Abstract
ARAP1 is a phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3)-dependent Arf GTPase-activating protein (GAP) with five PH domains that regulates endocytic trafficking of the epidermal growth factor receptor (EGFR). Two tandem PH domains are immediately N-terminal of the Arf GAP domain, and one of these fits the consensus sequence for PtdIns(3,4,5)P3 binding. Here, we tested the hypothesis that PtdIns(3,4,5)P3-dependent recruitment mediated by the first PH domain of ARAP1 regulates the in vivo and in vitro function of ARAP1. We found that PH1 of ARAP1 specifically bound to PtdIns(3,4,5)P3, but with relatively low affinity (≈1.6 μm), and the PH domains did not mediate PtdIns(3,4,5)P3-dependent recruitment to membranes in cells. However, PtdIns(3,4,5)P3 binding to the PH domain stimulated GAP activity and was required for in vivo function of ARAP1 as a regulator of endocytic trafficking of the EGFR. Based on these results, we propose a variation on the model for the function of phosphoinositide-binding PH domains. In our model, ARAP1 is recruited to membranes independently of PtdIns(3,4,5)P3, the subsequent production of which triggers enzymatic activity.
Pleckstrin homology (PH)2 domains are a common structural motif encoded by the human genome (1, 2). Approximately 10% of PH domains bind to phosphoinositides. These PH domains are thought to mediate phosphoinositide-dependent recruitment to membranes (1–3). Most PH domains likely have functions other than or in addition to phosphoinositide binding. For example, PH domains have been found to bind to protein and DNA (4–12). In addition, some PH domains have been found to be structurally and functionally integrated with adjacent domains (13, 14). A small fraction of PH domain-containing proteins (about 9% of the human proteins) have multiple PH domains arranged in tandem, which have been proposed to function as adaptors but have only been examined in one protein (15, 16). Arf GTPase-activating proteins (GAPs) of the ARAP family are phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3)-dependent Arf GAPs with tandem PH domains (17, 18). The function of specific PH domains in regulating Arf GAP activity and for biologic activity has not been described.
Arf GAPs are proteins that induce the hydrolysis of GTP bound to Arfs (19–23). The Arf proteins are members of the Ras superfamily of GTP-binding proteins (24–27). The six Arf proteins in mammals (five in humans) are divided into three classes based on primary sequence: Arf1, -2, and -3 are class 1, Arf4 and -5 are class 2, and Arf6 is class 3 (23, 24, 27–29). Class 1 and class 3 Arf proteins have been studied more extensively than class 2. They have been found to regulate membrane traffic and the actin cytoskeleton.
The Arf GAPs are a family of proteins with diverse domain structures (20, 21, 23, 30). ARAPs, the most structurally complex of the Arf GAPs, contain, in addition to an Arf GAP domain, the sterile α motif (SAM), five PH, Rho GAP, and Ras association domains (17, 18, 31, 32). The first and second and the third and fourth PH domains are tandem (Fig. 1). The first and third PH domains of the ARAPs fit the consensus for PtdIns(3,4,5)P3 binding (33–35). ARAPs have been found to affect actin and membrane traffic (21, 23). ARAP3 regulates growth factor-induced ruffling of porcine aortic endothelial cells (31, 36, 37). The function is dependent on the Arf GAP and Rho GAP domains. ARAP2 regulates focal adhesions, an actin cytoskeletal structure (17). ARAP2 function requires Arf GAP activity and a Rho GAP domain capable of binding RhoA·GTP. ARAP1 has been found to have a role in membrane traffic (18). The protein associates with pre-early endosomes involved in the attenuation of EGFR signals. The function of the tandem PH domains in the ARAPs has not been examined.
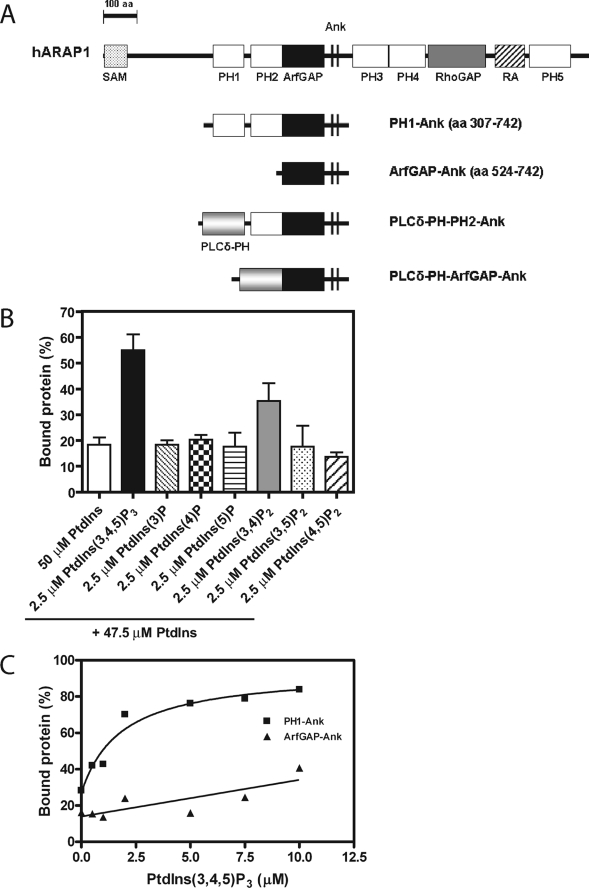
FIGURE 1.
ARAP1 binding to phospholipids. A, schematic of the recombinant proteins used in this study. Domain abbreviations: Ank, ankyrin repeat; PLCδ-PH, PH domain of phospholipase C δ; RA, Ras association motif; RhoGAP, Rho GTPase-activating domain. B, ARAP1 phosphoinositide binding specificity. 500 nm PH1-Ank recombinant protein was incubated with sucrose-loaded LUVs formed by extrusion through a 1-μm pore filter. LUVs contained PtdIns alone or PtdIns with 2.5 μm PtdIns(3,4,5)P3, 2.5 μm PtdIns(3)P, 2.5 μm PtdIns(4)P, 2.5 μm PtdIns(5)P, 2.5 μm PtdIns(3,4)P2, 2.5 μm PtdIns(3,5)P2, or 2.5 μm PtdIns(4,5)P2 with a total phosphoinositide concentration of 50 μm and a total phospholipid concentration of 500 μm. Vesicles were precipitated by ultracentrifugation, and associated proteins were separated by SDS-PAGE. The amount of precipitated protein was determined by densitometry of the Coomassie Blue-stained gels with standards on each gel. C, PtdIns(3,4,5)P3-dependent binding of ARAP1 to LUVs. 1 μm PH1-Ank or ArfGAP-Ank recombinant protein was incubated with 1 mm sucrose-loaded LUVs formed by extrusion through a 1-μm pore size filter containing varying concentration of PtdIns(3,4,5)P3. Precipitation of LUVs and analysis of associated proteins were performed as described in B. The average ± S.E. of three independent experiments is presented.
Here we investigated the role of the first two PH domains of ARAP1 for catalysis and in vivo function. The first PH domain fits the consensus sequence for PtdIns(3,4,5)P3 binding (33–35). The second does not fit a phosphoinositide binding consensus but is immediately N-terminal to the GAP domain. We have previously reported that the PH domain that occurs immediately N-terminal of the Arf GAP domain of ASAP1 is critical for the catalytic function of the protein (38, 39). We tested the hypothesis that the two PH domains of ARAP1 function independently; one recruits ARAP1 to PtdIns(3,4,5)P3-rich membranes, and the other functions with the catalytic domain. As predicted, PH1 interacted specifically with PtdIns(3,4,5)P3, and PH2 did not. However, both PH domains contributed to catalysis independently of recruitment to membranes. None of the PH domains in ARAP1 mediated PtdIns(3,4,5)P3-dependent targeting to plasma membranes (PM). PtdIns(3,4,5)P3 stimulated GAP activity, and the ability to bind PtdIns(3,4,5)P3 was required for ARAP1 to regulate membrane traffic. We propose that ARAP1 is recruited independently of PtdIns(3,4,5)P3 to the PM where PtdIns(3,4,5)P3 subsequently regulates its GAP activity to control endocytic events.
EXPERIMENTAL PROCEDURES
Plasmids
The cDNA fragments encoding the proteins described in Fig. 1 were cloned into NdeI and BamHI sites of the pET-19b bacterial expression vector (Novagen). The cDNA fragment encoding for PH1, PH2, PH1-PH2, PH3, PH4, or PH3-PH4 was cloned into the XhoI and BamHI sites of the mammalian expression vector pEGFP-C1 (Clontech). Point mutations were introduced using the QuikChange kit (Stratagene) following the manufacturer's instructions and confirmed by DNA sequencing.
An expression vector for a chimeric protein consisting of the PH domain of PLCδ (residues 1–134 (40)) followed by the PH2, ArfGAP, and Ank motifs of ARAP1 or the ArfGAP and Ank motifs of ARAP1 (see Fig. 1) was constructed by amplifying the open reading frame (ORF) encoding residues 427–742 or residues 524–742 of ARAP1 with NdeI and SalI restriction sites on the 5′-end of the ORF and BamHI site at the 3′-end. This DNA was ligated into pET-19b using the NdeI and BamHI sites. The ORF encoding residues 1–134 of PLCδ was amplified incorporating NdeI restriction sites on the 5′-end of the ORF and SalI and BamHI restriction sites on the 3′-end. This DNA was ligated into pET-19b using the NdeI and BamHI sites. After restriction digestion with SalI and BamHI, the ARAP1 fragments were ligated into the vector containing the PH domain of PLCδ digested with SalI and BamHI. A plasmid for the expression of GFP-tagged Rab5 in mammalian cells was a generous gift from Juan Bonifacino (NICHD, National Institutes of Health, Bethesda, MD).
