Abstract
Bax inhibitor-1 (BI-1) is a cell death suppressor protein conserved across a variety of organisms. The Arabidopsis atbi1-1 plant is a mutant in which the C-terminal 6 amino acids of the expressed BI-1 protein have been replaced by T-DNA insertion. This mutant BI-1 protein (AtBI-CM) produced in Escherichia coli can no longer bind to calmodulin. A promoter-reporter assay demonstrated compartmentalized expression of BI-1 during hypersensitive response, introduced by the inoculation of Pseudomonas syringae possessing the avrRTP2 gene, Pst(avrRPT2). In addition, both BI-1 knockdown plants and atbi1-1 showed increased sensitivity to Pst(avrRPT2)-induced cell death. The results indicated that the loss of calmodulin binding reduces the cell death suppressor activity of BI-1 in planta.
Bax inhibitor-1 (At5g47120, BI-1)2 is a highly conserved cell death suppressor protein that resides in the endoplasmic reticulum (ER) membranes of a range of organisms. BI-1 is important in the response of organisms to abiotic and biotic stresses. Down-regulation of BI-1 in tobacco suspension cells (BY2) induced sensitivity against starvation (1), whereas overexpression in barley induced the breakdown of mlo-mediated penetration resistance to the fungal pathogen, powdery mildew (Blumeria graminis) (2). Cultured rice cells overexpressing Arabidopsis BI-1 (AtBI-1) showed increased resistance to Magnaporthe grisea-induced hypersensitive response (HR)-like cell death, potentially confirming the role of BI-1 in HR regulation (3). Recent studies on animal and plant BI-1 indicated a close relationship with ER stress response (4–6). BI-1-deficient mice are hypersensitive to apoptosis induced by ER stress agents such as thapsigargin, tunicamycin, and brefeldin A (4). Such events correlate with decreased calcium release from the ER, and our previous study demonstrated an association of BI-1 with calcium signaling in stress-treated plant cells (7). However, the molecular mechanism by which BI-1 suppresses cell death is still unclear.
Recently, Watanabe et al. (5, 8) demonstrated that an Arabidopsis T-DNA-tagged mutant, atbi1-1, was more susceptible to fungal toxin-, heat-shock-, and tunicamycin-induced cell death. The atbi1-1 plant has T-DNA inserted into the AtBI-1 protein C-terminal region, which contains potential coiled-coil structures and is essential for inhibiting both Bax-induced lethality in yeast and oxidative stress-induced cell death in plant cells as we had demonstrated earlier (9). We also found that the C-terminal 14 amino acids of AtBI-1 were capable of binding to the calmodulin molecule, a mediator of calcium signaling (7). Here, the present study directly proved the functional interaction between the highly conserved calmodulin molecule and BI-1 using a genomic mutation of the AtBI-1 gene. Such a genomic mutant showed accelerated sensitivity against Pseudomonas-induced HR cell death. The results indicated that the C-terminal-less BI-1 protein, which lost the CaM binding, was associated with reduced cell death suppression activity in vivo.
EXPERIMENTAL PROCEDURES
Plant Materials
The Columbia (Col-0) ecotype of Arabidopsis thaliana (L.) Heynh was used as the wild-type plant in this study. Seeds of Arabidopsis were sterilized and either inoculated onto plates containing 0.2% Gerangam, 2% (w/v) sucrose, and 0.5× MS medium or grown on a Jiffy-7 pellet (Jiffy Products International AS, Stange, Norway). All plant material was grown at 23 °C under continuous light conditions (60 μmol m−2s−1). AtBI-1 knockdown plant (AtBI-RNAi strains) and GFP-containing control plant were described previously (7). The T-DNA-inserted line, atbi1-1, was obtained from Syngenta as Garlic_228_D08, and T-DNA insertion was confirmed by PCR and sequence determination.
Yeast Experiments
Saccharomyces cerevisiae wild-type strain W303-1A was used in the present study. The Yep51-Bax expression vector was provided by Dr. Reed (The Burnham Institute for Medical Research) (10). The pYX112-AtBI-CM expression vector was constructed by using the PCR-amplified EcoRI-tagged atbi1-1 cDNA fragment. The pYX112-AtBI-1 vector was described previously (11, 12). Yeast strains carrying Yep51-Bax were transformed with pYX112, pYX112-AtBI-1, or pYX112-AtBI-CM by the lithium acetate method. Yeast strains were cultured in YPD (1% yeast extract, 2% peptone, and 2% glucose) or synthetic dextrose medium with appropriate supplements at 30 °C. Ura−Leu− transformants were streaked either on synthetic dextrose-glucose (Glc) or on synthetic dextrose-galactose (Gal) plates and incubated at 30 °C for 3 (Glc) or 4 (Gal) days, respectively.
