Abstract
The Ca2+-binding protein calmodulin (CaM) has been shown to bind directly to cytoplasmic domains of some G protein-coupled receptors, including the dopamine D2 receptor. CaM binds to the N-terminal portion of the long third intracellular loop of the D2 receptor, within an Arg-rich epitope that is also involved in the binding to Gi/o proteins and to the adenosine A2A receptor, with the formation of A2A-D2 receptor heteromers. In the present work, by using proteomics and bioluminescence resonance energy transfer (BRET) techniques, we provide evidence for the binding of CaM to the A2A receptor. By using BRET and sequential resonance energy transfer techniques, evidence was obtained for CaM-A2A-D2 receptor oligomerization. BRET competition experiments indicated that, in the A2A-D2 receptor heteromer, CaM binds preferentially to a proximal C terminus epitope of the A2A receptor. Furthermore, Ca2+ was found to induce conformational changes in the CaM-A2A-D2 receptor oligomer and to selectively modulate A2A and D2 receptor-mediated MAPK signaling in the A2A-D2 receptor heteromer. These results may have implications for basal ganglia disorders, since A2A-D2 receptor heteromers are being considered as a target for anti-parkinsonian agents.
G-protein-coupled receptors are able to form homo- and hetero-oligomers with unique biochemical and functional characteristics (1–7), and they are easily detected in vitro by using biophysical techniques (8–10). Heteromers of adenosine A2A and dopamine D2 receptors were one of the first G-protein-coupled receptor heteromers to be described (11). A close physical interaction between both receptors was shown using co-immunoprecipitation and co-localization assays (11) and fluorescence and bioluminescence resonance energy transfer (FRET2 or BRET) techniques (12–14). At the biochemical level, two types of antagonistic A2A-D2 receptor interactions have been discovered that may explain the A2A-D2 receptor interactions described both at the neuronal and behavioral level (11, 15–18). First, by means of an allosteric interaction in the receptor heteromer, stimulation of A2A receptor decreases the affinity of D2 receptor for their agonists (12). Second, the stimulation of the Gi/o-protein-coupled D2 receptor inhibits the cAMP accumulation induced by the stimulation of the Gs/olf-protein-coupled A2A receptor (11, 17, 18). In view of the well known role of dopamine in Parkinson disease, schizophrenia, and drug addiction, it has been suggested that the A2A-D2 receptor interactions in the central nervous system may provide new therapeutic approaches to combat these disorders (16, 19).
An epitope-epitope electrostatic interaction between an Arg-rich epitope of the N terminus of the third intracellular loop (3IL) of the D2 receptor and an epitope containing a phosphorylated Ser localized in the distal part of the C terminus of the A2A receptor is involved in A2A-D2 receptor heteromer interface (14, 20, 21). The same Arg-rich epitope of the D2 receptor is able to interact with CaM (22–25). In the absence of phosphorylated residues, adjacent aspartates or glutamates, which are abundant in CaM, may also form non-covalent complexes with Arg-rich epitopes (26). Therefore, CaM can potentially convey a Ca2+ signal to the D2 receptor through direct binding to the 3IL of the D2 receptor (22). Mass spectrometry data have shown that bovine CaM can form multiple non-covalent complexes with an Arg-rich peptide corresponding to the N-terminal region of the 3IL of the D2 receptor (VLRRRRKRVN) (24) as well as a peptide from the proximal C terminus of the A2A receptor (24). This epitope, whose sequence is 291RIREFRQTFR300 in the human A2A receptor, also contains several Arg residues. Since the suspected interaction between the A2A receptor and CaM was awaiting confirmation by assays using complete proteins, the present study was undertaken to demonstrate the existence of interactions between the A2A receptor and CaM both in a recombinant protein expression cell system and in the brain. A proteomics approach was used for the discovery of protein-protein interactions between the A2A receptor and CaM in rat brain, whereas BRET in transfected cells demonstrated a direct interaction between CaM and this receptor. Furthermore, by using BRET and sequential resonance energy transfer (SRET) techniques and analyzing MAPK signaling in transfected cells, evidence was obtained for CaM-A2A-D2 receptor oligomerization and a selective Ca2+-mediated modulation of A2A and D2 receptor function in the A2A-D2 receptor heteromer.
EXPERIMENTAL PROCEDURES
Animals
Male Sprague-Dawley rats (250 g) were obtained from Harlan (Barcelona, Spain). Procedures involving animals were in accordance with the guidelines established by the normative of the European Council (86/609/EEC). The experimental design was approved by the Ethical Committee for Animal Testing of the University of Navarra (060/07).
Membrane Protein Purification
Animals were killed by decapitation, and the brains were rapidly removed. To obtain P2 membrane proteins, the striatum was dissected out and homogenized with a glass-Teflon homogenizer in ice-cold 0.32 m sucrose, 10 mm Hepes (pH 7,4), 2 mm EDTA, and complete protease inhibitors (Roche Applied Science). After spinning at 1,000 × g for 15 min, the supernatant was removed and centrifuged at 200,000 × g for 15 min to yield the crude membrane pellet (P2). The pellet was resuspended in homogenization buffer and spun again at 200,000 × g to yield washed crude membrane pellet (P2′).
Immunoprecipitation
The pellet containing membrane proteins (P2′) was solubilized in immunoprecipitation buffer (phosphate-buffered saline (pH 7.4), complete protease inhibitor mixture, and 1% Igepal Ca-630 (Sigma)) for 1 h at 4 °C. Protein concentration was determined by the BCA method (Pierce). A total amount of 0.5 mg of protein was used for immunoprecipitation. Samples were pretreated with 30 μl of protein A/G-agarose beads (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) with gentle rocking for 1 h at 4 °C and centrifuged at 10,000 × g to remove the beads. Membrane proteins were incubated with 5 μg of polyclonal anti A2A receptor (Affinity Bioreagents, bioNova Científica, Madrid, Spain) or with 5 μg of normal rabbit IgG (Upstate, Charlottesville, VA) for 90 min, and then 30 μl of Protein A/G beads were added and left for 16 h at 4 °C with gentle rocking. Immunoprecipitated proteins were washed three times with immunoprecipitation buffer.
Proteomic Analysis
Immunoprecipitated proteins were analyzed in the Proteomic Laboratory of CIMA. Briefly, proteins were eluted by incubation with 100 mm glycine-HCl (pH 2.5) for 10 min at room temperature with gentle rocking. The samples were centrifuged at 10,000 × g for 1 min, and the supernatant content was precipitated with 25% trichloroacetic acid for 20 min at 4 °C. After spinning at 10,000 × g for 15 min, the pellet was washed twice with precooled acetone, completely dried in a SpeedVac, and resuspended in 30 μl of 100 mm ammonium bicarbonate. Proteins were reduced with 10 mm dithiothreitol for 30 min at 56 °C, alkylated with 55 mm iodoacetamide for 20 min in darkness, and finally digested with trypsin 1:50 (v/v) for 16 h at 37 °C. Peptide mass fingerprinting was obtained from tryptic digests by liquid chromatography-electrospray ionization-tandem mass spectrometry analysis (27). A peak list was generated by ProteinLynx Global Server 2.1 (Waters, Milford, MA). A data base search was done with ProteinLynx Global Server 2.1 (Waters) and Phenyx for raw data and mzXML data, respectively. The data base used in this study was SwissProt.
