Abstract
The role of pleiotrophin in fetal lung development was investigated. We found that pleiotrophin and its receptor, protein-tyrosine phosphatase receptor β/ζ, were highly expressed in mesenchymal and epithelial cells of the fetal lungs, respectively. Using isolated fetal alveolar epithelial type II cells, we demonstrated that pleiotrophin promoted fetal type II cell proliferation and arrested type II cell trans-differentiation into alveolar epithelial type I cells. Pleiotrophin also increased wound healing of injured type II cell monolayer. Knockdown of pleiotrophin influenced lung branching morphogenesis in a fetal lung organ culture model. Pleiotrophin increased the tyrosine phosphorylation of β-catenin, promoted β-catenin translocation into the nucleus, and activated T cell factor/lymphoid enhancer factor transcription factors. Dlk1, a membrane ligand that initiates the Notch signaling pathway, was identified as a downstream target of the pleiotrophin/β-catenin pathway by endogenous dlk1 expression, promoter assay, and chromatin immunoprecipitation. These results provide evidence that pleiotrophin regulates fetal type II cell proliferation and differentiation via integration of multiple signaling pathways including pleiotrophin, β-catenin, and Notch pathways.
Fetal lung development is a complex biological process. Rat lung originates from the foregut endoderm as a bifurcation at embryonic day 10 (E10) and undergoes several generations of dichotomous branching morphogenesis to form a respiratory tree thereafter (1). The columnar epithelial cells differentiate into Clara cells and epithelial cells. Although surfactant protein C is detected as early as E13, fetal alveolar epithelial type II cells (fAEC II)3 are not fully differentiated until E18 when glycogen pool-enriched cells become cuboidal and differentiate into cells containing lamellar bodies. fAEC II can further differentiate and give rise to the alveolar epithelial type I cells (AEC I).
The regulation of fetal lung development includes coordinated regulation of molecular pathways as well as reciprocal interactions among mesenchymal cells, epithelial cells, and the extracellular matrix (2). Precise signals from mesenchymal cells regulate lung branching morphogenesis and lead to cell fate determination and the subsequent generation of cell type diversity in the lung epithelium. For example, mesenchymal cells secrete fibroblast growth factor 10 (3, 4). Fibroblast growth factor 10 in turn binds to its receptor, which is located on the surface of epithelial cells. The binding transduces a message to downstream signaling to regulate cell proliferation, differentiation, migration, and branching morphogenesis. Similarly, the epithelial cells also secrete signal molecules to interact with the mesenchyme. Sonic hedgehog (Shh) is growth factor expressed in the developing epithelium. Its receptor, Patched-1 (Ptc), is located on mesenchymal cells. The interaction between Shh and Ptc has been shown to be required for lung bud formation (5). Other growth factors such as platelet-derived growth factor, transforming growth factor-β, epidermal growth factor, and vascular endothelial growth factor also play active roles in mesenchyme-epithelium interactions (2).
Pleiotrophin (PTN), a heparin binding cytokine, is involved in cell transformation, growth, survival, migration, and angiogenesis (6). PTN is localized in the extracellular matrix and mesenchymal cells. It shares 50% sequence homology with midkine. PTN was first identified as a growth factor from the bovine uterus (7) and as a neurite outgrowth promoting factor in the neonatal rat brain (8). PTN expression peaks in the late stage of embryogenesis (9, 10) and postnatally in the nervous system (11, 12). PTN is also up-regulated in injured cells and is important for new tissue formation during recovery from injury (13). PTN levels are much lower in adult tissue than that in fetal tissue; however, PTN is overexpressed in a number of cancers including human breast cancers (14) and melanocytic tumors (15).
PTN signals through the cell surface receptor, protein-tyrosine phosphatase receptor (RPTP β/ζ). In U373-MG glioblastoma cells, the binding of PTN with RPTP β/ζ results in the dimerization and inactivation of the receptor, leading to an increase in tyrosine phosphorylation of β-catenin (16). β-Catenin is a transcription co-factor and an adaptor protein linking cadherins to the cytoskeleton. Because β-catenin is the central component of the Wnt signaling pathway, it may cross-talk between PTN and Wnt signaling pathways. PTN may also act via the receptor-tyrosine kinase, anaplastic lymphoma kinase receptor (17).
There is little information available regarding PTN in the lung. PTN is expressed in fetal lungs (10) and lung cancer cell lines (18). Our DNA microarray analysis of fetal lungs reveals that ptn is highly expressed during the late stage of fetal lung development (19). This is confirmed by real-time PCR and Western blot. However, whether PTN has a role in fetal lung development is unknown.
In this study we first examined the expression pattern of PTN and its receptor, RPTP β/ζ, during fetal lung development. Using primary fAEC II, we evaluated the role of PTN in cell proliferation, differentiation, and wound healing. We also determined the effects of PTN on fetal lung branching morphogenesis by knocking down the PTN expression with adenovirus-mediated small interfering RNA (siRNA). Finally, we dissected the signaling pathways initiated by PTN. Our results show that PTN regulates fetal lung epithelial cell proliferation and differentiation via β-catenin signaling pathway.
MATERIALS AND METHODS
Chemicals and Reagents
TRI reagents were purchased from Molecular Research Center, Inc. (Cincinnati, OH). Oligo(dT), random hexamers, and pGL3 firefly luciferase reporter vector were from Promega (Madison, WI). RNase-freee DNase was from Ambion (Austin, TX). Moloney murine leukemia virus reverse transcriptase, pENTR plasmid, LipofectamineTM 2000, minimal essential medium (MEM), and BGJb medium were from Invitrogen. SYBR Green I detection reagents were from Qiagen (Valencia, CA). NE-PER nuclear and cytoplasmic extraction reagents and M-PER mammalian protein extraction reagents were from Pierce. Protein A- or G-agarose beads were from Santa Cruz Biotechnology (Santa Cruz, CA). Matrigel, anti-RPTP β/ζ, and anti-β-catenin monoclonal antibodies were from BD Bioscience. Advantage 2 Polymerase Mix and Advantage GC Genomic Polymerase Mix were from Clontech (Mountain View, CA). EZ ChIP kit and anti-phosphotyrosine antibodies were from Upstate Biotechnology (Lake Placid, NY). Human recombinant pleiotrophin, collagenase, trypsin, and DNase I were from Sigma-Aldrich. Goat anti-pleiotrophin, rabbit anti-Dlk1, and mouse anti-bromodeoxyuridine (BrdUrd) antibodies were from Abcam (Cambridge, MA). Mouse anti-LB180 antibody was from Covance (Berkeley, CA). Alexa 568-conjugated anti-goat and Alexa 488-conjugated anti-mouse antibodies were from Molecular Probes (Eugene, OR). ABC reagents were from Vector Laboratories (Burlingame, CA). The TSATM-plus horseradish peroxidase system was from PerkinElmer Life Sciences. L2 cell line was from ATCC (Manassas, VA).
Real-time PCR
Total RNAs were isolated using TRI reagents and treated with RNase-freeTM DNase to remove DNA contamination. 1 μg of total RNAs was reverse-transcribed into cDNA using oligo(dT) and random hexamers and Moloney murine leukemia virus reverse transcriptase. The primers were designed using Primer Express® software (Applied Biosystems, Foster City, CA): PTN, 5′-ATACCAGCAGCAACGTCGAAA (forward) and 5′-GCACACACTCCATTGCCATT (reward); Dlk1, 5′-GTGAAGAACCATGGCAGTGTGT (forward) and 5′-GACAGTCCTTTCCAGAGAATCCA (reward); 18 S rRNA 5′-TCCCAGTAAGTGCGGGTCATA (forward) and 5′-CGAGGGCCTCACTAAACCATC (reward). Real-time PCR was performed on an ABI 7500 system using SYBR Green I detection as previously described (19). To eliminate the effects of primer dimers, we included a data acquisition step at a temperature of 2∼5 °C lower than the annealing temperature of the products. After the amplification, a dissociation curve was generated to check the specificity of the amplification. The data were normalized to 18 S rRNA.
