisms (10). About one-third of proMMPs contains a furin-recognition sequence and are activated in the secretion pathway by furin proprotein convertase cleavage of the prodomain. However, with the possible exception of proMMP2 activation by MMP14, the physiologic activation mechanism of most MMPs is not known (10).
Matrilysin (28 kDa zymogen, 19 kDa active enzyme) is expressed by mucosal epithelia and some macrophages and functions as a key effector of repair and immunity. Established functions of matrilysin include facilitating re-epithelialization (11, 12), cleaving Fas ligand to promote apoptosis (13, 14), shedding syndecan-1 to control neutrophil influx (15), and macrophage-mediated elastolysis (16).
In mice, matrilysin activates pro-α-defensins (procryptdins), a family of structurally similar 3–4 kDa antimicrobial peptides found in the granules of Paneth cells at the base of the crypts of Lieberkühn (17). Because of the lack of mature cryptdins, matrilysin-null (Mmp7−/−) mice have an impaired ability to battle enteric pathogens (4). Cryptdins are packaged as pro-proteins of 7–8 kDa and are cleaved at a conserved site by matrilysin within the secretion granules (4, 18). In resting Paneth cells, the steady-state levels of pro- and activated cryptdins are roughly equivalent. Upon stimulation, the balance of procryptdins is rapidly activated indicating efficient proteolysis by matrilysin within the secretory pathway (18, 19). However, in defined in vitro reactions containing just substrate and proteinase, activation of procryptdins by matrilysin is slow, with only 50% of the precursor cleaved in 8 h or longer (4). Furthermore, both pro- and active matrilysin are present in Paneth cells granules (4) indicating that this MMP is activated in vivo by prodomain cleavage. The inefficient cleavage of procryptdins in vitro, their rapid processing in vivo, and the presence of activated matrilysin in Paneth cell granules led us to hypothesize that other factors regulate both the activation of promatrilysin and its activity against physiologic substrates.
Yu et al. (20, 21) reported that heparin increases matrilysin activity about 2–4-fold in a transferrin zymogram assay, and they reported that matrilysin colocalizes to heparan sulfate molecules in tissue. However, transferrin is not a physiologic substrate of this MMP, and it is not known how heparin and other glycosaminoglycans affect matrilysin activity against established substrates, such as procryptdins. Therefore, we assessed matrilysin activity in vitro in the presence of various glycosaminoglycans (GAGs), and we found that both zymogen activation and activity against specific substrates are markedly enhanced by highly sulfated molecules. Our findings suggest that specific GAGs function to control matrilysin proteolysis.
EXPERIMENTAL PROCEDURES
Reagents
Recombinant human promatrilysin and active matrilysin were purchased from EMD Biosciences (San Diego, CA) or produced in our laboratory (below). Porcine intestinal heparin (16 kDa) and dermatan sulfate were purchased from Celsus Laboratories (Cincinnati, OH) or Sigma-Aldrich. Low molecular weight heparin (mean MW: 5.5 kDa) was from Sigma and dp16, a 16-mer oligosaccharide was prepared from controlled partial heparin lyase 1 digestion of bovine lung heparin (Sigma) followed by size fractionation (22). Chondroitin-4-sulfate (CS-A; bovine trachea) was purchased from Sigma and chondroitin-6-sulfate (CS-C), chondroitin-2,6-sulfate (CS-D; shark cartilage), chondroitin-4,6-sulfate (CS-E, squid cartilage) were from Seikagaku Corp (Tokyo, Japan). Dextran sulfate (MW: 500 kDa) and dextran were from Sigma, and heparan sulfate was purchased from Associates of Cape Cod (East Falmouth, MA).
Recombinant Proteins
Recombinant procryptdins expressed and purified from Hi-5 insect cells as described (4). Wild type and catalytically inactive mutant promatrilysin human cDNAs were subcloned into a pET23b expression vector (EMD Biosciences) and transformed into One Shot TOP10 (Invitrogen) competent bacteria. Colonies (5–8) were picked from each plate, and 1-ml overnight cultures were grown in LB media with 50 μg/ml carbenicillin. Plasmids were isolated using a Qiagen (Valencia, CA) miniprep kit, sequenced, and transfected into BL21-Codon-Plus (DE3)-RIL bacteria (Stratagene). At log-phase growth (A600 ∼ 0.4), 1 mm isopropyl-1-thio-β-d-galactopyranoside was added, and the cultures were grown for an additional 4 h. Bacteria were harvested, and the inclusion bodies were isolated using Bacterial Protein Extraction Reagent (B-PER) solution (Pierce). The isolated inclusion bodies were solubilized in 8 m urea containing 50 mm Tris, pH 7.4, 50 mm NaCl, 10 mm CaCl2, 0.1 mm zinc acetate, 0.05% Brij-35, 0.02% NaN3, and 0.01% Triton X-100).
To refold the recombinant proproteins, we modified a published protocol (23). The inclusion bodies in the 8 m urea solution were dialyzed against a series of buffers with decreasing amounts of urea (50 mm Tris, pH 7.4, 50 mm NaCl, 10 mm CaCl2, 0.1 mm zinc acetate, 0.05% Brij-35, 0.02% NaN3, and 0.01% Triton X-100; urea at 4 m, 2 m, 1 m, 0.5 m, and 0 m). Refolded proteins were centrifuged, filtered through 0.22-μm filter, and concentrated 8–10-fold using a Centricon centrifugal filter device (Millipore; 10,000 MWCO). Recombinant proteins were purified in 2 steps using an ÄKTA Fast Protein Liquid Chromatography system (GE Healthcare, Piscataway, NJ).
First, cation exchange chromatography was done using SP-Sepharose column. Samples were equilibrated and injected in 50 mm sodium acetate, pH 6.0, 10 mm CaCl2, 0.02% NaN3, and eluted in 1-ml fractions with a 0–1 m NaCl gradient. Aliquots of fractions were resolved by SDS-PAGE and immunoblotted with anti-hMMP-7-AB4 (Calbiochem) as described (24) to identify promatrilysin. Fractions containing promatrilysin were pooled, concentrated, and separated by size exclusion chromatography using a Superdex-75 column equilibrated in 50 mm Tris, pH 7.5, 10 mm CaCl2, 0.02% NaN3. Fractions (0.5 ml) were collected in 0.1 mm phenylmethylsulfonyl fluoride, screened by SDS-PAGE and staining with Gelcode Blue (Pierce) or SYPRO Ruby (Bio-Rad) and immunoblotting, pooled, and concentrated. Wild-type promatrilysin was activated by incubating with 1 mm aminophenylmercuric acetate (APMA, Sigma).
