Abstract
MyD88 couples the activation of the Toll-like receptors and interleukin-1 receptor superfamily with intracellular signaling pathways. Upon ligand binding, activated receptors recruit MyD88 via its Toll-interleukin-1 receptor domain. MyD88 then allows the recruitment of the interleukin-1 receptor-associated kinases (IRAKs). We performed a site-directed mutagenesis of MyD88 residues, conserved in death domains of the homologous FADD and Pelle proteins, and analyzed the effect of the mutations on MyD88 signaling. Our studies revealed that mutation of residues 52 (MyD88E52A) and 58 (MyD88Y58A) impaired recruitment of both IRAK1 and IRAK4, whereas mutation of residue 95 (MyD88K95A) only affected IRAK4 recruitment. Since all MyD88 mutants were defective in signaling, recruitment of both IRAKs appeared necessary for activation of the pathway. Moreover, overexpression of a green fluorescent protein (GFP)-tagged mini-MyD88 protein (GFP-MyD88-(27–72)), comprising the Glu52 and Tyr58 residues, interfered with recruitment of both IRAK1 and IRAK4 by MyD88 and suppressed NF-κB activation by the interleukin-1 receptor but not by the MyD88-independent TLR3. GFP-MyD88-(27–72) exerted its effect by titrating IRAK1 and suppressing IRAK1-dependent NF-κB activation. These experiments identify novel residues of MyD88 that are crucially involved in the recruitment of IRAK1 and IRAK4 and in downstream propagation of MyD88 signaling.
MyD88 was first discovered during studies addressing the differentiation of mouse myeloid cells in response to growth-inhibitory stimuli (1). Subsequent investigations revealed that MyD88 possesses a modular organization (2), with an amino-terminal death domain (DD),3 found in proteins involved in cell death (3, 4), and a carboxyl-terminal Toll-interleukin-1 receptor (TIR) domain, present in the intracytoplasmic tail of receptors belonging to the Toll-like receptor (TLR)/interleukin-1 receptor (IL-1R) superfamily (5). MyD88 also has an intermediate domain (ID) that is crucial in TLR signaling due to its interaction with IRAK4 (6). The role of MyD88 as a signal transducer was first shown in the pathways triggered by the activation of IL-1R (7, 8) and TLR4 (9). Further studies showed that all TLRs, with the sole exception of TLR3, and the IL-1R family utilize the adaptor protein MyD88 to initiate their signaling pathway (10).
By virtue of its modular organization, MyD88 critically bridges activated receptor complexes to downstream adaptors/effectors. Upon activation, MyD88 is recruited through its TIR domain by the homologous domain of the activated TLR/IL-1R (11, 12). MyD88, in turn, has been shown to interact with a family of downstream kinases, namely IRAK1 (13), IRAK2 (7), IRAK-M (15), and IRAK4 (16), through the interaction of its DD with the respective DDs present in the amino-terminal region of IRAKs (17). At this stage, this multimeric complex is competent to elicit the propagation of the signal downstream of the receptor(s). Although MyD88 recruits IRAK-1 via DD-DD interactions, its recruitment of IRAK-4 appears to be rather unusual. Burns et al. (6) first demonstrated that an alternatively spliced variant of MyD88 (MyD88s), lacking the ID domain, failed to interact with IRAK-4, suggesting that residues located in both the DD and ID of MyD88 are crucially involved in the recruitment of IRAK-4. Nevertheless, no information is available on the specific residues in the DD in MyD88 required for its interaction with either IRAK1 or IRAK4.
The DD was initially defined as the region of homology between the cytoplasmic tails of the FAS/Apo1/CD95 and TNF receptors required for their induction of cytotoxic signaling (18, 19). In analogy with other DD-containing proteins, this domain in MyD88 is also involved in the formation of homomeric and heteromeric interactions. Herein, we have undertaken an alanine-scanning mutational analysis to identify amino acids that are required for downstream signaling and might participate in the homomeric and heteromeric interactions. Our studies revealed that MyD88E52A and MyD88Y58A mutants are strongly impaired in the recruitment of both IRAK1 and IRAK4, whereas the MyD88K95A mutant is deficient in recruiting IRAK4. These findings identify residues within the DD of MyD88 crucially involved in the formation of higher order complexes containing IRAK1 and IRAK4 and required for the propagation of the TLR/IL1-R signaling pathways.