Protein Purification
To obtain myristoylated Arf5, human Arf5 was coexpressed with N-myristoyltransferase in Escherichia coli as described for Arf1 and Arf6 (41). MyrArf5 was purified as described for myrArf1 (39, 42), and the identity of the product was confirmed by mass spectrometry. MyrArf1 was purified as described above, and myrArf6 was purified as described previously (42, 43).
Non-myristoylated Arf5 and [L8K]Arf5 were purified as described previously (44). Because the preparations were contaminated with nucleotidase activity, the proteins were further purified using a Hiload 26/60 Superdex G75 column (GE Healthcare) developed in 20 mm Tris, pH 8.0, 100 mm NaCl, 1 mm MgCl2, 1 mm dithiothreitol, and 10% (v/v) glycerol and by fractionation on a hydroxylapatite column developed in a potassium Pi gradient from 20 mm to 500 mm. Non-myristoylated Arf1 and Arf6 were purified as described (44).
His10-tagged recombinant ARAP1 proteins, including PH1-Ank, ArfGAP-Ank, and chimeric PLCδ/ARAP1 proteins were purified as described for His10-ASAP1 (45, 46). Purification of GST-VHS-GATGGA3 was described previously (47, 48).
Large Unilamellar Vesicles (LUVs)
LUVs were composed of 40% phosphatidylcholine, 25% phosphatidylethanolamine, 15% phosphatidylserine, 7% PtdIns, 2.5% PtdIns(4,5)P2, 0.5% PtdIns(3,4,5)P3, and 10% cholesterol (unless indicated in the text) and were formed by extrusion as described (49). Lipids were purchased from Avanti Polar Lipids and, for experiments reported in Figs. 1B and 4, from Echelon.
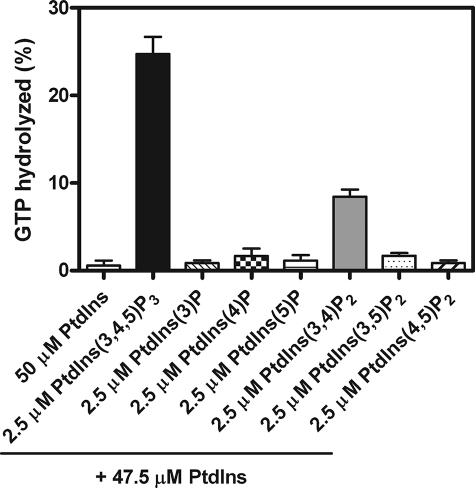
FIGURE 4.
Phosphoinositide dependence of PH1-Ank GAP activity. Reactions contained 0.03 nm PH1-Ank recombinant protein, myrArf5·GTP as a substrate, and LUVs comprising each of the different phosphorylated derivatives (2.5 μm) of phosphatidylinositol, a total phosphoinositide concentration of 50 μm, and a total phospholipid concentration of 500 μm. The averages from three experiments ± S.E. are presented.
Protein Association with LUVs
Protein association with LUVs was measured as described previously (49). Briefly, the purified recombinant proteins (500 nm or 1 μm) were incubated with sucrose-loaded LUVs containing 500 μm or 1 mm total phospholipids for 5 min at 30 °C. The LUVs were precipitated by ultracentrifugation at 75,000 rpm for 15 min at 4 °C. The proteins that precipitated with LUVs were separated by SDS-PAGE and visualized by Coomassie Blue staining. The signal was quantified by densitometry using Scion Image software.
Cell Culture and Transfection
HeLa and COS-7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in a 5% CO2 atmosphere at constant humidity. Cells were plated 1 day before transfection. For the experiments to examine protein localization, HeLa cells were transfected with plasmids using FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. COS-7 cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
For RNA interference assays, HeLa cells were transfected for 48 h with 19 nucleotide siRNA duplexes with 3′-UU overhangs specific for the 3′ untranslated region (5′-GUUCAGACCUCUUGGCCCAUU-3′) and the ORF (ON-TARGETplus SMARTpool) of human ARAP1 (GenBankTM accession number NM_139181) from Dharmacon. siCONTROL non-targeting siRNA 2 (Dharmacon) was used as a negative control. RNA interference transfections were performed using Oligofectamine (Invitrogen) according to the manufacturer's instructions. Endogenous ARAP1 was replaced with recombinant ARAP1 by first treating cells with siRNA and then, 48 h later, transfecting the cells with a plasmid for expression of ARAP1 using Lipofectamine 2000. The cells were used 24 h after the second transfection.
Immunofluorescence
Cells grown on glass coverslips coated with 10 μg/ml fibronectin (F1141, Sigma) were fixed with 3% paraformaldehyde for 20 min at room temperature or overnight at 4 °C. Cells were washed three times with phosphate-buffered saline (PBS), blocked, and lysed with 0.2% Triton X-100 and 10% fetal bovine serum in PBS for 20 min at room temperature. Cells were incubated with primary antibody anti-FLAG M5 monoclonal antibody (Sigma) diluted in 3% fetal bovine serum and 0.05% Tween 20 in PBS for 1 h at room temperature. Cells were washed twice with PBS, blocked for 5 min, and incubated with the appropriate secondary antibodies (Jackson ImmunoResearch), diluted as described previously, for 50 min. For EGF uptake experiment, HeLa cells were plated on glass coverslips in Dulbecco's modified Eagle's medium containing 10% fetal calf serum overnight and serum-starved for 1h at 37 °C in prewarmed uptake Dulbecco's modified Eagle's medium containing 20 mm HEPES, pH 7.4, and 1% bovine serum albumin. Cells were then incubated with1 μg/ml Alexa488-EGF (Molecular Probe) for 30 min on ice in ice-cold serum-free uptake medium and washed once in ice-cold PBS. Cells in prewarmed uptake medium were incubated for 10 min at 37 °C. The cells were cooled rapidly at 4 °C and incubated with ice-cold acetic acid buffer (pH 3.5) in ice-water for 2 min to remove fluorescence-conjugated reagents remaining at the cell surface. Cells were washed twice with ice-cold PBS before being fixed with 3% paraformaldehyde. All preparations were analyzed using a Zeiss LSM510-Meta laser-scanning confocal microscope with a ×63, 1.4 NA Plan Neofluar oil immersion lens. Images were processed using the Zeiss LSM 5 image software.
Pearson's coefficients were determined for 30 cells under each condition using Imarisx64 software. To determine the Pearson's coefficients for the images presented in Fig. 10, thresholds were set to exclude ARAP1 localized in the cytoplasm. The same threshold was applied between cells stimulated or not with EGF in one group. Association of protein with the cell edge was determined by visual inspection.
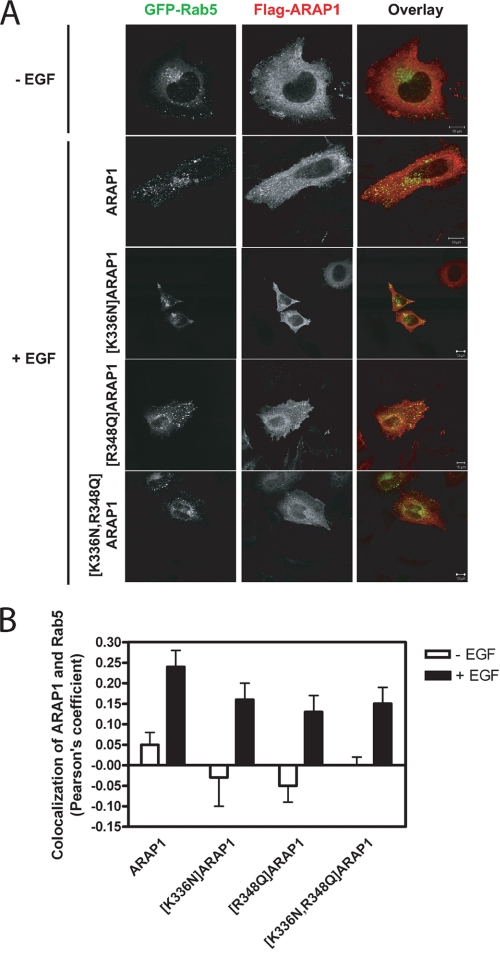
FIGURE 10.
Effect of EGF on association of ARAP1 and mutants with reduced PtdIns(3,4,5)P3 binding with a Rab5-positive endocytic compartment. A, images of representative cells expressing FLAG-ARAP1. HeLa cells expressing N-terminal FLAG-tagged ARAP1 or ARAP1 mutants and GFP-Rab5 were serum-starved overnight on fibronectin-coated coverslips (10 μg/ml) and treated with 100 ng/ml EGF for 5 min at 37 °C. Fixed cells were immunostained with monoclonal anti-FLAG antibody (red). Scale bar, 10 μm. B, quantification of colocalization of ARAP1 and mutants with Rab5-positive endosome. Effect of mutations, which affect binding to PtdIns(3,4,5)P3, in colocalization of ARAP1 with Rab5. Pearson's coefficients for colocalization of ARAP1 and Rab5 were determined as described under ”Experimental Procedures“ for at least 30 cells in each condition.
Live Cell Imaging
COS-7 cells were plated on glass coverslips (3 × 105 cells/35-mm dish) and 1 day after seeding were transfected with the indicated constructs (1 μg of DNA total/dish) using Lipofectamine 2000 for 24 h. Coverslips were mounted in metal Atto chambers (Invitrogen), and cells were incubated in a modified Krebs-Ringer buffer containing 120 mm NaCl, 4.7 mm KCl, 1.2 mm CaCl2, 0.7 mm MgSO4, 10 mm glucose, Na-Hepes 10 mm, pH 7.4. Confocal analysis was performed at 35 °C using a Zeiss LSM510-Meta confocal microscope. Post-acquisition processing was done using Adobe Photoshop to utilize the whole dynamic range of the image, but no change in the linearity was allowed.