Proteinase Activity Assays
Recombinant active human matrilysin was incubated with recombinant procryptdin-1, human E-cadherin/Fc fusion protein (hEcad/Fc, R&D Systems, Minneapolis, MN), or human fibronectin (BD Biosciences, Bedford MA) in 0.15 m NaCl, 10 mm Tris-HCl, pH 7.4, 5 mm CaCl2 (50 or 80 μl final volume) at 37 °C with or without the indicated GAGs. Reagent concentrations are listed in the figure legends. Sequencing of cleaved substrates was done by Edman degradation or tandem mass spectrometry. Promatrilysin autolytic activation assays were done in 100 μl in 10 mm HEPES, pH 7.4. 10 mm CaCl2 with or without GAGs. Reactions were stopped at the indicated times by adding sample buffer with 25 mm EDTA and were resolved through 15% Tris-Tricine or 4–12% Bis-Tris gels (Invitrogen, Carlsbad, CA). Gels were digitized using an UVP EpiChemi3 Gel Documentation System (Upland, CA). Procryptdin-1 cleavage data are presented as processing ratio, which was calculated as: [PCtx/(PCtx + Ctx)]/[PCt0/(Ct0 + PCt0)] and derived from band densities for procryptdin-1 (PCt0) and processed cryptdin-1 at time 0 (Ct0) and their densities at indicated sample times (PCtx, Ctx). Activation ratio for zymogen cleavage was similarly quantified.
MCA Substrate Assay
Recombinant active human matrilysin (13 nm) was preincubated with various GAGs (1.0 μg/ml for heparin, 0.1 μg/ml for other GAGs) in 50 mm HEPES, 10 mm CaCl2, 0.05% Brij-35, 10 μm ZnCl2, pH 7.0, on ice for 30 min, aliquoted in a 96-well plate in triplicate, and incubated at 37 °C for 30 min. OmniMMP MCA peptide (Mca-Ala-Pro-Lys(Dnp)-OH [Mca = (7-methoxycoumarin-4-yl)acetyl; Dnp = 2,4-dinitrophenyl]; Biomol International, Plymouth Meeting, MA) was added to a final concentration of 0, 5, 10, 15, or 20 μm (100 μl total volume). Samples were excited at 328 nm and fluorescence output was read at 393 nm in 30-s intervals for 30 min with a SpetraMax 250 plate reader (Molecular Devices, Sunnyvale, CA). Initial velocity (Vi) for each reaction was determined with using SoftmaxPro 4.6 plate reader software and was used to calculate Km and Vmax values. Data are presented as means and standard deviations from three separate experiments.
Surface Plasmon Resonance
To prepare biotinylated heparin and CS-E, heparin, or CS-E (2 mg) and amine-PEO3-Biotin (2 mg) were dissolved in 200 μl of H2O, and 10 mg of NaCNBH3 was added. The reaction mixture was heated at 70 °C for 24 h, after that a further 10 mg of NaCNBH3 was added, and the reaction was heated at 70 °C for another 24 h. After cooling to room temperature, the mixture was desalted with a spin column (3,000 MWCO). The biotinylated heparin and CS-E were immobilized onto Sensor SA (streptavidin) Chips following the manufacturer's protocol (GE Healthcare, Uppsala, Sweden) and confirmed by a 100–200 resonance unit (RU) increase in the sensor chip using a Biacore 3000 (GE Healthcare). Protein samples were diluted in 0.01 m HEPES, 0.15 m NaCl, 3 mm EDTA, 0.005% surfactant P20, pH 7.4, and different concentrations of pro- or active matrilysin were injected at a flow rate of 30 μl/min. The same buffer was then flowed over the sensor surface to facilitate dissociation, and the response (sensorgram) was monitored as a function of time at 25 °C. After a 2-min dissociation time, the sensor surface was regenerated with 30 μl of 2 m NaCl. Data were analyzed using BIAevaluate 4.0.1.
RESULTS
Sulfated GAGs Enhance Matrilysin Activity
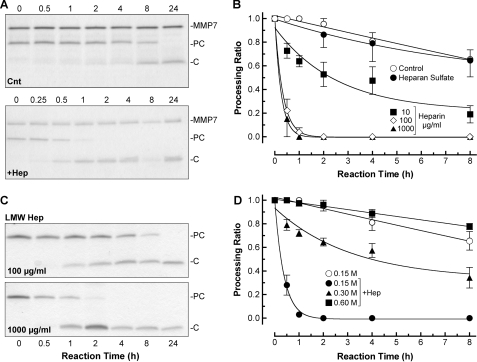
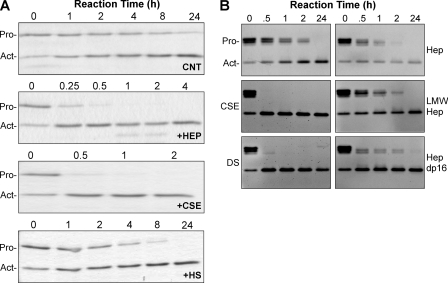
In reactions containing just active matrilysin and procryptdin-1, proteolytic cleavage of the substrate was slow, with 50% conversion (t½) in about 11 h (Fig. 1, A and B), if not longer (see Fig. 2), similar to previous observations (4). However, matrilysin processing of procryptdin-1 (7 kDa) to active cryptdin-1 (3.5 kDa) was markedly accelerated in the presence of heparin (16 kDa), which consistently reduced the t½ to <15 min, a greater than 44-fold increase in enzyme efficiency (Fig. 1, A and B). As we increased the concentration of heparin from 1 to 100 μg/ml, the rate of procryptdin-1 processing by matrilysin increased (Fig. 1B). Addition of 1.0 or 10 μg/ml of heparin did not affect the activity of matrilysin (data not shown). The matrilysin activity was also increased with low MW heparin (5.5 kDa) (Fig. 1C), but not as effectively (t½ > 60 min) as that seen with an equal mass of the larger polymer (t½ < 15 min). Heparin (or other GAGs) did not alter the P1′-P1 cleavage site (Ser39-Leu40) in procryptdin-1 (data not shown), which we mapped in earlier studies (4). As reported previously (18), matrilysin cleaves cryptdin prodomains into several smaller fragments not detectable in the gels.