Immunohistochemistry
Fetal lung tissues were fixed in 4% formaldehyde for 24 h, embedded in paraffin, and sectioned at a 4-μm thickness. The slides were dewaxed, rehydrated, boiled in citric buffer (pH 6.0) for 20 min for antigen retrieval, and permeabilized by a 10-min incubation in 0.3% Triton X-100. The slides were blocked in 10% normal horse serum and then incubated with goat anti-PTN (1:100) or mouse anti-RPTP β/ζ (1:200) at 4 °C overnight. After being rinsed with phosphate-buffered saline (PBS, pH 7.4), the slides were further incubated in corresponding secondary antibodies (1:100) for 1 h. The slides were rinsed again and incubated in ABC reagents for 30 min. The signal was amplified using the TSATM-plus horseradish peroxidase system. Signal was detected using diaminobenzidine substrate. The slides were counterstained by hematoxylin and viewed under a Nikon Eclipse E600 microscope with a Plan Fluor, ELWD 40×/0.6 or 20×/0.5 objective lenses. The images were captured with an Insight 2 color mosaic camera and the basic spot software (Diagnostic Instruments, Inc.).
Immunocytochemistry
fAEC II were fixed in 4% formaldehyde for 15 min followed by a 10-min incubation in 0.3% Triton X-100 for permeabilization. After being blocked in 10% fetal bovine serum (FBS) for 30 min, the cells were incubated with anti-LB180 (1:200) or anti-T1-α (1:250) antibodies at 4 °C overnight. The cells were rinsed with PBS and then incubated with Alexa 488-conjugated anti-mouse antibodies (1:250) for 1 h. Finally, cells were washed and viewed by a Nikon Eclipse TE2000-U inverted fluorescence microscope with a Plan Fluor, ELWD 40 ×/0.6 or 20 ×/0.5 objective lens. The images were captured with a Photometrics CoolSnap CCD camera (Roper Scientific) and the Metavue software (Universal Imaging Co.).
fAEC II Isolation and Culture
fAEC II were isolated as described (20). The Oklahoma State University Animal use and Care Committee approved all the procedures used in this study. Timed-pregnant Sprague-Dawley rats with a gestational day of 20 were sacrificed by carbon dioxide asphyxia. Fetuses were carefully separated from the uterus. Fetal lungs were excised from the fetuses and washed with cold Hanks' balanced salt solution. After removing the surrounding trachea, thymus, and heart, the lungs were chopped into 1-mm3 cubes and digested with MEM containing 1 mg/ml collagenase, 1 mg/ml trypsin, and 0.4 mg/ml DNase I for 10 min for 4 times. Released cells were filtered through 160- and 37-μm nylon gauze filters. The cells were then collected by centrifugation at 160 × g for 10 min and resuspended in MEM containing 20% FBS, 100 units/ml penicillin, and 10 μg/ml streptomycin at a final concentration of 1.7 × 106 cells/ml. To remove the fibroblasts, 40 ml of the cells were seeded in a 20-cm plastic dish for differential adhesion. After 4 rounds of a 45-min adherence, the cells were filtered through a 15-μm nylon gauze filter, centrifuged, and seeded on the plate for further culture. The resulting fAEC II had a purity of ∼90% and a viability of >95%.
For the trans-differentiation experiment, primary fAEC II were cultured in full MEM medium (FMEM: MEM supplemented with 10% FBS, 1% nonessential amino acids, and 1% penicillin and streptomycin) for 24 h. Media were changed to chemical defined serum-free medium (21) (DSFM, Dulbecco's modified Eagle's medium/F-12 1:1 supplemented with 1.25 mg/ml bovine serum albumin, 1% nonessential amino acids, and 1% penicillin and streptomycin) without or with 50 ng/ml human recombinant PTN. The cells were cultured for 4 more days, and media were changed every 2 days. Finally, the cells were fixed and used for immunofluorescence.
Wound Healing Assay
Primary fAEC II were cultured overnight in plastic dishes in FMEM. Culture media were changed to DSFM in the presence of different doses of PTN. Culture continued until a monolayer was formed (normally on day 2). The cell monolayer were then scratched with a yellow tip to mimic injury, and the gaps were photographed every 6 h. The wound width was measured, and the wound closure was defined as the wound width at a specific time minus the distance of the original wound width.
For the co-culture experiment, primary fibroblasts were collected during the differential adhesion stage when isolating fAEC II. 0.5 × 106 of fAEC II were seeded outside of a collagen-coated insert with 0.2-μm pore size, and fibroblasts (0.5 × 106) were seeded inside the insert. Both cells were cultured in FMEM. The insert was placed in each well of a 24-well plate. After 24 h the media were changed to DSFM, and the fAEC II monolayer was scratched for the wound healing assay.
Cell Proliferation Assay
Primary fAEC II were cultured in FMEM for 24 h. Then the media were changed to DSFM without or with 50 ng/ml PTN. The cells were cultured for additional 36 h. BrdUrd (10 μm) was added to the medium 12 h before collecting the cells. Finally, the cells were fixed in 4% paraformaldehyde and stained with anti-BrdUrd antibodies (1:100).
Western Blot
Cells were lysed in the M-PER mammalian protein extraction reagent with protease inhibitor mixture on ice for 10 min. The lysate was centrifuged at 12,000 × g for 10 min to clear the cell debris. Protein concentrations in the supernatant were determined the using the DC protein assay kit. The same amount of total protein was separated by 10% SDS-PAGE and then transferred onto a nitrocellulose membrane. The membrane was blocked with 5% dry milk in Tris-buffered saline for 1 h. Then the membrane was incubated with primary antibodies (anti-RPTP β/ζ, 1:500; anti-β-catenin, 1:1000; anti-γ-tubulin, 1:1000; anti-phosphotyrosine, 1:1000) overnight at 4 °C. After being washed with Tris-buffered saline containing 0.05% Tween 20, the membrane was incubated with corresponding secondary antibodies (1:2000) for 1 h. Finally, the membrane was developed with ECL reagents and exposed to x-ray film.
Construction of siRNA Adenoviral Vector
A highly efficient siRNA vector expressing four short hairpin RNAs under the control of mU6, hU6, H1, and 7SK promoters was previously developed in our laboratory (22). Four siRNA sequences specific to the coding region of PTN (549–569, 570–590, 607–627, and 909–929) were selected based on siRNA Design Software (SDS). A vector containing four unrelated siRNA sequences was used as a control. These siRNA sequences were cloned into a modified pENTR plasmid containing a cytomegalovirus-driven enhanced GFP protein for tracking the virus transduction. The siRNA and enhanced GFP sequences were transferred from the pENTR recombinant vector to the adenoviral vector, pAd/PL-DEST vector, by an LR recombination reaction using the Gateway technology. The plasmid from the positive clone was digested with PacI and transfected into a 293A producer cell line with the LipofectamineTM 2000 reagent. The adenovirus-containing cells were harvested, and the generated virus was used to infect 293A cells to amplify the viral stock. The titer was determined by counting the GFP-positive cells.
Fetal Lung Organ Culture
Timed-pregnant rats with a gestational day 14 (E14) were killed by CO2 asphyxia. Embryos were carefully removed from the uterus and placed in cold Hanks' balanced salt solution. Fetal lungs were isolated from the fetus by microdissection needles. Two or three lungs were kept on a 3-cm insert, which were placed in a 6-well plate with 1.5 ml of BGJb supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.2 mg/ml ascorbic acid. The fetal lungs were cultured for 1–3 days. The viruses were added to the lungs inside of the insert at day 0, moved outside of the insert after a 3-h treatment, and removed after 24 h. Medium was changed every day. The fetal lungs were photographed every 24 h to record the morphological changes. To determine the changes in lung-branching morphogenesis, the terminal buds of each lung were counted and normalized to those at day 0 of the same lung. The sizes of terminal and inside buds were determined with MetaviewTM (Universal Imaging Corp.). About 30 buds were evaluated for each lung.
Tyrosine Phosphorylation of β-Catenin
Primary fAEC II were cultured in FMEM for 1 day and then in DSFM for an additional day. The cells were treated with 50 ng/ml PTN for 0, 5, 10, 20, 60, or 120 min and lysed in the M-PER mammalian protein extraction reagent (supplemented with 1% Halt phosphatase inhibitor mixture and 1% protease inhibitor mixture and 1 mm sodium orthovanadate). The cell lysate (250 μg of protein) were incubated with anti-β-catenin antibodies (2 μg) at 4 °C for 2 h. 10 μl of protein A and 20 μl of protein G-agarose beads were added to the mixtures and incubated overnight at 4 °C by gentle end-to-end mixing. The agarose beads were washed six times with PBS. The proteins were eluted by boiling in 1× SDS sample buffer for detection of phosphorylation and β-catenin by Western blotting.