Cell Culture
HEK-293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 2 mm l-glutamine, 100 units/ml penicillin/streptomycin, and 5% (v/v) heat-inactivated fetal bovine serum (all supplements were from Invitrogen). Cells were maintained at 37 °C in an atmosphere of 5% CO2 and were passaged when they were 80–90% confluent (i.e. approximately twice per week).
Mutant A2A Receptors, Fusion Proteins, and Expression Vectors
The sequence R291IREFR296QTFR300 in the C-terminal domain of the human A2A receptor was mutated to A291IREFA296QTFA300 by site-directed mutagenesis (Cellogenetics, Ijamsville, MD). The human cDNAs for A2A, mutant A2A, which is denoted as A2ARAAA, D2, and A1 receptors, cloned into pcDNA3.1, were amplified without their stop codons using sense and antisense primers harboring unique EcoRI and BamHI sites to clone A2A receptor, mutant A2A receptor, or A1 receptor in Rluc vector and EcoRI and KpnI to clone D2 receptor in Rluc or EYFP vectors. The amplified fragments were subcloned to be in frame into restriction sites of pcDNA3.1Rluc and pEYFP-N1 (enhanced yellow variant of GFP; Clontech) vectors, resulting in the plasmids A2ARRluc, A2ARAAARluc, A1RRluc, D2RRluc, and D2RYFP. The cDNA encoding the human EYFP-CaM (CaMYFP) fusion protein was kindly provided by Dr. Carles Enrich (Hospital Clinic de Barcelona, Spain). CaM was subsequently subcloned into the pGFP2-C1 (BioSignal Packard, Montreal, Canada) using EcoRI and BamHI, resulting in the GFP2-CaM plasmid. The cDNA encoding the 5HT2BR-YFP fusion protein was kindly provided by Dr. Irma Nardi (University of Pisa, Italy). Expression of constructs was tested by confocal microscopy (see “Results”), and the receptor functionality was tested by second messengers, ERK1/2 phosphorylation, and cAMP production, as described previously (12, 14) (data not shown).
Transient Transfection and Protein Determination
HEK-293T cells growing in 6-well dishes were transiently transfected with the corresponding fusion protein cDNA by the polyethyleneimine (Sigma) method. Cells were incubated (4 h) with the corresponding cDNA together with polyethyleneimine (5 μl/μg cDNA of 10 μm polyethyleneimine) and 150 mm NaCl in a serum-free medium. After 4 h, the medium was changed to a fresh complete culture medium. Forty-eight hours after transfection, cells were washed twice in quick succession in HBSS with 10 mm glucose, detached by scraping, and resuspended in the same buffer unless otherwise indicated. To control the cell number, sample protein concentration was determined using a Bradford assay kit (Bio-Rad), using bovine serum albumin as the standard protein. Cell suspensions (20 μg of protein) in HBSS buffer containing 1.26 mm CaCl2 or, when indicated, in Ca2+-free HBSS buffer were distributed into 96-well microplates; black plates with a transparent bottom were used for fluorescence determinations, whereas white plates were used for BRET and SRET experiments.
BRET Assays
HEK-293T cells were transiently co-transfected with the indicated amounts of plasmid cDNAs corresponding to the indicated fusion proteins (see figure legends). To quantify fluorescence proteins, cells (20 μg of protein) were distributed in 96-well microplates (black plates with a transparent bottom), and fluorescence was read at 400 nm in a Fluo Star Optima Fluorimeter (BMG Labtechnologies, Offenburg, Germany) equipped with a high energy xenon flash lamp, using a 10-nm bandwidth excitation filter. Receptor fluorescence expression was determined as fluorescence of the sample minus the fluorescence of cells expressing protein-Rluc alone. For BRET measurements, the equivalent of 20 μg of cell protein was distributed in 96-well microplates (Corning 3600, white plates; Sigma), and 5 μm coelenterazine H (Molecular Probes, Inc., Eugene, OR) was added. After 1 min of adding coelenterazine H, the readings were collected using a Mithras LB 940, which allows the integration of the signals detected in the 485-nm short (440–500 nm) and the 530-nm long (510–590 nm) wavelength filters. To quantify receptor-Rluc expression, luminescence readings were performed after 10 min of adding 5 μm coelenterazine H. The same protocol was used to determine BRET with membranes obtained from cells (48 h post-transfection) disrupted using a Polytron homogenizer (PTA 20 TS rotor, setting 3; Kinematica, Basel, Switzerland). Disruption was performed for three 5-s periods in 10 volumes of free Ca2+ HBSS, pH 7.4, containing a proteinase inhibitor mixture (Sigma). Cell debris were eliminated, membranes were obtained by centrifugation at 105,000 × g (40 min, 4 °C), and the pellet was resuspended and recentrifuged under the same conditions. The net BRET is defined as (long wavelength emission/short wavelength emission) − Cf, where Cf corresponds to long wavelength emission/short wavelength emission for the Rluc construct expressed alone in the same experiment.
SRET Experiments
HEK-293T cells were transiently co-transfected with the indicated amounts of plasmid cDNAs, corresponding to the indicated fusion proteins (see the legend for Fig. 6). Using aliquots of transfected cells (20 μg of protein), three different determinations were performed in parallel. First, we performed quantification of protein-YFP expression by determination of the fluorescence due to protein-YFP. Cells distributed into 96-well microplates (black plates with a transparent bottom) were read in a Fluostar Optima fluorimeter (BMG Labtechnologies, Offenburg, Germany) equipped with a high energy xenon flash lamp, using an excitation filter at 485 nm and 10-nm bandwidth emission filters corresponding to 510 nm (506–515 nm) (Ch1) and 530 nm (527–536 nm) (Ch2). The contribution of the GFP2 and YFP proteins alone to the two detection channels (spectral signature) (28) was measured in experiments with cells expressing only one of these proteins and normalized to the sum of the signal obtained in the two detection channels. The spectral signatures of the different receptors fused to either GFP2 or YFP did not vary significantly from the determined spectral signatures of the fluorescent proteins alone. For protein-YFP expression quantification, linear unmixing was done taking into account the spectral signature as described by Zimmermann et al. (28, 29) to separate the two emission spectra; the sample fluorescence is the fluorescence calculated as described minus the fluorescence of cells expressing only protein-Rluc and protein-GFP2. Second, we performed quantification of protein-Rluc expression by determination of the luminescence due to protein-Rluc. Cells were distributed in 96-well microplates (Corning 3600), and luminescence was determined 10 min after the addition of 5 μm coelenterazine H in a Mithras LB 940 multimode reader (Berthold Technologies, DLReady, Germany). Third, we performed SRET2 measurements. Cells were distributed in 96-well microplates (Corning 3600), and 5 μm DeepBlueC (Molecular Probes) was added. The SRET2 signal was collected using a Mithras LB 940 reader with detection filters for short wavelength (400 nm (370–450 nm)) and long wavelength (530 nm (510–590 nm)). By analogy with BRET, net SRET is defined as (long wavelength emission/short wavelength emission) − Cf, where Cf corresponds to long wavelength emission/short wavelength emission for cells expressing protein-Rluc, protein-GFP2, and the other protein partner not fused to a fluorescence protein (similar values were obtained measuring Cf in cells expressing protein-Rluc only and protein-GFP2). Linear unmixing was done for SRET2 quantification, taking into account the spectral signature (28) to separate the two fluorescence emission spectra.