Total yeast RNA was extracted using the RNeasy plant miniprep kit (Qiagen, Hilden, Germany). Purified RNA (0.2 μg) was used for RT-PCR using specific primers for AtBI-1: a, 5′-ATGAGCATCCTTATCACTGCATT-3′; b, 5′-CTTCAACATTATGATGAGAAT-3′; c, 5′-GTTTCTCCTTTTCTTCTTC-3′; and d, 5′-CACCACAAAAGCGTCATTCTTCT-3′. PCR reactions were performed in 10 μl final volume using a direct amplification system (Ready-To-Go RT-PCR beads, Amersham Biosciences) as described in Oshima et al. (13).
Overlay Assay
The interaction of AtBI-1 with calmodulin was studied by overlay assay basically as described previously (7). The C-terminal 14 amino acids of AtBI-1, AtBI-CM, AtBI-29, and AtBI-30 were ligated into the pMAL vector (New England Biolabs) to express maltose-binding protein (MBP)-tagged proteins. The resultant plasmids were transformed into Escherichia coli BL21 strain, and MBP-tagged C-terminal proteins were purified according to the instructions provided by the manufacturer (New England Biolabs). The empty pMAL vector carrying in-frame β-galactosidase protein was used as a control (Gal). The His- and S-tagged Arabidopsis calmodulin 7 (AtCaM7) protein was also purified as described previously (7).
The purified C-terminal and Gal (control) proteins were subjected to SDS-PAGE, blotted onto polyvinylidene difluoride membrane, and immunodetected with anti-MBP antibody (New England Biolabs) as a control for protein loading. S-tagged AtCaM7 protein in 1× phosphate-buffered saline with 1 mm CaCl2 was used as the overlay. Binding was detected using horseradish peroxidase-conjugated S-protein (Novagen, Tokyo) and visualized by chemiluminescence (ECL kit, Amersham Biosciences). The coding regions of Arabidopsis calmodulin and calmodulin-related proteins (AtCaM3, AtCaM6, AtCaM8, AtCaM9, AtCML23, and AtCML12) were also amplified by PCR, ligated into the pMAL vector, and used in this assay. Primers used for PCR were as follows: AtCaM3, 5′-CACCATGGCGGATCAGCTCACCG-3′ and 5′-CTTAGCCATCATGACCTTAAC-3′; AtCaM6, 5′-CACCATGGCGGATCAGCTAACC-3′ and 5′-CTTTGCCATCATGACTTTG-3′; AtCaM8, 5′-CACCATGGAAGAAACAGCACTGAC-3′ and 5′-TCTTGACAAATCCATCATCCTTG-3′; AtCaM9, 5′-CACCATGGCGGATGCTTTCACAG-3′ and 5′-ATAAGAGGCAGCAATCATC-3′; At1g66400 (AtCML23), 5′-CACCATGTCGAAGAACGTTTCG-3′ and 5′-AGCACTACCATTAATCATC-3′; and At2g41100 (AtCML12), 5′-CACCATGGCGGATAAGCTCACTG-3′ and 5′-AGATAACAGCGCTTCGAAC-3′.
GUS Assay and Microscopic Analysis
The promoter-GUS reporter assay plasmid consisted of 2 kbp of AtBI-1 promoter region and the N-terminal 10 amino acids of AtBI-1 fused with the GUS reporter. The genomic fragment of AtBI-1 was amplified with 5′-GGTCGACCAAGTCAAAGACCTTCGATACAT-3′ and 5′-TCTAGACCAGCTTCTGCTACCAGGTTGAG-3′ by PCR and then introduced into the SalI/XbaI site of a cassette plasmid pHTS6.1 (14) containing the GUS reporter gene. The resultant plasmid was introduced into the Agrobacterium EHA105 strain, and transgenic Arabidopsis plants were obtained by Agrobacterium-mediated transformation. More than 10 of transgenic Arabidopsis lines possessing the AtBI-1-promoter-GUS construct were obtained. Among them, three lines, which show stable and typical GUS staining patterns, were chosen for further study. For the GUS assay, plant tissues were fixed in 90% acetone for 10 min on ice and vacuum-infiltrated with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide buffer (0.1 m NaPO4 buffer, pH 7.0, 10 mm EDTA, 3 mm potassium ferricyanide, 3 mm potassium ferrocyanide, 0.1% (v/v) Triton X-100, and 0.5 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-glucuronide). Leaves incubated overnight in the dark at 37 °C were decolored with acetic acid:ethanol (1:6) solution at room temperature. Transparentized tissues treated with chloride hydrate were observed and photographed. Autofluorescence induced by bacterial infection was monitored and imaged by fluorescence microscopy (MMRD; Leica, Wetzlar, Germany).
