Abstract
BAD is a proapoptotic member of the Bcl-2 protein family that is regulated by phosphorylation in response to survival factors. Although much attention has been devoted to the identification of phosphorylation sites in murine BAD, little data are available with respect to phosphorylation of human BAD protein. Using mass spectrometry, we identified here besides the established phosphorylation sites at serines 75, 99, and 118 several novel in vivo phosphorylation sites within human BAD (serines 25, 32/34, 97, and 124). Furthermore, we investigated the quantitative contribution of BAD targeting kinases in phosphorylating serine residues 75, 99, and 118. Our results indicate that RAF kinases represent, besides protein kinase A, PAK, and Akt/protein kinase B, in vivo BAD-phosphorylating kinases. RAF-induced phosphorylation of BAD was reduced to control levels using the RAF inhibitor BAY 43-9006. This phosphorylation was not prevented by MEK inhibitors. Consistently, expression of constitutively active RAF suppressed apoptosis induced by BAD and the inhibition of colony formation caused by BAD could be prevented by RAF. In addition, using the surface plasmon resonance technique, we analyzed the direct consequences of BAD phosphorylation by RAF with respect to association with 14-3-3 and Bcl-2/Bcl-XL proteins. Phosphorylation of BAD by active RAF promotes 14-3-3 protein association, in which the phosphoserine 99 represented the major binding site. Finally, we show here that BAD forms channels in planar bilayer membranes in vitro. This pore-forming capacity was dependent on phosphorylation status and interaction with 14-3-3 proteins. Collectively, our findings provide new insights into the regulation of BAD function by phosphorylation.
Apoptosis is a genetically programmed, morphologically distinct form of cell death that can be triggered by a variety of physiological and pathological stimuli (1–3). This form of cellular suicide is widely observed in nature and is not only essential for embryogenesis, immune responses, and tissue homeostasis but is also involved in diseases such as tumor development and progression. Bcl-2 family proteins play a pivotal role in controlling programmed cell death. The major function of these proteins is to directly modulate outer mitochondrial membrane permeability and thereby regulate the release of apoptogenic factors from the intermembrane space into the cytoplasm (for a recent review, see Ref. 4). On the basis of various structural and functional characteristics, the Bcl-2 family of proteins is divided into three subfamilies, including proteins that either inhibit (e.g. Bcl-2, Bcl-XL, or Bcl-w) or promote programmed cell death (e.g. Bax, Bak, or Bok) (5, 6). A second subclass of proapoptotic Bcl-2 family members, the BH32-only proteins, comprises BAD, Bik, Bmf, Hrk, Noxa, truncated Bid, Bim, and Puma (4). BH3-only proteins share sequence homology only at the BH3 domain. The amphipathic helix formed by the BH3 domain (and neighboring residues) associates with a hydrophobic groove of the antiapoptotic Bcl-2 family members (7, 8). Originally, truncated Bid has been reported to interact with Bax and Bak (9), suggesting that some BH3-only proteins promote apoptosis via at least two different mechanisms: inactivating Bcl-2-like proteins by direct binding and/or by inducing modification of Bax-like molecules. BAD (Bcl-2-associated death promoter, Bcl-2 antagonist of cell death) was described to promote apoptosis by forming heterodimers with the prosurvival proteins Bcl-2 and Bcl-XL, thus preventing them from binding with Bax (10). More recently, two major models have been suggested for how BH3-only proteins may induce apoptosis. In the direct model, all BH3-only proteins promote cell death by directly binding and inactivating their specific anti-death Bcl-2 protein partner (11, 12). In this model, the relative killing potency of different BH3-only proteins is based on their affinities for antiapoptotic proteins. Thus, the activation of Bax/Bak would be mediated through their release from antiapoptotic counterparts. Contrary to this model, Kim et al. (13) provided support for an alternative hierarchy model, in which BH3-only proteins are divided into two distinct subsets. According to this model, the inactivator BH3-only proteins, like BAD, Noxa, and some others, respond directly to survival factors, resulting in phosphorylation, 14-3-3 binding, and suppression of the proapoptotic function. In the absence of growth factors, these proteins engage specifically their preferred antiapoptotic Bcl-2 proteins. The targeted Bcl-2 proteins then release the other subset of BH3-only proteins designated the activators (truncated Bid, Bim, and Puma) that in turn bind to and activate Bax and Bak.
Non-phosphorylated BAD associated with Bcl-2/Bcl-XL is found at the outer mitochondrial membrane. Phosphorylation of specific serine residues, Ser-112 and Ser-136 of mouse BAD (mBAD) or the corresponding phosphorylation sites Ser-75 and Ser-99 of human BAD (hBAD), results in association with 14-3-3 proteins and subsequent relocation of BAD (14, 15). Phosphorylation of mBAD at Ser-155 (Ser-118 of hBAD) within its BH3 domain disrupts the association with Bcl-2 or Bcl-XL, promoting cell survival (16). Therefore, the phosphorylation status of BAD at these serine residues reflects a checkpoint for cell death or survival. Although the C-RAF kinase was the first reported BAD kinase (17), its target sites were not clearly defined. However, there is a growing body of evidence for direct participation of RAF in regulation of apoptosis via BAD (18, 19). In addition, Kebache et al. (20) reported recently that the interaction between adaptor protein Grb10 and C-RAF is essential for BAD-mediated cell survival. On the other hand, numerous reports suggest that PKA (21), Akt/PKB (22), PAK (18, 23, 24), Cdc2 (25), RSK (26, 27), CK2 (28), and PIM kinases (29) are involved in BAD phosphorylation as well. The involvement of c-Jun N-terminal kinase in BAD phosphorylation is controversially discussed. Whereas Donovan et al. (30) reported that c-Jun N-terminal kinase phosphorylates mBAD at serine 128, Zhang et al. (31) claimed that c-Jun N-terminal kinase is not a BAD-serine 128 kinase. On the other hand, it has been shown that c-Jun N-terminal kinase is able to suppress IL-3 withdrawal-induced apoptosis via phosphorylation of mBAD at threonine 201 (32). Thus, taken together, with respect to regulation of mBAD by phosphorylation, five serine phosphorylation sites (at positions 112, 128, 136, 155, and 170) and two threonines (117 and 201) have been identified so far. Intriguingly, only little data are available regarding the role of phosphorylation in regulation of hBAD protein, although significant structural differences between these two BAD proteins exist.
During apoptosis, some members of the Bcl-2 family of proteins, such as Bax or Bak, have been shown to induce permeabilization of the outer mitochondrial membrane, allowing proteins in the mitochondrial intermembrane space to escape into the cytosol, where they can initiate caspase activation and cell death (for a review, see Refs. 33 and 34). Despite intensive investigation, the mechanism whereby Bax and Bak induce outer membrane permeability remains controversial (34). Based on crystal structure (35), it became evident that Bcl-XL has a pronounced similarity to the translocation domain of diphtheria toxin (36), a domain that can form pores in artificial lipid bilayers. This discovery provoked the predominant view that upon commitment to apoptosis, the proapoptotic proteins Bax and Bak also form pores in the outer mitochondrial membrane (37). As expected from the structural considerations, Bcl-XL was found to form channels in synthetic lipid membranes (38). Since then, other Bcl-2 family members like Bcl-2, Bax, and the BH3-only protein Bid have been reported to have channel-forming ability. These pores can be divided into two different types: proteinaceous channels of defined size and ion specificity (38–42) and large lipidic pores that allow free diffusion of 2-megadalton macromolecules (43, 44). With respect to the BH3-only protein BAD, no pore-forming abilities have been reported so far, although human BAD has been found to possess per se high affinity for negatively charged phospholipids and liposomes, mimicking mitochondrial membranes (14).
The RAF kinases (A-, B-, and C-RAF) play a central role in the conserved Ras-RAF-MEK-ERK signaling cascade and mediate cellular responses induced by growth factors (45–47). Direct involvement of C-RAF in inhibition of proapoptotic properties of BAD established a link between signal transduction and apoptosis control (48, 49). However, the early works did not identify the exact RAF phosphorylation sites on BAD (17). Here we demonstrate that hBAD serves as a substrate of RAF isoforms. With respect to hBAD phosphorylation by PKA, Akt/PKB, and PAK1 in vivo, we observed different specificity compared with RAF kinases. hBAD phosphorylation by RAF was accompanied by reduced apoptosis in HEK293 cells (transformed human embryonic kidney cells) and NIH 3T3 cells (a mouse embryonic fibroblast cell line). Furthermore, we show that in vitro phosphorylation of hBAD by RAF at serines 75, 99, and 118 regulates the binding of 14-3-3 proteins and association with Bcl-2 and Bcl-XL. By use of mass spectrometry, we detected several novel in vivo phosphorylation sites of hBAD in addition to the established phosphorylation sites, serines 75, 99, and 118. Finally, we show here that hBAD forms channels in planar bilayer membranes in vitro. This pore-forming capacity was dependent on phosphorylation status and interaction with 14-3-3 proteins.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Immunoblotting
HEK293 cells and NIH 3T3 cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (heat-inactivated at 56 °C for 45 min), 2 mm l-glutamine, and 100 units/ml penicillin/streptomycin at 37 °C in humidified air with 5% CO2. U0126 (Promega), PD98059 (Promega), BAY 43-9006, forskolin (Alexis), isobutylmethylxanthine (Sigma), CI1040 (Axon Medchem), wortmannin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and LY294002 (Promega) were dissolved in DMSO (Sigma), whereas H-89 (Sigma) was dissolved in ethanol. NIH 3T3 fibroblasts were seeded at 2 × 105 cells/well of a 6-well plate and grown for 24 h before transfection by Lipofectamine (Invitrogen). HEK293 cells were seeded at 7.5 × 105 cells/well on a 6-well plate and grown for 24 h before they were transfected with expression plasmids by the calcium phosphate method (50). 16 h post-transfection, cells were washed twice with phosphate-buffered saline (PBS) and cultivated for the indicated time in medium supplemented with 0.3% serum to avoid activity of endogenous kinases. Cells were washed once in PBS, and equal amounts of cells were lysed by the direct addition of Laemmli buffer or in Nonidet P-40 buffer (10 mm Hepes, pH 7.5, 142.5 mm KCl, 5 mm MgCl2, 1 mm EGTA, and 0.2% Nonidet P-40 supplemented with a mixture of standard protease inhibitors). Protein concentration was determined by Bradford method. SDS-PAGE and immunoblotting were performed as described previously (14). The following primary antibodies were used: monoclonal anti-phospho-ERK antibody (catalog number 9106; Cell Signaling Technology), phosphospecific antibodies against mBAD phosphoserines 112 and 155 (catalog numbers 9295 and 9297; Cell Signaling Technology) and phosphoserine 136 (44-524Z; BIOSOURCE), polyclonal anti-actin (A2066; Sigma), polyclonal anti-Akt/PKB (catalog number 9272; Cell Signaling Technology), polyclonal anti-BAD antibody (sc-943 and sc-8044; Santa Cruz Biotechnology), anti-PAK antibody (sc-881; Santa Cruz Biotechnology), anti-B-RAF antibody (sc-166; Santa Cruz Biotechnology), anti-PKA antibody (sc-903; Santa Cruz Biotechnology), and pan-RAF antibody (30K; Institute for Medical Radiation and Cell Research).
Cell Survival Assay
For the analysis of cell survival, HEK293 cells were transiently transfected in triplicates. 16 h post-transfection, cells were washed twice with PBS and grown for 30 h in medium supplemented with 0.3% serum. Cell viability was assessed by staining cells in trypan blue (Sigma).
Colony Yield Assay
NIH 3T3 cells were transfected with the indicated plasmids using Lipofectamine. The day after transfection, cells were split, and around 50 cells of each set of transfection were seeded in 6-cm dishes. Colony assays were performed in triplicate by scoring the number of colonies (consisting of at least 20 cells) in the dishes grown for 14–18 days with appropriate antibiotic selection (450 μg/ml neomycin and 6 μg/ml puromycin; Calbiochem). To visualize the growing colonies, cells were washed once with PBS, fixed with methanol, and stained with Giemsa dye (Sigma).
DNA Expression Plasmids
Human BAD cDNA (kind gift of John Reed, La Jolla, CA) and cDNAs mutated at serine 75 (S75A), serine 99 (S99A), serine 118 (S118A), serines 75/99 (S75A/S99A), and serines 75, 99, and 118 (S75A/S99A/S118A) were cloned into pGEX-4T-1 (Amersham Biosciences). Deletion mutants of hBAD were PCR-amplified, with primer overhangs containing restriction sites and cloned into GST fusion vector pGEX-TT cleaved with the appropriate enzymes. Site-directed mutagenesis was performed by using QuikChange (Stratagene). To generate N-terminal His-tagged hBAD, the cDNA was released by EcoRI/NotI from pGEX-4T-1 and ligated in the EcoRI/NotI sites of pFastBac H1 (Invitrogen). Human Bcl-XL (John Reed, La Jolla, CA) was isolated from pcDNA3 using EcoRI and ligated with blunt ends in pFastBac-GST linearized by StuI. PAK1 constructs were from Jonathan Chernoff (Philadelphia, PA). Akt/PKB plasmids were kindly provided by Jakob Troppmair (Innsbruck, Austria). pGEX-Bcl-2 was kindly donated by John Reed.