FIGURE 6.
SRET for CaM, A2A receptor and D2 receptor in living cells. SRET saturation curves were performed in HEK-293 cells expressing A2AR-Rluc (0.75 μg of cDNA), CaMGFP2 (0.6 μg of cDNA), and increasing amounts of D2RYFP (0.5–5 μg of the cDNA). Net SRET was obtained by monitoring the YFP fluorescence emission after coelenterazine H addition, with subtraction of the value obtained with cells expressing the same amount of A2AR-Rluc and CaMGFP2. Significant net SRET was detected for A2ARRluc-CaMGFP2-D2RYFP coupling, whereas negligible or linear net SRET was obtained in cells expressing equivalent amounts (equivalent fluorescence and luminescence units) of A2ARRluc, CaMGFP2, and 5HT2BYFP or A1RRluc, CaMGFP2, and D2RYFP, which were used as negative controls. Values, expressed as net SRET, represent means ± S.E. of five independent experiments performed in triplicate. At the top, a scheme depicts the expressed proteins in SRET assays.
Immunostaining
For immunocytochemistry, transiently transfected HEK-293T cells were fixed in 4% paraformaldehyde for 15 min and washed with phosphate-buffered saline containing 20 mm glycine (buffer A) to quench the aldehyde groups. Then, after permeabilization with buffer A containing 0.2% Triton X-100 for 5 min, cells were treated with phosphate-buffered saline containing 1% bovine serum albumin. After 1 h at room temperature, protein-Rluc was labeled with the primary mouse monoclonal anti-Rluc antibody (1:100; Chemicon, Billerica, MA) for 1 h, washed, and stained with the secondary antibody Cy3 donkey anti-mouse (1:100; Jackson Immunoresearch Laboratories, West Grove, PA). CaMYFP was detected by its fluorescence properties. Samples were rinsed and observed in a Leica SP2 confocal microscope (Leica Microsystems, Mannheim, Germany).
ERK Phosphorylation Assay
Cells were grown in 25-cm2 flasks to 80% confluence and cultured in serum-free medium for 16 h before the addition of any agent. Cells resuspended in HBSS buffer containing 1.26 mm CaCl2 were treated or not with 1 μm ionomycin for 3 min before the addition of the following agonists: A2A receptor agonist CGS2168 (200 nm), the D2 receptor agonist quinpirole (1 μm), or a mixture of both ligands for 5 min. Cells were rinsed with ice-cold phosphate-buffered saline and lysed by the addition of 500 μl of ice-cold lysis buffer (50 mm Tris-HCl, pH 7.4, 50 mm NaF, 150 mm NaCl, 45 mm β-glycerophosphate, 1% Triton X-100, 20 μm phenyl-arsine oxide, 0.4 mm NaVO4, and protease inhibitor mixture). The cellular debris was removed by centrifugation at 13,000 × g for 5 min at 4 °C, and the protein was quantified by the bicinchoninic acid method using bovine serum albumin dilutions as a standard. To determine the level of ERK1/2 phosphorylation, equivalent amounts of protein (10 μg) were separated by electrophoresis on a denaturing 7.5% SDS-polyacrylamide gel and transferred onto polyvinylidene difluoride membranes. The membranes were then probed with a mouse anti-phospho-ERK1/2 antibody (1:2500; Sigma). In order to rule out the possibility that the differences observed were due to the application of unequal amounts of lysates, polyvinylidene difluoride blots were stripped and probed with a rabbit anti-ERK1/2 antibody that recognizes both phosphorylated and nonphosphorylated ERK1/2 (1:40,000; Sigma). Bands were visualized by the addition of anti-mouse horseradish peroxidase-conjugated (Dako, Glostrup, Denmark) or anti-rabbit horseradish peroxidase-conjugated (Sigma) secondary antibodies, respectively, and SuperSignal West Pico Chemiluminescent Substrate (Pierce). Band densities were quantified with a LAS-3000 imaging system (Fujifilm), and the level of phosphorylated ERK1/2 isoforms was normalized for differences in loading using the total ERK protein band intensities. Quantitative analysis of detected bands was performed by Image Gauge version 4.0 software. One-way analysis of variance and Student's t test for unpaired samples were used for statistical comparisons.
RESULTS
Identification of an Interaction between CaM and the A2A Receptor
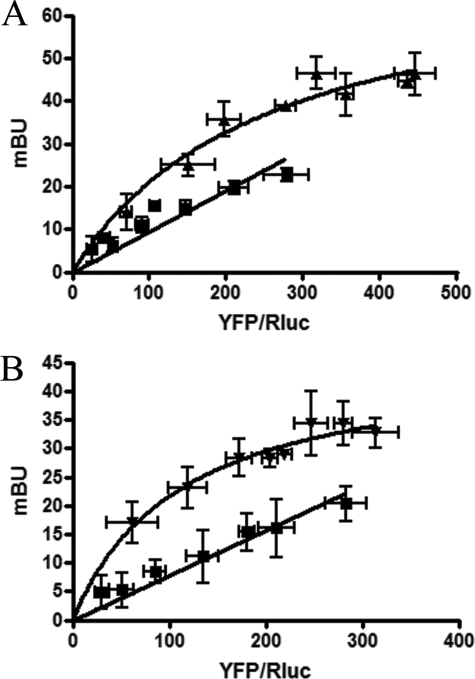
According to the CaM target data base (available on the World Wide Web), the sequence RIREFRQTFR, which is present in the proximal part of the C terminus of the A2A receptor, has a high probability to bind to CaM. In fact, we recently demonstrated that a synthetic peptide with the same sequence forms non-covalent complexes with bovine CaM (24). A proteomics approach was then used to find in situ evidence of direct interactions between the A2A receptor and CaM in the brain. Samples from rat striatal membranes were immunoprecipitated using an antibody directed against a synthetic peptide of the A2A receptor C terminus, SHGDMGLPDVELLSHELK, which does not overlap with the putative CaM-binding motif. Co-immunoprecipitates were digested with trypsin, and the resulting peptides were analyzed by mass spectrometry, as described under “Experimental Procedures.” Among other peptides, two corresponding to the sequence of rat CaM (P62161) were detected after co-immunoprecipitation with the specific polyclonal antibody but not with rabbit IgG (Table 1). The significance of the hits was confirmed by the low p values for both the 13-mer and the 9-mer peptides, which collectively covered 15% of the rat CaM sequence (Table 1). These results suggest that CaM and the A2A receptor may interact, but the existence of a third bridging protein cannot be ruled out. For this purpose, BRET assays were performed in cells transfected with the cDNAs for the A2ARRluc and CaMYFP fusion proteins. The hyperbola obtained upon increasing the YFP/Rluc ratio (Fig. 1A) indicates that a specific interaction between CaM and the A2A receptor can occur in living cells. Similar experiments were performed using D2RRluc and CaMYFP to confirm that these two proteins may establish direct molecular interactions (Fig. 1B). The specificity of the CaM and A2A receptor or CaM and D2 receptor interactions in HEK cells was confirmed by the nonspecific (linear) BRET signal obtained when assaying A1RRluc and CaMYFP (Fig. 1). To demonstrate that the A2A receptor sequence RIREFRQTFR acts as a CaM binding domain, BRET competition experiments were performed in cells transfected with the cDNAs for the A2ARRluc and CaMYFP fusion proteins (amounts adjusted to give values around the BRET50) and increasing amounts of cDNA corresponding to the wild type A2AR or to the A2ARAAA (RIREFRQTFR mutated to AIREFAQTFA). As expected, the A2A receptor did decrease BRET values by competing with A2ARRluc for its binding to CaMYFP, whereas A2ARAAA did not (Fig. 2A). Accordingly, in cells expressing A2ARAAARluc and CaMYFP, a linear and negligible BRET was detected in the absence or in the presence of ionomycin (Fig. 2B).