Infection with Pseudomonas
Pseudomonas syringae pv. tomato DC3000 strains containing avrRpt2, Pst(avrRpt2), or the empty plasmid vector Pst(vec) were grown for 1 day on King's B medium containing appropriate antibiotics (25 μg/ml rifampicin and 25 μg/ml kanamycin) at 30 °C. Leaves were inoculated with 5 × 108 colony-forming units/ml of P. syringae in 10 mm MgCl2, using 1-ml plastic syringes without needles. To detect ion leakage, three leaf discs obtained from plants inoculated with Pseudomonas were floated on autoclaved distilled water. Electrolyte leakage was monitored using an electrical conductivity meter (B-173; Horiba, Kyoto, Japan) at several time points and expressed as a relative value against total ion leakage, which was measured in the autoclaved samples. Bacterial growth was measured at 4 days after infiltration by extracting bacteria from leaf discs and plating a series of dilutions on the King's B medium supplemented with appropriate antibiotics.
RESULTS
Requirement of BI-1 C Terminus for Calmodulin Binding
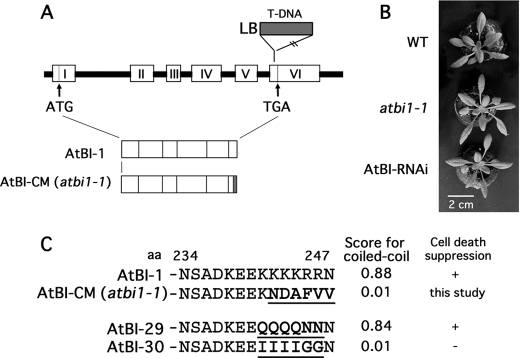
The AtBI-1 gene (At5G47120) consists of five introns and six exons. As reported previously, one of the T-DNA-tagged Arabidopsis lines, atbi1-1 (SAIL_228_D08), has a T-DNA insertion in exon 6. The atbi1-1 plants show accelerated progression of cell death upon infiltration of leaf with fungal toxin FB1, as well as increased sensitivity to heat-shock- and tunicamycin-induced cell death (5, 8). We also independently obtained the T-DNA-inserted plant (Garlic_228_D08, Syngenta), equivalent to SAIL_228_D08, and confirmed that this plant expresses mRNA containing the inserted boundary sequence of T-DNA encoding six replacement amino acids in-frame at the C terminus (Fig. 1). In addition, the plant showed no specific phenotype under normal growth conditions, similar to the AtBI-RNAi plant, in which AtBI-1 was down-regulated by RNAi method (Fig. 1B) (7).
FIGURE 1.
Schematic diagram of AtBI-1 mutants used in this study. A, genetic structure of the AtBI-1 and T-DNA-inserted mutant (AtBI-CM/atbi1-1). Exons are represented by numbered open boxes, and introns are represented by lines. LB, left border. B, representative phenotype of atbi1-1 and AtBI-RNAi plant. Plants were grown for 3 weeks under continuous light at 22 °C. WT, wild type. C, AtBI-1 mutants analyzed in the present study. The C-terminal 6 amino acids were replaced in mutant proteins (AtBI-CM, AtBI-29, and AtBI-30). Replaced amino acids are underlined. Calculated scores for coiled-coil structure (available through the COILS server) and cell death suppression activity in yeast analyzed previously (7, 9) are indicated.