FIGURE 1.
Cleavage of procryptdin-1 by active matrilysin is accelerated by heparin. A, active matrilysin (0.1 μg/μl; MMP7, 19 kDa) was incubated with 0.1 μg/μl recombinant procryptdin-1 (PC, 7 kDa; ∼3:1 substrate/enzyme molar ratio) with or without (Cnt) addition of 100 μg/ml heparin (+Hep), and reactions were stopped by placing aliquots in sample buffer at the indicated times (in hours). Shown are representative Coomassie Blue-stained gels. C, cleaved cryptdin-1, 3.5 kDa. B, heparin accelerated matrilysin cleavage of procryptdin-1 in a dose-dependent manner. Heparan sulfate (300 μg/ml for data shown) did not augment matrilysin activity. Data shown are the mean ± S.E. of processing ratios from four experiments. C, processing of procryptdin-1 by active matrilysin was also accelerated by low molecular weight heparin (LMW Hep). D, heparin-mediated (100 μg/ml) accelerated processing of procryptdin-1 by active matrilysin was inhibited in the presence of high NaCl concentration. Data shown are the processing ratios (mean ± S.E.) from three experiments.
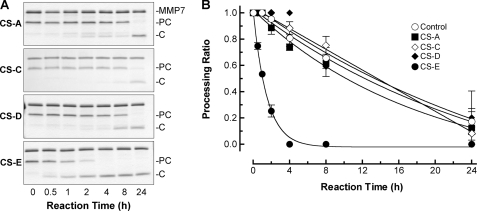
FIGURE 2.
CS-E selectively augments matrilysin activity. A, different classes of chondroitin sulfate (A, C, D, and E; 100 μg/ml each) were added to 80-μl reactions containing 0.1 μg/μl procryptdin-1 (PC; 7 kDa) and 0.1 μg/μl active matrilysin (MMP7; 19 kDa), and production of cryptdin-1 (C; 3.5 kDa) was assessed at the indicated times. B, data graphed are the procryptdin-1 processing ratios (mean ± S.E.) from three experiments.
Heparin is found only in mast cells (25), thus, it is not likely to affect matrilysin activity in vivo, which primarily comes from epithelial cells. Heparan sulfate is structurally similar to heparin but is less sulfated (26). Although the protein core of some heparin sulfate proteoglycans are substrates of matrilysin (15), heparan sulfate did not enhance matrilysin activity (Fig. 1B).
Using increasing salt concentrations, we assessed if electrostatic interactions drove heparin-mediated enhanced activity of matrilysin. Physiologic salt concentration (0.15 m) did not affect heparin-mediated acceleration of matrilysin activity (t½ ∼15 min; Fig. 1D). However, cleavage of procryptdin-1 was slowed in the presence of 0.3 m NaCl (t½ ∼ 4 h) and completely blocked with 0.6 m NaCl. This decrease was not due to salt inhibition of matrilysin catalytic activity directly. In the absence of heparin, high salt did not affect matrilysin activity to cleave procryptdin-1 or MCA peptide (data not shown). Lowering the pH from 7.4 to 5.2 also had no effect in these assays (data not shown).
Matrilysin Activity Promoted by Chondroitin-4,6-Sulfate
Chondroitin sulfates (CS), which are abundant on the cell surface, in secretion vesicles, and in the extracellular matrix, are comprised of a repeating disaccharide unit of N-acetylgalactosamine (GalNAc) and glucuronic acid (GlcA). There are several types of CS based on the extent and position of sulfate groups. Earlier nomenclature referred to these forms as A, C, D, or E (27). Yu et al. (20) reported that chondroitin 4-sulfate (CS-A) or chondroitin 6-sulfate (CS-C), which each have one sulfated GalNAc per disaccharide, did not affect matrilysin activity in a transferrin zymogram, and similarly we found that these GAGs did not significantly affect procryptdin-1 cleavage above control rates (Fig. 2; t½ > 14 h for control, >10 h for CS-A, and >13 h for CS-C). Chondroitin-2,6-sulfate (CS-D) has one sulfate on both the GlcA and GalNAc sugars and chondroitin-4,6-sulfate (CS-E) has two sulfates on the GalNAc sugar per disaccharide, but only CS-E significantly increased matrilysin activity (t½ ∼ 0.5 h; Fig. 2). The lack of activation by CS-D suggests that the presentation of the sulfur atom within the context of disaccharide unit, and not simply the degree of sulfation per disaccharide, is critical in affecting enzyme activity.
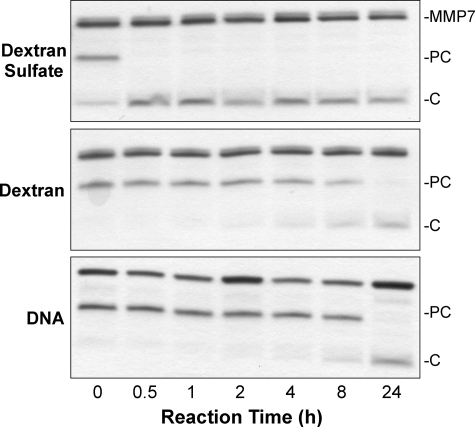
These data suggest that negative charge due to sulfation, within the appropriate context, is an important factor in modulating matrilysin activity. To assess this idea, we used dextran and dextran sulfate. Dextran sulfate is an O-sulfated polysaccharide, structurally different from heparin but with a similar degree of sulfation per unit sugar (28). Dextran sulfate accelerated procryptdin-1 processing by matrilysin similar to that mediated by CS-E and heparin (Fig. 3). In contrast, dextran, which is not sulfated, or heat-denatured DNA, a negatively charged nonsulfated polymer, had no effect (Fig. 3). These results demonstrate that highly sulfated GAGs enhance matrilysin catalytic activity.