TABLE 1.
Peptides identical to the rat calmodulin sequence identified by liquid chromatography-electrospray ionization-tandem mass spectrometry
| Sequence | Position | z | dm/z | z-Score | p value |
|---|---|---|---|---|---|
| ADQLTEEQIAEFK | 2–14 | 2 | −0.014 | 11.76 | 1.45E−30 |
| DTDSEEEIR | 79–87 | 2 | −0.035 | 4.16 | 4.8E−4 |
FIGURE 1.
Identification of CaM-A2A and CaM-D2 receptor oligomers by BRET experiments. BRET saturation curves were performed using HEK-293 cells co-expressing A2ARRluc and CaMYFP (triangles) (A) or D2RRluc and CaMYFP (inverted triangles) (B) or A1RRluc and CaMYFP (squares). Co-transfections were performed with increasing amounts of plasmid-YFP (0.1–1 μg of cDNA), whereas the plasmid-Rluc construct was maintained constant (0.6 μg of cDNA for A2ARRluc, 1 μg of cDNA for D2RRluc, and 0.7 μg of cDNA for A1RRluc). Both fluorescence and luminescence for each sample were measured before every experiment to confirm similar donor expressions (about 100,000 luminescent units) while monitoring the increase in acceptor expression. The relative amount of acceptor is given as the ratio between the fluorescence of the acceptor (YFP) and the luciferase activity of the donor (Rluc). BRET data are expressed as means ± S.D. of 4–8 different experiments grouped as a function of the amount of BRET acceptor.
FIGURE 2.
Identification of the A2A receptor CaM binding site. A, BRET competition experiments were performed with HEK-293 cells transfected with A2ARRluc (0.6 μg of cDNA), CaMYFP (0.6 μg of cDNA), and increasing amounts of cDNA corresponding to A2AR (white bars) or A2ARAAA (R291IREFR296QTFR300 mutated to A291IREFA296QTFA300; black bars). Both fluorescence and luminescence for each sample were checked to confirm similar donor and acceptor expression (about 100,000 luminescent units and 10,000 fluorescence units). A2AR and A2ARAAA receptor expression was monitored by Western blot (not shown). B, BRET saturation curves were performed with HEK-293 cells co-expressing A2ARAAARluc (0.6 μg of cDNA) and increasing amounts of CaMYFP (0.1–1 μg of cDNA) and untreated (white symbols) or treated (10 min; black symbols) with 1 μm ionomycin in HBSS buffer containing 1.26 mm CaCl2. Both fluorescence and luminescence of each sample were checked to confirm similar donor expressions (about 100,000 luminescent units) while monitoring the increase in acceptor expression. The relative amount of acceptor is given as the ratio between the fluorescence of the acceptor (YFP) and the luciferase activity of the donor (Rluc). BRET data are expressed as means ± S.D. of 4–6 different experiments grouped as a function of the amount of BRET acceptor. Results are compared with the curve obtained for A2ARRluc and CaMYFP (dotted line; see Fig. 1A).
CaM Interacts with A2A and D2 Receptors at the Plasma Membrane
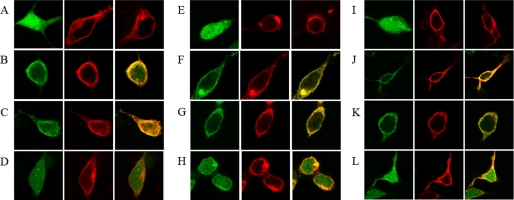
Immunolocalization of receptors and CaM were performed in co-transfected HEK cells. When expressed in the absence of receptors, CaM showed a cytosolic localization (Fig. 3). Co-expression of CaM and any of the two receptors did not modify the localization of the receptor. On the other hand, a significant membrane localization of CaM was only observed when the protein was co-expressed with either A2A or D2 receptors (Fig. 3). The translocation of CaM to the plasma membrane did not take place when co-expressed with the A1 receptor (Fig. 3), which is not able to establish molecular interactions with the protein (see above). Treatment of cells with a Ca2+-free buffer and EDTA or treatment with ionomycin did not modify colocalization of CaM and A2A or D2 receptors in cell membranes (Fig. 3).
FIGURE 3.
Membrane colocalization of CaM with A2A or D2 receptors. Co-transfected HEK-293 cells were washed and resuspended for 2.5 h in HBSS buffer containing 1.26 mm CaCl2 (A–D and I–L) or in a Ca+2-free HBSS buffer containing 1 mm EDTA (E–H). A, E, and I, confocal microscopy images of HEK-293T cells expressing (from left to right in each panel) CaMYFP (0.6 μg of cDNA), A2ARRluc (1 μg of cDNA), or D2RRluc (1 μg of cDNA). B–D, F–H, and J–L, confocal microscopy images of HEK-293T cells co-transfected with the above described amounts of cDNA for CaMYFP and A2ARRluc (B, F, and J), CaMYFP and D2RRluc (C, G, and K), or CaMYFP and A1RRluc (1 μg of cDNA) (D, H, and L). Ionomycin-treated cells (10 min prior to fixation) are shown in I–L. CaM was identified by YFP fluorescence (green images), and receptor-Rluc constructs were identified by immunocytochemistry (using a monoclonal anti-Rluc primary antibody and a cyanine-3-conjugated secondary antibody). Colocalization (yellow) is shown in the right panels.
Effect of Ca2+ Levels on the Interaction between CaM and A2A or D2 Receptors
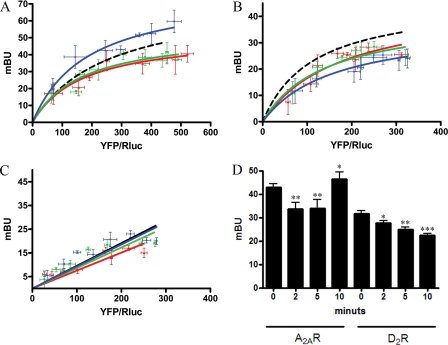
BRET experiments were performed to give some insight into the role of Ca2+ in the interaction of CaM with either A2A or D2 receptors. In cells co-expressing D2RRluc and CaMYFP, resuspended in a medium containing 1.26 mm CaCl2, ionomycin treatment led to significant decreases in BRET measurements obtained after periods of incubation of 2, 5, and 10 min (Fig. 4B). In cells co-expressing A2ARRluc and CaMYFP, ionomycin treatment also led to significant decreases in BRET measurements at 2 and 5 min of preincubation. In contrast, with a longer period of incubation (10 min), the BRET signal for A2ARRluc and CaMYFP increased significantly above that found in untreated cells (Fig. 4A). On the one hand, these data indicate that Ca2+ is not needed for CaM to be able to interact with A2A or D2 receptors. On the other hand, the ionophore-induced increase in intracellular Ca2+ concentration does not disrupt the oligomerization of CaM with the receptors but leads to conformational changes in the CaM molecule and/or the receptors that alter the distance between Rluc (fused to the C terminus of the D2 or A2A receptors) and YFP (fused to CaM). The observed effects are not due to the Ca+2 toxicity or changes in the membrane structure, since they were also observed in a cell-free system (Fig. 4D). Moreover, ionomycin did not induce significant changes in BRET measurements in cells co-expressing A1RRluc and CaMYFP (Fig. 4C).