Effects of ARAP1 on in Vivo Arf·GTP
COS-7 cells were transfected with plasmids directing expression of GFP-tagged Arf proteins and FLAG-tagged ARAP1 as indicated. 24 h after transfection, cellular levels of Arf·GTP proteins were determined as described (50). Briefly, purified recombinant GST-VHS-GATGGA3 (10 μg) were immobilized on glutathione-Sepharose 4B beads (GE Healthcare) for 1 h at room temperature. After three washes with ice-cold PBS, the immobilized GST fusion proteins were incubated for 1 h at 4 °C with equal amounts of lysates from COS-7 cells. The beads were collected by centrifugation and washed three times with ice-cold PBS. Proteins were eluted from the beads by boiling in SDS-PAGE sample buffer and detected by standard immunoblotting methods using mouse anti-GFP (Covance, MMS-118P) and rabbit anti-FLAG (Sigma, F7425) antisera. Signals from the immunoblots were quantified by densitometric scanning and plotted as the ratio of Arf·GTP level to the Arf·GTP level in control cells.
Miscellaneous
Arf GAP assays, in which hydrolysis of [α32P]GTP bound to Arf is measured, have been described previously (45, 51). Anti-Hsc70 antibody used to detect protein loading was from Santa Cruz Biotechnology. Protein concentrations were estimated using the Bio-Rad assay.
1,2-Dioctanoyl-sn-glycero-3-[phosphoinositol-3,4,5-trisphoshate] (dioctanoyl PtdIns(3,4,5)P3) and d-myo-inositol-1,3,4,5-tetraphosphate (IP4) were from Avanti Polar Lipids. Circular dichroism spectra were collected using a Jasco J720 CD spectropolarimeter for ARAP1 recombinant proteins diluted to 4 μm in PBS.
RESULTS
ARAP1 Binds to PtdIns(3,4,5)P3 through PH1
Bacterially expressed recombinant ARAP1 has not been characterized previously. We expressed and purified ARAP1-(307–742) (henceforth referred to as PH1-Ank), composed of the PH1, PH2, Arf GAP, and Ank repeats (see Fig. 1A for schematic of proteins used in this article) to study the function of the first set of tandem PH domains. LUVs containing the indicated phosphoinositides were incubated with PH1-Ank and rapidly separated from bulk solution by centrifugation. ARAP1 specifically associated with vesicles containing PtdIns(3,4,5)P3 and PtdIns(3,4)P2 (Fig. 1B). The Kd for the ARAP1·PtdIns(3,4,5)P3 complex, determined by titrating PtdIns(3,4,5)P3 into the binding reaction, was estimated to be 1.6 ± 0.3 μm (mean ± S.E., 13 experiments), which is similar to the concentration of PtdIns(3,4,5)P3 for half-maximal stimulation of GAP activity (Table 1). The binding of ARAP1-(524–742) (henceforth called ArfGAP-Ank), composed of the Arf GAP and Ank repeat domains and lacking the PH domains, to vesicles was not significantly affected by PtdIns(3,4,5)P3 (Fig. 1C). A recombinant protein that contained the second PH domain but not the first was not stable when expressed in bacteria.
TABLE 1.
ARAP1 PtdIns(3,4,5)P3-dependent Arf GAP activity and PtdIns(3,4,5)P3-ARAP1 association
PtdIns(3,4,5)P3, incorporated into LUVs with a total phospholipid concentration of 500 μm, was titrated into Arf GAP reactions containing 0.03 nm PH1-Ank, 0.03 nm mutant PH1-Ank, or 30 nm ArfGAP-Ank. For enzymatic activity, the reactions contained myrArf5¼GTP as a substrate. The average EC50 (concentration of PtdIns(3,4,5)P3 for half-maximal activity) ± S.E. from at least three independent experiments, estimated from the data in Fig. 5, is presented. The numbers in parentheses represent the number of experiments. For protein association, 1 μm protein was incubated with LUVs with a total phospholipid concentration of 1 mm. The Kd is estimated from the data in Fig. 2. The averages ± S.E. with the number of experiments in parentheses are presented.
| Recombinant protein | PtdIns(3,4,5)P3 dependence of enzymatic activity, EC50 | Lipid-protein association, Kd |
|---|---|---|
| μm | μm | |
| PH1-Ank | 1.6 ± 0.1 (7) | 1.6 ± 0.3 (13) |
| ArfGAP-Ank | >7.5 (3) | >10 (3) |
| [K336N]PH1-Ank | >7.5 (3) | >10 (4) |
| [K347N]PH1-Ank | 4.1 ± 1.7 (3) | 0.5 ± 0.2 (4) |
| [R348Q]PH1-Ank | >7.5 (3) | >10 (4) |
| [R351Q]PH1-Ank | 1.9 ± 0.4 (3) | 0.4 ± 0.1 (3) |
| [R450Q]PH1-Ank | 1.7 ± 0.4 (3) | 2.0 ± 0.4 (3) |
| [K453N]PH1-Ank | 2.7 ± 1.4 (3) | 1.9 ± 0.4 (3) |
| [K455N]PH1-Ank | 1.0 ± 0.2 (3) | 1.1 ± 0.1 (3) |
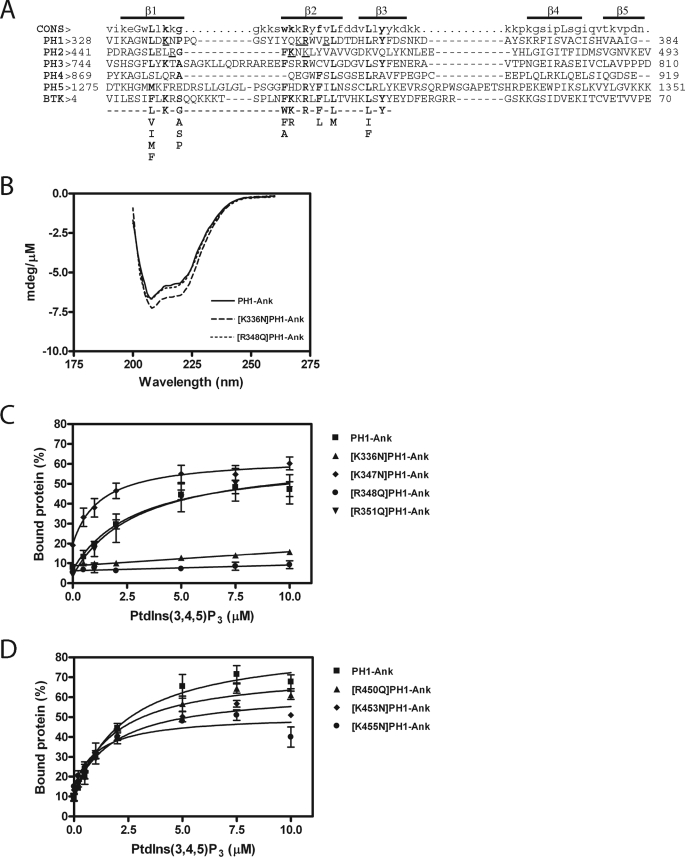
We examined the contribution of each PH domain to PtdIns(3,4,5)P3 binding. The PH domains of ARAP1 were aligned with PH domains known to bind PtdIns(3,4,5)P3 (Fig. 2A) to identify residues that might be involved in PtdIns(3,4,5)P3 binding. The effect of mutating the residues in PH1-Ank on binding to LUVs containing PtdIns(3,4,5)P3 was determined. Introduction of the mutations did not affect the circular dichroism spectra, indicating no gross changes in secondary structures (shown for [K336N] and [R348Q] in Fig. 2B). Two mutants, [K336N] and [R348Q], with changes in PH1 did not bind as efficiently as wild type PH1 (Fig. 2C and Table 1). Changes introduced into PH2 had less of an effect on binding to LUVs, although two mutations, [K453N] and [K455N], reduced the maximum protein binding by about 30% (Fig. 2D and Table 1).
FIGURE 2.
A, alignment of ARAP1 PH domains with the PH domain of the human Bruton's tyrosine kinase (GenBankTM accession number Q06187) and the consensus sequence for PtdIns(3,4,5)P3 binding PH domain-containing protein. Residues critical for binding to PtdIns(3,4,5)P3 are shown in bold type (33, 34). Residues that have been mutated in the first and second PH domains and tested in this study are underlined. The regions predicted to form β1, β2, β3, β4, and β5 strands are indicated. B, circular dichroism spectra of [K336N]PH1-Ank and [R348Q]PH1-Ank. CD spectra were determined for the indicated recombinant protein as described under ”Experimental Procedures.“ C and D, effect of mutations in PH1 (C) and PH2 (D) domains on PtdIns(3,4,5)P3-dependent binding. 1 μm PH1-Ank or mutant PH1-Ank was incubated with sucrose-loaded LUVs (total phospholipid concentration of 1 mm) formed by extrusion through a 1-μm pore size filter. LUVs contained the indicated concentrations of PtdIns(3,4,5)P3. The concentration of phosphatidylinositol was adjusted in each LUVs to maintain a total phosphoinositide concentration of 50 μm. Vesicles were precipitated by ultracentrifugation, and associated proteins were separated by SDS-PAGE. The amount of precipitated protein was determined by densitometry of the Coomassie Blue-stained gels with standards on each gel. The results presented are the average ± S.E. of at least three experiments.