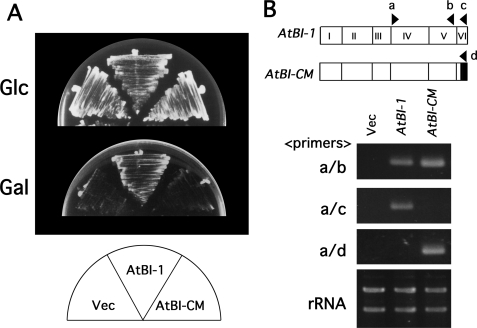
To test whether the atbi1-1 C-terminal mutant protein functions normally as a cell death suppressor, we assayed for rescue of Bax-induced yeast lethality. As reported previously, a C-terminal variant with a high coiled-coil score, AtBI-29, retained cell death suppression activity, whereas the AtBI-30 mutant with a reduced coiled-coil score failed to suppress Bax-induced lethality in yeast (Fig. 1C) (7). Another C-terminal variant, AtBI-CM (the resultant product of atbi1-1), had a coiled-coil score of 0.01 (Fig. 1C). As demonstrated in Fig. 2A, the yeast strain containing galactose-inducible proapoptotic protein Bax (Bax+vec) showed growth suppression on galactose-containing medium (Gal). Yeast cells expressing wild-type AtBI-1 recovered their growth activity on the Gal medium, whereas the AtBI-CM protein failed to suppress Bax-induced lethality. The expression of AtBI-1 and AtBI-CM mRNAs was confirmed by RT-PCR (Fig. 2B).
FIGURE 2.
AtBI-CM failed to suppress Bax-induced lethality in yeast cells. A, suppression of Bax-induced yeast lethality by AtBI-1 and AtBI-CM. Yeast cells possessing the galactose-inducible Bax with pYX112 (Bax+Vec), pYX112-AtBI-1 (Bax+AtBI-1), or pYX112-AtBI-CM (Bax+AtBI-CM) were streaked on Glc-containing medium (Glc) or Gal-containing medium (Gal). B, RT-PCR analysis of AtBI-1 expression in yeast strains possessing pYX112 (Vec), pYX112-AtBI-1 (AtBI-1), and pYX112-AtBI-CM (AtBI-CM). Exon numbers and the positions of the oligonucleotides (a–d) used for RT-PCR are indicated (upper panel). Ethidium bromide staining of rRNAs served as loading controls.
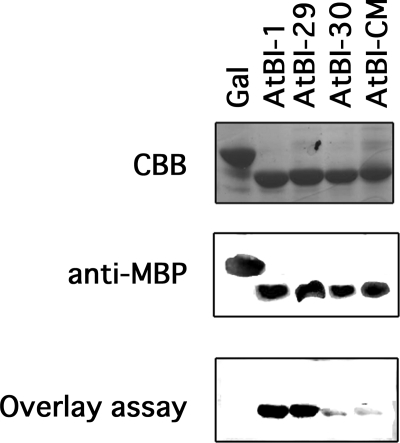
The C-terminal 14-residue domain of AtBI-1 interacts with calmodulin (7). Overlay binding assays were used to analyze whether calmodulin binding is coincident with the suppressor function of AtBI-1. Fusion proteins of MBP and the C terminus of AtBI-1 or the C-terminal mutants (AtBI-29, AtBI-30, and AtBI-CM) were expressed in E. coli, purified, and used for the overlay binding assay with AtCaM7 (Fig. 3). As expected from previous work (7), AtBI-1 interacted with AtCaM7. In addition, AtBI-29, which retained its cell death suppression activity, interacted with AtCaM7, whereas AtBI-30 and AtBI-CM showed reduced binding with calmodulin. These results suggest that calmodulin binding to the C-terminal region of AtBI-1 is in agreement with the suppressor function of the protein in yeast.
FIGURE 3.
Interaction of AtBI-1 C-terminal mutants with AtCaM7. The purified MBP-tagged C-terminal fragments of AtBI-1, AtBI-29, AtBI-30, AtBI-CM, and β-galactosidase (Gal, produced by empty vector) were separated by SDS-PAGE, immunoblotted, and examined by overlay assay. Coomassie Brilliant Blue staining (CBB), Western blotting with anti-MBP antibody, and overlay assay are shown.
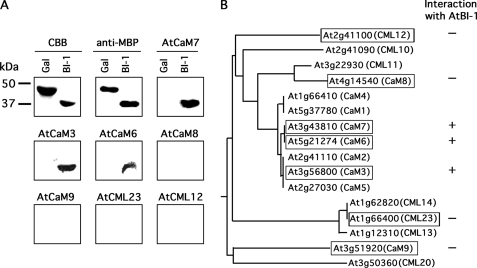
Plant species have evolved complex and intricate calcium signaling cascades. For instance, data base searching predicts more than 50 calmodulin-related proteins (CaMs and CMLs) encoded by the Arabidopsis genome. To define the calmodulin molecules that interact with AtBI-1, several classes of these proteins were expressed in E. coli, purified, and analyzed by overlay assay (Fig. 4). Only three of the calmodulins tested (CaM3, CaM6, and CaM7) interacted with AtBI-1, and all of these belong to the authentic calmodulin family gene group, which is highly conserved across a range of organisms.