Purification of Kinases, BAD, Bcl-XL, and 14-3-3 Proteins
Expression and purification of RAF kinases, PAK1, Akt/PKB, 14-3-3 proteins, and Bcl-XL from Sf9 insect cells (an insect cell line derived from pupal ovarian tissue of Spodoptera frugiperda) were performed as previously described (14, 51–53). For purification of phosphorylated His-hBAD, Sf9 cells were infected with baculoviruses at a multiplicity of infection of 5 and incubated for 48 h at 27 °C. The Sf9 cell pellet (2 × 108 cells) was lysed in 10 ml of lysis buffer containing 50 mm sodium phosphate, pH 8.0, 150 mm NaCl, 10 mm sodium pyrophosphate, 25 mm β-glycerophosphate, 25 mm NaF, 10% glycerol, 0.5% Nonidet P-40, and a mixture of standard proteinase inhibitors for 30 min with gentle rotation at 4 °C. The lysate was centrifuged at 27,000 × g for 30 min at 4 °C. The supernatant (10 ml) containing His-hBAD was incubated with 0.5 ml Ni2+-nitrilotriacetic acid-agarose for 2 h at 4 °C with rotation. After incubation, the beads were washed three times with lysis buffer containing 0.2% Nonidet P-40, 300 mm NaCl, and 20 mm imidazole, and hBAD was eluted with an imidazole step gradient. GST-tagged 14-3-3 proteins and GST-hBAD were expressed in Escherichia coli using pGEX-2T vector (Amersham Biosciences) and purified by glutathione-Sepharose affinity chromatography. For lipid bilayer experiments, GST-hBAD was additionally purified by ion exchange chromatography using an AKTA system (GE Healthcare). 14-3-3 proteins were released by thrombin (Sigma) cleavage using standard protocols. The purity of the proteins was assessed by SDS-PAGE and staining with Coomassie Blue. To enrich and separate phosphorylated His-hBAD from the non-phosphorylated fraction, His-hBAD purified from Sf9 cells (0.5 mg) was incubated with 0.5 mg of recombinant GST-14-3-3ζ on glutathione-Sepharose beads (300 μl) for 40 min at room temperature. After excessive washing with 10 mm Hepes, pH 7.4, 150 mm NaCl, and 0.01% Nonidet P-40, the phosphorylated hBAD fraction was released from the complex by 1% Empigen (Calbiochem).
Kinase Activity Assay
Human GST-BAD was incubated with purified preparations of RAF kinases, PKA, Akt/PKB, or PAK1 in 50 mm Hepes buffer, pH 7.6, in the presence of 10 mm MgCl2, 1 mm dithiothreitol, and 500 μm ATP. The mixture was incubated at 30 °C for 30 min, and the reaction was terminated by the addition of Laemmli buffer. The proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. The extent of BAD phosphorylation at serines 75, 99, and 118 was detected by phosphospecific antibodies. To inhibit kinase activity in vitro, purified kinases were preincubated with BAY 43-9006 or H-89 at room temperature for 30 min.
Mass Spectrometry Analysis of BAD Phosphorylation
Purified His-hBAD samples (about 100 pmol of each) were applied to SDS-PAGE. Proteins were visualized by Coomassie Blue staining, applying the method of Neuhoff et al. (54). In-gel reduction, acetamidation, and tryptic and/or GluC digestion were done according to Wilm et al. (55). After elution of the peptides, solutions were desalted using a Millipore C18 ZipTip according to the manufacturer's instructions. ESI-MS was performed on a Bruker APEX II FT-ICR mass spectrometer (Bruker Daltonic GmbH Bremen) equipped with an Apollo-Nano-ESI ion source in positive ion mode.
To determine the exact positions of phosphates within the peptides, the nano-LC-MS/MS analysis was carried out as follows. Purified hBAD samples were separated by SDS-PAGE, and protein bands were excised, washed, and digested in gel as described previously (56). Afterward, generated peptides were extracted using 15 μl of 0.1% trifluoroactic acid, and samples were analyzed by nano-LC-MS/MS on an LTQ Orbitrap XL mass analyzer (Thermo Scientific, Dreieich, Germany) coupled to an Ultimate 3000 LC/MS system (Dionex, Amsterdam, The Netherlands) using multistage activation as described previously (57). To this end, peptides were preconcentrated on a PepMap trapping column (100-μm inner diameter, 5-μm particle size, 100-Å pore size, 1-cm length; Dionex) and separated on a PepMap nano-LC column (75-μm inner diameter, 3-μm particle size, 100-Å pore size, 15-cm length; Dionex) at a flow rate of 270 nl/min, using a gradient ranging from 5 to 50% of 86% acetonitrile and 0.1% formic acid. Raw data were transformed into mgf format using extract_msn.exe as part of the Bioworks package, and generated peak lists were searched against an SGD data base with concatenated standard protein and BAD sequences (6319 entries) using Mascot version 2.2. The following search parameters were used: trypsin as protease with a maximum of one missed cleavage site, 10 ppm mass tolerance for MS, 0.5 Da for MS/MS, oxidation of Met (+15.99 Da) and phosphorylation of Ser/Thr/Tyr (+79.96 Da) as variable modifications, and 13C set to 1. All identified phosphopeptides were manually validated to correct for potentially false phosphorylation site assignments.
Lipid Bilayer Experiments
The channel-forming ability of proteins was assessed in artificial lipid bilayer membranes using a Teflon chamber as described previously (58, 59). Briefly, to form the membranes, a 1% (w/v) solution of diphytanoylphosphatidylcholine (Avanti Polar Lipids) in n-decane was used. Purified hBAD proteins (phosphorylated and non-phosphorylated) or Bcl-XL was added to the KCl buffer in both compartments of the chamber, and the single-channel conductance of the pores was measured after application of a fixed membrane potential. To test the effects of 14-3-3 proteins on the pore-forming abilities of hBAD, purified heterodimeric 14-3-3 protein (14-3-3ζ·14-3-3ϵ) was incubated with the phosphorylated form of BAD for 30 min at room temperature prior to channel formation. Samples were applied on both sides of the diphytanoylphosphatidylcholine membrane in KCl buffer, and single-channel formation was measured.
Biosensor Measurements
The biosensor measurements were carried out either on a BIAcore-X or BIAcore-J system (Biacore AB, Uppsala, Sweden) at 25 °C. The biosensor chip CM5 was loaded with anti-GST antibody using covalent derivatization according to the manufacturer's instructions. Purified and GST-tagged proteins (GST-BAD WT and substitution mutants) were immobilized in biosensor buffer (10 mm Hepes, pH 7.4, 150 mm NaCl, and 0.05% Nonidet P-40) at a flow rate of 10 μl/min, which resulted in a deposition of ∼1000 response units. Next, the purified analytes (14-3-3ζ or Bcl-XL) were injected at the indicated concentrations. The values for nonspecific binding measured in the reference cell were subtracted. To monitor the dissociation of Bcl-XL·GST-BAD complex induced by BAD phosphorylation the biosensor chip was treated with active C-RAF in the presence of 0.5 mm ATP. The modified biosensor buffer contained 5 mm MgCl2 and 0.5 mm dithiothreitol. The working temperature was set to 30 °C. The evaluation of kinetic parameters was performed by non-linear fitting of binding data using the BiaEvaluation version 2.1 analysis software. The apparent association (ka) and dissociation rates constant (kd) were evaluated from the differential binding curves (Fc2 − Fc1) assuming an A + B = AB association type for the protein-protein interaction. The dissociation constant KD was calculated from the equation, KD = kd/ka.
RESULTS
Detection of Novel in Vivo Phosphorylation Sites of Human BAD by Mass Spectrometry
Besides the highly conserved murine BAD phosphorylation sites at serines 112, 136, and 155 (corresponding to serines 75, 99, and 118 in human BAD) that are crucial for 14-3-3 binding and interaction with Bcl-2/Bcl-XL, four other murine phosphorylation sites (positioned at serines 128 and 170 and threonines 117 and 201) have been reported (for references, see the Introduction). Although human BAD compared with murine BAD shows striking differences in the length of its amino acid sequence (168 versus 204 residues) and in the number and locations of putative phosphorylation sites, no systematic analysis of in vivo phosphorylation of this proapoptotic protein has been performed.
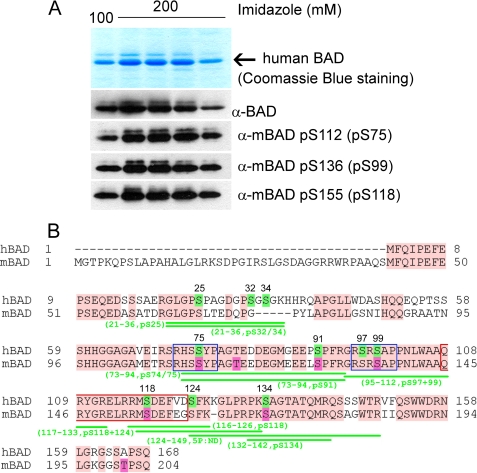
Since phosphospecific antibodies directed against the putative 14-3-3 binding sites (serines 75 and 99) and docking segment of Bcl-2/Bcl-XL proteins (surrounding serine 118) are available, we examined first the phosphorylation status at these established positions. As shown in Fig. 1A, all three of these sites were detectable in purified hBAD by use of phosphospecific antibodies, indicating that a fraction of hBAD expressed in Sf9 cells is associated with 14-3-3 proteins and decoupled from Bcl-2/Bcl-XL proteins. Such a complex is predicted to be excluded from mitochondria and, consequently, to act in an antiapoptotic manner (15).
FIGURE 1.
Analysis of in vivo phosphorylation of purified human BAD protein. A, purification of human BAD from Sf9 insect cells. The top panel shows Coomassie Blue staining after SDS-PAGE of human BAD obtained by elution from nickel-agarose beads by 100 and 200 mm imidazole, respectively. The phosphorylation of serines 75, 99, and 118 was verified by use of phosphospecific antibodies. B, sequence alignment of human and murine BAD protein and mass spectrometry analysis of phosphopeptides obtained by tryptic and GluC digestion of hBAD. For a detailed list of identified phosphopeptides, see also Table 1. The putative phosphorylation sites of human BAD are highlighted in green, and their positions within the sequence are indicated by numbers. The published phosphorylation sites of murine BAD are highlighted in magenta. The conserved regions between human and murine BAD are shown in pink. The putative 14-3-3 binding sites are indicated by blue rectangles, and the conserved BH3 domains are shown by red rectangles.
To analyze the complete phosphorylation status of human BAD, we performed a detailed mass spectrometry analysis of the protein purified from Sf9 cells. For that purpose, we digested purified hBAD by trypsin and/or GluC, and the selective detection of phosphopeptides was carried out by both ESI-MS and the nano-LC-MS/MS technique. Two independent measurements provided almost 89% coverage of the entire protein sequence. The combined results obtained for hBAD phosphorylation are summarized in Fig. 1B and Table 1, revealing several novel in vivo phosphorylation sites. To compare the mass spectrometry results obtained for human BAD with the known phosphorylation sites in murine BAD, we aligned the amino acid sequences of both BAD proteins (Fig. 1B). Remarkably, with exception of two novel phosphorylation sites localized closely to the N terminus (serines 25 and 32/34), most of the phosphorylated peptides obtained by mass spectrometry were localized in the C-terminal half of the protein. The last 20 residues at the very C-terminal sequence bear no phosphate molecules, whereby it should be noted that human BAD does not possess threonine 201, which was found to be phosphorylated in murine BAD. Two N-terminal phosphorylation sites were identified within the tryptic peptide 21–36, and the positions of the phosphates (25 and 32/34) were verified via fragmentation analysis using a nano-LC-MS/MS approach (see Table 1). Next, we identified a peptide corresponding to the hBAD 73–94 sequence carrying one phosphate. The fragmentation analysis of this peptide did not provide an unambiguous result; it suggested phosphorylation of either serine 74 or 75. We propose, however, that this phosphate could be ascribed to phosphoserine 75, since the phosphospecific antibody directed against the homologous site in mBAD identified also the hBAD protein (Fig. 1A). Interestingly, fragmentation analysis of the tryptic peptide 71–94 revealed phosphorylation at serine 91 but not at serine 75. Since the homologous serine in murine BAD (serine 128) has been reported to be phosphorylated, we can conclude that this serine in front of the putative 14-3-3 binding region may also play a regulatory role in human BAD signaling. The next two peptides (peptides 97–109 and 99–109) carrying one phosphate each cover the sequence of the putative 14-3-3 binding domain that has been described to be phosphorylated at serine 99. The antibody directed against the analogous site in mice recognized position Ser-99 within the purified hBAD protein (Fig. 1A). In accordance with this, the fragmentation analysis identified phosphoserine 99 within the tryptic peptide 99–109. Surprisingly, the peptide 97–109 carried the phosphate molecule at serine 97, revealing a novel phosphorylation site in hBAD. Of note, we identified also a peptide comprising the segment between residues 95 and 112 in which both serines (in positions 97 and 99) were phosphorylated (Table 1). This finding suggests a novel regulatory mechanism regarding 14-3-3 binding to their association partners. The next two peptides (peptides 116–126 and 117–126) carrying one phosphate each comprise the C-terminal part of BH3 domain of BAD, where serine 118 (corresponding to serine 155 in mBAD) is located. The phosphorylation of this residue regulates the interaction of BAD with Bcl-2/Bcl-XL proteins. The fragmentation analysis localized a phosphate molecule at serine 118 within both peptides. This finding documents that a fraction of BAD expressed in Sf9 cells does not appear to be associated with the antiapoptotic proteins Bcl-2/Bcl-XL. Importantly, the peptide 117–133 revealed two phosphorylation sites (serines 118 and 124). Because the number of phosphates within the peptide 117–133 corresponds to the number of phosphorylation possibilities, we conclude that besides the well characterized serine 118, serine 124 represents a novel regulatory site in hBAD. Phosphorylation of serine 124 may be of particular importance due to its proximity to the lipid binding domain of human BAD comprising the FKK motif that is located in close proximity to this serine (see Fig. 11).