PtdIns(3,4,5)P3-dependent Stimulation of ARAP1 GAP Activity Is, at Least in Part, Independent of Recruitment
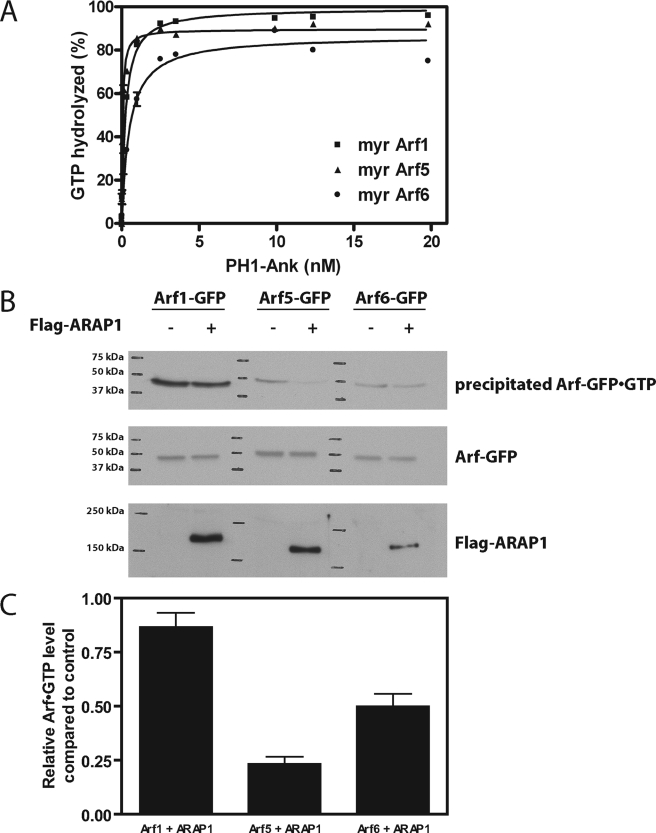
We further characterized the PtdIns(3,4,5)P3-dependent GAP activity of ARAP1. We started by determining the substrate specificity, which had not been examined previously for bacterially expressed recombinant ARAP1. Purified myrArf1·GTP, myrArf5·GTP, and myrArf6·GTP were used as representatives of class 1, 2, and 3 Arfs (Fig. 3A and Table 2). We also examined non-myristoylated Arfs (Table 2). PH1-Ank was titrated into the reaction to determine the amount required to induce hydrolysis of 50% of the bound GTP. We call this amount the C50, which is inversely proportional to the enzymatic power. Consistent with previous analyses using non-myristoylated Arfs and immunoprecipitated ARAP1, Arf5 was the preferred substrate (Fig. 3A and Table 2).
FIGURE 3.
Arf specificity. A, in vitro determination. PH1-Ank was titrated into reactions containing myrArf1·GTP, myrArf5·GTP, or myrArf6·GTP as substrate and LUVs containing 2.5 μm PtdIns(3,4,5)P3 and a total phospholipid concentration of 500 μm. Reactions were terminated after 3 min. Results averaged from three experiments are presented. The amount of enzyme to achieve 50% hydrolysis ± S.E. is given in Table 2. B, in vivo determination of Arf specificity. Arf1, Arf5, and Arf6 fused to GFP were coexpressed with N-terminal FLAG-tagged ARAP1 full-length wild type in COS-7 cells. Cells were lysed, and Arf1-GFP·GTP, Arf5-GFP·GTP, and Arf6-GFP·GTP were co-precipitated with GST-VHS-GATGGA3. The precipitated and overexpressed Arfs were detected by immunoblotting with monoclonal anti-GFP antibody, and the overexpressed ARAP1 was detected with polyclonal anti-FLAG antibody. C, quantification of in vivo results. The signals from the x-ray films were quantified by densitometry. The signal for Arf·GTP in cells expressing ARAP1 was divided by Arf·GTP in the control cells. The averages from three experiments ± S.E. are presented.
TABLE 2.
ARAP1 in vitro substrate specificity
C50 values for PH1-Ank recombinant protein were determined using Arf1, Arf5, and Arf6 as substrates in Arf GAP reactions containing 500 μm LUVs. Both myristoylated and non-myristoylated recombinant Arfs were used as indicated. The data presented are the average ± S.E. of three experiments. Numbers in parentheses represent the number of experiment.
| Modification | C50 |
||
|---|---|---|---|
| Arf1 | Arf5 | Arf6 | |
| nm | |||
| Non-myristoylated form | 0.028 ± 0.009 (3) | 0.019 ± 0.003 (3) | 0.33 ± 0.03 (3) |
| Myristoylated form | 0.24 ± 0.05 (3) | 0.049 ± 0.002 (3) | 0.55 ± 0.02 (3) |
We also examined substrate specificity in vivo. In these experiments, ARAP1 was coexpressed with Arf1-GFP, Arf5-GFP, or Arf6-GFP, and in vivo levels of the GTP bound forms of each Arf were determined. Consistent with previous reports (52), ARAP1 had no effect on Arf1-GFP, and it reduced Arf6-GFP·GTP levels. It also reduced Arf5-GFP·GTP levels (Fig. 3B and C).
The effect of phosphorylated derivatives of PtdIns on activity was determined using myrArf5 as a substrate and PH1-Ank at a concentration approximately that of the C50. PtdIns(3,4,5)P3 stimulated activity to a greater extent than other phosphoinositides (Fig. 4). The effect on activity was greater than the effect on vesicle binding (compare Fig. 1B with Fig. 4). PtdIns(3,4,5)P3 was titrated into the reaction (Fig. 5 and Table 1). The half-maximal effect (EC50) was achieved with 1.6 ± 0.1 μm PtdIns(3,4,5)P3 (mean ± S.E., n = 7), similar to the Kd for the ARAP1·PtdIns(3,4,5)P3 complex.
FIGURE 5.
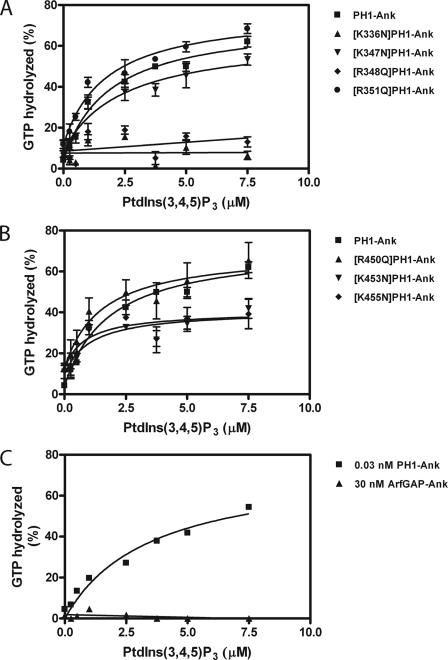
Role of the tandem PH domains of ARAP1 for PtdIns(3,4,5)P3-dependent GAP activity. 0.03 nm recombinant PH1-Ank and mutants in PH1 (A) and PH2 (B) and 30 nm recombinant ArfGAP-Ank (C) were added to a reaction mixture containing myrArf5·GTP and LUVs with the indicated concentration of PtdIns(3,4,5)P3, a total phosphoinositide concentration of 50 μm, and a total lipid concentration of 500 μm. Reactions were stopped after 3 min. The averages of three experiments ± S.E. are presented in A and B; a representative experiment of three is shown in C.
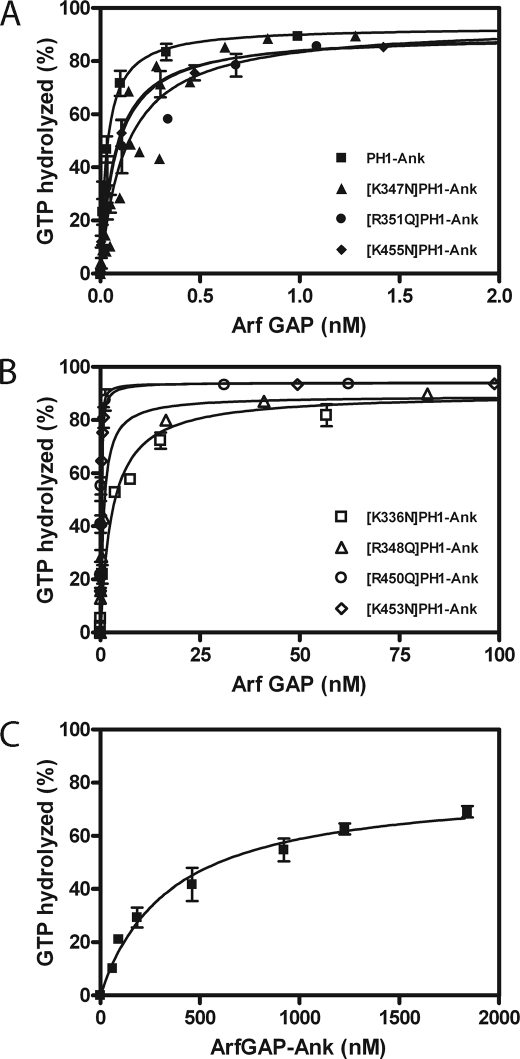
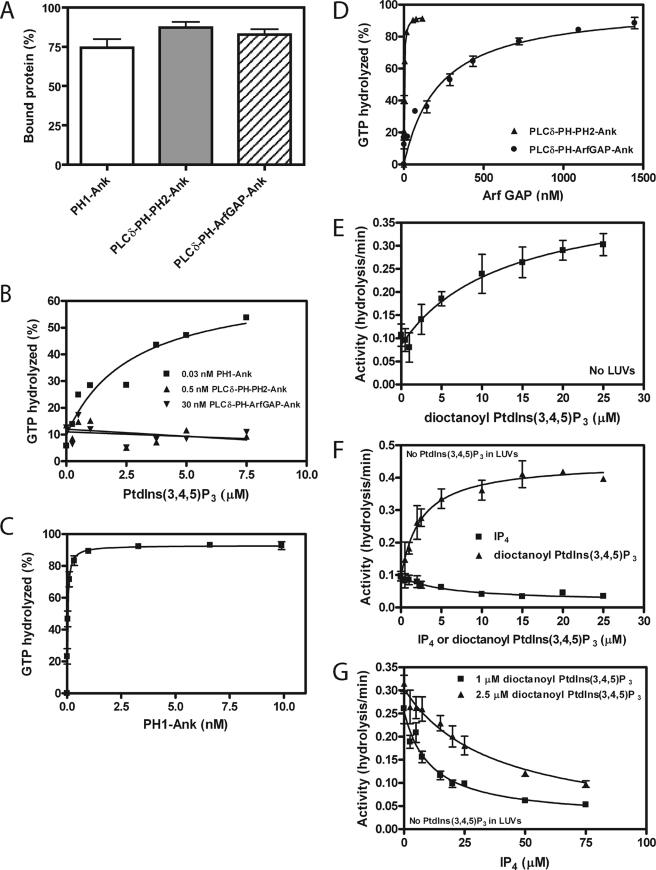
The role for the PH domain in controlling GAP activity was examined by comparing PH1-Ank, ArfGAP-Ank, and mutants of PH1-Ank with changes in PH1 and PH2. In the presence of PtdIns(3,4,5)P3, ArfGAP-Ank and point mutants in the PH1 with disrupted PtdIns(3,4,5)P3 binding ([K336N]PH1-Ank and [R348Q]PH1-Ank) had larger C50 values than PH1-Ank (Fig. 6, B and C, and Table 3). Mutations in PH2 and those in PH1 that did not affect PtdIns(3,4,5)P3 binding had less of an effect on the C50 (Fig. 6A and B and Table 3). We also compared the PtdIns(3,4,5)P3 dependence of GAP activity of the recombinant proteins. Proteins that did not bind PtdIns(3,4,5)P3 were not stimulated by PtdIns(3,4,5)P3 (Fig. 5 and Table 1). ArfGAP-Ank had less activity than PH1-Ank in the absence of PtdIns(3,4,5)P3 (Fig. 7A). The result was unexpected because neither protein was efficiently recruited to LUVs that lacked PtdIns(3,4,5)P3.