FIGURE 3.
Effect of other negatively charged polyanions. Dextran sulfate (MW = 500,000; 100 μg/ml) augmented the ability of active matrilysin (MMP7; 19 kDa) to cleave procryptdin-1 (PC; 7 kDa) to cryptdin-1 (C; 3.5 kDa). Dextran (MW = 500,000; 100 μg/ml) and DNA (100 μg/ml) did not affect matrilysin activity.
Other Substrates
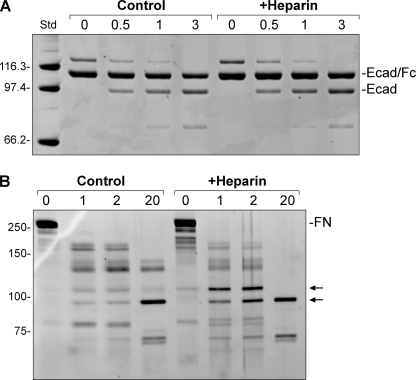
As demonstrated in cell-based models, matrilysin cleaves E-cadherin in its juxtamembrane stalk releasing the entire ectodomain (29, 30), and we reported that matrilysin sheds this junctional protein in vivo in response to tissue injury (12). To test if GAGs affect the ability of matrilysin to cleave this physiologic substrate, we used a fusion protein containing human E-cadherin ectodomain from the signal sequence up to the transmembrane domain (Met1-Ile707), a factor Xa cleavage site linker, and the Fc-portion (Pro100-Leu330) of human IgG1. Matrilysin cleaved the fusion protein producing an intact ectodomain fragment (Fig. 4A), and sequencing confirmed cleavage within the juxtamembrane stalk. In the absence of GAGs, matrilysin cleaved E-cadherin more efficiently than it cleaved procryptdin-1, requiring much less enzyme (5:1 substrate/enzyme molar ratio). Furthermore, heparin (Fig. 4) or any other GAG (not shown), including CS-E, did not change the efficiency or pattern of the cleavage.
FIGURE 4.
Effect on other substrates. A, recombinant human E-cadherin/Fc fusion protein (Ecad/Fc, 150 ng) was incubated with 5 ng (5:1 substrate/enzyme molar ratio) active matrilysin in 50-μl reactions that were preincubated on ice with or without 1 mg/ml heparin. B, human fibronectin (FN, 150 ng) was incubated with 30 ng of active matrilysin with or without 100 μg/ml heparin. Several degradation products were formed, but production of fragments around 100 kDa (arrows) were increased in the presence of heparin. Reactions in both panels were run at 37 °C and stopped at the indicated times (in hours) with addition of sample buffer, and the gels were stained with SYPRO Ruby. Std, molecular weight standards.
Fibronectin (250 kDa) is degraded by matrilysin into a range of fragments of 30 to 175 kDa (31, 32). Although heparin did not affect the overall degradation of fibronectin by matrilysin, it did selectively promote formation of fragments of about 100 kDa (Fig. 4B), which contain the cell binding domain (31).
The MCA peptide can be cleaved by essentially all MMPs. Heparin had a moderate, yet statistically significant (p < 0.05) effect on enhancing the rate of matrilysin cleavage of this test substrate, but other GAGs, including CS-E, did not (Table 1). Together, these data indicate that GAGs promote matrilysin activity to cleave or degrade some but not all substrates and that specific GAGs promote activity against different substrates.
TABLE 1.
Effect of GAGs on matrilysin cleavage of Mca substrate
Data are mean ± S.E. of triplicate determinations.
| GAG | Vmax | Km |
|---|---|---|
| RFU/s | μm | |
| None | 23.7 ± 7.7 | 11.6 ± 3.1 |
| Heparin | 18.9 ± 3.1 | 2.88 ± 1.6 |
| Heparan sulfate | 24.4 ± 5.0 | 10.9 ± 1.8 |
| CS-A | 28.6 ± 4.0 | 8.06 ± 2.7 |
| CS-C | 21.8 ± 3.3 | 12.1 ± 2.7 |
| CS-D | 28.0 ± 0.3 | 7.86 ± 2.0 |
| CS-E | 23.9 ± 1.5 | 6.70 ± 2.8 |
Sulfated GAGs Promote Autolytic Activation of Pro-matrilysin
We also assessed if sulfated GAGs promote the autolytic action of promatrilysin. By itself, promatrilysin will slowly autolytically convert itself to an active form, with a t½ of about 24 h (Fig. 5A) or less, depending on the activity of enzyme preparation being used. Similar to procryptdin-1 cleavage, autolytic activation of promatrilysin was markedly accelerated in the presence of heparin or CS-E (t½ < 0.5 h), whereas heparan sulfate (t½ ∼ 4 h) had only a moderate effect (Fig. 5, A and B). Dermatan sulfate, another sulfated GAG that is similar to CS (GlcA residues are epimerized into l-iduronic acid), also markedly promoted autolytic activation of promatrilysin (Fig. 5B). Low MW heparin and a 16-mer heparin oligosaccharide had intermediate effects.
FIGURE 5.
Sulfated GAGs accelerate autolytic activation of promatrilysin. A, promatrilysin (Pro; 0.1 μg/μl) was incubated in buffer alone (CNT) or with 100 μg/ml heparin (HEP), CSE, or heparan sulfate (HS), and autolytic processing to the active enzyme (Act) was assessed at the indicated times. B, similar studies were done with different batches of promatrilysin, heparin, and CS-E, as well as with dermatan sulfate (DS), low MW heparin (LMW Hep), or a 16-mer heparin oligosaccharide (Hep dp16). Promatrilysin (0.02 μg/μl) was incubated in buffer alone (upper left panel) or with 1 mg/ml heparin (Hep) or low MW heparin (LMW Hep) or 0.1 mg/ml CSE, dermatan sulfate, or heparin dp16 in 60-μl reactions. Reactions were stopped at the indicated time, 10-μl aliquots were resolved, and gels were stained with SYPRO Ruby.