TABLE 1.
Human BAD phosphopeptides obtained by trypsin and/or GluC digestion
The putative phosphorylation sites and the number of the phosphates detected in each of the peptides by mass spectrometry are indicated. ND, not determined.
| hBAD Peptide | Amino acid sequence | Phosphates/peptide | Phosphorylation site(s) |
|---|---|---|---|
| 21–36 | GLGPpSPAGDGPSGSGK | 1 | Ser-25 |
| 21–36 | GLGPSPAGDGPSGSGK | 1 | Ser-32 or Ser-34 |
| 73–94 | HSSYPAGTEDDEGMGEEPSPFR | 1 | Ser-74 or Ser-75 |
| 71–94 | SRHSSYPAGTEDDEGMGEEPpSPFR | 1 | Ser-91 |
| 73–94 | HSSYPAGTEDDEGMGEEPpSPFR | 1 | Ser-91 |
| 95–112 | GRpSRpSAPPNLWAAQRYGR | 2 | Ser-97 and Ser-99 |
| 97–109 | pSRSAPPNLWAAQR | 1 | Ser-97 |
| 97–109 | SRpSAPPNLWAAQR | 1 | Ser-99 |
| 99–109 | pSAPPNLWAAQR | 1 | Ser-99 |
| 116–126 | RMpSDEFVDSFK | 1 | Ser-118 |
| 117–126 | MpSDEFVDSFK | 1 | Ser-118 |
| 117–133 | MpSDEFVDpSFKKGLPRPK | 2 | Ser-118 and Ser-124 |
| 132–142 | PKpSAGTATQMR | 1 | Ser-134 |
| 134–142 | pSAGTATQMR | 1 | Ser-134 |
| 124–149 | SFKKGLPRPKSAGTATQMRQSSSWTR | 5 | ND |
FIGURE 11.
Amino acid sequences of the BAD fragment surrounding the BH3 domain from different mammalian species. Amino acid sequences were aligned using the ClustalW algorithm (available on the World Wide Web). The BH3 domain is highlighted in pink, and the vicinal FKK/FK regions are in turquoise. This alignment reveals that, in contrast to the highly conserved BH3 domain, the neighboring FKK lipid binding motif exists only in humans.
The alignment of human and murine BAD reveals that both BAD proteins contain the conserved segment RPKSAG, which represents an appropriate consensus sequence for some kinases. The phosphorylation of murine BAD at serine 170 within this segment has been previously reported (60). Our MS analysis revealed two tryptic peptides (132–142 and 134–143) carrying one phosphate each. This phosphorylation site was ascribed unambiguously to serine 134, which is homologous to murine serine 170. Finally, we detected a peptide that was cleaved by GluC and trypsin carrying five phosphates (Fig. 1B and Table 1). This peptide has been ascribed to the C-terminal BAD region located between residues 128 and 149. However, this sequence stretch bears up to eight possible phosphorylation sites. Unfortunately, we were not successful in specifying the exact positions of all of the phosphates detected by MS analysis within this fragment. Nevertheless, the phosphorylation of serines 124 and 134 is probable, since it has been detected within several peptides listed in Table 1.
Taken together, we present here for the first time a detailed analysis of human BAD phosphorylation. We have confirmed several phosphorylation sites that are common to both human and murine BAD and identified in addition a number of novel phosphorylation sites that are specific for human BAD. These novel sites may be involved in regulation of 14-3-3 binding and membrane anchoring of BAD.
Human BAD Is a Direct Substrate of RAF Kinases
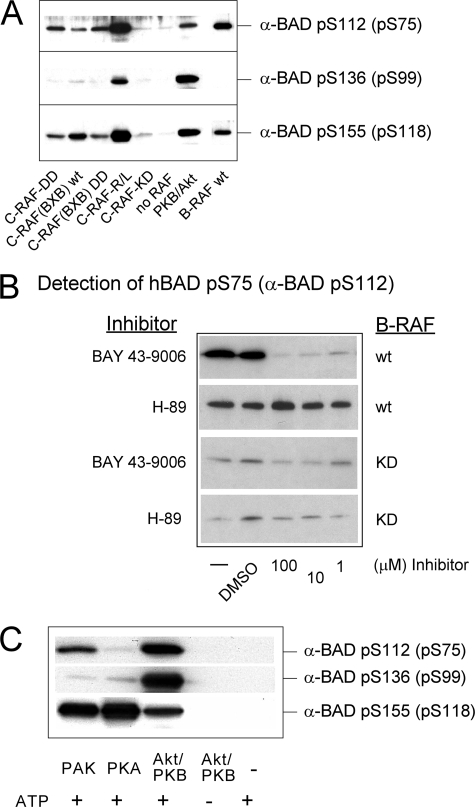
Next, we investigated whether RAF kinases are directly involved in phosphorylation of human BAD in vivo. It was previously shown that C-RAF phosphorylates hBAD (17). In these preliminary reports, the precise sites of RAF-mediated BAD phosphorylation have not been determined. More recent reports support the view that C-RAF participates in BAD phosphorylation either in conjunction with PAK (18) or the adaptor protein Grb10 (20). The role of other RAF isoforms (A- and B-RAF) has not been evaluated in this context. Particularly, the quantitative contribution of the RAF kinases in phosphorylating serine residues 112, 136, and 155 and the direct comparison of RAF isoforms with other BAD targeting kinases has not been performed so far.
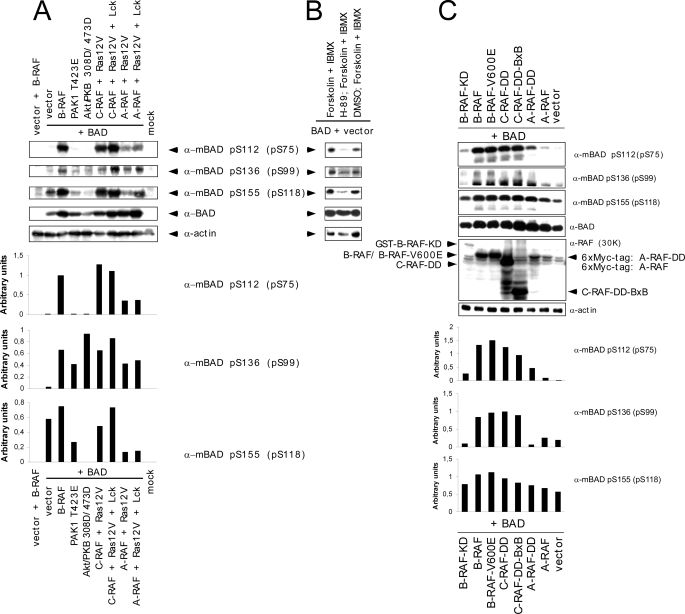
In order to address these open issues, we monitored the phosphorylation pattern on critical serine residues (serines 75, 99, and 118 of human BAD) by different RAF isoforms in vivo and in vitro and performed a comparative analysis involving other BAD-phosphorylating kinases, such as PAK, PKA, and/or Akt/PKB. Due to the fact that the sensitivity of the phosphospecific BAD antibodies is too low to analyze phosphorylation status of endogenous BAD in HEK293, NIH 3T3, or HeLa cells, we decided to overexpress hBAD. Analyzing co-transfected HEK293 cells we found that the exogenous BAD has been efficiently phosphorylated at the critical positions not only by active PAK1 and Akt/PKB mutants (PAK1-T423E and Akt/PKB-T308D/S473D) but also in the presence of activated RAF isoforms (Fig. 2A). In the absence of RAF kinases, Ras12V and Lck did not lead to significant phosphorylation of hBAD (data not shown). To exclude the participation of kinases other than RAF in BAD phosphorylation that may have been activated by overexpression of Ras12V and Lck, we co-expressed BAD with a variety of constitutively active A-, B-, and C-RAF mutants (Fig. 2C). Although inactive B-RAF-K483M (KD) or unstimulated A- and C-RAF failed to phosphorylate BAD significantly, mutants of RAF kinases with elevated kinase activity revealed the same phosphorylation pattern as presented in Fig. 2A. Similar results were obtained by stimulating endogenous PKA with forskolin in combination with isobutylmethylxanthine (Fig. 2B). Quantification of the results obtained by phosphospecific antibodies directed against serine 75, 99, and 118 revealed that active PAK1 and Akt/PKB were most efficient in the phosphorylation of the putative 14-3-3 binding site of BAD localized at serine 99 (see bar graph in Fig. 2A). Compared with these data, the activated or constitutively active forms of RAF kinases were more efficient in phosphorylation of serines 75 and 118. To investigate whether these results are HEK293 cell-specific, we co-transfected also NIH 3T3 and HeLa cells with BAD and activated forms of RAF, PAK1, and Akt/PKB. The results obtained with these cell lines were consistent among all three cell lines (data not shown). In contrast to in vivo data, more efficient phosphorylation of recombinant GST-BAD by activated PAK1 or Akt/PKB was observed using the intact cell lysates comprising the overexpressed active kinases from the experiment described in the legend to Fig. 2A (Fig. 3). Although cell lysate containing activated PAK1 was able to phosphorylate recombinant GST-BAD to low levels at serines 75 and 99 and to a higher extent at serine 118, activated Akt/PKB again showed efficient phosphorylation of BAD predominantly at serine 99 and more weakly at serine 118 and 75 (Fig. 3). These in vitro data are in accordance with findings published earlier (22, 23, 61). BAD phosphorylation by RAF kinases exhibited consistent data in vivo and in vitro.
FIGURE 2.
Comparative analysis of hBAD phosphorylation in HEK293 cells by PKA, Akt/PKB, PAK1, and RAF kinases. HEK293 cells were transiently transfected with the indicated expression vectors. A, C- and A-RAF kinases were activated in cells by co-transfecting Ras12V and Lck. In the case of Akt/PKB and PAK1, activating mutants (T308D/S473D and T423E, respectively) were used. In B, endogenous PKA was stimulated for 30 min with 25 μm forskolin and 500 μm isobutylmethylxanthine (IBMX). Pretreatment with 10 μm H-89 for 30 min prevented PKA activity. In C, B-RAF WT and various constitutively active mutants of RAF kinases (B-RAF-V600E, C-RAF-Y340D/Y341D (C-RAF-DD), and truncated C-RAF lacking N-terminal regulatory domains (BxB WT and BxB-DD)) were transfected. As a negative control, a kinase-dead B-RAF (B-RAF-K483A) mutant was chosen. 16 h post-transfection, cells were cultivated for an additional 30 h in medium supplemented with 0.3% serum. Total cell lysates were separated on a 15% SDS-polyacrylamide gel and blotted onto nitrocellulose membrane, and phosphorylation of human BAD at serines 75, 99, and 118 as well as BAD expression was analyzed. Endogenous actin was used as a loading control. The presence of different RAF proteins was verified with anti pan-RAF antibody. Representative blots from A and C were quantified by optical densitometry. These experiments were repeated three times with the same results. KD, kinase-dead.
FIGURE 3.
In vitro phosphorylation of recombinant GST-BAD by kinases overexpressed in HEK293 cells. HEK293 cells were transiently transfected with the indicated plasmids. Inactive mutants of PAK1 and Akt/PKB (K299R and K179A, respectively) were used as negative controls. 16 h post-transfection, cells were cultivated for an additional 30 h in medium supplemented with 0.3% serum. Afterward, cells were washed once in PBS and lysed by the direct addition of Nonidet P-40 buffer. For the kinase assay, 35 μg of the protein lysates containing the desired kinases were mixed with 1 μg of recombinant GST-BAD (purified from E. coli) in kinase buffer. Proteins were separated on a 12% SDS-polyacrylamide gel and blotted. Phosphorylation of BAD was visualized with phosphospecific BAD antibodies. Expression levels of RAF kinases, PAK1, and Akt/PKB are shown in the lower panels. This experiment was repeated three times with the same results.