FIGURE 6.
Effect of the tandem PH domains PH1 and PH2 on the enzymatic power of ARAP1. PH1-Ank, mutants of PH1-Ank, and ArfGAP-Ank were titrated into reactions containing myrArf5·GTP as a substrate and LUVs containing 2.5 μm PtdIns(3,4,5)P3. Reactions were terminated after 3 min. Averages ± S.E. from at least three experiments are presented. A, PH1-Ank, [K347N]PH1-Ank, [R351Q]PH1-Ank, and [K455N]PH1-Ank. B, [K336N]PH1-Ank, [R348Q]PH1-Ank, [R450Q]PH1-Ank, and [K453N]PH1-Ank. C, ArfGAP-Ank. The C50 values ± S.E. derived from these results are presented in Table 3.
TABLE 3.
Enzymatic activity of recombinant ARAP1s
The concentration of Arf GAP to hydrolyze 50% of the GTP bound to Arf (C50) for PH1-Ank, mutant PH1-Ank, ArfGAP-Ank, and chimeric ARAP1 proteins was determined using myristoylated Arf5 as substrate in reactions containing LUVs with 2.5 μm PtdIns(3,4,5)P3, a total phosphoinositide concentration of 50 μm, and a total phospholipid concentration of 500 μm. The data presented are the average ± S.E. The number of experiments is indicated in parentheses.
| Recombinant protein | C50 | -Fold change from PH1-Ank |
|---|---|---|
| nm | ||
| PH1-Ank | 0.03 ± 0.005 (4) | |
| ArfGAP-Ank | 370 ± 64 (3) | 12,300 |
| PLCδ-PH-PH2-Ank | 2.4 ± 0.2 (3) | 80 |
| PLCδ-PH-ArfGAP-Ank | 270 ± 63 (5) | 9,000 |
| [K336N]PH1-Ank | 2.73 ± 0.14 (3) | 91 |
| [K347N]PH1-Ank | 0.18 ± 0.004 (5) | 6 |
| [R348Q]PH1-Ank | 0.79 ± 0.01 (3) | 26 |
| [R351Q]PH1-Ank | 0.12 ± 0.04 (5) | 4 |
| [R450Q]PH1-Ank | 0.08 ± 0.01 (3) | 2.7 |
| [K453N]PH1-Ank | 0.16 ± 0.05 (3) | 5 |
| [K455N]PH1-Ank | 0.07 ± 0.01 (3) | 2 |
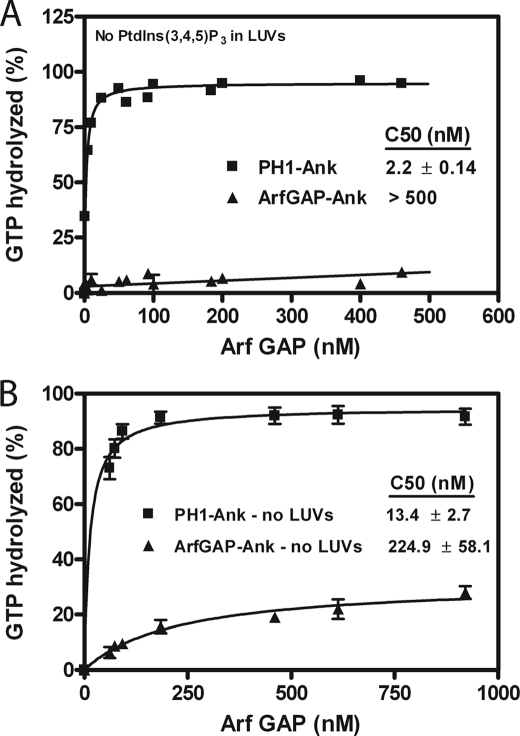
FIGURE 7.
Role of PH domains for PtdIns(3,4,5)P3-independent GAP activity. A, activity with LUVs lacking PtdIns(3,4,5)P3. PH1-Ank and ArfGAP-Ank were titrated into reaction mixtures containing myrArf5·GTP and 500 μm phospholipids but no PtdIns(3,4,5)P3. B, activity in the absence of lipids or detergents. PH1-Ank and ArfGAP-Ank were titrated into reactions containing [L8K]Arf5·GTP. The average ± S.E. from three experiments is presented.
To further examine the role of recruitment in regulating GAP activity, we compared the activity of PH1-Ank with ArfGAP-Ank using a soluble mutant of Arf5 ([L8K]Arf5) as a substrate in a reaction without lipids or detergent. If the function of the PH domain was to recruit the protein to membranes, the two recombinant proteins would have similar activity under these conditions. Contrary to the prediction, PH1-Ank was more active than ArfGAP-Ank (Fig. 7B). To further assess the relative role of recruitment to membranes for control of GAP activity, we constructed chimeric proteins. ARAP1 with a single PH domain deleted was not stable and, therefore, could not be examined. A fusion protein of the PH domain of PLCδ and PH2, Arf GAP and Ank repeat domains of ARAP1 (PLCδ-PH-PH2-Ank), was soluble and was as efficiently recruited to vesicles as PH1-Ank (Fig. 8A). Consistent with PtdIns(3,4,5)P3 binding to PH1 stimulating GAP activity, PtdIns(3,4,5)P3 did not affect the fusion protein (Fig. 8B). We determined the C50 for PLCδ-PH-PH2-Ank and found that it had less enzymatic power than PH1-Ank (Fig. 8, C and D, and Table 3). Similarly, ARAP1 with both PH1 and PH2 replaced with the PH domain of PLCδ (PLCδ-PH-ArfGAP-Ank) was recruited efficiently to vesicles (Fig. 8A) but was not activated by PtdIns(3,4,5)P3 (Fig. 8B) and had less enzymatic power than PH1-Ank or PLCδ-PH-PH2-Ank (Fig. 8, C and D, and Table 3).
FIGURE 8.
Role of membrane recruitment for ARAP1 GAP activity. A, binding of chimeric proteins to LUVs. 1 μm PLCδ-PH-PH2-Ank or PLCδ-PH-ArfGAP-Ank was incubated with 1 mm sucrose-loaded LUVs formed by extrusion through a 1-μm pore size filter. LUVs contained 12.5 μm PtdIns(4,5)P2 and 2.5 μm PtdIns(3,4,5)P3. Vesicles were precipitated by ultracentrifugation, and associated proteins were separated by SDS-PAGE. The amount of precipitated protein was determined by densitometry of the Coomassie Blue-stained gels with standards on each gel. B, PtdIns(3,4,5)P3 dependence of GAP activity of PLCδ-PH-Arf GAP-Ank and PLCδ-PH-PH2-Ank. 0.03 nm recombinant PH1-Ank, 0.5 nm PLCδ-PH-PH2-Ank, or 30 nm PLCδ-PH-ArfGAP-Ank was added to a reaction mixture containing myrArf5·GTP and LUVs with the indicated concentration of PtdIns(3,4,5)P3, a total phosphoinositide concentration of 50 μm, and a total lipid concentration of 500 μm. Reactions were stopped after 3 min. A representative experiment of three is shown. C and D, enzymatic power of chimeric recombinant proteins. PLCδ-PH-PH2-Ank, PLCδ-PH-ArfGAP-Ank, and PH1-Ank were titrated into reactions containing myrArf5·GTP and LUVs with 2.5 μm PtdIns(3,4,5)P3 and 12.5 μm PtdIns(4,5)P2. Average values from at least three experiments are presented; C50 values ± S.E. derived from the data are presented in Table 3. C, PH1-Ank. D, PLCδ-PH-PH2-Ank and PLCδ-PH-ArfGAP-Ank. E, effect of dioctanoyl PtdIns(3,4,5)P3 on GAP activity. 40 nm PH1-Ank recombinant protein was added to reaction mixtures containing [L8K]Arf5·GTP and the indicated concentrations of dioctanoyl PtdIns(3,4,5)P3 but no lipids or detergents. The results presented are the average ± S.E. from three experiments. F, Effect of IP4 and dioctanoyl PtdIns(3,4,5)P3 in presence of lipids. 1 nm PH1-Ank recombinant protein was incubated with the indicated concentration of IP4 or dioctanoyl PtdIns(3,4,5)P3 in the presence of LUVs containing 500 μm phospholipids but no PtdIns(3,4,5)P3. Myristoylated Arf5 was included as the substrate. The averages of three experiments ± S.E. are shown. G, inhibition of dioctanoyl PtdIns(3,4,5)P3-dependent GAP activity by IP4. IP4 was titrated in a reaction containing either 1 μm or 2.5 μm dioctanoyl PtdIns(3,4,5)P3 and LUVs. The average of three experiments ± S.E. is shown.