FIGURE 4.
Effect of intracellular Ca2+ on the interaction between CaM and A2A or D2 receptors detected by BRET experiments. A–C, BRET saturation curves were performed using HEK-293 cells co-expressing A2ARRluc and CaMYFP (A), D2RRluc and CaMYFP (B), or A1RRluc and CaMYFP as negative control (C) (see the legend to Fig. 1). In all cases, cells were treated with HBSS buffer containing 1.26 mm CaCl2 and 1 μm ionomycin at 10 (blue), 5 (green), or 2 (red) min before BRET determination. BRET saturation curves were compared with the one obtained in the absence of ionomycin (dotted line; see Fig. 1). The relative amount of acceptor is given as the ratio between the fluorescence of the acceptor (YFP) and the luciferase activity of the donor (Rluc). BRET data are expressed as means ± S.D. of 4–8 different experiments grouped as a function of the amount of BRET acceptor. D, HEK-293 cells were transfected with 0.6 μg of cDNA for A2ARRluc (A2AR) or 1 μg of cDNA for D2RRluc (D2R) and 1 μg of cDNA of CaMYFP, and membranes were obtained 48 h after transfection. Membranes were resuspended in free Ca2+ HBSS buffer, and 1.26 mm CaCl2 was added for the indicated times before BRET determination. BRET data are expressed as means ± S.E. of five different experiments. Significant differences over ionomycin-non-treated cells were calculated by one-way analysis of variance and Bonferroni test (*, p < 0.05; **, p < 0.01; ***, p < 0.005).
Interactions of CaM with the A2A-D2 Receptor Heteromer
By means of BRET competition experiments performed with cells expressing A2RRluc and CaMYFP (with expression levels adequate to give a suboptimal BRET value), the increase in D2 receptor expression (tested by Western blot experiments) did not modify the BRET signal due to A2RRluc and CaMYFP (Fig. 5A). Analogously, increasing the expression of CaM did not modify the BRET signal due to A2RRluc and D2RYFP (Fig. 5B). As indicated in the Introduction, CaM seems to interact with the Arg-rich epitope of the 3IL of the D2 receptor that is also involved in A2A-D2 receptor heteromerization (see the Introduction). In fact, competition assays demonstrated that increasing amounts of A2A receptor led to significant reduction in the BRET signal due to the interaction between the D2 receptor and CaM (Fig. 5C). However, a differential effect was obtained depending on the isoform of the D2 receptor (D2S or D2L). On the one hand, the BRET signal between D2LRRluc and CaMYFP increased with low quantities of A2A receptor but decreased dose-dependently with higher quantities of A2A receptor. On the other hand, using the pair D2SRRluc-CaMYFP, a dose-dependent decrease in BRET was found. These findings correlate with the potential ability of CaM and A2A receptors to bind to any of the two Arg-rich epitopes of the 3IL of the D2L receptor (one of which is not present in the D2S structure; see “Discussion”). The specificity of these effects was demonstrated by the independence of the BRET signal with increasing expression of the A1 receptor (Fig. 5C).
FIGURE 5.
CaM interaction with A2A-D2 receptor heteromers detected by BRET competition experiments. BRET competition experiments were performed with HEK-293 cells expressing donor and acceptor constructs to give submaximal BRET values. A, cells were co-transfected with the cDNA corresponding to A2ARRluc (0.6 μg) and CaMYFP (0.4 μg) and increasing amounts of D2R cDNA as competitor (0.3–2.5 μg), whose expression was monitored by Western blotting (bottom) mBU, milli-BRET units. B, cells were cotransfected with the cDNA corresponding to A2ARRluc (0.6 μg) and D2LRYFP (1.5 μg) and increasing amounts of cDNA of CaMGFP2 as competitor (0.2–1.5 μg), whose expression was monitored by measuring the GFP2 fluorescence (1,000–20,000 fluorescence units), as described under “Experimental Procedures.” To determine the YFP fluorescence in BRET experiments, the spectral signature was considered to control the GFP2 contribution to the detection channel (28, 29). Cells were cotransfected with the cDNA corresponding to D2LRRluc (1 μg; C) or D2SRRluc (0.8 μg; D) and CaMYFP (0.4 μg) and increasing amounts of cDNA of A2AR as competitor (black bars) or A1R as negative control (white bars), whose expression was monitored by dot blotting (results not shown). At the top of each panel, a scheme depicts the expressed proteins in BRET competition assays.
Oligomerization of CaM, A2A, and D2 Receptors
To test the possible existence of CaM-A2A-D2 receptor oligomerization, the recently described SRET technique was used (29). In SRET2, the oxidation of an Rluc substrate triggers GFP2 acceptor excitation by BRET2 and subsequent energy transfer to a YFP FRET acceptor. SRET2 was attempted using A2ARRluc, CaMGFP2, D2RYFP, and DeepBlueC as Rluc substrate. SRET2 would only occur if the two acceptor/donor pairs, A2ARRluc/CaMGFP2 and CaMGFP2/D2RYFP, are well oriented at a distance of less than 10 nm. A SRET saturation curve obtained by augmenting D2RYFP expression while maintaining the same A2ARRluc/CaMGFP2 ratio is shown in Fig. 6. To obtain optimal results (see Ref. 29), cells were transfected with an amount of cDNA (0.6 μg) for A2ARRluc giving readouts of about 100,000 luminescent units, an amount of cDNA (0.4 μg) for CaMGFP2 giving about 6,000 fluorescent units, and increasing amounts of cDNA (0.5–6 μg) for D2RYFP (giving a range of 500–8,000 fluorescence units). From the saturation curve (Fig. 6), apparent SRETmax (0.29 ± 0.03) and apparent SRET50 (0.010 ± 0.006) values were calculated. To prove whether SRET was possible when substituting A2A receptors by A1 receptors, cells were transfected with 0.7 μg of cDNA for A1RRluc (100,000 luminescent units), 0.4 μg of cDNA for CaMGFP2 (6,000 fluorescent units), and 0.5–6 μg of cDNA for D2RYFP (500–9,000 fluorescence units). The possible formation of a trimer in which the D2 receptor was substituted by the serotonin 5-HT2B receptor was tested using cells transfected with 0.6 μg of cDNA for A2ARRluc, 0.4 μg of cDNA for CaMGFP2, and 0.5–6 μg of cDNA for the 5-HT2B receptor (500–9,000 fluorescence units). Using these two combinations of fusion proteins, a very low and non-saturating (linear) SRET was observed, which indicates that oligomerization of CaM, A1, and D2 receptors or CaM, A2A, and 5-HT2B receptors does not occur.