EXPERIMENTAL PROCEDURES
Plasmids
Expression vectors for FLAG-tagged MyD88 or Myc-tagged MyD88, Myc-tagged IRAK1-kinase-dead (IRAK1KD) (K239S) and Myc-tagged IRAK4KD (K213A/K214A) were constructed by inserting PCR-generated cDNA fragments in the mammalian expression vectors p3X-FLAG and pCDNA3-N2-Myc, respectively. All site-directed mutations were inserted by PCR using oligonucleotides containing the mutated residue. Constructs for GFP-MyD88 fusion protein expression were obtained by subcloning cDNA encoding each protein into pEGFP-c1 vector. All constructs were confirmed by Cycle Sequencing (BMR Genomics, Padua, Italy). For the NF-κB reporter assays, the NF-κB luciferase and Renilla luciferase constructs were used according to the manufacturer's instructions (Promega Italia S.r.l., Milan, Italy).
Cell Culture and Transfections
The human embryonic kidney (HEK) 293T and HeLa cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen) and grown at 37 °C in a humidified atmosphere containing 95% air and 5% CO2. For co-immunoprecipitation of FLAG-MyD88/Myc-MyD88, HEK293T cells were first cultured in 10-cm diameter dishes and then transfected by the calcium phosphate method with 4–5 μg of the appropriate plasmids. To detect FLAG-MyD88 associated with Myc-IRAK1KD or Myc-IRAK4KD and GFP-MyD88-(27–72) associated with Myc-IRAK4KD, Myc-IRAK1KD, or Myc-MyD88, HEK293T cells were cultured in 6-cm diameter dishes and transfected by Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For immnunofluorescence analysis, HeLa cells were cultured in 3-cm diameter dishes and transfected with constructs for GFP or GFP-MyD88-(27–72) expression by FuGENE 6 reagent (Roche Applied Science) according to the manufacturer's instructions.
Co-immunoprecipitation Assay
HEK293T cells were collected 20 h after transfection, washed in ice-cold PBS, and lysed in buffer containing 50 mm Hepes, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 20 mm β-glycerophosphate, 2 mm dithiothreitol, 1 mm Na3VO4, and protease inhibitors. After incubating for 10 min on ice, cell lysates were centrifuged at 10,000 × g for 10 min at 4 °C, and cytosolic fractions were collected for immunoprecipitation. Cell extracts (1 mg of total proteins) were precleared by incubation for 1 h with protein G-Sepharose beads (Sigma) under constant shaking at 4 °C. After preclearing, cell extracts were incubated with 2 μg of mouse anti-FLAG M2 (Sigma) or rabbit anti-GFP antibodies (Molecular Probes) for 1 h under constant shaking at 4 °C. Concurrently, protein G-Sepharose beads or protein A-Sepharose beads were presaturated with 0.1% bovine serum albumin (Sigma) in PBS in the same conditions for 1 h. After incubation, the beads were washed twice with lysis buffer and then further incubated with cell extracts containing the antibodies for 1 h at 4 °C under constant shaking. Sepharose bead-bound immunocomplexes were washed three times in lysis buffer and eluted in SDS-PAGE sample buffer for Western blot analysis.
Western Blot Analysis
Cell extracts or immunoprecipitated proteins were diluted in SDS sample buffer, as described above, and boiled for 5 min. Proteins were separated on 8–10% SDS-PAGE gel and transferred to polyvinylidene fluoride Immobilon-P membranes (Millipore S.p.A., Vimodrone, Italy) using a semidry blotting apparatus (Bio-Rad). Membranes were saturated with 5% nonfat dry milk in PBS containing 0.1% Tween 20 for 1 h at room temperature and incubated overnight at 4 °C with the following primary antibodies: mouse anti-Myc and rabbit anti-Erk2 (1:1000 dilution; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), mouse anti-FLAG M2 and mouse anti-β tubulin (1:3000 and 1:1000 dilution, respectively; Sigma), and rabbit anti-GFP antibodies (1:1000 dilution; Molecular Probes). Secondary anti-mouse IgGs conjugated to horseradish peroxidase (Amersham Biosciences) were incubated with the membranes for 1 h at room temperature at a 1:10,000 dilution in PBS containing 0.1% Tween 20. Immunostained bands were detected by chemiluminescence (Santa Cruz Biotechnology).