To test for recruitment-independent activation of Arf GAP activity, the effect of the water-soluble PtdIns(3,4,5)P3 analog, dioctanoyl PtdIns(3,4,5)P3, and the PtdIns(3,4,5)P3 head group, IP4, on GAP activity using a water-soluble substrate, [L8K]Arf5, was examined. Dioctanoyl PtdIns(3,4,5)P3 increased activity by 3-fold (Fig. 8E). IP4 did not have a detectable effect by itself, nor did it inhibit dioctanoyl PtdIns(3,4,5)P3-dependent activity at concentrations up to 200 μm (not shown). We also examined the effect of dioctanoyl PtdIns(3,4,5)P3 and IP4 on ARAP1 activity in the presence of LUVs that did not contain PtdIns(3,4,5)P3. Dioctanoyl PtdIns(3,4,5)P3 stimulated Arf GAP activity by ∼4-fold, whereas IP4 inhibited activity by 85% (Fig. 8F). IP4 also inhibited dioctanoyl PtdIns(3,4,5)P3-stimulated GAP activity in the presence of LUVs (Fig. 8G). Taken together, our results indicate that PH1 of ARAP1 binds PtdIns(3,4,5)P3 leading to increased GAP activity by a mechanism that is, at least in part, independent of recruitment to membranes.
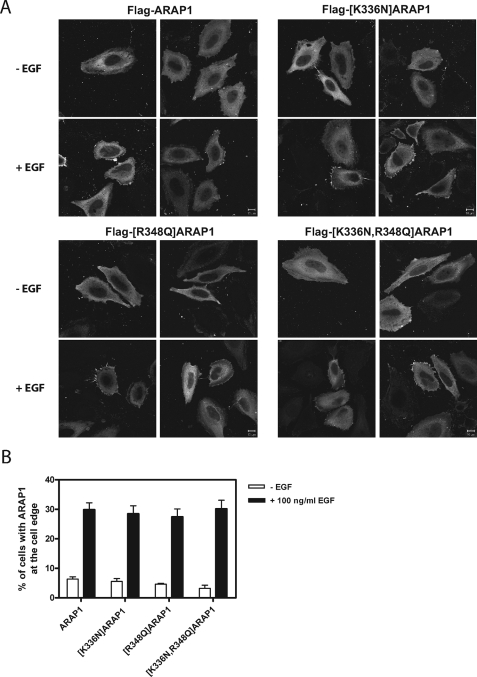
Role of PH Domains in Cellular Distribution of ARAP1
ARAP1 cycles among the Golgi apparatus, the cell edge, and a Rab5-positive endosome (53). Although recombinant ARAP1 does not associate efficiently with the Golgi apparatus, it is like endogenous ARAP1 in being recruited to the cell edge and the Rab5-positive endosomes upon treatment of the cells with EGF (53). We examined the cellular distribution of ARAP1 containing point mutations in PH1 and PH2. As reported previously, recombinant ARAP1 did not associate efficiently with the Golgi apparatus but did associate with the cell edge (Fig. 9) and, in EGF-treated cells, with a Rab5 compartment (Fig. 10). Mutant ARAP1 that does not bind PtdIns(3,4,5)P3 associated with the cell edge as efficiently as wild type ARAP1 (Fig. 9). Association of ARAP1 with the Rab5-positive pre-early endosome was most easily observed in cells expressing recombinant Rab5-GFP (53). Mutant ARAP1 that does not bind PtdIns(3,4,5)P3, also associated with the Rab5 positive pre-early endosome, to approximately the same extent as wild type ARAP1 in EGF-treated cells (Fig. 10).
FIGURE 9.
ARAP1 association with the cell edge. A, representative images. HeLa cells overexpressing N-terminal FLAG-tagged ARAP1 or mutant ARAP1 were serum-starved for 6 h on fibronectin-coated coverslips (10 μg/ml) and treated with 100 ng/ml EGF for 5 min at 37 °C. Fixed cells were immunostained with monoclonal anti-FLAG antibody. B, quantification of localization. More than 100 cells were scored for ARAP1 concentrated at the cell edge using visual inspection in three independent experiments. Images were captured with a Zeiss LSM510-Meta laser-scanning confocal microscope with a ×63, 1.4 NA Plan Neofluar oil immersion lens. Scale bar, 10 μm.
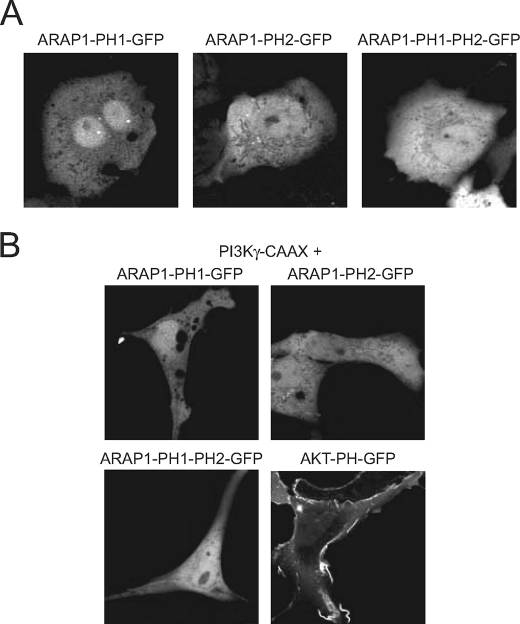
The localization of the mutant ARAP1 proteins was not consistent with predictions based on the typical role of PtdIns(3,4,5)P3-binding PH domains for targeting to the PM (1). To further assess the contribution of PH1 and PH2 to targeting to the PM, we prepared plasmids for the expression of GFP fused to PH1, PH2, or PH1-PH2 of ARAP1. The fusion proteins were expressed in COS-7 cells and were found to be distributed diffusely in the cytoplasm. EGF did not affect the distribution (data not shown). The distribution of the fusion proteins was also examined in cells expressing a membrane-targeted constitutively active PtdIns 3-kinase γ-CAAX construct (Fig. 11). None of the GFP-tagged PH domains were recruited to the PM. We obtained similar results with GFP fused to PH3, PH4, or PH3-PH4 (not shown). In contrast, GFP-PHAkt was efficiently recruited to the PM by expression of constitutively active PtdIns 3-kinase.
FIGURE 11.
Cellular distribution of ARAP1-PH domains expressed in COS-7 cells. The GFP-tagged versions of ARAP1-PH1, ARAP1-PH2, and ARAP1-PH1-PH2 tandem were expressed either separately (A) or together with a membrane-targeted PtdIns 3-kinase γ (PtdIns 3-K γ-CAAX) (B). Live cell imaging was then performed 24 h later by laser confocal microscopy. Note that none of the ARAP1 PH domains localized to the PM either in quiescent cells (indicating no recruitment by PtdIns(4,5)P2) or in the presence of PtdIns 3-kinase γ-CAAX (when PtdIns(3,4,5)P3 is generated). The plasma membrane localization of the PtdIns(3,4,5)P3 reporter Akt-PH domain is apparent when expressed with the constitutively active PtdIns 3-kinase γ. Note that the cells expressing PtdIns 3-Kγ-CAAX become quite elongated allowing identification of the cells expressing the active enzyme.
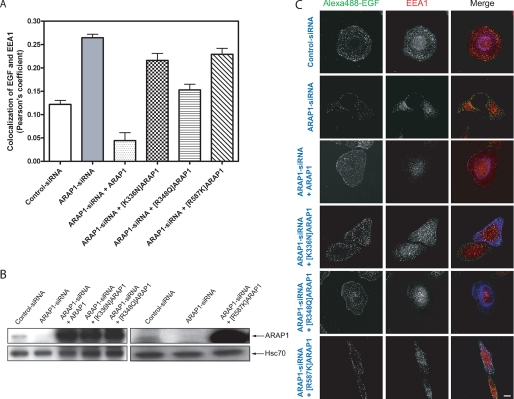
The Ability of PH1 to Bind PtdIns(3,4,5)P3 Is Necessary for the Regulation of Endocytic Trafficking of EGF by ARAP1
ARAP1 has been found to affect trafficking of the EGF receptor (53). Reducing ARAP1 expression accelerates EGF association with EEA1-positive endosomes. We determined whether mutant ARAP1s could replace endogenous ARAP1 to regulate the kinetics of EGF-EEA1 association. Overexpression of wild type ARAP1 completely reversed the effect of reduced expression of endogenous ARAP1, reducing EGF-EEA1 colocalization to levels that were lower than observed in cells expressing endogenous ARAP1. Two mutants of ARAP1 with reduced PtdIns(3,4,5)P3 binding were less effective than the wild type ARAP1 (Fig. 12). Because of the effect of PtdIns(3,4,5)P3 on GAP activity, we also examined an ARAP1 mutant that was deficient in Arf GAP activity ([R587K]ARAP1). Similar to the PH domain mutants [R587K]ARAP1 was less effective than wild type recombinant ARAP1 (Fig. 12).
FIGURE 12.