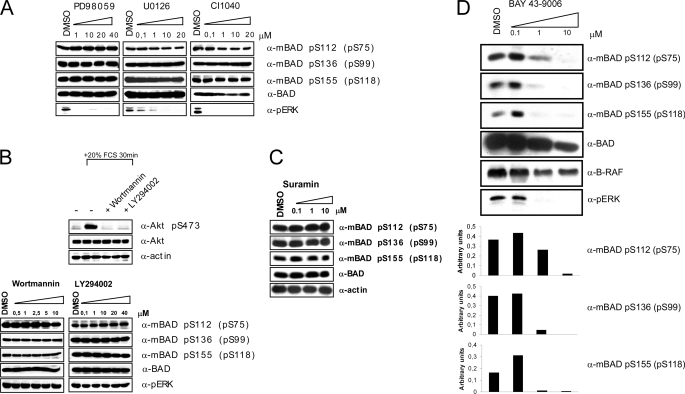
To demonstrate that BAD is a direct substrate of RAF kinases, several inhibitors were applied to exclude the possible influence of endogenous kinases. Importantly, we observed no reduction of BAD phosphorylation after treatment of HEK293 cells co-transfected with hBAD and B-RAF using three different MEK inhibitors (U0126, PD98059, and CI1040) or the PI3K inhibitors wortmannin and LY294002 (Fig. 4, A and B). An involvement of autocrine loops could be excluded, since treatment with suramin, an inhibitor blocking the activation of growth factor receptors at the plasma membrane through interference with receptor-ligand binding (62, 63), did not prevent phosphorylation of BAD by B-RAF (Fig. 4C). Importantly, the RAF kinase inhibitor BAY 43-9006 reduced BAD phosphorylation at all three serine residues already at 1 μm (Fig. 4D). The results presented here (using RAF and MEK inhibitors) are in accordance with data published by Jin et al. (18). Finally, the PKA inhibitor H-89 reduced considerably the phosphorylation degree at all three sites (Fig. 2B).
FIGURE 4.
Kinase inhibitors indicate direct involvement of RAF kinases in BAD phosphorylation. HEK293 cells were transiently transfected with BAD and B-RAF expression vectors. 16 h post-transfection, cells were cultivated in medium supplemented with 0.3% serum. During that time, cells were additionally treated with various concentrations of MEK inhibitors PD98059, U0126, and CI1040 (A), with PI3K inhibitors wortmannin and LY294002 (B), with suramin (C), and with RAF inhibitor (BAY 43-9006) (D) for 22 h, respectively. Phosphorylation of BAD was detected with phosphospecific BAD antibodies. The efficiency of the PI3K inhibitors has been verified by abolition of Akt-S473 phosphorylation in stimulated HEK293 cells (B). Treatment with the RAF inhibitor BAY 43-9006 prevented both BAD and ERK phosphorylation in a concentration-dependent manner. These experiments were repeated three times with the same results.
In Vitro Phosphorylation of BAD by Purified B- and C-RAF Kinases
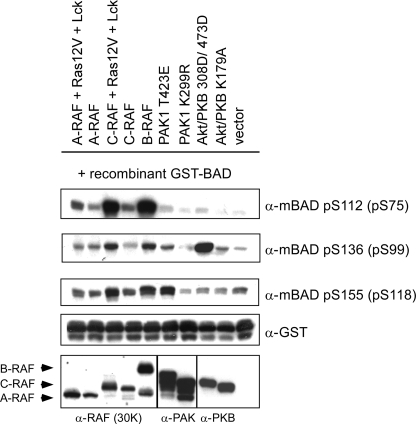
To further explore whether BAD is a direct substrate of RAF, we analyzed phosphorylation of recombinant BAD by purified RAF kinases in vitro. For C-RAF, we tested several activated forms, including constitutively active C-RAF (C-RAF-Y340D/Y341D, termed C-RAF-DD), highly activated C-RAF (co-expressed in the presence of Ras12V and Lck, called here C-RAF-R/L), and truncated C-RAF lacking N-terminal regulatory domains (BxB WT and BxB-DD). In experiments with B-RAF, the WT kinase was chosen because of its high basal activity compared with A- and C-RAF WT. As illustrated in Fig. 5A, all active RAF preparations phosphorylated hBAD at Ser-75 and Ser-118. Only C-RAF-R/L additionally phosphorylated BAD at Ser-99. Of note, kinase-dead mutants of C- and B-RAF (C-RAF-K375M and B-RAF-K483M) failed to phosphorylate hBAD in vitro (Fig. 5, A and B). In the presence of Akt/PKB, we observed phosphorylation at Ser-75, -99, and -118, whereas Ser-118 was the main target of PKA. PAK phosphorylated Ser-75 and Ser-118 (Fig. 5C). In accordance with in vivo data obtained by use of RAF inhibitor BAY 43-9006, we also observed in vitro effective inhibition of RAF-mediated hBAD phosphorylation by BAY 43-9006 (Fig. 5B).
FIGURE 5.
In vitro phosphorylation of recombinant GST-BAD by purified PKA, PAK1, Akt/PKB, and RAF kinases. A–C, phosphorylation of non-phosphorylated BAD purified from E. coli was carried out as described under “Experimental Procedures.” Recombinant GST-BAD (20 pmol) was phosphorylated by purified B- and C-RAF and corresponding RAF mutants (2 pmol each) as indicated, catalytic subunit of PKA, constitutively active Akt/PKB, or PAK1 (4 pmol each). B, whereas the RAF inhibitor BAY 43-9006 prevented BAD phosphorylation in the concentration range between 1 and 100 μm, the PKA inhibitor H-89 had no effect on BAD phosphorylation, demonstrating that purified B-RAF was not associated with PKA. Following SDS-PAGE and immunoblotting, BAD phosphorylation was visualized by phosphospecific antibodies directed against phosphoserines 112, 136, and 155 of mouse BAD (corresponding to phosphoserines 75, 99, and 118 of human BAD). These experiments were repeated five times with comparable results. KD, kinase-dead.
BAD Phosphorylation by RAF Promotes Survival of Mammalian Cells
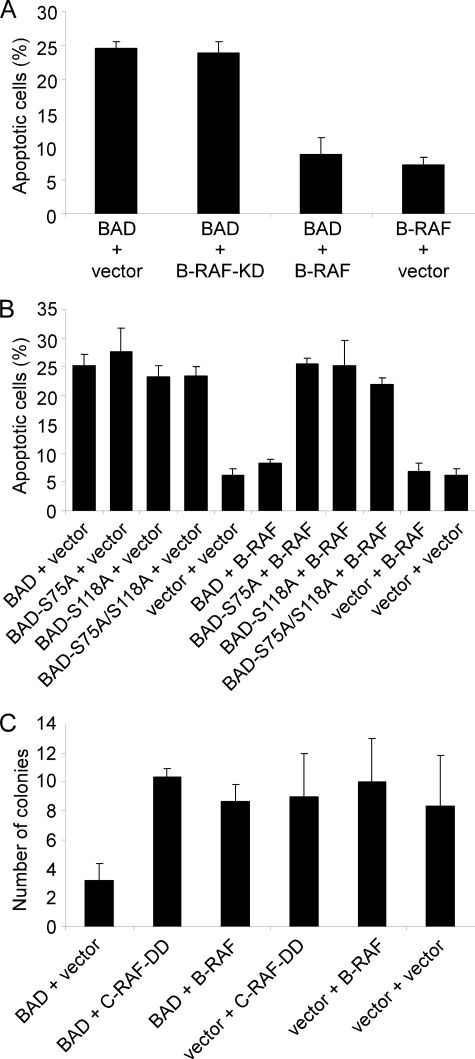
BAD protein has to be phosphorylated in order to neutralize its proapoptotic properties (2, 64). To demonstrate that active RAF prevents BAD-mediated apoptosis, we performed different cell survival assays using HEK293 and NIH 3T3 cells. HEK293 cells were transiently transfected with the indicated expression plasmids and starved for 30 h in medium supplemented with 0.3% serum. The number of apoptotic cells was determined by trypan blue exclusion (Fig. 6A). As demonstrated in Fig. 6A, BAD-induced apoptosis was effectively inhibited by B-RAF overexpression. About 24% of BAD overexpressing cells were apoptotic, whereas co-expression of B-RAF reduced the percentage of apoptotic cells to a level comparable with control cells. Under starvation conditions, substitution mutants of human BAD, S75A, S118A, or S75A/S118A, induce apoptosis to the same extend as was observed for wild type. However, in contrast to co-expression with BAD WT, B-RAF did not inhibit BAD-mediated apoptosis, when it was co-expressed with BAD-S75A, BAD-S118A, or BAD-S75A/S118A mutants (Fig. 6B). Of importance, expression of the kinase-dead form of B-RAF did not prevent the BAD-induced apoptosis of the cells as well (Fig. 6A).
FIGURE 6.
Expression of B- and C-RAF delays BAD-mediated apoptotic death of HEK293 cells following growth factor removal. A and B, HEK293 cells were transiently transfected in triplicates with the indicated expression vectors (ratio 1:1). 16 h post-transfection, cells were cultivated for an additional 30 h in medium supplemented with 0.3% serum. Cell viability was assessed by trypan blue staining. Mean and S.D. values are shown. The experiment was repeated twice. C, colony formation assay shows reproductive survival of triplicate cultures of NIH 3T3 cells stably expressing either empty vectors, BAD alone, BAD in combination with B-RAF or activated C-RAF (C-RAF-DD), and B-RAF and C-RAF-DD alone. BAD phosphorylation by RAF protected cells from apoptosis and led to an increased number of colonies.
In order to corroborate the importance of B- and C-RAF-mediated BAD phosphorylation for the survival of mammalian cells, we additionally performed colony yield assays (Fig. 6C). BAD-overexpressing cells seeded at low density in Petri dishes have difficulties in forming clonogenic colonies, clear evidence that they are not protected against its proapoptotic activity. On the other hand, co-expression with active B- or C-RAF is expected to improve colony yields. In our experimental set, we generated stable expression of the transfected DNA by double selection with different antibiotics. As a result, we found that the number of colonies formed by control cells and cells continuously expressing BAD in combination with C-RAF-DD or B-RAF was at least 3 times higher compared with cells expressing BAD only (Fig. 6C).
The Associations of BAD with 14-3-3 Proteins and Bcl-2/Bcl-XL Is Affected by RAF Kinases
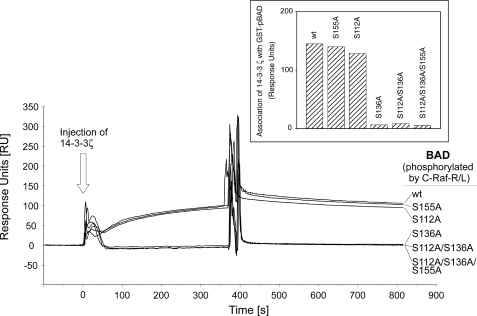
Following prosurvival signaling, BAD becomes phosphorylated, which enables complex formation with 14-3-3 proteins. To analyze the putative 14-3-3 binding site(s) of hBAD, particularly with respect to phosphorylation by RAF, purified hBAD and its mutants (S75A, S99A, S118A, S75A/S99A, and S75A/S99A/S118A) have been treated in an in vitro kinase assay with active C-RAF-R/L, as described in the legend to Fig. 5A. The real time association with 14-3-3 proteins has been performed by use of the BIAcore technique. For that purpose, phosphorylated GST-BAD samples were captured on a chip surface, and interactions with purified 14-3-3ζ were monitored. In contrast to S75A and S118A mutants, no 14-3-3ζ binding was observed for BAD-S99A mutant, indicating that the domain surrounding Ser(P)-99 represents the major 14-3-3 binding site. The double mutant (S75A/S99A) and the triple mutant (S75A/S99A/S118A) did not interact with 14-3-3 as well (Fig. 7). GST-BAD WT revealed the highest 14-3-3 binding efficiency, indicating that a further binding site may enhance or stabilize this association. These data suggest that in vivo interaction of BAD with active C-RAF may result in 14-3-3 association and depletion of the BAD-14-3-3 complex from mitochondrial membranes.
FIGURE 7.
Phosphorylation of BAD wild type and BAD mutants by C-RAF promotes association with 14-3-3 proteins. Interactions of BAD with 14-3-3ζ were monitored by the surface plasmon resonance technique. GST-BAD samples (200 pmol) were phosphorylated by highly activated C-RAF-R/L (10 pmol), as described under “Experimental Procedures.” Approximately 1000 response units of phosphorylated BAD were captured onto an anti-GST chip. 14-3-3ζ (200 nm) was injected, and the association-dissociation curves were monitored. For better understanding, we used here (and in Fig. 8) the mouse nomenclature.