NF-κB Reporter Assay
HeLa cells (1 × 105) were cultured in 12-well plates and transfected with 0.5 μg of an NF-κB-dependent luciferase reporter gene and Renilla luciferase reporter gene (4 ng) as an internal control together with the constructs for expression of MyD88 mutants or GFP-MyD88 fusion proteins by using the FuGENE 6 reagent according to the manufacturer's instructions. Twenty-four h after transfection, the cells were stimulated or not with 30 ng/ml IL-1β (R&D Systems, Minneapolis, MN) or 100 μg/ml poly(I-C) (Invivogen, San Diego, CA) for 6 h. After rinsing with PBS, the cells were harvested in two tubes and lysed separately. For biocounter luminometer analysis, the cells were lysed in 250 μl of passive lysis buffer (dual luciferase reporter assay system; Promega Italia S.r.l., Milan, Italy) for 15 min at room temperature. Cell lysates were cleared for 30 s by centrifugation at top speed in a refrigerated microcentrifuge and transferred to a fresh tube prior to reporter enzyme analysis. Ten μl of cell lysates were mixed with 100 μl of luciferase assay reagent II (Promega), and the NF-κB-firefly luciferase activity was determined using a biocounter luminometer. For the assessment of Renilla luciferase activity, 100 μl of Stop & Glo® reagent were added to the same sample. For all samples, the reporter data were normalized for transfection efficiency by dividing firefly luciferase activity by that of the Renilla luciferase. Data are expressed as mean -fold induction ± S.D. from a minimum of three separate experiments. To analyze the expression of the MyD88 mutants or GFP-MyD88 fusion proteins, HeLa cells were lysed in 30 μl of buffer (50 mm HEPES, pH 7.4, 15 mm MgCl2, 150 mm NaCl, 15 mm EGTA, 10% glycerol, 1% Triton X-100, protease inhibitor mixture (Sigma), 20 mm β-glycerophosphate, 2 mm dithiothreitol, 1 mm Na3VO4). Cells were centrifuged at 10,000 × g for 10 min, and the resulting supernatants were diluted in SDS sample buffer for Western blot analysis of levels of wild type or mutated MyD88 or GFP-MyD88 fusion proteins, respectively.
Immunofluorescence Microscopy
HeLa cells were cultured in 3-cm diameter dishes and transfected with constructs for GFP or GFP-MyD88-(27–72) expression by FuGENE 6 reagent (Roche Applied Science) according to the manufacturer's instructions. Twenty h after transfection, HeLa cells expressing GFP or GFP-MyD88-(27–72) were stimulated with 20 ng/ml IL-1β for 20 min at 37 °C. The cells were fixed at room temperature for 10 min with 4% paraformaldehyde in PBS and permeabilized at room temperature for 30 min with 0.5% Tween 20, 10% goat serum, 1% bovine serum albumin in PBS. After two washes in PBS, the samples were incubated overnight at 4 °C with anti-p65 NF-κB antibodies (1:500 dilution in PBS with 1% goat serum; Santa Cruz Biotechnology). Cells were washed three times with PBS and incubated for 1 h at room temperature with secondary antibodies (1: 500 dilution in PBS; Jackson ImmunoResearch Laboratories). Hoechst dye (0.1 μg/ml; Sigma) was added during the last 10 min of incubation to stain the nuclei. Slides were mounted in Mowiol 4-88 reagent (Calbiochem).
Densitometry and Statistical Analysis
Densitometric analysis of the Western blots was performed using underexposed images from 3–5 experiments and analyzed by ImageQuant version 5.0 software. Statistical significance was determined using the two-tailed Student's t test.