Translocation of β-Catenin
Primary fAEC II were cultured in FMEM for 24 h and in DSFM for another 24 h. The cells were treated with 50 or 100 ng/ml PTN for 12 h. The nuclear and cytoplasmic fractions were separated using the NE-PER nuclear and cytoplasmic extraction reagents. The amounts of β-catenin in the cytoplasmic and nuclear fractions were determined using Western blot.
fAEC II Culture on Matrigel
Matrigel was thawed fully at 4 °C overnight and kept on ice until use. 200 μl of Matrigel was added to each well of 12-well plate and incubated for 1 h at room temperature to solidification. Primary fAEC II was seeded at a density of 5 × 105 cells/well in FMEM. After 24 h media were changed to DSFM, and the cells were treated with various concentrations of Wnt3a or LiCl for 48 h. The cells were collected in 1 ml of Tri-reagent for isolation of total RNA.
TOPflash Assay
Freshly isolated fAEC II (5 × 106) were transfected with phRL-TK (1.14 μg) and TOPflash or FOPflash (1.3 μg) by electroporation using the Amaxa NHBE Nucleofector kit for normal human bronchial epithelial cells and the Program T-13. The cells (2.5 × 106/well) were seeded in a 48-well plate and cultured in 10% FBS-Dulbecco's modified Eagle's medium for 24 h. The fAEC II were then treated with 100 ng/ml PTN for 24 h and harvested for a dual-luciferase assay.
Dlk1 Promoter Assay
The Dlk1 promoter (−1471 to 190) was amplified from mouse genomic DNA by overlap PCR. Three sets of primers were designed to amply Dlk1 promoter from mouse genomic DNA. The first fragment, A (631 bp) ranging from −1471 to −848, was amplified by Advantage 2 Polymerase Mix with the primers (5′-CGTGCTAGCCAAAGGAGCTATGTCAATGAC-3′ and 5′-AGCACTGATTCTTTCCAACATG-3′). The second fragment, B (624 bp) ranging from −894 to −271, and the third fragment, C (503 bp) ranging from −305 to 190, were amplified by Advantage GC Genomic Polymerase Mix with the primer sets of 5′-CATACGTGTTGCTGCGAGGTGTGTACATGTT-3′ and 5′-GGCCCGCTTAGCGCAAGTCTCAGGAACCAA-3′ and of 5′-CTGGCTTGGTTCCTGAGACTTGCGCTAA-3′ and 5′-GCCAAGCTTGGAGGGCTCCGGTCGCGATCATCTC-3′). Using the gel-purified three Dlk1 fragments mixture as template, the full-length Dlk1 promoter was obtained by overlap PCR with two outside primers (5′-CGTGCTAGCCAAAGGAGCTATGTCAATGAC-3′ and 5′-GCCAAGCTTGGAGGGCTCCGGTCGCGATCATCTC-3′). The purified PCR products were digested by Nhe1 and Hind3 and cloned into the pGL3 firefly luciferase reporter vector to replace the SV40 promoter through Nhe1-Hind3 sites. The amplified Dlk1 promoter was verified by DNA sequencing. The Dlk1 promoter-driven firefly luciferase vector was named as pDlk1-Fluc.
Freshly isolated fAEC II (4 × 106) was transfected with 1 μg of pDlk1-Fluc, 1.14 μg of Renilla luciferase vector phRL-TK, 100 or 500 ng of wild type β-catenin, or constructively activated mutant ΔGSK β-catenin using the Amexa Nucleofector kit for normal human bronchial epithelial cells and the Program T-13. The cells (2 × 106/well) were seeded into a 48-well plate and cultured in 10% FBS-Dulbecco's modified Eagle's medium for 24 h. The medium was changed with fresh 10% FBS-Dulbecco's modified Eagle's medium with or without PTN (100 ng/ml), and the cells were harvested for dual luciferase assay after a 24-h treatment.
Chromatin Immunoprecipitation (ChIP)
The EZ ChIP kit was used for the CHIP assay. fAEC II were fixed in 1% formaldehyde in PBS for 10 min at room temperature. Then glycine was added at a final concentration of 0.125 m to stop the cross-linking. 6 × 106 cells were washed by cold PBS 2 times and then sonicated in 300 μl of SDS lysis buffer (Branson Sonifier 450, Output power 7, 20 s for 20 times). For each sample chromatin from 1.5 × 106 cells was immunoprecipitated with 2 μg of normal mouse IgG or mouse monoclonal antibody for β-catenin as described in the kit manual. After reversal of the cross-linking by adding 200 mm NaCl and incubating at 65 °C overnight, 2 μl of purified immuno-selected DNAs were used for PCR (32 cycles, 94 °C, 30 s; 56 °C, 30 s; 72 °C, 30 s). 5 μl of the reaction products were run on 1.5% agarose gel. Two pairs of primers were used to amplify two putative TCF/LEF binding sites and the following flanking sequences: Dlk1-BS1, forward, 5′-TGGAGATTAAATTCAAGCTGTCAG-3′, and reverse, 5′-GCAGCCAAACTTGAGTTTGATC-3′; Dlk1-BS2, forward, 5′-CATTTGACRGTGAACATATTGG-3′ (R = A/G), and reverse, 5′-GCCCAGWCCCCAAATCTGTC-3′ (W = A/T). The degenerated primers were designed to amplify both rat and mouse Dlk1 promoter sequences. CCDN1 promoter fragment (−173 to +117) was amplified as a positive control for TCF/LEF binding using a primer pair: forward, 5′-TTCTCTGCCCGGCTTTGAT-3′, and reverse, 5′-CACAGGAGCTGGTGTTCCATG-3′. Glyceraldehyde-3-phosphate dehydrogenase promoter, which has no TCF/LEF binding site, served as a negative control and was amplified using a primer pair: forward, 5′-GTGCAAAAGACCCTGAACAATG-3′, and reverse, 5′-GAAGCTATTCTAGTCTGATAACCTCC-3′.
RESULTS
PTN and RPTP β/ζ Expression in the Developing Lung
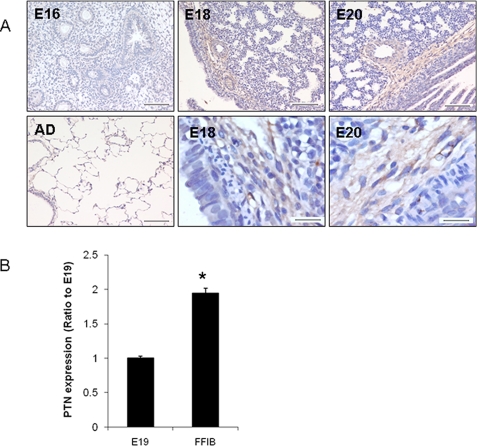
We have previously shown that PTN mRNA and protein are highly expressed in the late stage of fetal lung development by analyzing fetal lungs at different gestational days as well as the lungs of newborn and adult rats using DNA microarray analysis, real-time PCR, and Western blots (19). We further investigated temporal and spatial expression of PTN during rat lung development by ABC immunostaining. PTN was barely detectable in the embryonic day (E16) lungs (Fig. 1A). A strong staining in the mesenchyme adjacent to the developing epithelium and endothelium of the E18 and E20 lungs was seen. Little PTN signals were observed in adult lungs. No positive signals were found in the negative controls without primary antibodies (data not shown). Real-time PCR revealed that PTN was enriched in fetal lung fibroblasts in comparison with fetal lung tissues (Fig. 1B).
FIGURE 1.
PTN expression during fetal lung development. A, immunostaining of PTN in the developing lungs. Embryonic days 16, 18, and 20 (E16, E18, E20) and adult (AD) lung tissue sections were stained using anti-PTN antibodies and the Vectastain ABC kit. Scale, 100 μm. The lower right two panels are high magnifications (scale bar, 20 μm). B, real-time PCR analysis of PTN mRNA expression in E19 fetal lung tissue (E19) and isolated E19 fetal fibroblasts (FFIB). The results were normalized to 18 S rRNA and expressed as a ratio of lung tissue. Data shown are the means ± S.E.; *, p < 0.05 versus lung tissue (n = 3 cell preparations, assayed in duplicate).