Next, we examined in more detail the interaction between BAD and Bcl-2 or Bcl-XL, respectively. In a direct comparison of association-dissociation kinetics (supplemental Table S1), we have measured that the Bcl-XL-BAD complex (KD = 0.65 nm) was more stable than Bcl-2-BAD (KD = 2.16 nm). Notably, similar values have been reported for interaction between Bcl-XL and synthetic peptide representing the BH3 domain of BAD (65). In contrast, the affinity of BAD-BH3 peptide to Bcl-2 was much lower (KD = 15 nm), indicating that the binding parameters using full-length proteins may differ considerably from those using peptide samples (65). Due to the higher binding affinity, we used in the following experiments Bcl-XL as a binding partner of BAD.
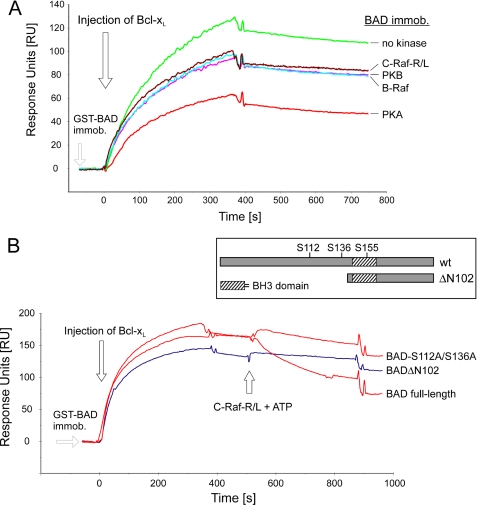
To investigate the relevance of phosphorylation of hBAD with respect to Bcl-XL association, we first phosphorylated hBAD (purified from E. coli) in vitro with various kinases, such as PKA, Akt/PKB, B-RAF, and C-RAF-R/L. Next, the phosphorylated hBAD samples were immobilized on the Biosensor chip, and interactions with Bcl-XL were monitored. As depicted in Fig. 8A, phosphorylation by C-RAF-R/L, B-RAF, and Akt/PKB led to a ∼25% decrease in Bcl-XL association. Incubation with PKA resulted in a ∼50% reduction. This result is in agreement with earlier reports, suggesting that PKA phosphorylates BAD preferentially at serine 155 (16, 66).
FIGURE 8.
Phosphorylation of recombinant BAD inhibits complex formation between BAD and Bcl-XL and disrupts preexisting complex. A, purified GST-BAD (200 pmol) was phosphorylated by purified and active B- and C-RAF, Akt/PKB, and PKA (20 pmol each) as described under “Experimental Procedures.” Association of phosphorylated BAD with full-length Bcl-XL was monitored using the surface plasmon resonance technique. Approximately 800 response units of GST-BAD phosphorylated with respective kinases were immobilized by anti-GST-coated surface. Bcl-XL (200 nm) was injected, and the association-dissociation curves were monitored. B, approximately 1000 response units of GST-tagged BAD WT, BAD-S75A/S99A (here termed as BAD-S112A/S136A), and BAD(ΔN102) were immobilized, and Bcl-XL (200 nm) was injected. After saturation, the formed complexes were treated with C-RAF-R/L in the presence of ATP. The structure of BAD samples used in this assay is illustrated in the inset.
Finally, we asked whether RAF-mediated phosphorylation of BAD reported above is able to trigger the dissociation of the preexisting Bcl-XL-BAD complex. For that purpose, different complexes of Bcl-XL with the following hBAD variants were used (see Fig. 8B, inset): BAD WT, mutated BAD-S75A/S99A, and truncated BAD(ΔN102) consisting of the C-terminal fragment harboring the BH3 domain. We used BAD(ΔN102) because it lacks phosphorylation sites Ser-75 and Ser-99 that are involved in 14-3-3 binding. The complexes were formed on the surface of the biosensor chip (Fig. 8B). To monitor the effect of hBAD phosphorylation by RAF, C-RAF-R/L was injected in the presence of ATP. Whereas the complex with wild type BAD dissociated readily, the complex with the C-terminal part of BAD(ΔN102) remained stable. The double mutant (S75A/S99A) dissociated slowly, indicating that phosphorylation sites other than S75A and S99A are involved (Fig. 8B). From these experiments, we conclude that RAF kinases do not only prevent the formation of the complex between BAD and Bcl-XL but also possess the ability to mediate the disruption of existing BAD-Bcl-XL complexes.
Channel-forming Activity of Human BAD in Planar Lipid Bilayers Is Influenced by Phosphorylation and 14-3-3 Proteins
Most Bcl-2 family proteins contain a C-terminal hydrophobic transmembrane domain, indicating that these proteins may exist as integral membrane proteins (67–69). Some of them, including pro- and antiapoptotic members as well as BH3-only protein Bid, show the ability to form pores in lipid bilayers (38–44).
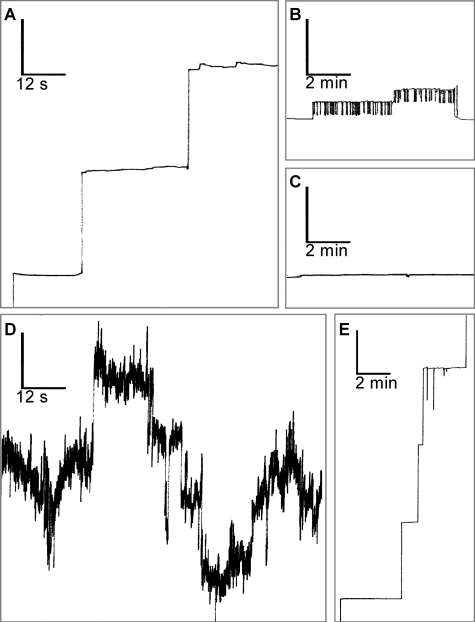
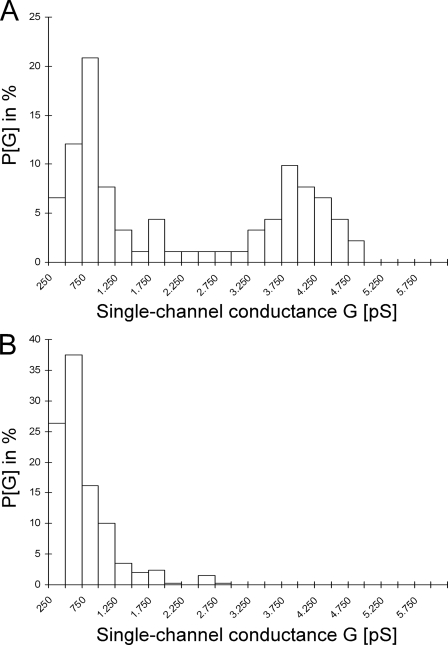
Regarding the BH3-only protein BAD, Hekman et al. (14) reported pronounced lipophilic properties and identified two lipid binding regions (LBD1 and LBD2) in hBAD. Therefore, we supposed that BAD, like several other Bcl-2 family members, may possess channel-forming ability. To test this issue, we investigated whether human BAD is able to form pores in artificial lipid bilayers. Here we used a planar bilayer configuration, where two Teflon chambers are separated by a septum having a small (800-μm diameter) aperture in which the membrane bilayer is formed (58, 59). This or similar configurations are commonly used for the characterization of channels formed by Bcl-2 family proteins, mitochondrial porins, or bacterial toxins (38–42, 70–72). It has been previously shown that Bcl-XL forms channels in synthetic lipid membranes (38). Thus, we applied this measurement as a positive control (Fig. 9E). For the measurements with hBAD, we used two different preparations that differ in their phosphorylation states. Besides the complete dephosphorylated protein (produced and purified from E. coli), we utilized hBAD expressed in Sf9 cells that was also analyzed by mass spectrometry and was shown here to be phosphorylated at several serine residues (Fig. 1). In the case of dephosphorylated hBAD, we did not detect any pore formation (Fig. 9C). In contrast, ∼7 min after injection of phosphorylated hBAD into both chambers, steplike current fluctuations representing channel openings to various discrete conductance states were observed. These channels can be classified in two groups: small channels that show a flickering behavior between a closed and an open state (Fig. 9B) and larger permanent open pores (Fig. 9A). The small flickering pores had a single-channel conductance of about 500 picosiemens. The large permanent open pores occur to a lesser frequency and showed single-channel conductance states of about 0.75 and 3.75 nanosiemens. Two histograms of the probability P(G) for the occurrence of specific conductivity states for 91 (Fig. 10A) and 341 (Fig. 10B) single-conductance events that were observed within the same experiment with phosphorylated hBAD are shown in Fig. 10. It has been reported that phosphorylation of serines 112 and 136 of mBAD (corresponding to serines 75 and 99 of hBAD) leads to cytoplasmic sequestration by 14-3-3 proteins (15). To test the functional consequence of this interaction with respect to hBAD pore-forming abilities, phosphorylated hBAD was preincubated with heterodimeric 14-3-3 protein (14-3-3ζ·14-3-3ϵ) before analyzing the channel activity. The consequence was a disruption of the assembly of hBAD into the lipid membrane. Furthermore, 14-3-3 proteins cause removal or closing of existing BAD channels (Fig. 9D). 14-3-3 proteins alone had no measurable effect on the lipid bilayer (data not shown).
FIGURE 9.
Channel-forming activity of hBAD. Single-channel recordings of purified hBAD or Bcl-XL (30 ng/ml, respectively) in a diphytanoylphosphatidylcholine membrane were monitored. The aqueous phase contained 1 m KCl. The temperature was 20 °C, and the applied voltage was 20 mV. Phosphorylated hBAD forms large permanent open pores (A) as well as small channels that show a flickering behavior between a closed and an open state (B). Non-phosphorylated hBAD has no pore-forming ability (C). Preincubation of phosphorylated hBAD with the 14-3-3ζ·14-3-3ϵ heterodimer (1:2, mol/mol) for 30 min at 22 °C resulted in a disruption of hBAD assembly into the lipid membrane and caused removal or closing of existing BAD channels (D). 14-3-3 proteins alone had no measurable effect on the lipid bilayer (data not shown). E, Bcl-XL was used as a positive control. The vertical bars indicate conductance of 2.5 nanosiemens. These measurements were repeated three times with the same results.
FIGURE 10.
Histogram of the probability for the occurrence of a given conductivity unit. The histogram was observed with membranes formed using 1% diphytanoylphosphatidylcholine in the presence of 30 ng of phosphorylated hBAD. P(G) is the probability that a given conductance increment G is observed in the single-channel experiments. It was calculated by dividing the number of fluctuations with a given conductance increment by the total number of conductance fluctuations. A, large and permanent open pores (91 single-channel events were integrated). B, small channels that show a flickering behavior between a closed and an open state (341 single-channel events were considered).
DISCUSSION
The phosphorylation of BAD provides an important connection between cell survival signaling and the apoptotic death machinery. A current model of BAD function implicates phosphorylation of at least three serine residues (using mouse nomenclature, these are serines 112, 136, and 155). The consequence of phosphorylation of thee sites is complex formation of BAD with 14-3-3 proteins and removal of prosurvival Bcl-2 members at the outer mitochondrial membrane. Besides these highly conserved phosphorylation domains, four other phosphorylation sites of murine BAD (positioned at serines 128 and 170 and threonines 117 and 201) have been identified. Remarkably, although much attention has been devoted to the phosphorylation-mediated regulation of murine BAD function, with some exceptions, the regulation of human BAD by phosphorylation has so far not been investigated. Therefore, we performed a systematic analysis of in vivo phosphorylation sites in human BAD by combined use of phosphospecific antibodies and mass spectrometry.
MS Analysis of Human BAD Phosphorylation and Identification of Novel in Vivo Phosphorylation Sites
As shown in Fig. 1A phosphorylation of all three highly conserved serines in purified hBAD were detectable by use of phosphospecific antibodies, indicating that a fraction of human BAD expressed in Sf9 cells is associated with 14-3-3 proteins. Such a complex is predicted to be cytosolic or relocated to lipid rafts and not to be associated with Bcl-2/Bcl-XL proteins (14, 15). To investigate whether hBAD is phosphorylated on more than the three established phosphorylation sites (serines 75, 99, and 118), the purified hBAD samples were analyzed by both ESI-MS and nano-LC-MS/MS technique. The results obtained by MS analysis revealed numerous novel phosphorylation sites (Fig. 1B and Table 1). Interestingly, with the exception of serines 25 and 32/34, most of the phosphorylated peptides are clustered within a 75-amino acid stretch comprising also the BH3-like domain. In contrast, the last 20 residues at the very C-terminal sequence bear no phosphate molecules.