Effect of ARAP1 on EGF traffic to EEA1-positive endosomes. HeLa cells were transfected either with control-siRNA or ARAP1-specific siRNA. After 48 h, cells were transfected with plasmids directing the expression of FLAG-ARAP1, [K336N]ARAP1, [R348Q]ARAP1, and [R587K]ARAP1. Twenty-four h later, cells were serum-starved overnight and incubated with 1 μg/ml fluorescent Alexa488-EGF for 10 min at 37 °C. The cells were fixed and immunostained with mouse anti-EEA1 (red) and rabbit anti-FLAG (blue). Pearson's coefficients for signals from EGF and EEA1 were determined for 25 cells captured with a Zeiss LSM510-Meta laser-scanning confocal microscope with a ×63, 1.4 NA Plan Neofluar oil immersion lens under each condition using Zeiss LSM 5 image software. The average and range from two experiments is shown in A. An immunoblot to determine ARAP1 levels in lysates from cells treated with siRNAs or expression plasmids, as indicated, is shown in B. Antiserum (1153) raised against ARAP1 was used to determine the ARAP1 levels, and anti-Hsc70 antibody was used as a protein loading control. Representative micrographs are shown in C. Scale bar, 10 μm.
DISCUSSION
We have investigated the role of a tandem of two PH domains for the function of ARAP1. The first PH domain of the tandem has the consensus sequence of a PtdIns(3,4,5)P3-binding PH domain and, as predicted, did bind PtdIns(3,4,5)P3, which recruited the protein to lipid vesicles in vitro and stimulated GAP activity. The ability to bind PtdIns(3,4,5)P3 was also important for the in vivo function of ARAP1 in EGF endocytic traffic. However, the effect of both PH domains on GAP activity was partly independent of recruitment. Furthermore, mutant ARAP1s, which bind PtdIns(3,4,5)P3 poorly, were efficiently recruited to the cell edge, and the isolated PH domains were not recruited by PtdIns(3,4,5)P3 to the cell edge. Based on these results, we concluded that PtdIns(3,4,5)P3-dependent recruitment is not the primary function of PH1-PtdIns(3,4,5)P3 association in ARAP1.
The first PH domain of ARAP1 appears to function in a different capacity than other PtdIns(3,4,5)P3-binding PH domains. Proteins such as Btk are recruited to the PM by PtdIns(3,4,5)P3 binding to the PH domain (4, 54). The isolated PH domain of ARAP1 was not recruited by PtdIns(3,4,5)P3, and ARAP1 with mutations in the PH domain abrogating PtdIns(3,4,5)P3 binding was recruited to the cell edge on treatment of cells with EGF. Mutant ARAP1 also translocated to the Rab5-positive pre-early endosome as efficiently as did wild type protein. PH1 of ARAP1 was nevertheless important for ARAP1 function. The mutant ARAP1 proteins did not efficiently replace endogenous ARAP1 to regulate EGF trafficking to the EEA1-positive endosome. PH1 and PtdIns(3,4,5)P3 binding also stimulated GAP activity by a mechanism that was at least in part independent of membrane recruitment. We propose that PH1 of ARAP1 is not a recruitment module. Instead, ARAP1 is recruited to the membrane by a mechanism independent of PH1. The PH domains of other Arf GAPs may be similar. ASAP1 has a PH domain that binds specifically PtdIns(4,5)P2; however, ASAP1 is recruited to specific cellular sites independent of the PH domain (55). ASAP1 is recruited to focal adhesions by binding to focal adhesion kinase through the SH3 domain of ASAP1 (56) and binding to Crk through the proline-rich motif of ASAP1 (57). Recruitment to circular dorsal ruffles is similarly independent of the PH domain (55), although specific molecular interactions driving association with the ruffles have not been identified.
Although we did not observe recruitment by a PtdIns(3,4,5)P3-PH1 domain interaction, our results do not exclude the possibility that one PH domain from ARAP1 mediates recruitment. Our preliminary examination did not support the idea that any of the PH domains were involved in PtdIns(3,4,5)P3-dependent targeting, but we did not examine PH3, PH4, or PH5 in detail. Furthermore, the PH domains could mediate recruitment by binding to a protein. AGAP1 and -2 are Arf GAPs with PH domains that bind to clathrin adaptor proteins, which mediate recruitment to endosomes (6, 5). Of the five PH domains in ARAP1, two are not strong candidates for phosphoinositide binding, leading us to consider that these may mediate protein interactions. ARAP1 has additional domains that could function in membrane recruitment including the SAM domain and the Ras association domain. Another plausible targeting mechanism is association with Arf·GTP through the Arf GAP domain or association with RhoA·GTP through the Rho GAP domain.
PH1 and PH2 are integral to the function of the Arf GAP domain. Based on analogy with other PH domain containing Arf GAPs, the recruitment-independent contribution of PH2 to Arf GAP activity was anticipated. The PH domain of ASAP1 has limited contact with the Arf GAP domain and, like the PH domains in DH-PH proteins, may bind and orient the substrate GTP-binding protein (14, 58, 59). Similarly, PH2 may form an interface with the Arf GAP domain and/or bind the substrate Arf·GTP. The effects of mutations in PH2 on GAP activity are consistent with this model. The recruitment independent function of PH1 was not anticipated. However, PH2 did not fold independently of a second PH domain (we were unable to prepare isolated PH2 as a soluble protein in bacteria), which is consistent with the idea that PH1 and PH2 are integrally folded, explaining the recruitment independent effects of PH1 on GAP activity. Determination of the molecular basis of regulation by PH domain will depend on structural studies. Although there is some information about how PH domains fold together with other protein domains, e.g. BAR, there is limited information about how PH domains fold with Arf GAP domains or other PH domains that occur within the same protein.
The effect of water-soluble PtdIns(3,4,5)P3 analogs on ARAP1 Arf GAP activity is consistent with the idea that PtdIns(3,4,5)P3 binding leads to an activating conformational change in ARAP1. Dioctanoyl PtdIns(3,4,5)P3 stimulated activity in the presence or absence of LUVs. In contrast, IP4 inhibited activity. Differences in the effects of IP4 and PtdIns(3,4,5)P3 and between phosphoinositides with different acyl groups in the 2-position of the glycerol backbone have been reported for other proteins (55, 60). The differences may indicate that the glycerol backbone and acyl groups also make contact with the PH domain-containing protein and may contribute to conformational changes.
The molecular basis of ARAP1-dependent regulation of endocytosis of the EGFR or other membrane traffic events remains to be discovered. For instance, it is not known whether the Arf and Rho GAP domains are dependently or independently regulated, whether ARAP1 has a function in addition to GAP activity, and what might be the role of Arf proteins in EGFR traffic. A possible mechanism that should be considered includes ARAP1 function as an effector for Rho, with Rho binding to the Rho GAP domain controlling the hydrolysis of GTP on Arf by the Arf GAP domain. A similar model has been proposed for ARAP2. Our results provide a framework on which to base further investigations into the molecular basis of ARAP1 function. We propose that ARAP1 is recruited to the PM where it may be incorporated into a forming pre-early endosome under the control of PtdIns(3,4,5)P3-dependent hydrolysis of GTP on Arf. Once incorporated into the vesicle, ARAP1 controls the rate of entry of the receptor into the degradative pathway.
In summary, our results contradict the idea that the first PH domain of ARAP1 functions as a recruitment module. We propose, instead, that the PH domain functions as a binding site for an allosteric modifier.
Acknowledgment
We thank Mark Lemmon for discussion and teaching PR about data base analysis.
This work was supported, in whole or in part, by National Institutes of Health grants from the NCI Intramural Program and NICHD.
- PH
- pleckstrin homology
- GAP
- GTPase-activating protein
- PtdIns
- phosphatidylinositol
- EGF
- epidermal growth factor
- EGFR
- epidermal growth factor receptor
- PLC
- phospholipase C
- ORF
- open reading frame
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- LUV
- large unilamellar vesicle
- siRNA
- small interfering RNA
- PBS
- phosphate-buffered saline
- dioctanoyl PtdIns(3,4,5)P3
- 1,2-dioctanoyl-sn-glycero-3-[phosphoinositol-3,4,5-trisphoshate]
- IP4
- d-myo-inositol-1, 3,4,5-tetraphosphate
- SAM
- sterile α-motif
- PM
- plasma membranes.