FIGURE 4.
AtBI-1 interacts with authentic CaM proteins. A, Arabidopsis calmodulin and calmodulin-related proteins (AtCaM3, AtCaM6, AtCaM7, AtCaM8, AtCaM9, AtCML23, and AtCML12) fused with His and S-tags were expressed in E. coli, and purified proteins were used for the overlay assay with AtBI-1. Coomassie Brilliant Blue staining (CBB) and Western blotting with anti-MBP antibody (anti-MBP) were also examined as controls for protein loading. Molecular size markers (in kDa) are indicated on the left. B, phylogenic relationships of Arabidopsis calmodulin-related proteins and interaction with AtBI-1. A phylogenic tree of CaMs and selected CMLs was constructed by the ClustalW program. Proteins analyzed in this study are denoted by boxes with the identified interaction (+ or −). The designation of genes was in accordance with McCormack et al. (18).
AtBI-1 Is Associated with the Pathogen Response
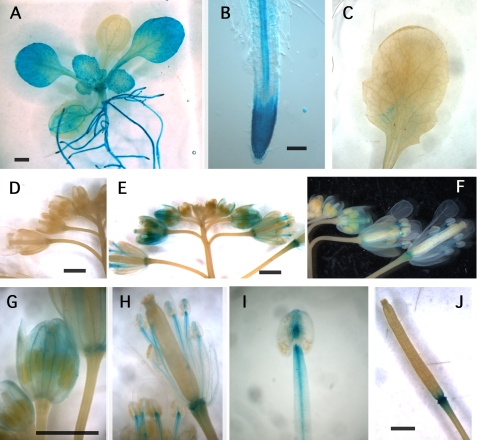
Rice cells overexpressing AtBI-1 show resistance to the HR-like cell death induced by an elicitor derived from M. grisea (3). To further characterize the tissue specificity of AtBI-1 expression, a GUS reporter gene was cloned under the control of the AtBI-1 promoter region (−2 kbp). After the successive transformation, we obtained more than 10 of single GUS gene-inserted lines. Among them, three lines, which show stable and typical GUS staining patterns, were chosen for further study. Whole mount GUS staining demonstrated high expression levels in young leaves and roots of 2-week-old seedlings (Fig. 5A). The highest expression in roots was seen at the root tip and in vascular tissue (Fig. 5B). Strong staining was also found in flower tissues, including stamens and sepals, and in the base of siliques (Fig. 5, D and E–J). In contrast, a mature leaf did not show clear GUS activity (Fig. 5C). Thus, the mature leaves (third to fifth leaves of 3-week-old plants) were used for followed experiments.
FIGURE 5.
Histochemical analysis of GUS expression in AtBI-1 promoter-GUS transgenic plants. The GUS reporter gene was expressed in wild-type Arabidopsis plants under the control of the AtBI-1 promoter region (2 kbp). A, young seedling (12-day-old). B, root. C, mature leaf (fourth leaf in 3-week-old seedling). D, flowers of control plant without transgene. E and F, flower buds of GUS-containing plant. G, magnified flower buds. H, blooming flower. I, stamen and anther. J, silique. Bar = 1 mm.
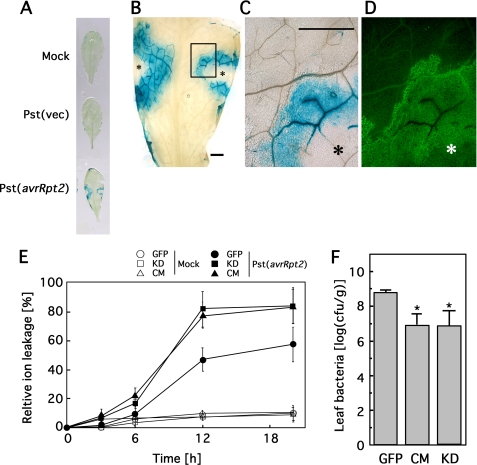
Although AtBI-1 gene expression is known to be stress-responsive (15), the tissue specificity is not known. To evaluate AtBI-1 expression at the tissue level, mature leaves of transgenic plants containing the GUS reporter gene under the control of an AtBI-1 promoter were infected with Pseudomonas containing either empty vector or avrRpt2 (16). As shown in Fig. 6A, site-specific GUS expression was observed in leaves inoculated for 16 h with Pst(avrRpt2), whereas mock-inoculated (Mock) and vector (Pst(vec)) controls showed only non-tissue-specific expression of a lower magnitude. Leaf cells that underwent HR demonstrated autofluorescence, and as shown in Fig. 6, B–D, the GUS activity was largely coincident with autofluorescence. Furthermore, Arabidopsis plants that exhibited altered AtBI-1 expression (overexpressed line, knockdown, and C-terminal mutant) were inoculated with P. syringae expressing avrRpt2. Electrolyte leakage, measured as an increase in conductivity of the solution containing the treated leaf discs, is often used to assay irreversible membrane damage during HR (17). No clear leakage was detected during the first 3 h of incubation (Fig. 6E), but 6 h after inoculation, the C-terminal mutant (CM/atbi1-1) and knockdown (KD) lines exhibited elevated sensitivity to HR cell death. We also assessed the bacterial growth of Pst(avrRpt2) in infected leaves. The C-terminal mutant (CM/atbi1-1) and RNAi (KD) plants demonstrated reduced bacterial growth, suggesting that AtBI-1 is associated with Pst(avrRpt2)-triggered immunity.