Three peptides (peptide 95–112, 97–109, and 99–109) carrying one or two phosphates comprise the sequence of the putative 14-3-3 binding domain RSRS99AP. By use of phosphospecific antibodies, the serine 99 has been found to be phosphorylated in the hBAD sample (see Fig. 1A). Surprisingly, in addition to serine 99, the peptide 95–112 was phosphorylated also at serine 97, indicating a novel regulatory mechanism regarding association of 14-3-3 proteins with BAD. Possibly, the phosphorylation of the second serine in the position 97 within the 14-3-3 binding motif (RS97RSAP) inhibits the association of BAD with 14-3-3 proteins. Similar accumulation of phosphates has been observed within the C-terminal 14-3-3 binding motif of A-RAF kinase (RS580AS582EP), where both serines 580 and 582 were found to be phosphorylated (73). We have proposed that the multiple phosphorylation of the C-terminal 14-3-3 binding region in A-RAF may be one of the reasons for the relative low activity of this RAF isoform. On the other hand, perturbations within the internal 14-3-3 binding domain of C-RAF (RSTS259TP) have been reported to be a reason for severe cardio-facio-cutaneous disorders called Noonan and Leopard syndrome (74, 75). Displacement of the serine 259 by phenylalanine abolished the autoinhibitory mechanism of C-RAF, resulting in a permanent active kinase form. In conclusion, we suggest that phosphorylation of the serine in position −2 relative to the obligatory phosphorylated serine within the 14-3-3 binding motif (e.g. serine 99 in hBAD) is sufficient to displace 14-3-3 from BAD. In this scenario, previous dephosphorylation of the crucial serine within the 14-3-3 binding domain would be dispensable.
The binding motif surrounding serine 75 (RHSS75YP) fulfills the criteria for a typical 14-3-3 binding site as well (76). In human BAD, phosphorylation of serine 75 has been detected by use of phosphospecific antibody (see Fig. 1A). Also, the fragmentation of the tryptic peptide 73–94 suggests phosphorylation of this residue (see Table 1). However, binding data presented in Fig. 7 obtained by mutated BAD proteins do not support a significant contribution of this domain for association of 14-3-3 proteins. We propose that phosphorylated serine 75 is functioning as a gatekeeper for the association to dimeric 14-3-3 molecules. In other words, the heterodimeric 14-3-3 proteins may occupy both functional 14-3-3 binding sites, with a region surrounding serine 99 representing the high affinity binding site. Besides displacement of 14-3-3 protein from the internal binding site, also other functions of the phosphoserine 75 should be taken into account. Indeed, Fueller et al. (77) reported recently that the transient phosphorylation of serine 75 in hBAD protein mediated by the activated catalytic domain of C-RAF promotes polyubiquitylation of hBAD and increases the turnover of this protein by proteosomal degradation. The alignment of the amino acid sequences of several mammalian BAD proteins reveals two PEST regions, which constitute a marker for proteins that undergo proteosomal degradation. Interestingly, one of these PEST regions overlap with the 14-3-3 binding domain surrounding phosphoserine 75, thus indicating a competition between 14-3-3 binding and the ubiquitylation machinery.
Four other phosphopeptides (114–127, 116–126, 117–126, and 117–133) carrying either one or two phosphates partially cover the BH3 domain where the serine 118 (corresponding to serine 155 in mBAD) is located. Phosphorylation of this residue regulates the interaction of BAD with Bcl-2/Bcl-XL proteins. Importantly, within the peptide 117–133, two phosphates were detected (see Fig. 1B and Table 1). Because the number of phosphates within the peptide 117–133 corresponds to the number of phosphorylation possibilities, both serine residues (serines 118 and 124) within this peptide appear to be phosphorylated in vivo. Thus, we propose that besides the well characterized serine 118, the serine 124 represents a novel phosphorylation site in hBAD, possibly regulating the interaction with membrane lipids. In our previous attempts to characterize the translocation of BAD to mitochondria, we made an unexpected observation that BAD associates with the same efficiency with non-treated and protein-depleted mitochondria (14). By use of plasmon resonance-based measurements, two lipid-binding domains (termed LBD1 and LBD2) were identified in C-terminal region of human BAD. Although LBD2 overlaps with helix-5 localized at the very C terminus, LBD1 encompasses the C-terminal half of BH3 helix. The LBD1 segment covers also the short FKK motif that has been shown to be necessary for lipid binding of hBAD, because mutations within this motif led to dysfunction of BAD and reduced significantly the association with liposomes (14). Therefore, we suggest that the phosphorylation of serine 124 may be of regulatory importance due to its close proximity to the FKK motif (S124FKK127). It is reasonable to speculate that the negative charge of the phosphate molecule may serve to neutralize the influence of the positively charged lysines 126 and 127, with the consequence of reduced affinity of BAD for certain membrane lipids or membrane lipid microdomains. Importantly, the proposed regulation of BAD function by phosphorylation of serine 124 seems to be unique for human BAD protein, since most of the mammalian homologues do not contain the complete FKK motif (see also the sequence alignment of several mammalian species illustrated in Fig. 11).
Finally, we detected a peptide carrying five phosphates overlapping partially with the peptide 117–133 (see Fig. 1B). This peptide has been ascribed to the C-terminal BAD region located between residues 128 and 149. At present, we cannot definitively specify the exact positions of all of the phosphates found by MS analysis. Nevertheless, the phosphorylation of serine 124 is very probable, since it has already been detected within the peptide 117–133. Also, the phosphorylation of murine BAD at serine 170 (corresponding to serine 134 in hBAD) has been previously reported (60). Since the alignment of human and murine BAD shows that both BAD proteins contain the conserved segment RPKS134/170AG, it appears probable that this position is phosphorylated in both proteins. Indeed, two other phosphopeptides (132–142 and 134–142) confirmed this assumption. Nevertheless, the role of multiple phosphorylations at the C-terminal end of the BAD protein remains unclear. Such an accumulation of negative charged residues may support the 14-3-3 depletion of BAD from mitochondria.
Inhibition of BAD-mediated Apoptosis by RAF Kinases
The proapoptotic protein BAD has been reported to be a substrate for a broad spectrum of kinases. Here, we demonstrate that BAD is phosphorylated in vivo and in vitro by RAF kinases as well. However, the role of RAF kinases in BAD phosphorylation is still controversial (15, 18, 20, 21). B-RAF has so far not been considered as a BAD-phosphorylating kinase. Our results here indicate that RAF kinases (particularly B- and C-RAF) play an active role in BAD phosphorylation and regulation of apoptosis. Due to quality limitations of the phosphospecific BAD antibodies, it was almost impossible to analyze the phosphorylation status of endogenous BAD in cell lines like HEK293, NIH 3T3, or HeLa. To avoid immunoprecipitation experiments where phosphorylated BAD fractions are highly enriched in vitro, we decided to perform forced expression experiments. An advantage of this method is that it allows direct testing of RAF kinases, Akt/PKB, or PAK1 on BAD phosphorylation and avoids broad activation of effector kinases that normally occurs after stimulation with growth factors like epidermal growth factor or platelet-derived growth factor.
It has been previously demonstrated that active C-RAF is involved in BAD phosphorylation (17). However, the exact position of BAD phosphorylation by RAF has not been elucidated. Here we resolved BAD phosphorylation mediated by all three RAF kinases and compared it with data obtained by PKA, Akt/PKB, and PAK1. In vitro experiments are in accordance with previously published reports (16, 21, 23) that PKA, Akt/PKB, and PAK1 phosphorylate BAD to different extents at serines 75, 99, and 118 (Figs. 3 and 5). Under the same conditions, B-RAF phosphorylated BAD predominantly at Ser-75 and Ser-118, whereas highly active C-RAF-R/L phosphorylated BAD at all of these positions (Fig. 5). In contrast, in vivo experiments show that RAF kinases and PKA possess the ability to phosphorylate BAD at all three crucial serines (75, 99, and 118), whereas Akt/PKB and PAK1 were more efficient in phosphorylation of serine 99, which is involved in association of 14-3-3 proteins (Fig. 2). These findings indicate that besides kinase specificity, intracellular localization may be important for substrate recognition.
Blocking of autocrine loops by suramin (e.g. NF-κB pathway or stress kinase cascades) (Fig. 4) to down-regulate BAD phosphorylation suggests that these pathways do not play an essential role in BAD regulation. Cultivating cells with three different MEK inhibitors (U0126, PD98059, and CI1040), we observed no differences in BAD phosphorylation in the presence of B-RAF (Fig. 4). In contrast, cells grown in the presence of RAF inhibitor BAY 43-9006 or PKA inhibitor H-89 showed significant reduction of BAD phosphorylation in vivo and in vitro, suggesting that RAF kinases and PKA are involved directly in suppression of BAD-mediated apoptosis. Although BAY 43-9006 was initially developed as a RAF kinase inhibitor, it can additionally target the MAP kinase p38 and several tyrosine kinases, including VEGFR-2, Flt-3, and c-Kit, but none of the reported BAD kinases (78, 79). Importantly, Jin et al. (18) showed, in accordance with our results, that C-RAF/PAK-mediated BAD phosphorylation could be effectively inhibited in vivo in the presence of 2 μm RAF inhibitor BAY 43-9006. In contrast, the use of MEK inhibitor PD98059 (20 μm) did not prevent BAD phosphorylation. Also, consistent with our data, it has been shown by using the same cell line that Akt/PKB phosphorylates BAD efficiently at serine 136 (22). Collectively, we compare here in the same experiment BAD phosphorylation by RAF and other kinases and show that Akt/PKB and PAK1 phosphorylate BAD with different specificity compared with RAF and PKA. Furthermore, we demonstrated that BAD-induced apoptosis can be inhibited by B- and C-RAF and showed that this inhibition is dependent on the phosphorylation of serines 75 and 118 of hBAD (Fig. 6).
Based on data presented here, we suggest that in vivo phosphorylation of BAD by RAF kinases represents an important pathway in the phosphorylation of BAD domains that are involved either in 14-3-3 protein association or mediate coupling/decoupling of BAD with Bcl-2 and Bcl-XL proteins. To corroborate these findings, we performed binding studies with purified components by use of the BIAcore technique. In Fig. 7, we demonstrate that in vitro phosphorylation of BAD by activated C-RAF promotes association of BAD with 14-3-3ζ. We used 14-3-3ζ, since of the seven 14-3-3 isoforms analyzed, this isoform bound phosphorylated BAD most efficiently (14). Whereas serines 75 and 118 are essentially not required for 14-3-3 binding, the domain surrounding serine 99 represents the preferential 14-3-3 binding site. However, a second binding site may enhance or stabilize this association. Quantitative interactions of BAD with truncated Bcl-2 and Bcl-XL had previously been investigated only using peptide samples derived from the BH3 domain of BAD (65, 80). Consistent with these data, we detected a preference for the interaction between BAD and full-length Bcl-XL, suggesting that Bcl-XL is the preferential binding partner of BAD (supplemental Table S1).
C-RAF has been found to colocalize with mitochondria markers, indicating a high proportion of C-RAF located at mitochondria (81). The presence of activated C-RAF at mitochondria is also supported by the contribution of Jin et al. (18), who demonstrated that PAK mediates C-RAF activation and its subsequent translocation to the mitochondria. At present, we cannot completely exclude the possibility that RAF kinases and PKA act simultaneously or synergistically, since it has been reported that C-RAF and PKA form a complex in vivo (82). However, the C-RAF-PKA complex was found to be stable only in non-stimulated cells. It is possible that Akt/PKB and C-RAF also act as a complex in vivo. In this scenario, C-RAF would phosphorylate mBAD mainly at serines 112 and 155, and Akt/PKB might be responsible for Ser-136 phosphorylation. These combined phosphorylations would enable effective association of 14-3-3 with BAD and separation from the BAD-Bcl-XL. She et al. (83) proposed that BAD might represent the convergence point of the RAF- and the PI3K/Akt kinase pathway. According to this report, BAD protein acts as a switch that integrates the anti-apoptotic effects of the epidermal growth factor receptor/MAPK and PI3K/Akt pathways (as detected in MDA-468 cancer cells). This model is further supported by the observation that BAD can associate with PKB and B-RAF in conjunction with the co-chaperone BAG-1 on the mitochondrial level (84).
Pore-forming Activity of Human BAD Is Regulated by Phosphorylation and 14-3-3 Proteins
Within the Bcl-2 family of proteins, Bcl-XL, Bcl-2, Bax, Bak, and the BH3-only protein Bid have been reported to possess channel-forming ability in artificial lipid bilayers (38–42). In addition, it was observed by confocal and electron microscopy that Bax and Bak coalesce during apoptosis into large clusters on the surface of mitochondria (85). Here we present biophysical evidence that the proapoptotic BH3-only protein BAD forms channels in artificial membranes. To form pores, such proteins must contain helices that are long enough to span the membrane bilayer, and these helices must be largely devoid of charged residues (70). Since an average lipid bilayer has a hydrophobic cross-section of ∼30 Å (86), the α-helix needs to be ∼20 residues long in order to span a membrane bilayer and to be able to participate in channel formation (70). A helix probability plot of human BAD exhibited a C-terminal region of about 20 residues with a high probability of a helical structure and only two charged residues (14). This region is surrounded by positively charged residues, which may additionally facilitate the association of the protein with membranes. Although one helix is insufficient to form a channel, some molecules could come together, each contributing their hydrophobic helix to create a pore. Furthermore, in the vicinity of this putative C-terminal helix, a second lipid binding domain in human BAD comprising the FKK motif has been identified (14).