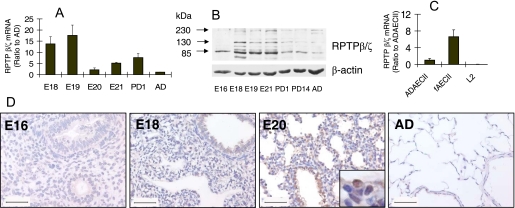
We also examined the expression of RPTP β/ζ, one of the PTN receptors, during fetal lung development. As shown by real-time PCR, RPTP β/ζ mRNA was highly expressed in E18 and E19 fetal lungs (Fig. 2A). Western blot revealed 3 bands with molecular masses of 230, 130, and 85 kDa (Fig. 2B), which correspond to 3 alternatively spliced species of RPTP β/ζ (16). The RPTP β/ζ protein was highly expressed in E18–21 fetal lungs but was relatively low in postnatal day 1, day 14, and adult lung tissues. A high expression of RPTP β/ζ protein in the E21 lungs in comparison with mRNA may be because of its stability. The time-restricted RPTP β/ζ expression corresponds with the PTN expression, indicating that the PTN pathway may be active during the late stage of fetal lung development. To further identify which types of cells express RPTP β/ζ, real-time PCR was used to examine the RPTP β/ζ mRNA expression levels in fAEC II, adult type II cells (ADAEC II), and L2 cells (a rat lung epithelial cell line derived from the adult rat lung). RPTP β/ζ expressed highly in fAEC II but lowly in ADAEC II (Fig. 2C). L2 had no expression of the receptor. RPTP β/ζ cellular localization was further viewed using ABC staining in E16, E18, E20 and adult lung tissues. As shown in Fig. 2D, RPTP β/ζ staining was strong in E18 and E20 lungs but was not detectable in E16 or adult lungs. Consistent with the real-time PCR results, RPTP β/ζ was localized in airway epithelial cells.
FIGURE 2.
RPTP β/ζ expression during fetal lung development. A, mRNA levels of RPTP β/ζ in lung tissues. B, protein levels of RPTP β/ζ in lung tissues. C, mRNA levels of RPTP β/ζ in various cells. mRNA and protein levels were determined by real-time PCR and Western blot. E18-E21, fetal lungs; PD1 and PD14: postnatal day 1 and 14 lungs; AD, adult lungs; ADAEC II, adult type II cells; fAEC II, E20 primary fetal type II cells; L2, a cell line derived from adult rat lungs. The mRNA levels were normalized to 18 S rRNA and expressed as a percentage of AD or ADAEC II. Data shown are the means ± S.E. (n = 6). D, immunostaining of E16, -18, -20, and AD lungs with anti-RPTP β/ζ antibodies. Scale bar, 50 μm.
PTN Arrests fAEC II Trans-differentiation into AEC I
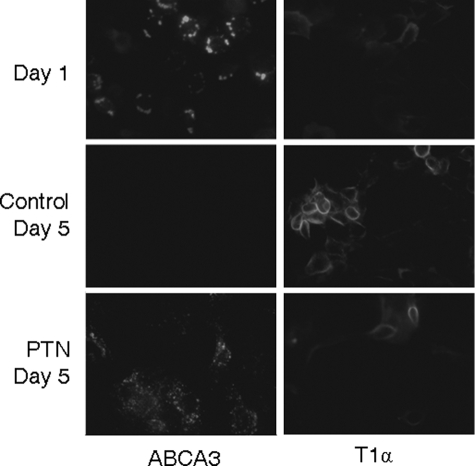
When AEC II were cultured on plastic dishes, the cells gradually lost the AEC II phenotype and trans-differentiated into AEC I. Using this in vitro system, we examined the effect of PTN on fAEC II trans-differentiation. Primary fAEC II were cultured on plastic dishes overnight and then treated with 50 ng/ml PTN. After being cultured for four more days, AEC II cells were stained with antibodies against ABCA3 (AEC II-specific marker) and T1α (AEC I-specific marker). At day 1 the cells had strong ABCA3 staining but weak T1α staining (Fig. 3). After day 5 of culture, the control cells had no ABCA3 staining and a clear T1α staining, indicating that fAEC II trans-differentiated into AEC I. However, in the PTN-treated fAEC II, strong ABCA3 staining still remained. The results suggest that PTN can arrest the fAEC II trans-differentiation into AEC I.
FIGURE 3.
Effect of PTN on fAEC II trans-differentiation. Primary fAEC II were cultured for 1 or 5 days on plastics. PTN (50 ng/ml) was added at day 1. ABCA3 (type II cell marker) and T1α (Type I cell marker) were stained by immunofluorescence.
PTN Increases fAEC II Proliferation
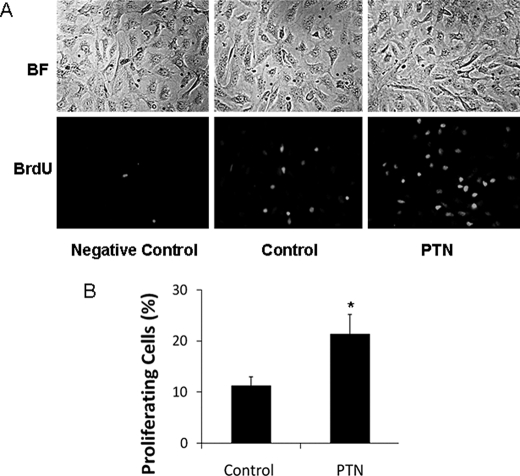
To determine whether PTN can promote fAEC II proliferation, primary fAEC II were treated with PTN and labeled with BrdUrd. The cells were then fixed and stained with anti-BrdUrd antibodies to view the proliferating cells. As shown in Fig. 4, the PTN-treated cells contained about twice as many proliferating cells as the control cells without any treatment.
FIGURE 4.
PTN stimulates fAEC II proliferation. Primary fAEC II were cultured in serum-free medium on plastic dishes. The cells were treated with 50 ng/ml PTN at day 1. After 36 h, the cells were collected and stained with anti-BrdUrd (BrdU) antibodies to view the proliferating cells. A, representative images. BF, brightfield. B, quantitation. The proliferating cells (BrdUrd-positive cells) were expressed as a percentage of total cells (means ± S.E.). *, p < 0.05 versus control, n = 5.
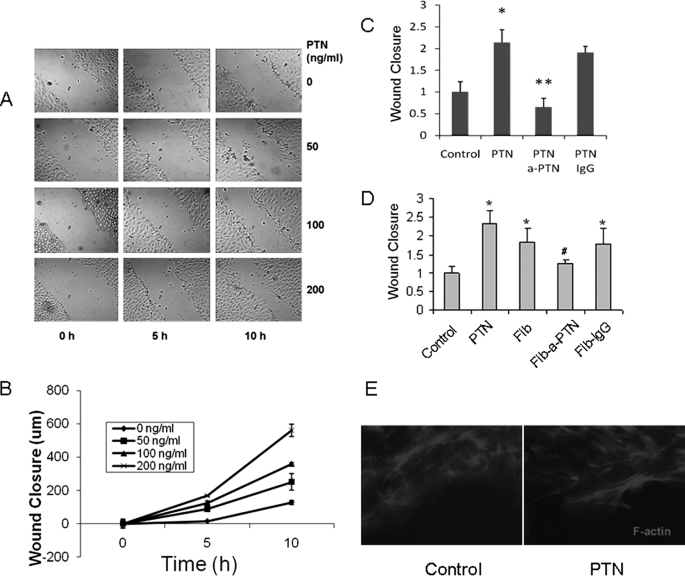
PTN Promotes Wound Healing
We determined whether PTN enhances alveolar epithelial cell repair. fAEC II were cultured in serum-free medium with PTN. After washing, a linear wound was created using a pipette tip, and the bright field images were taken at various time points. PTN significantly increased the wound closure of fAEC II in a dose-dependent manner (Fig. 5A). The wound closure was further quantified as the gap width at 0 h minus the gap width at late time points (Fig. 5B). When anti-PTN antibody was added together with PTN, the wound closure induced by PTN was blocked (Fig. 5C). The control IgG had no effects. Because PTN was enriched in fetal lung fibroblasts, we further examined whether it was the PTN secreted from the fibroblasts that are responsible for fAEC II repair. fAEC II were co-cultured in a 6-well plate with fetal lung fibroblasts in a Transwell insert placed in the plate, and thus, fibroblasts may secrete growth factors into the AEC II culture medium. Fibroblasts significantly promoted the fAEC II wound closure. This effect was again blocked by PTN antibodies but not the control IgG. These results indicate that PTN is secreted from fibroblasts and acts on fAEC II to promote wound healing.
FIGURE 5.