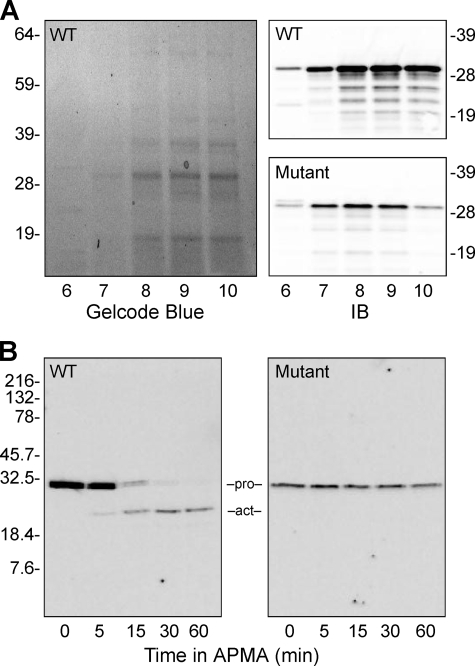
In these reactions, the autolytic activation of promatrilysin was likely mediated by the presence of active matrilysin in the commercial enzyme preparations (Fig. 5; the Act band in 0-h lanes). Thus, to generate the zymogen in the absence of any active form, we expressed and purified recombinant human promatrilysin produced in bacteria under conditions designed to preclude spontaneous autoactivation during the purification process. We did not add tags to bar any potential steric artifacts caused by additional motifs. Proteins in exclusion bodies were purified by 2-step chromatography, which yielded highly purified pro-enzymes with no contaminating active forms (Fig. 6A). In the presence of APMA, the refolded wild-type enzyme was able to undergo autolytic cleavage and activation, whereas the catalytically inactive mutant was not (Fig. 6B). (The gradual disappearance of active wild-type enzyme is a consequence of matrilysin degrading itself, which it tends to do in the absence of other substrates.)
FIGURE 6.
Recombinant wild-type and catalytically inactive mutant matrilysin. A, bacterially expressed wild-type (WT) and catalytically inactive mutant human promatrilysins were purified by cation exchange chromatography. Shown is a Gelcode-stained gel of fractions 6–10 of the pro-wild-type product; the pro-mutant fractions were similar. Gels were immunoblotted (IB) for matrilysin protein to identify fractions containing promatrilysin. B, pooled cation exchange fractions were further purified by size exclusion chromatography. Fractions containing promatrilysin proteins were screened by immunoblotting and pooled, and purity was assessed by SYBRO Ruby-stained gels (shown). Incubation with 1 mm APMA mediated autolytic activation of wild type but not mutant promatrilysin.
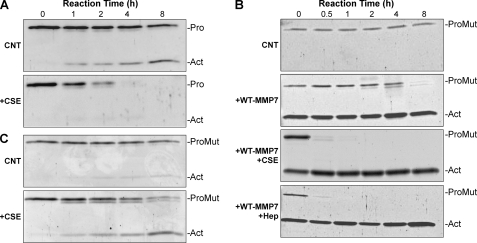
In buffer alone, wild-type promatrilysin activated slowly, but in the presence of CS-E, activation was markedly accelerated (Fig. 7A). CS-E and heparin also markedly accelerated the ability of active wild-type matrilysin to cleave inactive promatrilysin (Fig. 7, B and C). In buffer alone, mutant promatrilysin was quite stable, and with addition of active matrilysin, it was slowly cleaved (Fig. 7B). (In Figs. 7, B and C and 9C, Act indicates the cleaved form of mutant matrilysin. This protein, of course, does not gain enzymatic activity.)
FIGURE 7.
Intra- and intermolecular activation of promatrilysin. A, purified recombinant promatrilysin (Pro; 0.04 μg/μl) was incubated with or without 100 μg/ml CSE in 100 μl. Reactions were stopped at the indicated times (in hours) by adding sample buffer. Once converted to an active enzyme (Act), matrilysin has a propensity to degrade itself, which, along with activation of the zymogen, was accelerated with CSE. B, recombinant inactive mutant promatrilysin (ProMut, 0.04 μg/μl) was incubated alone (CNT) or with active wild-type matrilysin (WT-MMP7; 0.08 μg/μl; 1:3 Pro:active molar ratio) plus 100 μg/ml CSE or 100 μg/ml heparin (Hep), and reactions were stopped at the indicated times. Note that the active form present at 0 h corresponds to the added wild-type matrilysin. C, recombinant inactive mutant promatrilysin (Mut, 0.06 μg/μl) was incubated alone (CNT) or with active matrilysin (0.004 μg/ml; 1:10 Pro:active molar ratio) plus 100 μg/ml CSE. Reactions were stopped at the times indicated in A.
GAG-Matrilysin Interaction
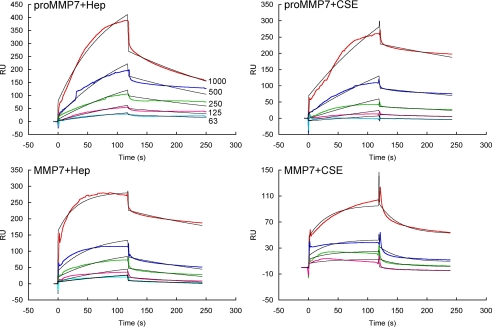
We used surface plasmon resonance (SPR) to assess in real time the direct interaction of heparin and CS-E with promatrilysin and active matrilysin (Fig. 8 and Table 2). Promatrilysin associated rapidly with both heparin and CS-E and dissociated slowly from both GAGs (Table 2). The KD of promatrilysin-GAG interactions was 400 nm and 630 nm for heparin and CS-E, respectively. Active matrilysin also bound heparin with similar on-off kinetics and affinity but it interacted weakly with CS-E. These data suggest that CS-E interacts preferentially with the prodomain. Heparan sulfate interacted weakly with promatrilysin and negligibly with active matrilysin. Procryptdin protein also bound to CSE with high affinity (KD, 420 nm) and about one-log weaker to heparin (KD, 5.3 μm).
FIGURE 8.