RESULTS
Identification of Residues within the DD of MyD88 That Are Required for Its Activity
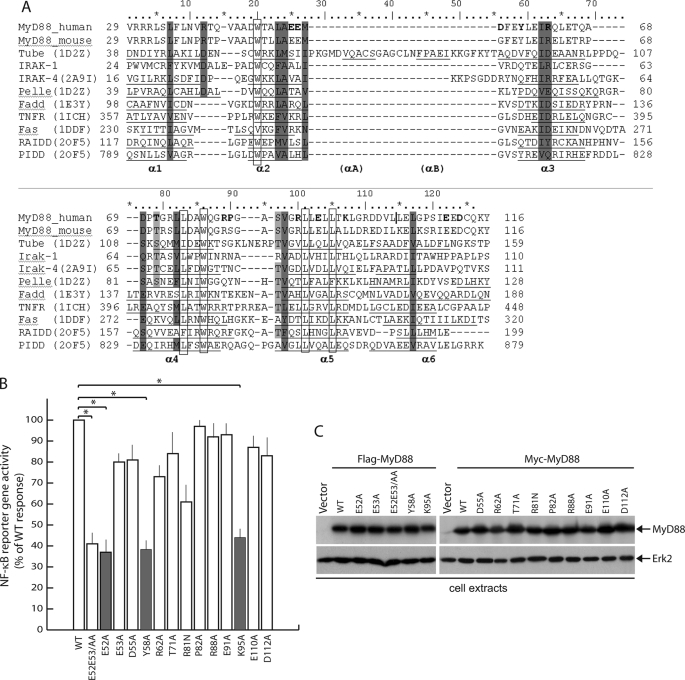
In order to delineate the relevant residues within the DD of MyD88 required for self-association and for interaction with downstream components of the pathway, we performed site-directed mutagenesis of selected residues. Previous mutagenesis studies carried out on FADD DD (20) and Pelle DD (21) highlighted the importance of polar/charged amino acids located on helices α2, α3, and α5 and loops α2-α3 and α4-α5 for the association with their partners Fas DD and Tube DD. For the alignment of human and mouse MyD88 and IRAK1 DD with homologous protein domains, we used the ClustalW algorithm (22) (Fig. 1A). A structural alignment of the DD of homologous proteins was performed using the available crystallographic/NMR data and the Swiss-Pdb viewer (23). The amino acids shown in boldface type (Fig. 1) in human MyD88 were substituted with alanine or asparagine (Arg81), and the effect on protein function was tested by assaying downstream activation of NF-κB caused by overexpression of MyD88 in the HeLa human cell line. Co-transfection of wild type or mutant MyD88 with a reporter construct containing NF-κB-responsive elements upstream of the firefly luciferase gene confirmed that wild type MyD88 triggers activation of NF-κB even in the absence of exogenous stimuli (Fig. 1B) (24). On the other hand, several mutations strongly affected MyD88 function. The E52A/E53A, Y58A, and K95A mutations reduced NF-κB activation by almost 60% (Fig. 1B, gray bars), whereas the R81N mutation was slightly less effective. The other MyD88 mutants analyzed activated NF-κB to a similar extent as wild type MyD88. All of the MyD88 proteins tested were expressed at comparable levels (Fig. 1C). Remarkably, single substitution of Glu52 or Glu53 to alanine indicated that the first glutamic acid residue (Glu52) is the one required for full function of MyD88, whereas E53A behaved similarly to the wild type protein. These results indicate that residues Glu52 (predicted in helix α2), Tyr58 (predicted in helix α3), and Lys95 (predicted in helix α5) are required for MyD88 activity.
FIGURE 1.
Effect of mutations in MyD88 on NF-κB signaling in HeLa cells. A, alignment of MyD88 DD with homologous protein domains. Conserved amino acids of the hydrophobic core are shown in open boxes; similar polar or hydrophobic residues are displayed in gray, whereas conserved Ser/Thr residues are shown in light gray; underlined residues form α-helical motifs according to NMR or x-ray structures; residues in boldface type are the ones selected for mutagenesis studies. B, NF-κB activity monitored by luciferase reporter assay. HeLa cells were transiently transfected with wild type (WT) or mutant MyD88 constructs together with NF-κB reporter constructs expressing firefly luciferase and a control plasmid expressing Renilla luciferase. After 24 h, the cells were harvested, and luciferase activity was measured in soluble extracts as described under “Experimental Procedures.” Data were normalized for transfection efficiency by dividing firefly luciferase activity by that of the Renilla luciferase. Data are expressed as a percentage of wild type ± S.D. from a minimum of three separate experiments. Statistical significance was determined by Student's t test. *, p < 0.01; **, p < 0.05. C, expression levels of wild type or mutated MyD88 in transfected HeLa determined by Western blotting with either anti-FLAG- or anti-Myc-specific antibodies. Cell extract loading was normalized with anti-Erk2 antibodies (lower panel).