PTN promotes wound healing of fAEC II monolayer. A, fAEC II were isolated from E20 lungs and cultured for 2 days. Different doses of PTN were added at day 1. On day 2 fAEC II monolayers were scratched, and the generated gaps were photographed every 5 h. B, wound closure was measured as the difference between the original wound width and the final wound width. C, effect of anti-PTN antibodies. PTN (100 ng/ml) was added at day 1 with or without anti-PTN (a-PTN) antibodies or IgG (2 μg/ml). Wound closure was measured 16 h after the scratching and expressed as a percentage of control. D, fAEC II were co-cultured with fibroblasts on Transwell inserts. Anti-PTN antibodies or IgG (2 μg/ml) were added at day 1. fAEC II cultured in the insert alone in the absence (control) or presence of PTN (100 ng/ml) were used as negative and positive controls. Wound closure was measured 16 h after the scratching. Fib, fibroblasts; Fib-a-PTN, fibroblasts plus anti-PTN antibodies; Fib-IgG, fibroblasts plus IgG. All data shown are the means ± S.E. *, p < 0.05 versus control; **, p < 0.05 versus PTN; #, p < 0.05 versus fibroblasts, n = 3 independent cell preparations. E, F-actin staining. The control and PTN-treated fAEC II at the end of wound healing assay were fixed in 4% formaldehyde for 30 min, permeabilized in 1% Triton X-100 for 10 min, and blocked in 10% FBS and 1% bovine serum albumin for 15 min. The cells were then incubated with 1 μm Alexa Fluor 568 phalloidin for 1.5 h at 4 °C. After washing, the fluorescence images were viewed under a Nikon inverted fluorescence microscope.
Previously, PTN was reported to be able to stimulate the epithelial-mesenchymal-like transition by altering actin cytoskeleton organization (23). We examined whether PTN influences F-actin distribution. fAEC II treated with PTN were stained with Alexa Fluor 568-tagged phalloidin. The cells that were away from the scratched gap did not show much difference in stimulated and nonstimulated cells (data not shown). However, the PTN-treated cells on the edge of the wound gap had sharper and longer F-actin staining compared with untreated control cells (Fig. 5E). The redistribution of the actin cytoskeleton would allow the cells to migrate.
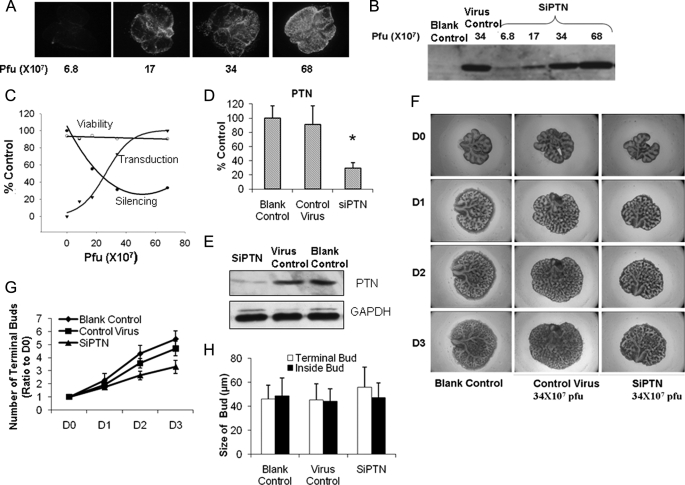
PTN Promotes Fetal Lung Branching Morphogenesis
Because PTN has been shown to play an important role in renal branching morphogenesis (24), we sought to determine whether PTN plays a role in fetal lung branching morphogenesis. We silenced PTN in a fetal lung organ culture by using the pK4-shRNA vector (22) and examined the effects of the treatment on lung branching morphogenesis. An adenoviral vector carrying four siRNAs targeted to the different regions of rat PTN was constructed to knock down PTN, and another vector with four non-relevant siRNAs was used as a control. The vectors contained cytomegalovirus promoter-driven GFP for monitoring transduction efficiency. The silencing efficiency of the PTN adenovirus was more than 90%, as tested in fetal lung fibroblasts (data not shown). E14 fetal lungs were cultured for 3 days on 30-mm Millicell inserts (Millipore) placed in 6-well plates with 1.5 ml of BGJb medium in each well in the presence of various doses of viruses. The transduction efficiency gradually increased as a function of virus dose as seen by GFP fluorescence (Fig. 6A). Western blot analysis also revealed an increase in GFP expression with increasing amounts of viruses (Fig. 6B). The quantitation of transduction efficiency as GFP expression level was shown in Fig. 6C. The silencing efficiency increased as measured by PTN mRNA levels when the dose increased and reached a maximum level at 30 × 107 plaque-forming units (Fig. 6, C and D). The PTN protein level was also decreased by the PTN siRNA (Fig. 6E). The control virus had no effect on PTN mRNA and protein level expression (Fig. 6, D and E). Cell viability was not significantly changed as determined by measuring lactate dehydrogenase release (Fig. 6C). In lungs infected with siRNA adenovirus, the counts of terminal buds decreased (Fig. 6, F and G). The control virus had no significant effects on terminal bud numbers. There were no significant differences on the sizes of terminal buds on the surface and inside of the lungs between siRNA-treated and control virus-treated or nonvirus-treated groups (Fig. 6H).
FIGURE 6.
Effect of PTN on fetal lung branching morphogenesis. A–C, E14 fetal lungs were isolated and treated with various doses of a PTN siRNA adenovirus (SIPTN) or a virus control. GFP fluorescence was taken at day 3 (A), and fetal lung tissues were analyzed by Western blot using anti-GFP antibodies (B). Pfu, plaque-forming units. The transduction efficiency, silencing efficiency, and cell viability were monitored and expressed as a percentage of untreated lungs or a maximal value (C). Transduction efficiency was measured by determining GFP expression levels. Silencing efficiency was measured by determining ptn mRNA levels using real-time PCR. mRNA levels were normalized to 18 S rRNA. Viability was measured by determining lactate dehydrogenase release. D–H, E14 fetal lungs were treated with a PTN siRNA adenovirus or a virus control (34 × 107 plaque-forming units) for 3 days. PTN mRNA (D) and protein (E) levels were determined by real-time PCR and Western blot. Photographs were taken at day 0 to day 3 (D0–D3) (F). Terminal bud numbers (G) were counted and normalized to day 0. The sizes of terminal buds on surface and inside of the lungs were also measured at day 3 (H). Data shown are the means ± S.E.; n = 6 from 3 different pregnant rats.
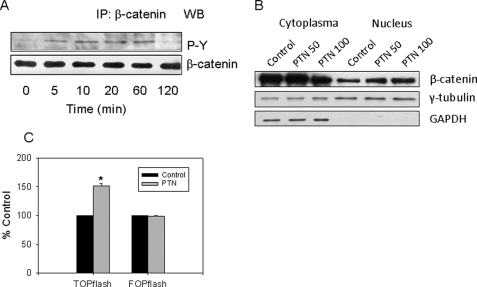
PTN Signals through the RPTP β/ζ and β-Catenin Pathway
We hypothesized that the binding of PTN to the RPTP β/ζ receptor would lead to its inactivation and an increase in β-catenin tyrosine phosphorylation, causing the translocation of β-catenin into nuclei. To examine whether PTN stimulates tyrosine phosphorylation of β-catenin, fAEC II were serum-starved for 24 h and incubated with PTN (50 ng/ml) for 0–120 min. At the end of incubation, the cells were lysed and immunoprecipitated with anti-β-catenin antibodies followed by Western blots using anti-phosphotyrosine antibodies. As shown in Fig. 7A, PTN increased tyrosine phosphorylation of β-catenin in a time-dependent manner with a peak at 10–60 min. The tyrosine phosphorylation of β-catenin decreased after a 2-h treatment.
FIGURE 7.
PTN initiates signaling pathway through RPTP β/ζ and β-catenin. A, tyrosine phosphorylation. fAEC II were serum-starved overnight and stimulated with 50 ng/ml PTN for 0–120 min. β-Catenin was immunoprecipitated (IP) with anti-β-catenin antibodies and probed by Western blot (WB) with anti-phosphotyrosine (P-Y). B, β-catenin translocation. fAEC II were cultured on plastic dishes for 2 days and then treated with 50 or 100 ng/ml PTN for 6 h in serum-free medium. β-Catenin amounts in the cytoplasm and nucleus were examined using Western blot. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and γ-tubulin were used as cytoplasmic and nuclear markers as well as loading controls. C, TOPflash assay for TCF/LEF activation. fAEC II were transfected with TOPflash or FOPflash and phRL-TK Renilla luciferase and then treated with PTN (100 ng/ml) for an additional 24 h. The firefly and Renilla luciferase activities were determined. The results were expressed as a percentage of control (untreated cells). Data shown are the means ± S.E. *, p < 0.05 versus control, n = 9, 3 cell preparations assayed in triplicate.