Surface plasmon resonance of GAG-matrilysin interaction. Promatrilysin or active matrilysin at the nanomolar concentrations indicated were interacted with heparin (Hep) or CSE immobilized to streptavidin sensor chips. The colored lines are the raw data, and the black curves are the fitted lines using models from BIAevaluate 4.0.1.
TABLE 2.
Matrilysin-GAG Interactions-surface plasmon resonance
| Components | kon | koff | KD |
|---|---|---|---|
| 1/ms | 1/s | m | |
| proMMP7 + Heparin | 8.2 × 103 | 3.2 × 10−3 | 4.0 × 10−7 |
| proMMP7 + CS-E | 2.8 × 103 | 1.8 × 10−3 | 6.3 × 10−7 |
| proMMP7 + Heparan sulfate | 79 | 0.05 | 6.0 × 10−4 |
| MMP7 + Heparin | 2.8 × 104 | 4.1 × 10−3 | 1.5 × 10−7 |
| MMP7 + CS-E | 505 | 0.03 | 6.0 × 10−5 |
| MMP7 + Heparan sulfate | NDa | ND | ND |
| proCryptdin + Heparin | 9.2 × 103 | 0.48 × 10−3 | 5.3 × 10−6 |
| proCryptdin + CS-E | 4.3 × 104 | 0.18 × 10−3 | 4.2 × 10−7 |
a ND, no detectable interaction.
DISCUSSION
Our in vitro data demonstrate that highly sulfated GAGs promote both the activation of promatrilysin and the ability of the active proteinase to cleave specific substrates. We propose that GAGs serve two broad functions in regulating matrilysin activity and possibly that of other MMPs. First, they act as allosteric modulators of matrilysin catalysis, particularly in promoting zymogen activation via autolytic (either inter- or intramolecular) cleavage of the prodomain. Second, GAGs provide an anchor on the cell surface or within secretion granules or the pericellular environment to compartmentalize matrilysin proteolysis to specific substrates within defined locations. In the case of procryptdin activation, our binding data suggest that GAGs, and specifically chondroitin-4,6-sulfate, also can interact with the substrate possibly in trimeric complex. As for most protein-protein interactions, MMP specificity may be driven by (at least) a third component, and identifying the nature of these anchors will be a key advance toward identifying activation mechanisms and substrates.
Compartmentalization, that is where and how in the pericellular environment an MMP is released and localized, is a critical process for regulating the specificity of proteolysis, possibly more so than the affinity of enzyme-substrate interactions. Analogous to MMPs with a transmembrane domain, the so-called “secreted” MMPs, such as matrilysin, are likely confined to precise compartments, thereby maintaining both a locally high enzyme concentration and targeting their catalytic activity to specific substrates. Indeed, several examples of specific cell-MMP interactions have been reported, such as the binding of MMP1 to the α2β1 integrin (24, 33) and MMP9 to CD44 (34).
Matrilysin has been reported to bind heparan sulfate (20, 21), cholesterol sulfate (32, 35), and CD151, a tetraspanin (36, 37). Although these macromolecules may provide different cell surface anchors directing proteolysis to specific substrates, it is not clear if these diverse interactions all affect activation and/or catalytic activity per se. For example, matrilysin interacts with cell surface heparan sulfate proteoglycans, such as CD44v3 (20, 21), and sheds others, such as syndecan-1 (15); but we found that heparan sulfate did not enhance either matrilysin activity or its autolytic activation. CD151 can bind and presumably activate pro-matrilysin (36), but CD151-dependent pro-enzyme activation is incomplete and occurs only in the presence of transferrin, a nonphysiologic substrate. Furthermore, siRNA ablation of CD151 reduced matrilysin expression (38), suggesting that CD151 influences enzyme activity via an effect on production. Yamamoto et al. (35) reported that active matrilysin binds cholesterol sulfate, but an effect on proteinase activity was shown by indirect means.
We speculate that matrilysin interacts with different macromolecules to direct its activity to specific substrates. Indeed, whereas cleavage of procryptdin-1 was markedly enhanced with sulfated GAGs, matrilysin proteolysis of E-cadherin was quite efficient without added GAGs. Thus, activated matrilysin may interact with a surface molecule, such as CD151 or cholesterol sulfate, such as when directed to shedding E-cadherin, but when cleaving other substrates in other compartments, such as procryptdins in secretion vesicles, the enzyme may be controlled by other interactions, specifically with sulfated GAGs.
In our studies, heparin and chondroitin-4,6-sulfate, as well as dextran sulfate, were consistently the most effective at promoting autolytic activation of promatrilysin and activity of the active proteinases. These findings indicate that anionic charge due to sulfation is critical in potentiating matrilysin activation and activity. Heparan sulfate is less sulfated than heparin or chondroitin-4,6-sulfate (39) and was only weakly effective at promoting autolytic activation or ineffective at enhance matrilysin activity. Chondroitin-2,6-sulfate (CS-D), which has one sulfate on each monosaccharide oriented on different sides of the molecule, also did not enhance matrilysin activity. In contrast, chondroitin-4,6-sulfate (CS-E), which has two sulfates on each GalNAc sugar oriented on the same side of the molecule, potently affected both activation and activity. The different effect between these two GAGs suggests that the presentation of sulfates is more important than the overall degree of sulfation. Similarly, active matrilysin binds cholesterol sulfate but not unsulfated cholesterol on colon cancer cells (35). Interestingly, structural modeling predicts that both heparin (20) and cholesterol sulfate (32) bind the surface of matrilysin opposite to the catalytic cleft, thereby minimizing any interference with substrate interactions.