Effects of Intracellular Ca2+ on CaM-A2A-D2 Receptor Oligomerization
BRET was measured in the presence and in the absence of ionomycin in cells co-transfected with A2ARRluc and D2RYFP. The calcium ionophore led to qualitative changes in the BRET curves (Fig. 7A) similar to those in the experiments with cells co-expressing A2ARRluc and CaMYFP (see Fig. 4A). Thus, the BRET signal for A2ARRluc and D2RYFP decreased at shorter times of incubation with ionomycin, whereas it increased at longer times (Fig. 7A). These results suggest that Ca2+ binding to CaM induces conformational changes in the A2A receptor such that the distance between the donor Rluc (fused to the C terminus of the A2A) and the acceptor YFP is modified. Taking all of the results into account, the most probable scenario is the existence of a CaM-A2A-D2 receptor oligomeric complex, Ca2+ binding to CaM being unable to disrupt A2A-D2 receptor heteromers. To further test this possibility, SRET2 for A2ARRluc-CaMGFP2-D2RYFP was measured in cells treated with ionomycin. As shown in Fig. 6B, a positive SRET2 was detected in the presence of ionomycin with an apparent SRETmax of 0.24 ± 0.08 and an apparent SRET50 of 0.051 ± 0.003. Therefore, calcium binding to CaM led to a decrease in the apparent SRETmax and an increase in the apparent SRET50 values. CaM-A2A-D2 receptor oligomers seem to be stable complexes that are not disrupted by calcium binding to CaM.
FIGURE 7.
Effect of intracellular Ca2+ on the molecular interactions between CaM, A2A, and D2 receptors. A, effect of Ca2+ levels on the A2A-D2 receptor heteromerization detected by BRET. BRET saturation curves were performed with HEK-293 cells co-expressing A2ARRluc (0.6 μg of cDNA) and increasing amounts of D2RYFP (0.3–4 μg of cDNA). Cells in HBSS buffer containing 1.26 mm CaCl2 were untreated (black) or treated with 1 μm ionomycin for 2 (red), 5 (green) or 10 (blue) min before BRET determination. Both fluorescence and luminescence for each sample were measured to confirm similar donor expressions (about 100,000 luminescent units) while monitoring the increase acceptor expression (1,000–15,000 fluorescent units). The relative amount of acceptor is given as the ratio between the fluorescence of the acceptor (YFP) and the luciferase activity of the donor (Rluc). BRET data are expressed as means ± S.D. of four different experiments grouped as a function of the amount of BRET acceptor. mBU, milli-BRET units. B, effect of Ca2+ levels on the CaM-A2A-D2 receptor oligomerization detected by SRET. SRET saturation curves were performed in HEK-293 cells expressing A2ARRluc, CaMGFP2, and increasing amounts of D2RYFP, as described in the legend to Fig. 6. Cells in HBSS buffer containing 1.26 mm CaCl2 were treated with 1 μm ionomycin for 10 min before SRET determination as described in the legend to Fig. 6, and the SRET saturation curve was compared with the one obtained in the absence of ionomycin (dotted line; see Fig. 6).
Effects of Intracellular Ca2+ on the Function of A2A-D2 Receptor Heteromers
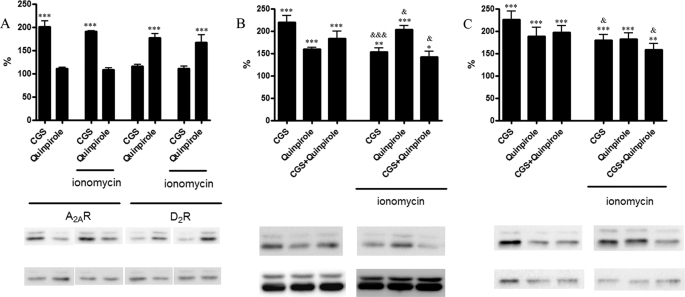
To test whether calcium binding to CaM-A2A-D2 receptor oligomers modifies receptor functionality, MAPK pathway signaling in cells expressing CaM and A2AR, D2R, or both receptors was analyzed. ERK1/2 phosphorylation was determined in cells treated with a 200 nm concentration of the A2A receptor agonist CGS21680, and/or with 1 μm D2 receptor agonist quinpirole in the presence and in the absence of 1 μm ionomycin. As shown in Fig. 8A, activation of A2A or D2 receptors, in cells transfected with low amounts of cDNA for CaM (0.3 μg) and expressing A2A or D2 receptors, induced similar ERK1/2 phosphorylation in the absence or in the presence of ionomycin. On the other hand, in cells transfected with 0.3 μg of cDNA for CaM and expressing the two receptors, a different effect was obtained in the absence or in the presence of ionomycin. In this case, ionomycin significantly reduced or potentiated ERK1/2 phosphorylation induced by activation of A2A or D2 receptors (Fig. 8B), respectively. The well known antagonistic interaction between A2A and D2 receptors was enhanced upon treatment with ionomycin. Results similar to those in Fig. 8, A and B, were obtained if cells were expressing higher amounts of CaM (1 μg of cDNA; results not shown), indicating that moderate amounts of exogenous CaM are already able to modulate receptor operation. In contrast, the effects of ionomycin were not observed in cells expressing only endogenous CaM (Fig. 8C). Therefore, the effects of ionomycin were dependent on the intracellular levels of CaM. Results similar to those in Fig. 8, A and B, were also obtained using cells in suspension (not adherent cells) (i.e. in the same conditions as those used for BRET and SRET experiments) (results not shown). Overall, the results suggest a correlation between structural changes in the oligomer and the modulation of its functionality exerted by Ca+2 binding.
FIGURE 8.
Effect of intracellular Ca2+ on A2A receptor-, D2 receptor-, and A2A-D2 receptor heteromer-mediated ERK1/2 phosphorylation. HEK-293 cells expressing A2A receptor (1.2 μg of cDNA) or D2 receptor (1 μg of cDNA) and CaM (0.3 μg of cDNA) (A); A2A receptor (1.2 μg of cDNA), D2R (1 μg of cDNA), and CaM (0.3 μg of cDNA) (B); or A2A receptor (1.2 μg of cDNA) and D2R (1 μg of cDNA) (C) were placed in HBSS buffer containing 1.26 mm CaCl2 and treated 3 min with vehicle or with 1 μm ionomycin before the addition of the A2A receptor agonist CGS21680 (200 nm; CGS), the D2 receptor agonist quinpirole (1 μm), or both. A representative Western blot from ERK1/2 phosphorylation assays, which were performed as described under “Experimental Procedures,” is shown. No significant differences in the basal levels were detected by the presence of CaM or CaM plus ionomycin. The immunoreactive bands from 4–6 independent experiments were quantified, and values represent the mean ± S.E. of percentage of increase of phosphorylation over the basal levels (100%) found in ionomycin-untreated or -treated cells. Significant differences of agonist-treated versus basal (*) or agonist-treated in the presence versus the absence of ionomycin (&) were calculated by one-way analysis of variance and Bonferroni test (* and &, p < 0.05; ** and &&, p < 0.01; *** and &&&, p < 0.005).