We next examined whether PTN can cause β-catenin translocation into the nuclei. fAEC II were treated with PTN for 6 h. The cells were then collected, and the nuclei were isolated. As shown in Fig. 7B, PTN increased the nuclear β-catenin content.
To further examine whether the cellular translocation of β-catenin subsequently activates TCF/LEF transcription factors, we used a reporter gene TOPflash assay to determine the activity of TCF/LEF transcription factors. The TOPflash plasmid contains multiple copies of the TCF/LEF binding site and a thymidine kinase minimal promoter-driven firefly luciferase. We transfected primary fAEC II with the Amaxa Nucleofector because of its high transfection efficiency on primary cells. We achieved >60% transfection efficiency using the NHBE Nucleofector kit for normal human bronchial epithelial cells and the Program T-13 as tested by a cytomegalovirus-GFP vector. As shown in Fig. 7C, PTN increased the luciferase activity. However, PTN had little effect on FOPflash, a negative control that has a mutated TCF site in the TOPflash. The results suggest that PTN activates TCF/LEF transcription factors.
Dlk1 Is a Downstream Target of PTN Signaling
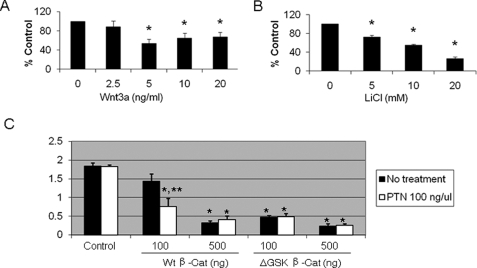
β-Catenin/TCF can be activated by LiCl or Wnt ligand. We determined whether LiCl or Wnt3a affects Dlk1 expression in primary fAEC II. E20 fAEC II were cultured on Matrigel for 24 h and treated with Wnt3a (0–20 ng/ml) or LiCl (0–20 mm) for 2 days. As shown in Fig. 8, both LiCl and Wnt3a decreased Dlk1 mRNA expression. The maximal inhibition was 44 and 73% for Wnt3a and LiCl, respectively.
FIGURE 8.
Effects of the activation of β-catenin pathway on Dlk1 expression. fAEC II were cultured on Matrigel with various concentrations of Wnt3a (A) or LiCl (B). dlk1 mRNA level was determined by real-time PCR and normalized to 18 S rRNA. Data shown are the means ± S.E.; *, p < 0.05 versus untreated control cells (0), n = 6, assayed in duplicate from 3 independent cell preparations. C, effect of β-catenin on Dlk1 promoter activity. fAEC II (3 × 106) were co-transfected with pDlk1-FLluc, phRL-TK, and wild type (Wt) β-catenin or ΔGSK β-catenin mutant by the Amaxa Nucleofector. After a 24-h culture, the cells were incubated with PTN for 24 h. The Dlk1 promoter activity was expressed as a ratio of Firefly over Renilla luciferase activities. Data shown are the means ± S.E. *, p < 0.05 versus control (without wt β-catenin and PTN treatment); **, p < 0.05 versus minus PTN and 100 ng of β-catenin; n = 3 independent cell preparations.
We further examined whether β-catenin affected Dlk1 promoter activity. We co-transfected freshly isolated E20 fAEC II with pDlk1-FLluc, phRL-TK Renilla luciferase, and wild type β-catenin. After overnight culture the cells were stimulated with PTN (100 ng/ml). As shown in Fig. 8C, co-transfection of β-catenin decreased Dlk1 promoter activity in a dose-dependent manner. The addition of PTN further reduced the Dlk1 promoter activity. ΔGSK-β-catenin is a constitutively active form of β-catenin which has four point mutations in the GSK-3β phosphorylation sites of β-catenin (Ser/Thr → Ala) and, thus, cannot be phosphorylated by GSK-3β (25). ΔGSK-β-catenin had more profound effects on Dlk1 promoter activity. The results suggest that PTN and β-catenin regulate Dlk1 gene expression at the transcriptional level. It is worthy to note that PTN did not influence the Dlk1 promoter activity in the absence of co-transfection of β-catenin, indicating that the co-activation of Wnt/β-catenin pathway is required for the PTN signaling.
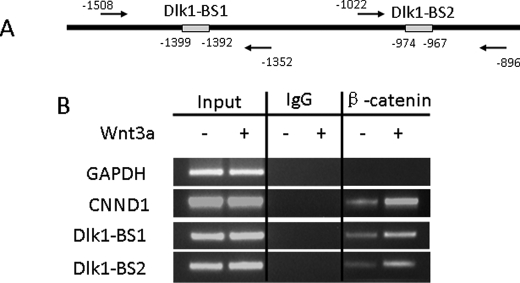
To determine whether Dlk1 is a direct target of TCF/LEF, we performed a ChIP assay using the EZ ChIP kit. E20 fAEC II (1.5 × 106) were treated with 20 ng/ml Wnt3a for 8 h. The chromatin was immunoprecipitated with 2 μg of normal mouse IgG or mouse monoclonal antibodies against β-catenin. The immuno-selected DNAs were amplified using the primers for two putative TCF/LEF binding sites (Dlk1-BS1 and Dlk1-BS2) and the flanking sequences of the Dlk1 promoter (Fig. 9A). Wnt3a increased the binding of β-catenin to the promoter of cyclin D1 (CNND1), a known target of TCF/LEF/β-catenin (Fig. 9B). β-Catenin did not bind to the promoter of glyceraldehyde-3-phosphate dehydrogenase, which does not contain the TCF/LEF binding site. β-Catenin bound to both TCF/LEF binding sites on the Dlk1 promoter, and this binding was enhanced by Wnt3a (Fig. 9B). The negative control using normal mouse IgG did not have any bands. These results suggest that Dlk1 is a direct target of TCF/LEF.
FIGURE 9.
ChIP assay showing that Dlk1 is a direct target of TCF/LEF in fAEC II. A, two putative TCF/LEF binding sites (Dlk1-BS1, −1399 to −1392, and Dlk1-BS2, −974 to −967) of the Dlk1 promoter. The arrows indicate primers used for PCR amplification. B, the immunoprecipitated DNAs using anti-β-catenin antibodies were PCR-amplified using the primer pairs for Dlk1-BS1 and Dlk1-BS2. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and cyclin D1 (CNND1) served as negative and positive controls, respectively. Normal IgG was used as an immunoprecipitation control.
DISCUSSION
In this study we investigated the role of PTN in fetal epithelial cell proliferation and differentiation and underlying mechanisms. PTN and its receptor, RPTP β/ζ, were found in the mesenchymal and epithelial cells of the fetal lungs at the late stages of development. PTN promoted fAEC II proliferation and epithelial repair but inhibited fAEC II trans-differentiation into AEC I. The knockdown of PTN in E14 fetal lung organ culture decreased lung branching morphogenesis. Furthermore, PTN increased the tyrosine phosphorylation of β-catenin and translocation of β-catenin into the nucleus and activated the TCF/LEF transcription factors, leading to transcriptional depression of Dlk1.
Fetal lung development are divided into five stages: embroyonic (E0–13), pseudoglandular (E13–18), canalicular (E18–20), sacular (E20-term), and alveolar stages (term-adult). Branching morphogenesis occurs at the pseudoglandular stage and is accomplished by inducing the formation of buds from the outmost periphery of the epithelial tubes. Alveolar epithelial cell differentiation is a two-step process. At the canalicular stage, the columnar epithelial cells or pre-type II cells differentiate into mature type II cells, which in turn trans-differentiate into type I cells in the secular stage. PTN has multiple roles in fetal lung development at various stages. PTN is necessary for lung branching morphogenesis at the early stage of development; PTN also promotes epithelial type II cell proliferation and migration but suppresses the trans-differentiation of type II cells into type I cells at the late stage of development.
At 50 ng/ml, PTN has a marked effect on cell proliferation and differentiation. However, this dose appears to have a modest effect on wound healing, and a higher dose is required for this assay. It is, therefore, likely that there are different effective doses for different functional consequences caused by PTN.