Although autolytic activation of promatrilysin was enhanced by sulfated GAGs, there was a significant increase in the activation when active matrilysin was included in the reaction. Furthermore, active wild-type matrilysin cleaved the prodomain of wild-type pro-matrilysin about as efficiently as it cleaved the prodomain from mutant promatrilysin (Fig. 7). These in vitro observations suggest that the bulk of autolytic activation proceeds via intermolecular cleavage and that the interaction of active matrilysin with promatrilysin is facilitated by GAGs. Chondroitin-4,6-sulfate bound promatrilysin but not active matrilysin, suggesting that an allosteric interaction with positively charged amino acids in the prodomain drives intermolecular zymogen activation. However, when considering how the zymogen would be activated in vivo, this proposed mechanism leads logically to an egg-and-chicken augment. Assuming no other proteinases are involved, at least one promatrilysin molecule must be activated by intramolecular cleavage to ignite activation of other zymogen molecules. Our in vitro data with purified promatrilysin suggest that chondroitin-4,6-sulfate can promote such a series of reactions, but other factors, such as zymogen concentration, may also be important. In vivo, such as in the microenvironment of secretion vesicles of Paneth cells, locally high concentrations of promatrilysin combined with the right GAG may create an environment and the needed allosteric interactions to permit both intra- and intermolecular activation. During protein refolding and purification, we found that promatrilysin began to convert to its active form when the concentration exceeded 0.1 mg/ml. Interesting, we have found that Paneth cells also contain serglycin,4 a chondroitin-4,6-sulfate proteoglycan found in secretion vesicles of various leukocytes (40–42). Consistent with the idea that this secretion vesicle proteoglycan may function to control matrilysin activity, in mice lacking serglycin, leukocyte proteases are missorted and have reduced activity (42–44). We are currently assessing if serglycin functions to control matrilysin activity in vivo.
In summary, we propose that sulfated GAGs play important roles in controlling both the activation and activity of matrilysin. Through direct interaction with the zymogen, possibly to the prodomain, sulfated GAGs, such as chondroitin-4,6-sulfate, act as allosteric modulators promoting the autolytic activation of the proteinase. Once activated, GAGs may facilitate proteolysis of certain substrates by interacting with the substrate, the enzyme, or both. An implication of our data is that the specificity of proteolysis requires more than just enzyme and substrate, and that an allosteric interaction with at least one other macromolecule is needed to confine proteolysis to targeted substrates within defined compartments. Indeed, the specificity of most protein-protein reactions requires them to interact within higher order complexes. Heparan sulfate proteoglycans regulate several serine proteases, the coagulation enzymes, mast cell tryptase, and tripeptidyl-peptidase. The autolytic activation of proMMP2 and activity of MMP1 are enhanced by heparin (45, 46), suggesting that sulfated GAGs may have wide roles in controlling MMP proteolysis. The observations that heparin did not enhance cleavage of E-cadherin may indicate that distinct accessory molecule function to spatially confine matrilysin activity to selective substrates. Thus, identifying how (and if) MMPs are anchored to specific microenvironments may provide new mechanisms to target in manipulating enzyme activity.
Acknowledgments
We thank Dr. Andre J. Ouellette, University of Southern California, for procryptdins, Dr. Lijuan Zhang, WA University in St. Louis, for providing chondroitin sulfates and technical advice, and Thomas Broekelmann and Robert Mecham, WA University, for help with Edman sequencing. This work was supported in part by the University of Washington's Proteomics Resource (UWPR95794). Some of the initial work for these studies was done while some authors (H. J. R., C. L. W., and W. C. P.) were at Washington University in St. Louis.
This work was supported, in whole or in part, by National Institutes of Health Grants HL029594 (to W. C. P.), GM038060 (to R. J. L.), and F32-HL083665 (to S. H. B.).
W. C. Parks, unpublished observations.
- MMP
- matrix metalloproteinase
- CS-A
- chondroitin-4-sulfate
- CS-C
- chondroitin-6-sulfate
- CS-D
- chondroitin-2,6-sulfate
- CS-E
- chondroitin-4,6-sulfate
- DS
- dermatan sulfate
- GAG
- glycosaminoglycan
- MCA
- Mca-Ala-Pro-Lys(Dnp)-OH
- Mca
- (7-methoxycoumarin-4-yl)acetyl
- Dnp
- 2,4-dinitrophenyl
- SPR
- surface plasmon resonance
- RFU
- relative fluorescence units.
REFERENCES
- 1.Parks W. C., Wilson C. L., López-Boado Y. S. (2004) Nat. Rev. Immunol. 4, 617–629 [DOI] [PubMed] [Google Scholar]
- 2.Page-McCaw A., Ewald A. J., Werb Z. (2007) Nat. Rev. Mol. Cell Biol. 8, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manicone A. M., McGuire J. K. (2008) Semin. Cell Dev. Biol. 19, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson C. L., Ouellette A. J., Satchell D. P., Ayabe T., López-Boado Y. S., Stratman J. L., Hultgren S. J., Matrisian L. M., Parks W. C. (1999) Science 286, 113–117 [DOI] [PubMed] [Google Scholar]
- 5.Wilson C. L., Schmidt A. P., Pirilä E., Valore E. V., Ferri N., Sorsa T., Ganz T., Parks W. C. (2009) J. Biol. Chem. 284, 8301–8311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McQuibban G. A., Gong J. H., Tam E. M., McCulloch C. A., Clark-Lewis I., Overall C. M. (2000) Science 289, 1202–1206 [DOI] [PubMed] [Google Scholar]