Mutations in MyD88 DD Affect Recruitment of IRAK1 and IRAK4
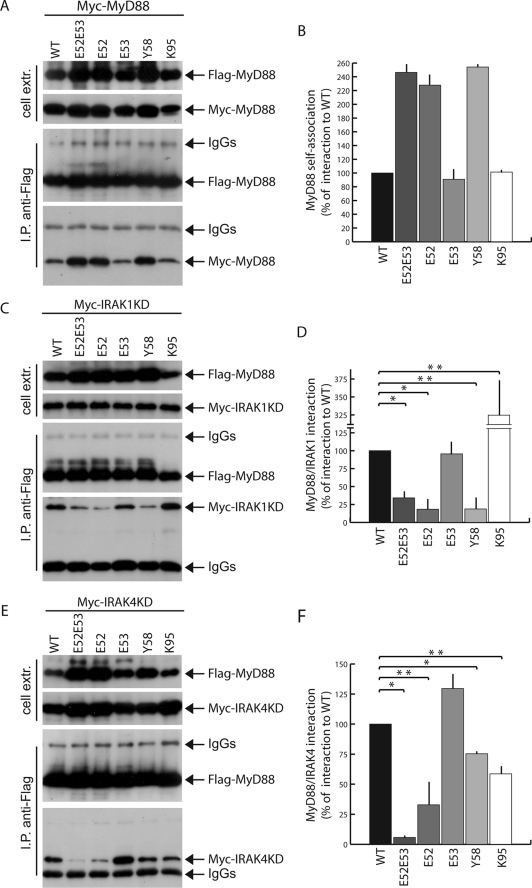
Death domains have six anti-parallel α-helices, arranged in a Greek key structure (25). The DD can fold to offer different surfaces of homomeric interactions (26). In the case of MyD88, the DD self-associates and also binds to the DDs of IRAKs (7, 8, 16). To determine which of these functions were disrupted in the MyD88 loss-of-function mutants, we first tested their ability to recruit wild type MyD88 by a co-immunoprecipitation assay (28). HEK293T cells were co-transfected with FLAG-tagged wild type or mutated MyD88 and wild type Myc-tagged MyD88. Cell extracts were immunoprecipitated with anti-FLAG antibodies, and association with the wild type protein was tested by detecting Myc-MyD88 in the immunoprecipitates. Remarkably, none of the mutations impaired MyD88 self-association (Fig. 2, A and B), demonstrating that the Glu52, Tyr58, and Lys95 residues are not required for MyD88 self-association.
FIGURE 2.
Mutations in MyD88 DD affect protein self-association and recruitment of IRAK1 and IRAK4. A, HEK293T cells were transfected with Myc-MyD88 in combination with wild type (first lane) or mutated (second through sixth lanes) FLAG-MyD88. Twenty h after transfection, cells were collected, and self-association of MyD88 was assessed by co-immunoprecipitation. Cell extracts were immunoprecipitated (I.P.) with anti-FLAG antibodies, and the immunoprecipitated proteins were then analyzed by Western blotting with either anti-FLAG- or anti-Myc-specific antibodies to detect association. The mutations in FLAG-MyD88 increase MyD88 self-association about 2.5 times in comparison with the wild type protein. B, densitometric analysis of the effect of MyD88 mutants used in A on MyD88 dimerization. C–E, HEK293T cells were transfected with either Myc-IRAK1KD (C) or Myc-IRAK4KD (E) in combination with wild type (first lane in C and E) or mutated (second through sixth lanes in C and E) FLAG-MyD88. Twenty h after transfection, the cells were harvested, and the interaction of MyD88 with IRAKs was determined by co-immunoprecipitation. Cell extracts were immunoprecipitated with anti-FLAG antibodies, and the immunoprecipitated proteins were then analyzed by Western blotting with either anti-FLAG- or anti-Myc-specific antibodies to reveal the interaction. MyD88 mutants (E52A/E53A, E52A, and Y58A) strongly interfere with recruitment of IRAK1 and IRAK4 by MyD88 (densitometric analysis, D and F). Densitometric data in B, D, and F are expressed as a percentage of the wild type ± S.D. from a minimum of three separate experiments. Statistical significance was determined by Student's t test. *, p < 0,01; **, p < 0.05.
Next, we tested whether the signaling-defective MyD88 mutants were impaired in their ability to associate with IRAK1 and IRAK4. Since the interaction between MyD88 and these kinases is rapid and transient, it can be reproducibly detected only by expressing kinase-dead IRAK1 and IRAK4 (IRAK1KD and IRAK4KD) (8, 16). Hence, wild type or mutant FLAG-tagged MyD88 proteins were co-expressed with either Myc-tagged IRAK1KD (Fig. 2C) or IRAK4KD (Fig. 2E), and cell extracts were immunoprecipitated with anti-FLAG antibodies. We observed a strong interaction between wild type MyD88 and IRAK1KD or IRAK4KD (lane 1 in Fig. 2, C and E). By contrast, MyD88E52A/E53A, MyD88E52A, and MyD88Y58A were impaired in the recruitment of both IRAK1KD (Fig. 2C) and IRAK4KD (Fig. 2E). In line with its ability to elicit NF-κB activation, MyD88E53A behaved like the wild type protein in the recruitment of both IRAKs (Fig. 2, C and E). Interestingly, mutation of the Lys95 residue exerted a different effect on MyD88 interaction with the two kinases. Indeed, whereas it strongly bound to IRAK1KD (Fig. 2, C and D), MyD88K95A interacted less efficiently than the wild type protein with IRAK4KD (Fig. 2, E and F). These findings highlight novel amino acid residues, within the DD of MyD88, that are crucial for the recruitment of IRAK1 and IRAK4 but not for its self-association.