PTN has been reported to be highly expressed during the late stages of embryogenesis (9–11). Our previous real-time PCR and Western blot results have shown that PTN mRNA and protein expression levels were high at the late stage of fetal lung development (19). The current detailed immunohistochemical studies of fetal lungs at different stages confirmed that PTN was mainly expressed in mesenchymal cells surrounding the epithelium at E18 to E20. Similar analyses of the PTN receptor, RPTP β/ζ, revealed its localization in columnar and alveolar epithelial cells and its high expression at E20. Therefore, both PTN and its receptor RPTP β/ζ had the highest expression during the stage when alveolar epithelial cells proliferation and differentiation occur. The restricted temporal and spatial expression of these two genes suggests that PTN is a mediator of mesenchymal-epithelial interactions during fetal lung development, possibly reflecting their roles in fAEC II proliferation and differentiation.
It is well known that epithelial-mesenchymal interaction is essential for normal growth, morphogenesis, and cell differentiation in the developing lung (2). Signals from the mesenchymal cells determine the patterning of the epithelium as well as the differentiated phenotype of the epithelium. Removal of the mesenchyme from the developing lung impairs lung branching morphogenesis and stops the epithelium from proliferating and differentiating (26). During the late stage of fetal lung development, fAEC II differentiate from progenitor cells and further trans-differentiate into AEC I. Many factors secreted from the mesenchyme have been identified to regulate fAEC II differentiation including fibroblast growth factor 10 (27). Our current study identified PTN as an additional such factor.
PTN increased fAEC II proliferation and inhibited fAEC II trans-differentiation. The regulation of cell proliferation is coupled with that of cell differentiation. The cells that enter the differentiation pathway normally go into G0 phase and stop proliferating (28–30). The cell proliferation and differentiation are independent but well coupled processes that can be precisely regulated by many factors. In normal development the progenitor cells generate enough cells for differentiation and then exit from cell cycle and start to differentiate into mature cells (29). In myogenic precursor cells this process is achieved by up-regulating P21, which inhibits the cyclin-dependent kinase activities (30). Reversely, it has been demonstrated that the overexpression of the cell cycle protein cyclin D1 blocks the terminal differentiation (31). Collectively, PTN may act as a link between fAEC II proliferation and differentiation. It may achieve this by keeping the fAEC II in a proliferating state and maintaining its phenotype. Because coordinated cell proliferation and differentiation are essential for normal organogenesis. PTN may play an essential role in fetal lung development.
Recently, PTN was found to play a role in epithelial-mesenchymal transition (23). PTN-treated U373 epithelial cells exhibited fibroblast-like cells with elongated shape, increased cell migration, and disappeared cell polarity. We found that PTN increased wound healing, possibly because of an increased cell migration. This is supported by the observation that PTN-treated fAEC II had fibroblast-like F-actin distribution on the edge of the wound area. Furthermore, a canonical Wnt signaling pathway is important for the cancer cell invasion and migration (32). Our results showed that PTN activated the Wnt/β-catenin signaling. The effect of PTN on fAEC II wound healing may also be partly because of the cell proliferation or migration.
PTN has been identified as an important factor on renal branching morphogenesis (24). Glial-derived neurotrophic factor, a peptide growth factor that is required for ureteric bud outgrowth, induces the branching in isolated ureteric bud explants at the presence of PTN. However, purified PTN alone also induces ureteric bud outgrowth. This result suggests that PTN is necessary for glial-derived neurotrophic factor signaling. Additionally, using whole embryonic kidney organ culture, PTN induces the ureteric bud outgrowth. Similar to renal branching, the lung undergoes branching morphogenesis during the development. Knockdown of PTN expression decreased the number of terminal buds, indicating its role in lung branching morphogenesis. However, how PTN regulates lung branching morphogenesis remains to be determined.
We found that PTN increased the tyrosine phosphorylation of β-catenin in fAEC II, resulting in β-catenin accumulation in the nucleus and, thus, increase LEF/TCF transcriptional activity. Tyrosine phosphorylation of β-catenin at Tyr-654 and Tyr-142 reduces the binding affinity of β-catenin to E-cadherin and α-catenin, respectively (33–35). Several lines of evidence have demonstrated that tyrosine phosphorylation of β-catenin abolishes adhesion and increases the transcription activity (36). Thus, PTN may act as a key regulator in switching the role of β-catenin from cell adhesion to transcription. Because PTN enhanced TCF/LEF activity as assessed by the TOPflash assay, we for the first time integrated the PTN/RPTP β/ζ pathway with the canonical β-catenin/LEF-TCF pathway.
Wnt regulates lung morphogenesis through the canonical β-catenin/LEF-TCF pathway (37). β-Catenin signaling is essential for cell fate determination in epithelium and mesenchyme in the developing lung (38, 39). In the absence of Wnt signal, β-catenin is phosphorylated at several serine-threonine sites by glycogen synthase kinase 3β and casein kinase and is subsequently marked for proteasomal degradation (36, 40, 41). In the presence of Wnts, β-catenin is stabilized and accumulates in the nucleus to activate the transcription of its downstream genes such as N-myc, bone morphogenetic protein 4, and fibroblast growth factor. In U373 cells, PTN increases tyrosine phosphorylation of β-catenin and releases β-catenin from the membrane (23). However, subsequent β-catenin nuclear accumulation is not reported. In the present studies we observed that PTN not only increased tyrosine phosphorylation of β-catenin but also resulted in the β-catenin nuclear translocation and the activation of TCF/LEF in fAEC II. Our results suggest that PTN may enhance the Wnt/β-catenin signaling and, thus, regulate lung development.
Both canonical Wnt and Notch signaling pathway is indispensable for stem and progenitor cell fate determination and subsequent differentiation during embryonic development and adult life (42–46). These two pathways are reciprocally regulated. Abolishment of Wnt signaling leads to unstable notch activities during somitogenesis (47). These results suggested that Wnt pathway may act as an upstream regulator of Notch pathway. Dlk-1 is a ligand of Notch receptor. Another member of this family, Dll1, was identified as a direct target of TCF/LEF transcription factors (48, 49). The following evidence supports that PTN/Wnt-β-catenin signaling pathways negatively regulate Dlk1 in fAEC II. (i) β-Catenin decreased Dlk1 promoter activity in a dose-dependent manner, and the addition of PTN further reduced the Dlk1 promoter activity. ΔGSK-β-catenin, a constitutively activated β-catenin mutant, had more profound effects on the Dlk1 promoter activity. (ii) ChIP assays showed that β-catenin bound to both TCF/LEF binding sites on the Dlk1 promoter, and this binding was enhanced by Wnt3a. (iii) Dlk1 was down-regulated in Wnt3a- or LiCl-treated fAEC II. Together with the fact that PTN activated the Wnt signaling pathway, these results suggest a novel molecular link between Notch and Wnt signaling pathways during fetal lung development.
Acknowledgments
We thank Dr. Angela Barth (Stanford University) for β-catenin and ΔGSK β-catenin constructs and Dr. Mary Williams (Boston University) for T1α antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL-052146, R01 HL-071628, and R01 HL-083188 (to L. L.).
- fAEC II
- fetal alveolar epithelial type II cells
- ADAEC
- adult AEC
- PTN
- pleiotrophin
- FMEM
- full MEM
- BrdUrd
- bromodeoxyuridine
- PBS
- phosphate-buffered saline
- FBS
- fetal bovine serum
- siRNA
- small interfering RNA
- MEM
- minimal essential medium
- FBS
- fetal bovine serum
- GSK
- glycogen synthase kinase 3
- ChIP
- chromatin immunoprecipitation
- RPTP
- protein-tyrosine phosphatase receptor
- GFP
- green fluorescent protein
- TCF/LEF
- T cell factor/lymphoid enhancer factor.