- 7.Van Lint P., Libert C. (2007) J. Leukocyte Biol. 82, 1375–1381 [DOI] [PubMed] [Google Scholar]
- 8.Cauwe B., Van den Steen P. E., Opdenakker G. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 113–185 [DOI] [PubMed] [Google Scholar]
- 9.Van Wart H. E., Birkedal-Hansen H. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5578–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ra H. J., Parks W. C. (2007) Matrix Biol. 26, 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunsmore S. E., Saarialho-Kere U. K., Roby J. D., Wilson C. L., Matrisian L. M., Welgus H. G., Parks W. C. (1998) J. Clin. Invest. 102, 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGuire J. K., Li Q., Parks W. C. (2003) Am. J. Pathol. 162, 1831–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powell W. C., Fingleton B., Wilson C. L., Boothby M., Matrisian L. M. (1999) Curr. Biol. 9, 1441–1447 [DOI] [PubMed] [Google Scholar]
- 14.Mitsiades N., Yu W. H., Poulaki V., Tsokos M., Stamenkovic I. (2001) Cancer Res. 61, 577–581 [PubMed] [Google Scholar]
- 15.Li Q., Park P. W., Wilson C. L., Parks W. C. (2002) Cell 111, 635–646 [DOI] [PubMed] [Google Scholar]
- 16.Filippov S., Caras I., Murray R., Matrisian L. M., Chapman H. A., Jr., Shapiro S., Weiss S. J. (2003) J. Exp. Med. 198, 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouellette A. J., Selsted M. E. (1996) FASEB J. 10, 1280–1289 [DOI] [PubMed] [Google Scholar]
- 18.Ayabe T., Satchell D. P., Pesendorfer P., Tanabe H., Wilson C. L., Hagen S. J., Ouellette A. J. (2002) J. Biol. Chem. 277, 5219–5228 [DOI] [PubMed] [Google Scholar]
- 19.Ayabe T., Satchell D. P., Wilson C. L., Parks W. C., Selsted M. E., Ouellette A. J. (2000) Nat. Immunol. 1, 113–118 [DOI] [PubMed] [Google Scholar]
- 20.Yu W. H., Woessner J. F., Jr. (2000) J. Biol. Chem. 275, 4183–4191 [DOI] [PubMed] [Google Scholar]
- 21.Yu W. H., Woessner J. F., Jr., McNeish J. D., Stamenkovic I. (2002) Genes Dev. 16, 307–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pervin A., Gallo C., Jandik K. A., Han X. J., Linhardt R. J. (1995) Glycobiology 5, 83–95 [DOI] [PubMed] [Google Scholar]
- 23.Itoh F., Yamamoto H., Hinoda Y., Imai K. (1996) Cancer 77, 1717–1721 [DOI] [PubMed] [Google Scholar]
- 24.Dumin J. A., Dickeson S. K., Stricker T. P., Bhattacharyya-Pakrasi M., Roby J. D., Santoro S. A., Parks W. C. (2001) J. Biol. Chem. 276, 29368–29374 [DOI] [PubMed] [Google Scholar]
- 25.Humphries D. E., Wong G. W., Friend D. S., Gurish M. F., Qiu W. T., Huang C., Sharpe A. H., Stevens R. L. (1999) Nature 400, 769–772 [DOI] [PubMed] [Google Scholar]
- 26.Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. (1999) Annu. Rev. Biochem. 68, 729–777 [DOI] [PubMed] [Google Scholar]
- 27.Silbert J. E., Sugumaran G. (2002) IUBMB Life 54, 177–186 [DOI] [PubMed] [Google Scholar]
- 28.Alter S. C., Metcalfe D. D., Bradford T. R., Schwartz L. B. (1987) Biochem. J. 248, 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies G., Jiang W. G., Mason M. D. (2001) Clin. Cancer Res. 7, 3289–3297 [PubMed] [Google Scholar]
- 30.Noë V., Fingleton B., Jacobs K., Crawford H. C., Vermeulen S., Steelant W., Bruyneel E., Matrisian L. M., Mareel M. (2001) J. Cell Sci. 114, 111–118 [DOI] [PubMed] [Google Scholar]
- 31.von Bredow D. C., Nagle R. B., Bowden G. T., Cress A. E. (1995) Exp. Cell Res. 221, 83–91 [DOI] [PubMed] [Google Scholar]
- 32.Higashi S., Oeda M., Yamamoto K., Miyazaki K. (2008) J. Biol. Chem. 283, 35735–35744 [DOI] [PubMed] [Google Scholar]
- 33.Stricker T. P., Dumin J. A., Dickeson S. K., Chung L., Nagase H., Parks W. C., Santoro S. A. (2001) J. Biol. Chem. 276, 29375–29381 [DOI] [PubMed] [Google Scholar]
- 34.Yu Q., Stamenkovic I. (2000) Genes Dev. 14, 163–176 [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto K., Higashi S., Kioi M., Tsunezumi J., Honke K., Miyazaki K. (2006) J. Biol. Chem. 281, 9170–9180 [DOI] [PubMed] [Google Scholar]
- 36.Shiomi T., Inoki I., Kataoka F., Ohtsuka T., Hashimoto G., Nemori R., Okada Y. (2005) Lab. Invest. 85, 1489–1506 [DOI] [PubMed] [Google Scholar]
- 37.Fujita Y., Shiomi T., Yanagimoto S., Matsumoto H., Toyama Y., Okada Y. (2006) Arthritis Rheum. 54, 3233–3243 [DOI] [PubMed] [Google Scholar]
- 38.Hasegawa M., Furuya M., Kasuya Y., Nishiyama M., Sugiura T., Nikaido T., Momota Y., Ichinose M., Kimura S. (2007) Lab. Invest. 87, 882–892 [DOI] [PubMed] [Google Scholar]
- 39.Capila I., Linhardt R. J. (2002) Angew. Chem. Int. Ed. Engl. 41, 391–412 [DOI] [PubMed] [Google Scholar]
- 40.Schick B. P., Jacoby J. A. (1995) J. Cell. Physiol. 165, 96–106 [DOI] [PubMed] [Google Scholar]
- 41.Toyama-Sorimachi N., Kitamura F., Habuchi H., Tobita Y., Kimata K., Miyasaka M. (1997) J. Biol. Chem. 272, 26714–26719 [DOI] [PubMed] [Google Scholar]
- 42.Zernichow L., Abrink M., Hallgren J., Grujic M., Pejler G., Kolset S. O. (2006) J. Biol. Chem. 281, 26792–26801 [DOI] [PubMed] [Google Scholar]
- 43.Abrink M., Grujic M., Pejler G. (2004) J. Biol. Chem. 279, 40897–40905 [DOI] [PubMed] [Google Scholar]
- 44.Grujic M., Braga T., Lukinius A., Eloranta M. L., Knight S. D., Pejler G., Abrink M. (2005) J. Biol. Chem. 280, 33411–33418 [DOI] [PubMed] [Google Scholar]
- 45.Crabbe T., Ioannou C., Docherty A. J. (1993) Eur. J. Biochem. 218, 431–438 [DOI] [PubMed] [Google Scholar]
- 46.Crabbe T., O'Connell J. P., Smith B. J., Docherty A. J. (1994) Biochemistry 33, 14419–14425 [DOI] [PubMed] [Google Scholar]