FIGURE 6.
AtBI-1 expression associated with HR caused by P. syringae pv. tomato DC3000 carrying avrRpt2. A, histochemical expression analysis of AtBI promoter-GUS in HR caused by Pst(avrRpt2) inoculation. The third to fifth leaves of Arabidopsis plants carrying the AtBI-promoter-GUS construct were inoculated with 10 mm MgCl2 (Mock), Pst(vec), or Pst(avrRpt2). After 1 day, leaves were stained for GUS activity. B, magnified image of leaf inoculated with Pst(avrRpt2). Asterisks indicate the inoculated sites. C and D, magnified view of boxed region in B and autofluorescence observed by fluorescence microscopy, respectively. Bars = 1 mm. E, electrolyte leakage induced by HR cell death. The electrical conductivity was measured in the Pst(avrRPT2)-inoculated control (GFP), AtBI-1 KD, and atbi1-1 (CM) plants as described under “Experimental Procedures.” Data are mean ± S.E. (error bars) of five plants each. F, bacterial growth in control (GFP) and C-terminal mutant. Arabidopsis plants (GFP, CM, and KD) were inoculated with Pst(avrRpt2), and bacterial growth was measured at 4 days after inoculation. *, significant difference (p < 0.05) between the control (GFP) and CM or KD (n = 5). Error bars indicate S.E.
DISCUSSION
The Arabidopsis genome harbors seven calmodulins and about 50 calmodulin-like genes (18, 19). Despite their potential importance in mediating plant calcium signaling, the physiological functions of these calmodulin-related genes remain largely unknown due to redundancy. Only seven of the Arabidopsis calmodulin genes encode proteins with high identity to vertebrate calmodulins. These belong to a class of plant calmodulins that activates NAD kinase in vitro and binds both kinesin-like motor proteins and cyclic nucleotide-gated ion channels (20–22). We have demonstrated the interaction between BI-1 and calmodulin using a split ubiquitin two-hybrid system and have demonstrated similar binding in plant cells by BiFC analysis (7). Calmodulin binds to the C-terminal cytoplasmic tail of BI-1, which is relatively conserved among BI-1 family members. In this study, we demonstrated that genomic BI-1 mutant (AtBI-CM) lacked the calmodulin binding activity, which in turn caused increased sensitivity to Pst(avrRpt2). Overlay assays performed in this study to determine which type of calmodulin-related protein binds BI-1 in vitro demonstrated that the authentic conserved type of calmodulin binds to the C terminus of AtBI-1. Microarray expression data that are available from several on-line sources (23) enabled us to estimate which calmodulin-related molecule is co-expressed with AtBI-1 (see supplemental data). The AtBI-1, CaMs, and CMLs were expressed in almost tissues, at least at the basal level, and did not show typical tissue specificity. However, the expression data in response to different stresses revealed distinct patterns of co-expression. Interestingly, the CaM mRNA co-expressed with AtBI-1 during the response to Pst(avrRPT2) was At3g43810 (CaM7), which is the first identified ABI-1-binding protein reported in our previous study (7). In contrast, CaMs co-expressing under abiotic stresses (cold, osmosis, UV-B, wounding, and heat) were At3g56800 (CaM3) and At2g41110 (CaM2). These data are in agreement with the present study, which demonstrates that AtBI-1 binds authentic CaM proteins, not CML molecules. To clarify the functional partner for AtBI-1 under each stress condition, further analysis using single or multigene disruptants of CaM will be useful. The basic structure of Arabidopsis calmodulins is highly similar to those of animal calmodulins in both the Ca2+-binding loops and the EF hands. Thus, it would be also interesting to establish whether the function of mammalian BI-1 also requires calmodulin binding.