REFERENCES
- 1.Hilfer S. R. (1996) Annu. Rev. Physiol. 58, 93–113 [DOI] [PubMed] [Google Scholar]
- 2.Shannon J. M., Hyatt B. A. (2004) Annu. Rev. Physiol. 66, 625–645 [DOI] [PubMed] [Google Scholar]
- 3.Bellusci S., Grindley J., Emoto H., Itoh N., Hogan B. L. (1997) Development 124, 4867–4878 [DOI] [PubMed] [Google Scholar]
- 4.Sekine K., Ohuchi H., Fujiwara M., Yamasaki M., Yoshizawa T., Sato T., Yagishita N., Matsui D., Koga Y., Itoh N., Kato S. (1999) Nat. Genet. 21, 138–141 [DOI] [PubMed] [Google Scholar]
- 5.Litingtung Y., Lei L., Westphal H., Chiang C. (1998) Nat. Genet. 20, 58–61 [DOI] [PubMed] [Google Scholar]
- 6.Deuel T. F., Zhang N., Yeh H. J., Silos-Santiago I., Wang Z. Y. (2002) Arch. Biochem. Biophys. 397, 162–171 [DOI] [PubMed] [Google Scholar]
- 7.Milner P. G., Li Y. S., Hoffman R. M., Kodner C. M., Siegel N. R., Deuel T. F. (1989) Biochem. Biophys. Res. Commun. 165, 1096–1103 [DOI] [PubMed] [Google Scholar]
- 8.Rauvala H. (1989) EMBO J. 8, 2933–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herradon G., Ezquerra L., Nguyen T., Vogt T. F., Bronson R., Silos-Santiago I., Deuel T. F. (2004) Biochem. Biophys. Res. Commun. 324, 1041–1047 [DOI] [PubMed] [Google Scholar]
- 10.Vanderwinden J. M., Mailleux P., Schiffmann S. N., Vanderhaeghen J. J. (1992) Anat. Embryol. 186, 387–406 [DOI] [PubMed] [Google Scholar]
- 11.Li Y. S., Milner P. G., Chauhan A. K., Watson M. A., Hoffman R. M., Kodner C. M., Milbrandt J., Deuel T. F. (1990) Science 250, 1690–1694 [DOI] [PubMed] [Google Scholar]
- 12.Silos-Santiago I., Yeh H. J., Gurrieri M. A., Guillerman R. P., Li Y. S., Wolf J., Snider W., Deuel T. F. (1996) J. Neurobiol. 31, 283–296 [DOI] [PubMed] [Google Scholar]
- 13.Yeh H. J., He Y. Y., Xu J., Hsu C. Y., Deuel T. F. (1998) J. Neurosci. 18, 3699–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wellstein A., Fang W. J., Khatri A., Lu Y., Swain S. S., Dickson R. B., Sasse J., Riegel A. T., Lippman M. E. (1992) J. Biol. Chem. 267, 2582–2587 [PubMed] [Google Scholar]
- 15.Seykora J. T., Jih D., Elenitsas R., Horng W. H., Elder D. E. (2003) Am. J. Dermatopathol. 25, 6–11 [DOI] [PubMed] [Google Scholar]
- 16.Meng K., Rodriguez-Peña A., Dimitrov T., Chen W., Yamin M., Noda M., Deuel T. F. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2603–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoica G. E., Kuo A., Aigner A., Sunitha I., Souttou B., Malerczyk C., Caughey D. J., Wen D., Karavanov A., Riegel A. T., Wellstein A. (2001) J. Biol. Chem. 276, 16772–16779 [DOI] [PubMed] [Google Scholar]
- 18.Jäger R., Noll K., Havemann K., Pflüger K. H., Knabbe C., Rauvala H., Zugmaier G. (1997) Int. J Cancer 73, 537–543 [DOI] [PubMed] [Google Scholar]
- 19.Weng T., Chen Z., Jin N., Gao L., Liu L. (2006) Am. J. Physiol. Lung Cell Mol. Physiol. 291, L1027–L1037 [DOI] [PubMed] [Google Scholar]
- 20.Batenburg J. J., Otto-Verberne C. J., Ten Have-Opbroek A. A., Klazinga W. (1988) Biochim. Biophys. Acta 960, 441–453 [DOI] [PubMed] [Google Scholar]
- 21.Fraslon C., Rolland G., Bourbon J. R., Rieutort M., Valenza C. (1991) In Vitro Cell Dev. Biol. 27A, 843–852 [DOI] [PubMed] [Google Scholar]
- 22.Gou D., Weng T., Wang Y., Wang Z., Zhang H., Gao L., Chen Z., Wang P., Liu L. (2007) J. Gene Med. 9, 751–763 [DOI] [PubMed] [Google Scholar]
- 23.Perez-Pinera P., Alcantara S., Dimitrov T., Vega J. A., Deuel T. F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17795–17800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakurai H., Bush K. T., Nigam S. K. (2001) Development 128, 3283–3293 [DOI] [PubMed] [Google Scholar]
- 25.Barth A. I., Stewart D. B., Nelson W. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4947–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon J. M., Nielsen L. D., Gebb S. A., Randell S. H. (1998) Dev. Dyn. 212, 482–494 [DOI] [PubMed] [Google Scholar]
- 27.Maeda Y., Davé V., Whitsett J. A. (2007) Physiol. Rev. 87, 219–244 [DOI] [PubMed] [Google Scholar]
- 28.Heller H., Gredinger E., Bengal E. (2001) J. Biol. Chem. 276, 37307–37316 [DOI] [PubMed] [Google Scholar]
- 29.Sachs L. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 6152–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh K., Perlman H. (1997) Curr. Opin. Genet. Dev. 7, 597–602 [DOI] [PubMed] [Google Scholar]
- 31.Rao S. S., Chu C., Kohtz D. S. (1994) Mol. Cell Biol. 14, 5259–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neth P., Ries C., Karow M., Egea V., Ilmer M., Jochum M. (2007) Stem Cell Rev. 3, 18–29 [DOI] [PubMed] [Google Scholar]
- 33.Piedra J., Miravet S., Castaño J., Pálmer H. G., Heisterkamp N., García de Herreros A., Duñach M. (2003) Mol. Cell. Biol. 23, 2287–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lilien J., Balsamo J. (2005) Curr. Opin. Cell Biol. 17, 459–465 [DOI] [PubMed] [Google Scholar]
- 35.Roura S., Miravet S., Piedra J., García de Herreros A., Duñach M. (1999) J. Biol. Chem. 274, 36734–36740 [DOI] [PubMed] [Google Scholar]
- 36.Piedra J., Martinez D., Castano J., Miravet S., Dunach M., de Herreros A. G. (2001) J. Biol. Chem. 276, 20436–20443 [DOI] [PubMed] [Google Scholar]
- 37.Pongracz J. E., Stockley R. A. (2006) Respir. Res. 7, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mucenski M. L., Nation J. M., Thitoff A. R., Besnard V., Xu Y., Wert S. E., Harada N., Taketo M. M., Stahlman M. T., Whitsett J. A. (2005) Am. J. Physiol. Lung Cell Mol. Physiol. 289, L971–L979 [DOI] [PubMed] [Google Scholar]
- 39.De Langhe S. P., Carraro G., Tefft D., Li C., Xu X., Chai Y., Minoo P., Hajihosseini M. K., Drouin J., Kaartinen V., Bellusci S. (2008) PLoS. ONE. 3, e1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eberhart C. G., Argani P. (2001) Pediatr. Dev. Pathol. 4, 351–357 [DOI] [PubMed] [Google Scholar]
- 41.Wodarz A., Nusse R. (1998) Annu. Rev. Cell Dev. Biol. 14, 59–88 [DOI] [PubMed] [Google Scholar]
- 42.Aoyama K., Delaney C., Varnum-Finney B., Kohn A. D., Moon R. T., Bernstein I. D. (2007) Stem Cells 25, 2488–2497 [DOI] [PubMed] [Google Scholar]
- 43.Eijken M., Meijer I. M., Westbroek I., Koedam M., Chiba H., Uitterlinden A. G., Pols H. A., van Leeuwen J. P. (2008) J. Cell Biochem. 104, 568–579 [DOI] [PubMed] [Google Scholar]
- 44.Cheng P., Gabrilovich D. (2008) Immunol. Res. 41, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen E. D., Tian Y., Morrisey E. E. (2008) Development 135, 789–798 [DOI] [PubMed] [Google Scholar]
- 46.Osborne B. A., Minter L. M. (2007) Nat. Rev. Immunol. 7, 64–75 [DOI] [PubMed] [Google Scholar]
- 47.Aulehla A., Wehrle C., Brand-Saberi B., Kemler R., Gossler A., Kanzler B., Herrmann B. G. (2003) Dev. Cell 4, 395–406 [DOI] [PubMed] [Google Scholar]
- 48.Galceran J., Sustmann C., Hsu S. C., Folberth S., Grosschedl R. (2004) Genes Dev. 18, 2718–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofmann M., Schuster-Gossler K., Watabe-Rudolph M., Aulehla A., Herrmann B. G., Gossler A. (2004) Genes Dev. 18, 2712–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]