We show here that human BAD is able to form ion channels, which exhibit multiple conductance states with complex opening kinetics. Similar properties have been also reported for other Bcl-2 family members, including Bcl-XL, Bcl-2, Bax, and Bid (38, 39, 41, 42, 87). The presence of three different channel activities with progressively greater conductances (∼500, ∼750, and ∼3750 picosiemens) but occurring with progressively lesser frequency raises the possibility of stepwise oligomerization of BAD protein molecules in planar bilayers. The BAX channel progresses within 2–4 min of its initial appearance (42). This includes an early low conductance channel, followed by a transition phase with multiple subconductance levels and finally achieves an apparently stable ohmic pore of large conductance. Our findings that the lower conductive hBAD channels are flickering between a closed and an open state and the higher conductive hBAD channels persist open raises the question of what factors may control opening and closing of hBAD channels in vivo. Although it was demonstrated that phosphorylation of BAD does not affect membrane binding (14), dephosphorylated hBAD fails to form discrete channels in lipid bilayers. Possibly, some specific phosphorylation patterns of hBAD are responsible for the formation of particular conductance states. Furthermore, we observed that 14-3-3 proteins disrupt the assembly of hBAD into the lipid membrane and that 14-3-3 is able to remove or close existing hBAD channels. Based on these data, we propose that the formation of hBAD pores is a reversible process that is regulated by phosphorylation and 14-3-3 proteins. This fits well to the suggested model that BAD is a membrane-associated protein that has the hallmarks of a receptor rather than a ligand, which shuttles in a phosphorylation-dependent manner between mitochondria and other membranes with 14-3-3 as a key regulator of this relocation (14). Additionally, our results emphasize that phosphorylation alone is insufficient to release BAD from membranes, because the depletion process depends on 14-3-3 proteins. In vivo, the formation of hBAD pores may also be affected by other proteins that have been reported to interact with BAD. Two candidates are Akt/PKB and B-RAF, which were demonstrated to co-immunoprecipitate with BAD (84). It is possible that these kinases affect the pore-forming ability of hBAD besides the BAD-phosphorylating activity. Another open issue is the putative influence of other pore-forming members of the Bcl-2 family of proteins, like Bcl-2 and Bcl-XL, that have been shown to interact with BAD (10, 38, 41). Do they merely shut off their own and hBAD pore-forming activity by heterodimerization, or do they alternatively form counteracting pores? Our preliminary data suggest that Bcl-XL does not abolish pore formation of hBAD (data not shown). Similar observations were reported with respect to the effects of Bax on the pore-forming ability of Bcl-2. Although it was recently demonstrated that Bax interacts preferentially with the membrane-inserted form of Bcl-2 (88), it was reported that Bax does not merely abrogate pore formation of Bcl-2 (41). These authors suggested that Bcl-2 allows the transport across membranes in a direction that is cytoprotective, whereas Bax does the opposite. Bcl-XL and hBAD may also be involved in controlling such a homeostasis. In this regard, it should be mentioned that hBAD forms pores in its phosphorylated and non-apoptotic state. Therefore, it is possible that it cooperates with anti-apoptotic proteins instead of counteracting them.
Results presented here raised the question, what could be the physiological role of BAD channels? BAD has been found to localize at the outer mitochondrial membrane and cholesterol-rich rafts at the plasma membrane (14, 89). Therefore, the ion-conducting BAD channels could contribute to the regulation of the mitochondrial permeability transition (90), a process that has been suggested to be critical in apoptosis (91, 92). Recently, it was demonstrated that BAD targets the permeability transition pore and sensitizes it to Ca2+ in a phosphorylation-dependent manner (93). It is also possible that hBAD conducts molecules other than ions, such as cytochrome c or further apoptogenic factors. Also, Bax was originally described to form ion-conducting channels (42), whereas cell-free studies on isolated mitochondria demonstrated that it can accelerate the release of cytochrome c (94). Another function of BAD channels could be to influence metabolic processes apart from apoptosis. It was already suggested that mitochondrial ion channels such as those regulated by Bcl-2 family members may control the export of metabolites, including ATP, under normal physiological conditions (95–97). An example for a Bcl-2 family member that may regulate a non-apoptotic process by the formation of pores is the naturally occurring proteolytic cleavage fragment of Bcl-XL (Bcl-XS). There is evidence that during hypoxia, Bcl-XS results in the formation of large conductance channels (98), which contribute to the run-down of synaptic transmission in the squid presynaptic terminal (99).
Other interesting aspects that could unravel the function of pore formation by Bcl-2 proteins are observations that link apoptosis to mitochondrial morphogenesis. Bax/Bak were found to colocalize with mitochondrial fission sites and dynamin family GTPases, Drp1 and Mfn2 (85). Additionally, it was found that during apoptosis close in time to Bax translocation and cytochrome c release, mitochondria fragment into small units (100). This indicates that cytochrome c release may occur through Bax-modified mitochondrial scission machinery. It is also feasible that the insertion of hBAD into membranes is of greater importance to its function as adaptor or docking protein than for pore formation, as has been suggested for Bcl-2, Bax, and Bcl-XL (70).
Conclusions
Generally, BH3-only proteins are proposed to function as sentinels of the cellular health status. Data presented by She et al. (83) indicate that BAD might represent the convergence point of the RAF- and the PI3K/Akt kinase pathway. According to this report, BAD protein acts as a switch integrating the antiapoptotic effects of the central mitogenic and PI3K/Akt pathways. This model is further supported by the observation that BAD can associate with both PKB and RAF kinases at the mitochondrial level. Data presented here suggest also that there is interplay between RAF and the PKB pathway and that BAD can function as a node of these two signaling pathways. Thus, C-RAF might coordinate the assembly, recruitment, and possibly the activity of other kinases, resulting in the correct signaling output. Identification of novel hBAD phosphorylation sites indicates another type of regulation for human BAD compared with murine (and some other mammalian) BAD proteins. The finding that human BAD exhibits pore-forming activities opens new insights into the regulation of apoptotic mechanisms mediated by Bcl-2 family of proteins.
Supplementary Material
Acknowledgments
We thank Barbara Bauer, Nadine Schubert, and Elke Maier for excellent technical assistance.
This work was supported by the Ministerium für Innovation, Wissenschaft, Forschung, und Technologie des Landes Nordrhein-Westfalen and by the Bundesministerium für Bildung und Forschung. This work was also supported by Deutsche Forschungsgemeinschaft Grant SFB 487, Project C3.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- BH3
- Bcl-2 homology domain 3
- mBAD
- mouse BAD
- hBAD
- human BAD
- R/L
- activated by co-transfection with Ras12V and Lck
- LBD
- lipid binding domain
- PKA
- protein kinase A
- PKB
- protein kinase B
- ERK
- extracellular signal-regulated kinase
- PBS
- phosphate-buffered saline
- GST
- glutathione S-transferase
- ESI
- electrospray ionization
- MS
- mass spectrometry
- LC
- liquid chromatography
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase.
REFERENCES
- 1.Reed J. C., Doctor K. S., Godzik A. (2004) Sci. STKE 2004, re9. [DOI] [PubMed] [Google Scholar]
- 2.Danial N. N., Korsmeyer S. J. (2004) Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 3.Letai A. (2006) Mol. Cell 21, 728–730 [DOI] [PubMed] [Google Scholar]
- 4.Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 5.Adams J. M., Cory S. (1998) Science 281, 1322–1326 [DOI] [PubMed] [Google Scholar]
- 6.Gross A., McDonnell J. M., Korsmeyer S. J. (1999) Genes Dev. 13, 1899–1911 [DOI] [PubMed] [Google Scholar]
- 7.Fesik S. W. (2000) Cell 103, 273–282 [DOI] [PubMed] [Google Scholar]
- 8.Petros A. M., Olejniczak E. T., Fesik S. W. (2004) Biochim. Biophys. Acta 1644, 83–94 [DOI] [PubMed] [Google Scholar]
- 9.Wei M. C., Lindsten T., Mootha V. K., Weiler S., Gross A., Ashiya M., Thompson C. B., Korsmeyer S. J. (2000) Genes Dev. 14, 2060–2071 [PMC free article] [PubMed] [Google Scholar]
- 10.Yang E., Zha J., Jockel J., Boise L. H., Thompson C. B., Korsmeyer S. J. (1995) Cell 80, 285–291 [DOI] [PubMed] [Google Scholar]
- 11.Galonek H. L., Hardwick J. M. (2006) Nat. Cell Biol. 8, 1317–1319 [DOI] [PubMed] [Google Scholar]
- 12.Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 13.Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 14.Hekman M., Albert S., Galmiche A., Rennefahrt U. E., Fueller J., Fischer A., Puehringer D., Wiese S., Rapp U. R. (2006) J. Biol. Chem. 281, 17321–17336 [DOI] [PubMed] [Google Scholar]
- 15.Zha J., Harada H., Yang E., Jockel J., Korsmeyer S. J. (1996) Cell 87, 619–628 [DOI] [PubMed] [Google Scholar]
- 16.Datta S. R., Katsov A., Hu L., Petros A., Fesik S. W., Yaffe M. B., Greenberg M. E. (2000) Mol. Cell 6, 41–51 [PubMed] [Google Scholar]
- 17.Wang H. G., Rapp U. R., Reed J. C. (1996) Cell 87, 629–638 [DOI] [PubMed] [Google Scholar]
- 18.Jin S., Zhuo Y., Guo W., Field J. (2005) J. Biol. Chem. 280, 24698–24705 [DOI] [PubMed] [Google Scholar]
- 19.Panka D. J., Wang W., Atkins M. B., Mier J. W. (2006) Cancer Res. 66, 1611–1619 [DOI] [PubMed] [Google Scholar]
- 20.Kebache S., Ash J., Annis M. G., Hagan J., Huber M., Hassard J., Stewart C. L., Whiteway M., Nantel A. (2007) J. Biol. Chem. 282, 21873–21883 [DOI] [PubMed] [Google Scholar]
- 21.Harada H., Becknell B., Wilm M., Mann M., Huang L. J., Taylor S. S., Scott J. D., Korsmeyer S. J. (1999) Mol. Cell. 3, 413–422 [DOI] [PubMed] [Google Scholar]
- 22.Datta S. R., Dudek H., Tao X., Masters S., Fu H., Gotoh Y., Greenberg M. E. (1997) Cell 91, 231–241 [DOI] [PubMed] [Google Scholar]
- 23.Schürmann A., Mooney A. F., Sanders L. C., Sells M. A., Wang H. G., Reed J. C., Bokoch G. M. (2000) Mol. Cell. Biol. 20, 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gnesutta N., Qu J., Minden A. (2001) J. Biol. Chem. 276, 14414–14419 [DOI] [PubMed] [Google Scholar]
- 25.Konishi Y., Lehtinen M., Donovan N., Bonni A. (2002) Mol. Cell 9, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 26.Shimamura A., Ballif B. A., Richards S. A., Blenis J. (2000) Curr. Biol. 10, 127–135 [DOI] [PubMed] [Google Scholar]
- 27.She Q. B., Ma W. Y., Zhong S., Dong Z. (2002) J. Biol. Chem. 277, 24039–24048 [DOI] [PubMed] [Google Scholar]
- 28.Klumpp S., Mäurer A., Zhu Y., Aichele D., Pinna L. A., Krieglstein J. (2004) Neurochem. Int. 45, 747–752 [DOI] [PubMed] [Google Scholar]
- 29.Macdonald A., Campbell D. G., Toth R., McLauchlan H., Hastie C. J., Arthur J. S. (2006) BMC Cell Biol. 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donovan N., Becker E. B., Konishi Y., Bonni A. (2002) J. Biol. Chem. 277, 40944–40949 [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., Liu J., Yu C., Lin A. (2005) Cancer Res. 65, 8372–8378 [DOI] [PubMed] [Google Scholar]