Cytosolic calcium increases in response to a range of diverse stimuli, including environmental, hormonal, and developmental prompts. This elevation is sensed by calmodulin and transduced into altered target activity to trigger a cellular response. Calcium plays a major role in initiating the HR, which in turn limits pathogen growth by promoting plant cell wall reinforcement or by eliminating host functions necessary for pathogen multiplication (24). The HR of plants to avirulent pathogens effectively restricts pathogen growth and includes a characteristic programmed cell death at the site of pathogen invasion (25). Such events typically include the induction of reactive oxygen species, callose deposition, and expression of several defense-related genes (26). This study focused on HR mediated by the Arabidopsis R gene RPS2 to infection by the bacterial pathogen P. syringae pv. tomato DC3000 (Pst) carrying the type III effector gene avrRpt2. The type III effector was first identified based on its ability to trigger pathogen recognition in resistant host plants (27), and interaction between the products of the avrRpt2 gene and the RPS2 resistance gene is a well characterized example of HR-induced cell death (28). Sanchez et al. (29) demonstrated that AtBI-1 expression is rapidly up-regulated during wounding or pathogen challenge. A reporter gene linked to the promoter from AtBI-1 provided direct evidence that the infection by Pseudomonas possessing the avrRpt2 gene can regulate AtBI-1 transcription, with the GUS signal surrounding the lesioned region. The ion leakage occurs at the late stage of HR. The loss of plasma membrane semipermeability and the protoplast shrinkage cause ion leak (30). Cells dying via the HR usually become autofluorescent and brown, attributed to the accumulation of phenolic compounds (31). The autofluorescence is observed in the cell walls of both dying cells and adjacent living cells in close contact with the infected site. Many defense genes are induced with the dying cells and in their adjacent living neighbors (30). Thus, the HR associates with both cell death and defense. The expression of AtBI-1 in the adjacent neighbor of infected region may suggest the role of BI-1 in defense response with unknown function. Recently, we reported that cytochrome b5, which is an electron transfer protein in lipid metabolism in ER, is another AtBI-1-binding protein in Arabidopsis (32). One possible speculation is that AtBI-1 would be associated with membrane remodification of adjacent neighbor of infected site.
Regarding AtBI-1-overexpressing plants, we did not see any phenotypes on the ion leakage or bacterial growth (data not shown). This is in agreement with the previous studies (8). AtBI-1 mutant showed higher sensitivity against FB1 or heat shock treatment; however, the overexpression line did not show any differences. We assume that the native level of AtBI-1 expression may be sufficient or saturated to execute its function as a cell death suppressor in wild-type Arabidopsis plants.
Elucidating the biological function of BI-1 protein is a significant future challenge for the field of plant cell death regulation. For one thing, it would help to clarify the cell protection mechanism operating in plants. Furthermore, understanding the relationship between BI-1 and calmodulin might reveal how the versatile nature of calcium signaling facilitates the dynamic behaviors and environmental adaptability typical of plants.
Supplementary Material
Acknowledgments
We thank Dr. J. C. Reed for kindly providing the described material. We also thank Ms. Y. Takahashi and Ms. M. Sakamoto for technical assistance.
This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan and a grant from CREST, JST, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- BI
- Bax Inhibitor
- CaM
- calmodulin
- CML
- CaM-like
- HR
- hypersensitive response
- MBP
- maltose-binding protein
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein
- RNAi
- RNA interference
- RT-PCR
- reverse transcription-PCR
- Vec
- vector
- KD
- knockdown.