DISCUSSION
Ca2+ plays an important role in the physiology of higher order organisms and is involved in the regulation of many cellular events. Various stimuli, such as membrane depolarization or binding of ligands to plasma transmembrane receptors, trigger Ca2+ channel opening, which results in a significant influx of Ca2+ into the cytosol. Then Ca2+-binding proteins act as sensors and mediators of the initial Ca2+ signal. CaM is a highly conserved, soluble, intracellular 17-kDa Ca2+-binding protein, regarded as a major transducer of Ca2+ signals in mammalian cells (30). CaM has been shown to bind directly to cytoplasmic domains of plasma membrane proteins, including G-protein-coupled receptors, such as opioid μ, serotonin 5-HT1A, acetylcholine muscarinic, and dopamine D2 receptors (21, 31–33). In the present work, we also provide clear evidence for the binding of CaM to adenosine A2A receptors. It was demonstrated that the sequence RIREFRQTFR, which is present in the proximal part of the C terminus of the A2A receptor, is the CaM binding domain in these receptors. Furthermore, we provide evidence for CaM-A2A-D2 receptor oligomerization and for a specific Ca2+-dependent modulation of A2A-D2 receptor heteromer function. Previous studies have shown that an Arg-rich domain of the N-terminal portion of the 3IL of the D2 receptor is involved in the binding to Gi/o proteins, CaM, and the A2A receptor (14, 20, 22, 24, 34). The binding of CaM to the D2 receptor disrupts Gi/o coupling and, therefore, its ability to inhibit the activity of adenylyl cyclase (22). We here report competition of CaM and A2A receptors for the same domain in the D2 receptor. This competition was particularly evident when the short isoform of the D2 receptor was assayed. The two isoforms of the D2 receptor (short and long, D2S and D2L, respectively) are generated by alternative splicing (35). D2L, the isoform used in most of the experiments of the present study, contains 29 additional amino acid residues within the middle portion of the 3IL. The Arg-rich epitope of the D2 receptor (VLRRRRKRVN) that is able to interact with the A2A receptor is common to both D2 receptor isoforms and is localized in the N-terminal portion of their long 3IL (14, 20). However, there is an additional Arg-rich epitope in the 3IL of D2L (VNRRRVEAA), which is not present in D2S and which can potentially interact with CaM or with the A2A receptor (21). The VNRRRVEAA peptide interacts in vitro with CaM and with a phosphorylated Ser-containing epitope of the C terminus of the A2A receptor that is involved in A2A-D2 receptor heteromerization (data not shown). In BRET competition experiments using D2SRRluc-CaMYFP, the increase of A2A receptor expression produced a significant decrease of the BRET signal. On the other hand, when co-transfecting the A2A receptor, the BRET signal between D2LRRluc and CaMYFP first increased, whereas higher expression levels led to a reduced BRET signal. It is possible that, in the D2L receptor, CaM binds preferentially to the VLRRRRKRVN epitope and that increasing quantities of transfected A2A receptor induce, first, translocation of CaM to the VNRRRVEAA epitope, thus leading to an enhancement of the BRET signal. Upon further increase of receptor expression, A2AR may also bind to the VNRRRVEAA epitope, with the consequent displacement of CaM from the D2L receptor. The results from BRET competition experiments demonstrating that increasing expression of D2 receptor did not modify the BRET signal due to A2RRluc and CaMYFP and that increasing expression of CaM did not modify the BRET signal due to A2RRluc and D2RYFP were also compatible with the possibility of CaM-A2A-D2 receptor oligomerization with A2A receptors able to bind simultaneously to CaM and to the D2 receptor. This was demonstrated with the use of the recently introduced sequential SRET techniques, which allow the demonstration of direct interaction of three protein molecules (29).
The results not only indicated that CaM may interact with A2A receptors in cells that express A2A-D2 receptor heteromers but also that the conformation and function of A2A-D2 receptor heteromers are modulated by Ca2+. In fact ionomycin, an ionophore able to increase intracellular Ca2+, did affect the energy transfer between A2ARRluc and D2RYFP. Agonist-induced conformational modifications in a receptor heteromer can be demonstrated by using resonance energy transfer techniques (36). In the case of the A2A-D2 receptor heteromer and by means of the same techniques used in the present study, no energy transfer variations were found after exposure to A2A or D2 receptor agonists (12). Therefore, the ability of BRET techniques to detect Ca2+-triggered conformational changes within the CaM-A2A receptor, CaM-D2 receptor, and CaM-A2A-D2 receptor oligomers suggests that Ca2+ exerts a strong CaM-dependent control of the function of A2A and D2 receptors and A2A-D2 receptor heteromers. Interestingly, Ca2+ exerted a selective modulation of A2A-D2 receptor heteromer-mediated activation of the MAPK pathway. On the other hand, Ca2+ binding to CaM did not individually modulate the receptor-mediated signaling toward the MAPK pathway, and Ca2+ significantly modified ERK1/2 phosphorylation induced by activation of A2A or of D2 receptors in cells co-expressing the two receptors. Furthermore, the previously described (18) antagonistic interaction between A2A and D2 receptors was regulated by ionomycin-mediated variation in the intracellular calcium concentration.
The present study demonstrates the existence of oligomeric intermolecular interactions between CaM, A2A, and D2 receptors. When not expressed together, A2A or D2 receptors may potentially bind CaM, but in cells expressing the A2A-D2 receptor heteromer, CaM binds preferentially to the A2A receptor. Furthermore, CaM transduces Ca2+-dependent changes of MAPK signaling in the CaM-A2A-D2 receptor oligomer. This work adds new information about the function of A2A-D2 receptor heteromers, which are considered as a target for the development of anti-parkinsonian agents (37).
Acknowledgments
We acknowledge the technical help of Drs. Laura Sesma, Fernando Corrales, and Carmen Miqueo (Proteomics Laboratory, CIMA, Universidad de Navarra), Carmen Molina (Laboratory of Basal Ganglia Neuromorphology, CIMA, Universidad de Navarra), and Jasmina Jiménez (Molecular Neurobiology Laboratory, Barcelona University).
This work was supported, in whole or in part, by the National Institutes of Health, NIDA, Intramural Research Program (to S. F. and A. S. W.). This work was also supported by Spanish Ministry of Education and Science Grants SAF2008-00146 (to E. I. C.) and SAF2006-05481 (to R. F.) and Fundació La Marató de TV3 Grant 060110 (to E. I. C.). The Proteomics Laboratory at CIMA belongs to the network of the Spanish National Institute of Proteomics Facilities, ProteoRed.
- FRET
- fluorescence resonance energy transfer
- BRET
- bioluminescence resonance energy transfer
- CaM
- calmodulin
- SRET
- sequential resonance energy transfer
- 3IL
- third intracellular loop
- MAPK
- mitogen-activated protein kinase
- GFP
- green fluorescent protein
- YFP
- yellow fluorescent protein
- EYFP
- enhanced yellow fluorescent protein
- HBSS
- Hanks' balanced salt solution
- ERK
- extracellular signal-regulated kinase.