Identification of the MyD88 DD Region Required for the Recruitment of IRAK1
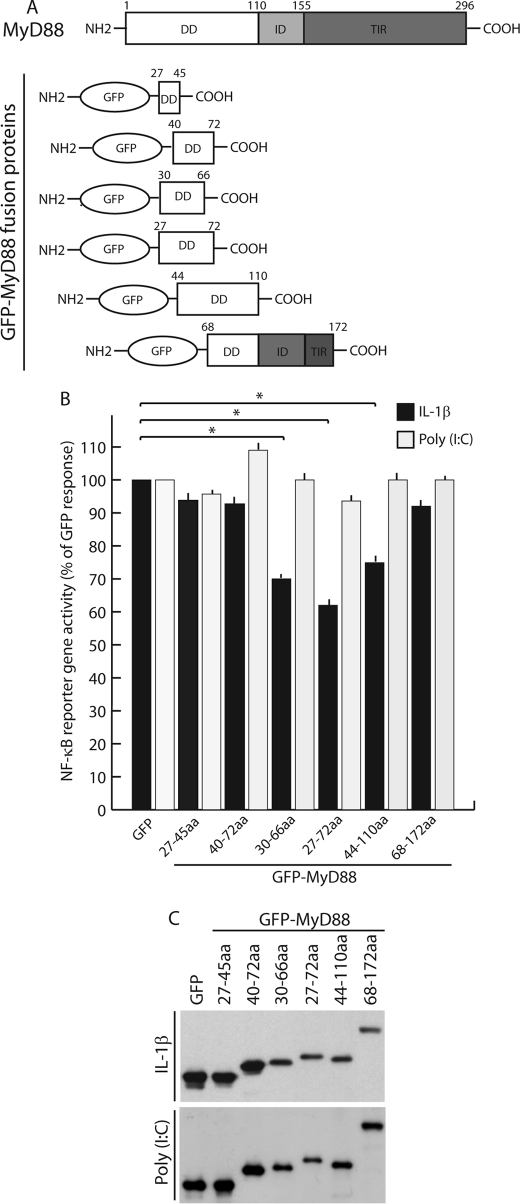
Our results suggested that residues 52–95 define a subregion of the MyD88 DD domain required for the interaction with IRAK1 and IRAK4. To determine whether expression of different regions of MyD88 DD affected IL1-R signal transduction, we produced a series of GFP-MyD88 fusion proteins containing portions of this domain, as schematically represented in Fig. 3A. We reasoned that these chimeric proteins might exert a dominant-negative effect on the endogenous MyD88 by titrating out IRAKs without triggering NF-κB activation. Using a reporter gene assay, we tested whether overexpressed GFP or GFP-MyD88 fusion proteins were able to interfere with IL-1-mediated activation of NF-κB. We found that GFP-MyD88-(27–72), GFP-MyD88-(30–66), and GFP-MyD88-(44–110) significantly reduced IL-1-dependent activation of NF-κB (by approximately 40, 30 and 25%, respectively; p < 0.01; Fig. 3B). Remarkably, the effect of the GFP-MyD88 DD proteins was specific, since they did not inhibit activation of NF-κB by the MyD88-independent (29) poly(I-C)/TLR3 pathway (Fig. 3B). By contrast, GFP-MyD88-(27–45), MyD88-(40–72), and GFP-MyD88-(68–172) did not exert significant effects on either IL-1-dependent or poly(I-C)-dependent activation of NF-κB (Fig. 3B). Similar amounts of chimeric proteins were expressed in the samples (Fig. 3C). Since GFP-MyD88-(27–72) exerted the strongest inhibition of NF-κB transcriptional activity, which relies on its translocation from cytoplasm to the nucleus (30), we also tested whether overexpression of GFP-MyD88-(27–72) interfered with the subcellular localization of the p65/NF-κB protein. In non-stimulated cells, p65 was localized exclusively in the cytoplasm (Fig. 4, A and C). Stimulation with IL-1β for 20 min caused the translocation of p65 into the nucleus of GFP-transfected cells (Fig. 4B). However, in cells expressing GFP-MyD88-(27–72) (Fig. 4, G and H), we found that significant amounts of p65 remained in the cytoplasm after IL-1β treatment (indicated by the arrows in Fig. 4D). Taken together, these results suggest that GFP-MyD88-(27–72) attenuates MyD88-dependent activation of NF-κB, probably by exerting a dominant negative effect on MyD88 function in live cells.