- 32.Yu C., Minemoto Y., Zhang J., Liu J., Tang F., Bui T. N., Xiang J., Lin A. (2004) Mol. Cell 13, 329–340 [DOI] [PubMed] [Google Scholar]
- 33.Zamzami N., Kroemer G. (2003) Curr. Biol. 13, R71–R73 [DOI] [PubMed] [Google Scholar]
- 34.Antignani A., Youle R. J. (2006) Curr. Opin. Cell Biol. 18, 685–689 [DOI] [PubMed] [Google Scholar]
- 35.Muchmore S. W., Sattler M., Liang H., Meadows R. P., Harlan J. E., Yoon H. S., Nettesheim D., Chang B. S., Thompson C. B., Wong S. L., Ng S. L., Fesik S. W. (1996) Nature 381, 335–341 [DOI] [PubMed] [Google Scholar]
- 36.Choe S., Bennett M. J., Fujii G., Curmi P. M., Kantardjieff K. A., Collier R. J., Eisenberg D. (1992) Nature 357, 216–222 [DOI] [PubMed] [Google Scholar]
- 37.Martinou J. C., Green D. R. (2001) Nat. Rev. Mol. Cell Biol. 2, 63–67 [DOI] [PubMed] [Google Scholar]
- 38.Minn A. J., Vélez P., Schendel S. L., Liang H., Muchmore S. W., Fesik S. W., Fill M., Thompson C. B. (1997) Nature 385, 353–357 [DOI] [PubMed] [Google Scholar]
- 39.Antonsson B., Conti F., Ciavatta A., Montessuit S., Lewis S., Martinou I., Bernasconi L., Bernard A., Mermod J. J., Mazzei G., Maundrell K., Gambale F., Sadoul R., Martinou J. C. (1997) Science 277, 370–372 [DOI] [PubMed] [Google Scholar]
- 40.Schendel S. L., Azimov R., Pawlowski K., Godzik A., Kagan B. L., Reed J. C. (1999) J. Biol. Chem. 274, 21932–21936 [DOI] [PubMed] [Google Scholar]
- 41.Schendel S. L., Xie Z., Montal M. O., Matsuyama S., Montal M., Reed J. C. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5113–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlesinger P. H., Gross A., Yin X. M., Yamamoto K., Saito M., Waksman G., Korsmeyer S. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11357–11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basañez G., Nechushtan A., Drozhinin O., Chanturiya A., Choe E., Tutt S., Wood K. A., Hsu Y., Zimmerberg J., Youle R. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 5492–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., Newmeyer D. D. (2002) Cell 111, 331–342 [DOI] [PubMed] [Google Scholar]
- 45.Daum G., Eisenmann-Tappe I., Fries H. W., Troppmair J., Rapp U. R. (1994) Trends Biochem. Sci 19, 474–480 [DOI] [PubMed] [Google Scholar]
- 46.Wellbrock C., Karasarides M., Marais R. (2004) Nat. Rev. Mol. Cell Biol. 5, 875–885 [DOI] [PubMed] [Google Scholar]
- 47.Rapp U. R., Götz R., Albert S. (2006) Cancer Cell 9, 9–12 [DOI] [PubMed] [Google Scholar]
- 48.Rapp U. R., Rennefahrt U., Troppmair J. (2004) Biochim. Biophys. Acta 1644, 149–158 [DOI] [PubMed] [Google Scholar]
- 49.Troppmair J., Rapp U. R. (2003) Biochem. Pharmacol. 66, 1341–1345 [DOI] [PubMed] [Google Scholar]
- 50.Chen C., Okayama H. (1987) Mol. Cell. Biol. 7, 2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hekman M., Wiese S., Metz R., Albert S., Troppmair J., Nickel J., Sendtner M., Rapp U. R. (2004) J. Biol. Chem. 279, 14074–14086 [DOI] [PubMed] [Google Scholar]
- 52.Fischer A., Baljuls A., Reinders J., Nekhoroshkova E., Sibilski C., Metz R., Albert S., Rajalingam K., Hekman M., Rapp U. R. (2009) J. Biol. Chem. 284, 3183–3194 [DOI] [PubMed] [Google Scholar]
- 53.Hekman M., Hamm H., Villar A. V., Bader B., Kuhlmann J., Nickel J., Rapp U. R. (2002) J. Biol. Chem. 277, 24090–24102 [DOI] [PubMed] [Google Scholar]
- 54.Neuhoff V., Arold N., Taube D., Ehrhardt W. (1988) Electrophoresis 9, 255–262 [DOI] [PubMed] [Google Scholar]
- 55.Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., Mann M. (1996) Nature 379, 466–469 [DOI] [PubMed] [Google Scholar]
- 56.Reinders J., Wagner K., Zahedi R. P., Stojanovski D., Eyrich B., van der Laan M., Rehling P., Sickmann A., Pfanner N., Meisinger C. (2007) Mol. Cell Proteomics 6, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 57.Zahedi R. P., Lewandrowski U., Wiesner J., Wortelkamp S., Moebius J., Schütz C., Walter U., Gambaryan S., Sickmann A. (2008) J. Proteome Res. 7, 526–534 [DOI] [PubMed] [Google Scholar]
- 58.Benz R., Maier E., Thinnes F. P., Götz H., Hilschmann N. (1992) Biol. Chem. Hoppe Seyler 373, 295–303 [DOI] [PubMed] [Google Scholar]
- 59.Benz R. (1994) Biochim. Biophys. Acta 1197, 167–196 [DOI] [PubMed] [Google Scholar]
- 60.Dramsi S., Scheid M. P., Maiti A., Hojabrpour P., Chen X., Schubert K., Goodlett D. R., Aebersold R., Duronio V. (2002) J. Biol. Chem. 277, 6399–6405 [DOI] [PubMed] [Google Scholar]
- 61.Tang Y., Zhou H., Chen A., Pittman R. N., Field J. (2000) J. Biol. Chem. 275, 9106–9109 [DOI] [PubMed] [Google Scholar]
- 62.Hosang M. (1985) J. Cell. Biochem. 29, 265–273 [DOI] [PubMed] [Google Scholar]
- 63.Huang R. R., Dehaven R. N., Cheung A. H., Diehl R. E., Dixon R. A., Strader C. D. (1990) Mol. Pharmacol. 37, 304–310 [PubMed] [Google Scholar]
- 64.Green D. R., Kroemer G. (2005) J. Clin. Invest. 115, 2610–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petros A. M., Medek A., Nettesheim D. G., Kim D. H., Yoon H. S., Swift K., Matayoshi E. D., Oltersdorf T., Fesik S. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3012–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Virdee K., Parone P. A., Tolkovsky A. M. (2000) Curr. Biol. 10, 1151–1154 [DOI] [PubMed] [Google Scholar]
- 67.del Mar Martínez-Senac M., Corbalán-García S., Gómez-Fernández J. C. (2000) Biochemistry 39, 7744–7752 [DOI] [PubMed] [Google Scholar]
- 68.del Mar Martínez-Senac M., Corbalán-García S., Gómez-Fernández J. C. (2001) Biochemistry 40, 9983–9992 [DOI] [PubMed] [Google Scholar]
- 69.Martínez-Senac Mdel M., Corbalán-García S., Gómez-Fernández J. C. (2002) Biophys. J. 82, 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schendel S. L., Montal M., Reed J. C. (1998) Cell Death Differ. 5, 372–380 [DOI] [PubMed] [Google Scholar]
- 71.Le Mellay V., Troppmair J., Benz R., Rapp U. R. (2002) BMC Cell Biol. 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manich M., Knapp O., Gibert M., Maier E., Jolivet-Reynaud C., Geny B., Benz R., Popoff M. R. (2008) PLoS ONE 3, e3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baljuls A., Schmitz W., Mueller T., Zahedi R. P., Sickmann A., Hekman M., Rapp U. R. (2008) J. Biol. Chem. 283, 27239–27254 [DOI] [PubMed] [Google Scholar]
- 74.Pandit B., Sarkozy A., Pennacchio L. A., Carta C., Oishi K., Martinelli S., Pogna E. A., Schackwitz W., Ustaszewska A., Landstrom A., Bos J. M., Ommen S. R., Esposito G., Lepri F., Faul C., Mundel P., López Siguero J. P., Tenconi R., Selicorni A., Rossi C., Mazzanti L., Torrente I., Marino B., Digilio M. C., Zampino G., Ackerman M. J., Dallapiccola B., Tartaglia M., Gelb B. D. (2007) Nat. Genet. 39, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 75.Razzaque M. A., Nishizawa T., Komoike Y., Yagi H., Furutani M., Amo R., Kamisago M., Momma K., Katayama H., Nakagawa M., Fujiwara Y., Matsushima M., Mizuno K., Tokuyama M., Hirota H., Muneuchi J., Higashinakagawa T., Matsuoka R. (2007) Nat. Genet. 39, 1013–1017 [DOI] [PubMed] [Google Scholar]
- 76.Aitken A. (2002) Plant Mol. Biol. 50, 993–1010 [DOI] [PubMed] [Google Scholar]
- 77.Fueller J., Becker M., Sienerth A. R., Fischer A., Hotz C., Galmiche A. (2008) Biochem. Biophys. Res. Commun. 370, 552–556 [DOI] [PubMed] [Google Scholar]
- 78.Fabian M. A., Biggs W. H., 3rd, Treiber D. K., Atteridge C. E., Azimioara M. D., Benedetti M. G., Carter T. A., Ciceri P., Edeen P. T., Floyd M., Ford J. M., Galvin M., Gerlach J. L., Grotzfeld R. M., Herrgard S., Insko D. E., Insko M. A., Lai A. G., Lélias J. M., Mehta S. A., Milanov Z. V., Velasco A. M., Wodicka L. M., Patel H. K., Zarrinkar P. P., Lockhart D. J. (2005) Nat. Biotechnol. 23, 329–336 [DOI] [PubMed] [Google Scholar]
- 79.Wilhelm S. M., Carter C., Tang L., Wilkie D., McNabola A., Rong H., Chen C., Zhang X., Vincent P., McHugh M., Cao Y., Shujath J., Gawlak S., Eveleigh D., Rowley B., Liu L., Adnane L., Lynch M., Auclair D., Taylor I., Gedrich R., Voznesensky A., Riedl B., Post L. E., Bollag G., Trail P. A. (2004) Cancer Res. 64, 7099–7109 [DOI] [PubMed] [Google Scholar]
- 80.Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., Huang D. C. (2005) Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 81.Galmiche A., Fueller J., Santel A., Krohne G., Wittig I., Doye A., Rolando M., Flatau G., Lemichez E., Rapp U. R. (2008) J. Biol. Chem. 283, 14857–14866 [DOI] [PubMed] [Google Scholar]
- 82.Dumaz N., Marais R. (2003) J. Biol. Chem. 278, 29819–29823 [DOI] [PubMed] [Google Scholar]
- 83.She Q. B., Solit D. B., Ye Q., O'Reilly K. E., Lobo J., Rosen N. (2005) Cancer Cell 8, 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Götz R., Wiese S., Takayama S., Camarero G. C., Rossoll W., Schweizer U., Troppmair J., Jablonka S., Holtmann B., Reed J. C., Rapp U. R., Sendtner M. (2005) Nat. Neurosci. 8, 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karbowski M., Lee Y. J., Gaume B., Jeong S. Y., Frank S., Nechushtan A., Santel A., Fuller M., Smith C. L., Youle R. J. (2002) J. Cell Biol. 159, 931–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Montal M., Mueller P. (1972) Proc. Natl. Acad. Sci. U.S.A. 69, 3561–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dejean L. M., Martinez-Caballero S., Kinnally K. W. (2006) Cell Death Differ. 13, 1387–1395 [DOI] [PubMed] [Google Scholar]
- 88.Dlugosz P. J., Billen L. P., Annis M. G., Zhu W., Zhang Z., Lin J., Leber B., Andrews D. W. (2006) EMBO J. 25, 2287–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fleischer A., Ghadiri A., Dessauge F., Duhamel M., Cayla X., Garcia A., Rebollo A. (2004) Mol. Cancer Res. 2, 674–684 [PubMed] [Google Scholar]
- 90.Zoratti M., Szabò I. (1995) Biochim. Biophys. Acta 1241, 139–176 [DOI] [PubMed] [Google Scholar]
- 91.Zamzami N., Marchetti P., Castedo M., Hirsch T., Susin S. A., Masse B., Kroemer G. (1996) FEBS Lett. 384, 53–57 [DOI] [PubMed] [Google Scholar]
- 92.Zamzami N., Susin S. A., Marchetti P., Hirsch T., Gómez-Monterrey I., Castedo M., Kroemer G. (1996) J. Exp. Med. 183, 1533–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roy S. S., Madesh M., Davies E., Antonsson B., Danial N., Hajnóczky G. (2009) Mol. Cell 33, 377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jürgensmeier J. M., Xie Z., Deveraux Q., Ellerby L., Bredesen D., Reed J. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4997–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gottlieb E., Armour S. M., Thompson C. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12801–12806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vander Heiden M. G., Thompson C. B. (1999) Nat. Cell Biol. 1, E209–216 [DOI] [PubMed] [Google Scholar]
- 97.Jonas E. A., Hoit D., Hickman J. A., Brandt T. A., Polster B. M., Fannjiang Y., McCarthy E., Montanez M. K., Hardwick J. M., Kaczmarek L. K. (2003) J. Neurosci. 23, 8423–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jonas E. A., Hickman J. A., Chachar M., Polster B. M., Brandt T. A., Fannjiang Y., Ivanovska I., Basañez G., Kinnally K. W., Zimmerberg J., Hardwick J. M., Kaczmarek L. K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13590–13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jonas E. A., Hickman J. A., Hardwick J. M., Kaczmarek L. K. (2005) J. Biol. Chem. 280, 4491–4497 [DOI] [PubMed] [Google Scholar]
- 100.Frank S., Gaume B., Bergmann-Leitner E. S., Leitner W. W., Robert E. G., Catez F., Smith C. L., Youle R. J. (2001) Dev. Cell 1, 515–525 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.