REFERENCES
- 1.Agnati L. F., Ferré S., Lluis C., Franco R., Fuxe K. (2003) Pharmacol. Rev. 55, 509–550 [DOI] [PubMed] [Google Scholar]
- 2.Pin J. P., Neubig R., Bouvier M., Devi L., Filizola M., Javitch J. A., Lohse M. J., Milligan G., Palczewski K., Parmentier M., Spedding M. (2007) Pharmacol. Rev. 59, 5–13 [DOI] [PubMed] [Google Scholar]
- 3.Lee S. P., O'Dowd B. F., George S. R. (2003) Life Sci. 74, 173–180 [DOI] [PubMed] [Google Scholar]
- 4.Rashid A. J., So C. H., Kong M. M., Furtak T., El-Ghundi M., Cheng R., O'Dowd B. F., George S. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prinster S. C., Holmqvist T. G., Hall R. A. (2006) J. Pharmacol. Exp. Ther. 318, 974–981 [DOI] [PubMed] [Google Scholar]
- 6.Prinster S. C., Hague C., Hall R. A. (2005) Pharmacol. Rev. 57, 289–298 [DOI] [PubMed] [Google Scholar]
- 7.Ferré S., Baler R., Bouvier M., Caron M. G., Devi L. A., Durroux T., Fuxe K., George S. R., Javitch J. A., Lohse M. J., Mackie K., Milligan G., Pfleger K. D., Pin J. P., Volkow N. D., Waldhoer M., Woods A. S., Franco R. (2009) Nat. Chem. Biol. 5, 131–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milligan G., Bouvier M. (2005) FEBS J. 272, 2914–2925 [DOI] [PubMed] [Google Scholar]
- 9.Pfleger K. D., Eidne K. A. (2006) Nat. Methods 3, 165–174 [DOI] [PubMed] [Google Scholar]
- 10.Kocan M., See H. B., Seeber R. M., Eidne K. A., Pfleger K. D. (2008) J. Biomol. Screen. 13, 888–898 [DOI] [PubMed] [Google Scholar]
- 11.Hillion J., Canals M., Torvinen M., Casado V., Scott R., Terasmaa A., Hansson A., Watson S., Olah M. E., Mallol J., Canela E. I., Zoli M., Agnati L. F., Ibanez C. F., Lluis C., Franco R., Ferre S., Fuxe K. (2002) J. Biol. Chem. 277, 18091–18097 [DOI] [PubMed] [Google Scholar]
- 12.Canals M., Marcellino D., Fanelli F., Ciruela F., de Benedetti P., Goldberg S. R., Neve K., Fuxe K., Agnati L. F., Woods A. S., Ferré S., Lluis C., Bouvier M., Franco R. (2003) J. Biol. Chem. 278, 46741–46749 [DOI] [PubMed] [Google Scholar]
- 13.Kamiya T., Saitoh O., Yoshioka K., Nakata H. (2003) Biochem. Biophys. Res. Commun. 306, 544–549 [DOI] [PubMed] [Google Scholar]
- 14.Ciruela F., Burgueño J., Casadó V., Canals M., Marcellino D., Goldberg S. R., Bader M., Fuxe K., Agnati L. F., Lluis C., Franco R., Ferré S., Woods A. S. (2004) Anal. Chem. 76, 5354–5363 [DOI] [PubMed] [Google Scholar]
- 15.Ferre S., von Euler G., Johansson B., Fredholm B. B., Fuxe K. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7238–7241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferré S., Fredholm B. B., Morelli M., Popoli P., Fuxe K. (1997) Trends Neurosci. 20, 482–487 [DOI] [PubMed] [Google Scholar]
- 17.Ferré S., Ciruela F., Quiroz C., Luján R., Popoli P., Cunha R. A., Agnati L. F., Fuxe K., Woods A. S., Lluis C., Franco R. (2007) Scientific WorldJournal 7, 74–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferré S., Quiroz C., Woods A. S., Cunha R., Popoli P., Ciruela F., Lluis C., Franco R., Azdad K., Schiffmann S. N. (2008) Curr. Pharm. Des. 14, 1468–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altamura A. C., Bassetti R., Cattaneo E., Vismara S. (2005) World J. Biol. Psychiatry 6, Suppl. 2, 23–30 [DOI] [PubMed] [Google Scholar]
- 20.Woods A. S., Ferré S. (2005) J. Proteome Res. 4, 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woods A. S., Ciruela F., Fuxe K., Agnati L. F., Lluis C., Franco R., Ferré S. (2005) J Mol. Neurosci. 26, 125–132 [DOI] [PubMed] [Google Scholar]
- 22.Bofill-Cardona E., Kudlacek O., Yang Q., Ahorn H., Freissmuth M., Nanoff C. (2000) J. Biol. Chem. 275, 32672–32680 [DOI] [PubMed] [Google Scholar]
- 23.Liu Y., Buck D. C., Macey T. A., Lan H., Neve K. A. (2007) J. Recept. Signal. Transduct. Res. 27, 47–65 [DOI] [PubMed] [Google Scholar]
- 24.Woods A. S., Marcellino D., Jackson S. N., Franco R., Ferré S., Agnati L. F., Fuxe K. (2008) J. Proteome Res. 7, 3428–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woods A. S. (2004) J. Proteome Res. 3, 478–484 [DOI] [PubMed] [Google Scholar]
- 26.Woods A. S., Huestis M. A. (2001) J. Am. Soc. Mass Spectrom. 12, 88–96 [DOI] [PubMed] [Google Scholar]
- 27.Muñoz J., Fernández-Irigoyen J., Santamaría E., Parbel A., Obeso J., Corrales F. J. (2008) Proteomics 8, 1898–1908 [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann T., Rietdorf J., Girod A., Georget V., Pepperkok R. (2002) FEBS Lett. 531, 245–249 [DOI] [PubMed] [Google Scholar]
- 29.Carriba P., Navarro G., Ciruela F., Ferré S., Casadó V., Agnati L., Cortés A., Mallol J., Fuxe K., Canela E. I., Lluís C., Franco R. (2008) Nat. Methods 5, 727–733 [DOI] [PubMed] [Google Scholar]
- 30.Vetter S. W., Leclerc E. (2003) Eur. J. Biochem. 270, 404–414 [DOI] [PubMed] [Google Scholar]
- 31.Belcheva M. M., Szùcs M., Wang D., Sadee W., Coscia C. J. (2001) J. Biol. Chem. 276, 33847–33853 [DOI] [PubMed] [Google Scholar]
- 32.Turner J. H., Gelasco A. K., Raymond J. R. (2004) J. Biol. Chem. 279, 17027–17037 [DOI] [PubMed] [Google Scholar]
- 33.Lucas J. L., Wang D., Sadée W. (2006) Pharm. Res. 23, 647–653 [DOI] [PubMed] [Google Scholar]
- 34.Senogles S. E., Heimert T. L., Odife E. R., Quasney M. W. (2004) J. Biol. Chem. 279, 1601–1606 [DOI] [PubMed] [Google Scholar]
- 35.Missale C., Nash S. R., Robinson S. W., Jaber M., Caron M. G. (1998) Physiol. Rev. 78, 189–225 [DOI] [PubMed] [Google Scholar]
- 36.Vilardaga J. P., Nikolaev V. O., Lorenz K., Ferrandon S., Zhuang Z., Lohse M. J. (2008) Nat. Chem. Biol. 4, 126–131 [DOI] [PubMed] [Google Scholar]
- 37.Müller C. E., Ferré S. (2007) Recent Pat. CNS Drug Discov. 2, 1–21 [DOI] [PubMed] [Google Scholar]