FIGURE 3.
Chimeric GFP-MyD88 proteins differentially affect NF-κB signaling. A, GFP-MyD88 fusion proteins are schematically depicted. B, NF-κB activity monitored by luciferase reporter assay. HeLa cells were transfected with GFP or GFP-MyD88 fusion protein constructs together with NF-κB reporter constructs expressing firefly luciferase and a control plasmid expressing Renilla luciferase. Twenty-four h after transfection, the cells were left untreated or were stimulated with either 30 ng/ml IL-1β (dark bars) or 100 μg/ml poly(I-C) (gray bars) for an additional 6 h. At the end of incubation, the cells were harvested, and luciferase activity was measured in soluble extracts as described under “Experimental Procedures.” NF-κB-driven firefly luciferase reporter activity was normalized to the control Renilla luciferase activity, as described above. Activities are plotted as percentage of GFP ± S.D. of at least three experiments for each fusion protein. GFP-MyD88-(27–72), GFP-MyD88-(30–66), and GFP-MyD88-(44–110) significantly reduced IL-1-dependent activation of NF-κB. *, p < 0.01 (n = 3). C, expression levels of GFP-MyD88 fusion proteins in transfected HeLa cells as determined by Western blot with anti-GFP antibodies. aa, amino acids.
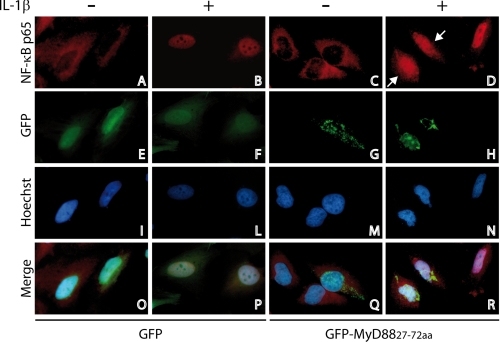
FIGURE 4.
GFP-MyD88-(27–72) interferes with the subcellular localization of the p65/NF-κB protein. Immunofluorescence analysis of the effect of GFP-MyD88-(27–72) on nuclear translocation of endogenous NF-κB p65. HeLa cells were transfected with GFP or GFP-MyD88-(27–72) (panels E and F and panels G and H, respectively). Twenty h after transfection, the cells were left untreated (panels A, E, I, and O and panels C, G, M, and Q) or treated with 20 ng/ml IL-1β for 20 min (panels B, F, L, and P and panels D, H, N, and R). Cells were fixed, blocked, and stained with anti-NF-κB p65 antibodies (red) and Hoechst (blue) for nuclear staining. Nuclear NF-κB p65 is observed 20 min after treatment with IL-1β in the cells expressing GFP, but it is held back in the cytoplasm in those expressing GFP-MyD88-(27–72) (indicated by an arrow in D).