REFERENCES
- 1.Bolduc N., Brisson L. F. (2002) FEBS Lett. 532, 111–114 [DOI] [PubMed] [Google Scholar]
- 2.Hückelhoven R., Dechert C., Kogel K. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5555–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsumura H., Nirasawa S., Kiba A., Urasaki N., Saitoh H., Ito M., Kawai-Yamada M., Uchimiya H., Terauchi R. (2003) Plant J. 33, 425–434 [DOI] [PubMed] [Google Scholar]
- 4.Chae H. J., Kim H. R., Xu C., Bailly-Maitre B., Krajewska M., Krajewski S., Banares S., Cui J., Digicaylioglu M., Ke N., Kitada S., Monosov E., Thomas M., Kress C. L., Babendure J. R., Tsien R. Y., Lipton S. A., Reed J. C. (2004) Mol. Cell 15, 355–366 [DOI] [PubMed] [Google Scholar]
- 5.Watanabe N., Lam E. (2008) J. Biol. Chem. 283, 3200–3210 [DOI] [PubMed] [Google Scholar]
- 6.Lisbona F., Rojas-Rivera D., Thielen P., Zamorano S., Todd D., Martinon F., Glavic A., Kress C., Lin J. H., Walter P., Reed J. C., Glimcher L. H., Hetz C. (2009) Mol. Cell 33, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ihara-Ohori Y., Nagano M., Muto S., Uchimiya H., Kawai-Yamada M. (2007) Plant Phys. 143, 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe N., Lam E. (2006) Plant J. 45, 884–894 [DOI] [PubMed] [Google Scholar]
- 9.Kawai-Yamada M., Ohori Y., Uchimiya H. (2004) Plant Cell 16, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Q., Reed J. C. (1998) Mol. Cell 1, 337–346 [DOI] [PubMed] [Google Scholar]
- 11.Kawai-Yamada M., Jin L., Yoshinaga K., Hirata A., Uchimiya H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12295–12300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawai M., Pan L., Reed J. C., Uchimiya H. (1999) FEBS Lett. 464, 143–147 [DOI] [PubMed] [Google Scholar]
- 13.Oshima R., Yoshinaga K., Ihara-Ohori Y., Fukuda R., Ohta A., Uchimiya H., Kawai-Yamada M. (2007) FEBS Lett. 581, 4627–4632 [DOI] [PubMed] [Google Scholar]
- 14.Pan L., Kawai M., Yano A., Uchimiya H. (2000) Plant Phys. 122, 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hückelhoven R. (2004) Apoptosis 9, 299–307 [DOI] [PubMed] [Google Scholar]
- 16.Yu I., Fengler K. A., Clough S. J., Bent A. F. (2000) Mol. Plant Microbe. Interact. 13, 277–286 [DOI] [PubMed] [Google Scholar]
- 17.Pike S. M., Zhang X. C., Gassmann W. (2005) Plant Phys. 138, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCormack E., Tsai Y. C., Braam J. (2005) Trends Plant Sci. 10, 383–389 [DOI] [PubMed] [Google Scholar]
- 19.McCormack E., Braam J. (2003) New Phytol. 159, 585–598 [DOI] [PubMed] [Google Scholar]
- 20.Liao B., Gawienowski M. C., Zielinski R. E. (1996) Arch. Biochem. Biophys. 327, 53–60 [DOI] [PubMed] [Google Scholar]
- 21.Reddy V. S., Safadi F., Zielinski R. E., Reddy A. S. N. (1999) J. Biol. Chem. 274, 31727–31733 [DOI] [PubMed] [Google Scholar]
- 22.Köhler C., Neuhaus G. (2000) FEBS Lett. 471, 133–136 [DOI] [PubMed] [Google Scholar]
- 23.Winter D., Vinegar B., Nahal H., Ammar R., Wilson G. V., Provart N. J. (2007) Plos. One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon R. A., Harrison M. J., Lamb C. J. (1994) Annu. Rev. Phytopathol. 32, 479–501 [Google Scholar]
- 25.Lam E., Kato N., Lawton M. (2001) Nature 411, 848–853 [DOI] [PubMed] [Google Scholar]
- 26.Gómez-Gómez L., Boller T. (2002) Trends Plant Sci. 7, 251–256 [DOI] [PubMed] [Google Scholar]
- 27.Dong X., Mindrinos M., Davis K. R., Ausubel F. M. (1991) Plant Cell 3, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackey D., Belkhadir Y., Alonso J. M., Ecker J. R., Dangl J. L. (2003) Cell 112, 379–389 [DOI] [PubMed] [Google Scholar]
- 29.Sanchez P., de Torres , Zabala M., Grant M. (2000) Plant J. 21, 393–399 [DOI] [PubMed] [Google Scholar]
- 30.Heath M. C. (2000) Plant Mol. Biol. 44, 321–334 [DOI] [PubMed] [Google Scholar]
- 31.Heath M. C. (1998) New Phytol. 138, 251–263 [DOI] [PubMed] [Google Scholar]
- 32.Nagano M., Ihara-Ohori Y., Imai H., Inada N., Fujimoto M., Tsutsumi N., Uchimiya H., Kawai-Yamada M. (2009) Plant J. 58, 122–134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.