REFERENCES
- 1.Lemmon M. A. (2003) Traffic 4, 201–213 [DOI] [PubMed] [Google Scholar]
- 2.Lemmon M. A., Ferguson K. M. (1998) Curr. Top. Microbiol. Immunol. 228, 39–74 [DOI] [PubMed] [Google Scholar]
- 3.Várnai P., Lin X., Lee S. B., Tuymetova G., Bondeva T., Spät A., Rhee S. G., Hajnóczky G., Balla T. (2002) J. Biol. Chem. 277, 27412–27422 [DOI] [PubMed] [Google Scholar]
- 4.Balla T. (2005) J. Cell Sci. 118, 2093–2104 [DOI] [PubMed] [Google Scholar]
- 5.Nie Z., Fei J., Premont R. T., Randazzo P. A. (2005) J. Cell Sci. 118, 3555–3566 [DOI] [PubMed] [Google Scholar]
- 6.Nie Z., Boehm M., Boja E. S., Vass W. C., Bonifacino J. S., Fales H. M., Randazzo P. A. (2003) Dev. Cell 5, 513–521 [DOI] [PubMed] [Google Scholar]
- 7.Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., Matteis M. A. (2004) Nat. Cell Biol. 6, 393–404 [DOI] [PubMed] [Google Scholar]
- 8.Cohen L. A., Honda A., Varnai P., Brown F. D., Balla T., Donaldson J. G. (2007) Mol. Biol. Cell 18, 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gervais V., Lamour V., Jawhari A., Frindel F., Wasielewski E., Dubaele S., Egly J. M., Thierry J. C., Kieffer B., Poterszman A. (2004) Nat. Struct. Mol. Biol. 11, 616–622 [DOI] [PubMed] [Google Scholar]
- 10.VanDemark A. P., Blanksma M., Ferris E., Heroux A., Hill C. P., Formosa T. (2006) Mol. Cell 22, 363–374 [DOI] [PubMed] [Google Scholar]
- 11.Fürst J., Schedlbauer A., Gandini R., Garavaglia M. L., Saino S., Gschwentner M., Sarg B., Lindner H., Jakab M., Ritter M., Bazzini C., Botta G., Meyer G., Kontaxis G., Tilly B. C., Konrat R., Paulmichl M. (2005) J. Biol. Chem. 280, 31276–31282 [DOI] [PubMed] [Google Scholar]
- 12.Prehoda K. E., Lee D. J., Lim W. A. (1999) Cell 97, 471–480 [DOI] [PubMed] [Google Scholar]
- 13.Zhu G., Chen J., Liu J., Brunzelle J. S., Huang B., Wakeham N., Terzyan S., Li X., Rao Z., Li G., Zhang X. C. (2007) EMBO J. 26, 3484–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossman K. L., Worthylake D. K., Snyder J. T., Siderovski D. P., Campbell S. L., Sondek J. (2002) EMBO J. 21, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowler S., Currie R. A., Downes C. P., Alessi D. R. (1999) Biochem. J. 342, 7–12 [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas C. C., Dowler S., Deak M., Alessi D. R., Van Aalten D. M. (2001) Biochem. J. 358, 287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon H. Y., Miura K., Cuthbert E. J., Davis K. K., Ahvazi B., Casanova J. E., Randazzo P. A. (2006) J. Cell Sci. 119, 4650–4666 [DOI] [PubMed] [Google Scholar]
- 18.Miura K., Jacques K. M., Stauffer S., Kubosaki A., Zhu K., Hirsch D. S., Resau J., Zheng Y., Randazzo P. A. (2002) Mol. Cell 9, 109–119 [DOI] [PubMed] [Google Scholar]
- 19.Nie Z. Z., Randazzo P. A. (2006) J. Cell Sci. 119, 1203–1211 [DOI] [PubMed] [Google Scholar]
- 20.Inoue H., Randazzo P. A. (2007) Traffic 8, 1465–1475 [DOI] [PubMed] [Google Scholar]
- 21.Randazzo P. A., Inoue H., Bharti S. (2007) Biol. Cell 99, 583–600 [DOI] [PubMed] [Google Scholar]
- 22.Randazzo P. A., Hirsch D. S. (2004) Cell. Signal. 16, 401–413 [DOI] [PubMed] [Google Scholar]
- 23.Gillingham A. K., Munro S. (2007) Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 24.D'Souza-Schorey C., Chavrier P. (2006) Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 25.Donaldson J. G. (2003) J. Biol. Chem. 278, 41573–41576 [DOI] [PubMed] [Google Scholar]
- 26.Randazzo P. A., Nie Z., Miura K., Hsu V. (2000) Sci. STKE 2000 59, RE1. [DOI] [PubMed] [Google Scholar]
- 27.Logsdon J. M., Kahn R. A. (2003) in Arf Family GTPases ( Kahn R. A. ed) pp. 1–21, Kluwer Academic Publishers, Dordrecht [Google Scholar]
- 28.Moss J., Vaughan M. (1998) J. Biol. Chem. 273, 21431–21434 [DOI] [PubMed] [Google Scholar]
- 29.Kahn R. A., Cherfils J., Elias M., Lovering R. C., Munro S., Schurmann A. (2006) J. Cell Biol. 172, 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahn R. A., Bruford E., Inoue H., Logsdon J. M., Jr., Nie Z., Premont R. T., Randazzo P. A., Satake M., Theibert A. B., Zapp M. L., Cassel D. (2008) J. Cell Biol. 182, 1039–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krugmann S., Anderson K. E., Ridley S. H., Risso N., McGregor A., Coadwell J., Davidson K., Eguinoa A., Ellson C. D., Lipp P., Manifava M., Ktistakis N., Painter G., Thuring J. W., Cooper M. A., Lim Z. Y., Holmes A. B., Dove S. K., Michell R. H., Grewal A., Nazarian A., Erdjument-Bromage H., Tempst P., Stephens L. R., Hawkins P. T. (2002) Mol. Cell 9, 95–108 [DOI] [PubMed] [Google Scholar]
- 32.I, S. T., Nie Z., Stewart A., Najdovska M., Hall N. E., He H., Randazzo P. A., Lock P. (2004) J. Cell Sci. 117, 6071–6084 [DOI] [PubMed] [Google Scholar]
- 33.Lietzke S. E., Bose S., Cronin T., Klarlund J., Chawla A., Czech M. P., Lambright D. G. (2000) Mol. Cell 6, 385–394 [DOI] [PubMed] [Google Scholar]
- 34.Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., Lemmon M. A. (2000) Mol. Cell 6, 373–384 [DOI] [PubMed] [Google Scholar]
- 35.Park W. S., Heo W. D., Whalen J. H., O'Rourke N. A., Bryan H. M., Meyer T., Teruel M. N. (2008) Mol. Cell 30, 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krugmann S., Williams R., Stephens L., Hawkins P. T. (2004) Curr. Biol. 14, 1380–1384 [DOI] [PubMed] [Google Scholar]
- 37.Krugmann S., Andrews S., Stephens L., Hawkins P. T. (2006) J. Cell Sci. 119, 425–432 [DOI] [PubMed] [Google Scholar]
- 38.Luo R., Miller Jenkins L. M., Randazzo P. A., Gruschus J. (2008) Cell. Signal. 20, 1968–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo R., Ahvazi B., Amariei D., Shroder D., Burrola B., Losert W., Randazzo P. A. (2007) Biochem. J. 402, 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Várnai P., Balla T. (1998) J. Cell Biol. 143, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Randazzo P. A., Kahn R. A. (1995) Methods Enzymol. 250, 394–405 [DOI] [PubMed] [Google Scholar]
- 42.Ha V. L., Thomas G. M., Stauffer S., Randazzo P. A. (2005) Methods Enzymol. 404, 164–174 [DOI] [PubMed] [Google Scholar]
- 43.Randazzo P. A., Fales H. M. (2002) in GTPase Protocols: The Ras Superfamily ( Manser E., Leung T. eds) Vol. 189, pp. 169–179, Humana Press, Totowa, NJ [Google Scholar]
- 44.Randazzo P. A., Weiss O., Kahn R. A. (1992) Methods Enzymol. 219, 362–369 [DOI] [PubMed] [Google Scholar]
- 45.Che M. M., Nie Z. Z., Randazzo P. A. (2005) Methods Enzymol. 404, 147–163 [DOI] [PubMed] [Google Scholar]
- 46.Randazzo P. A., Miura K., Jackson T. R. (2001) Methods Enzymol 329, 343–354 [DOI] [PubMed] [Google Scholar]
- 47.Hirsch D. S., Stanley K. T., Chen L. X., Jacques K. M., Puertollano R., Randazzo P. A. (2003) Traffic 4, 26–35 [DOI] [PubMed] [Google Scholar]
- 48.Jacques K. M., Nie Z., Stauffer S., Hirsch D. S., Chen L. X., Stanley K. T., Randazzo P. A. (2002) J. Biol. Chem. 277, 47235–47241 [DOI] [PubMed] [Google Scholar]
- 49.Nie Z., Hirsch D. S., Luo R., Jian X., Stauffer S., Cremesti A., Andrade J., Lebowitz J., Marino M., Ahvazi B., Hinshaw J. E., Randazzo P. A. (2006) Curr. Biol. 16, 130–139 [DOI] [PubMed] [Google Scholar]
- 50.Yoon H. Y., Bonifacino J. S., Randazzo P. A. (2005) Methods Enzymol. 404, 316–332 [DOI] [PubMed] [Google Scholar]
- 51.Randazzo P. A., Kahn R. A. (1994) J. Biol. Chem. 269, 10758–10763 [PubMed] [Google Scholar]
- 52.Cuthbert E. J., Davis K. K., Casanova J. E. (2008) Am. J. Physiol. Cell Physiol. 294, C263–C270 [DOI] [PubMed] [Google Scholar]
- 53.Yoon H. Y., Lee J. S., Randazzo P. A. (2008) Traffic 9, 2236–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemmon M. A., Ferguson K. M. (2000) Biochem. J. 350, 1–18 [PMC free article] [PubMed] [Google Scholar]
- 55.Kam J. L., Miura K., Jackson T. R., Gruschus J., Roller P., Stauffer S., Clark J., Aneja R., Randazzo P. A. (2000) J. Biol. Chem. 275, 9653–9663 [DOI] [PubMed] [Google Scholar]
- 56.Liu Y., Loijens J. C., Martin K. H., Karginov A. V., Parsons J. T. (2002) Mol Biol. Cell 13, 2147–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oda A., Wada I., Miura K., Okawa K., Kadoya T., Kato T., Nishihara H., Maeda M., Tanaka S., Nagashima K., Nishitani C., Matsuno K., Ishino M., Machesky L. M., Fujita H., Randazzo P. (2003) J. Biol. Chem. 278, 6456–6460 [DOI] [PubMed] [Google Scholar]
- 58.Che M. M., Boja E. S., Yoon H. Y., Gruschus J., Jaffe H., Stauffer S., Schuck P., Fales H. M., Randazzo P. A. (2005) Cell. Signal. 17, 1276–1288 [DOI] [PubMed] [Google Scholar]
- 59.Rossman K. L., Cheng L., Mahon G. M., Rojas R. J., Snyder J. T., Whitehead I. P., Sondek J. (2003) J. Biol. Chem. 278, 18393–18400 [DOI] [PubMed] [Google Scholar]
- 60.Stokoe D., Stephens L. R., Copeland T., Gaffney P. R., Reese C. B., Painter G. F., Holmes A. B., McCormick F., Hawkins P. T. (1997) Science 277, 567–570 [DOI] [PubMed] [Google Scholar]