GFP-MyD88-(27–72) Binds IRAK1 and Interferes with Recruitment of IRAK1 and IRAK4 by MyD88
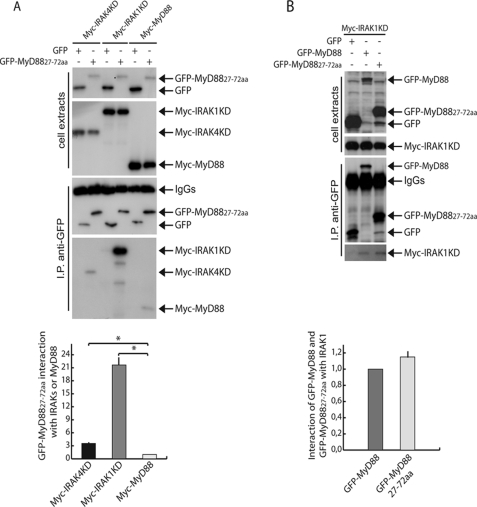
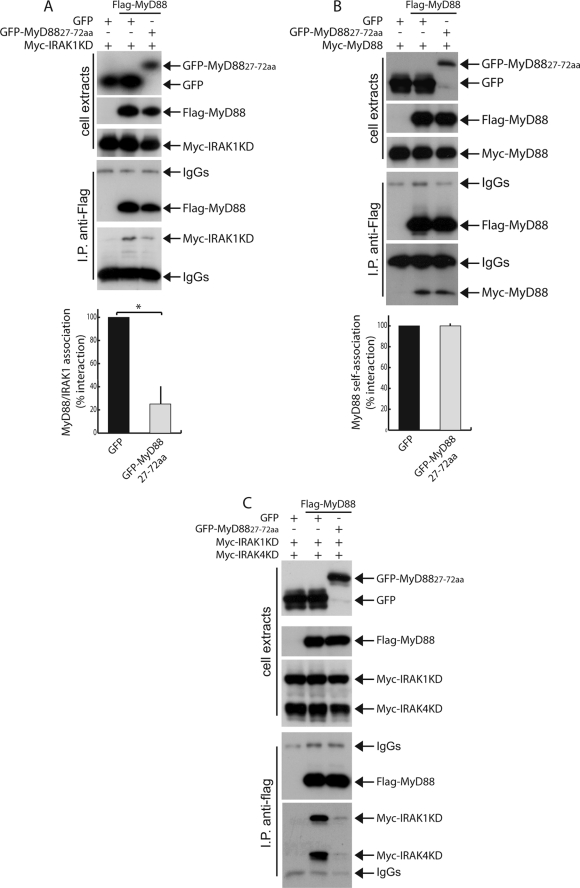
To further investigate the inhibitory effect of GFP-MyD88-(27–72) on MyD88 signaling, we analyzed its ability to bind full-length MyD88 and IRAK1/4 by co-immunoprecipitation assays. GFP or GFP-MyD88-(27–72) were co-expressed in HEK293T cells with MyD88, IRAK1KD, or IRAK4KD. Western blot analysis showed that GFP-MyD88-(27–72) strongly interacted with IRAK1KD (Fig. 5A), whereas it bound very weakly to either IRAK4KD (Fig. 5A) or full-length MyD88 (Fig. 5A). The association of GFP-MyD88-(27–72) with IRAK1KD was comparable with that of full-length MyD88 (Fig. 5B), suggesting that the miniprotein folded correctly and maintained the original structure. This result suggests that this region of MyD88 is sufficient to associate with IRAK1. To test whether the inhibitory effect of GFP-MyD88-(27–72) was due to its ability to titrate out IRAK1, we performed co- immunoprecipitation assays between MyD88 and IRAK1KD in HEK293T cells expressing either GFP or GFP-MyD88-(27–72). As shown in Fig. 6A, GFP-MyD88-(27–72) was able to significantly inhibit (p < 0.01) IRAK1KD recruitment by MyD88 by ∼70%. This effect was specific, because in similar experiments, GFP-MyD88-(27–72) did not interfere with MyD88 self-association (Fig. 6B). To test whether inhibition of IRAK1 recruitment also affected the interaction of MyD88 with IRAK4, we co-expressed all components of the complex together with GFP or GFP-MyD88-(27–72). Co-immunoprecipitation assays of MyD88 with IRAK1KD and IRAK4KD in HEK293T cells showed that GFP-MyD88-(27–72) also prevents the recruitment of IRAK4 by MyD88 (Fig. 6C).
FIGURE 5.
GFP-MyD88-(27–72) interacts with IRAK1. A, HEK293T cells were transfected with GFP or GFP-MyD88-(27–72) in combination with Myc-IRAK4KD (lanes 1 and 2) or Myc-IRAK1KD (lanes 3 and 4) or Myc-MyD88 (lanes 5 and 6). Twenty h after transfection, the cells were collected, and the interaction of GFP-MyD88-(27–72) with Myc-IRAK4KD, Myc-IRAK1KD, or Myc-MyD88 was assessed by co-immunoprecipitation. Cell extracts were immunoprecipitated (IP) with anti-GFP antibodies, and immunoprecipitated proteins were analyzed by Western blot with either anti-GFP or anti-Myc antibodies to detect association. GFP-MyD88-(27–72) strongly interacts with IRAK1 (lane 4). Densitometric analysis of the degree of interaction of GFP-MyD88-(27–72) with Myc-IRAK4KD, Myc-IRAK1KD, or Myc-MyD88, respectively, is shown and is expressed as -fold induction with respect to MyD88. Statistical significance of the effects observed is indicated as follows. *, p < 0.01 (n = 3). B, HEK293T cells were transfected with Myc-IRAK1KD in combination with GFP (lane 1), GFP-MyD88 (lane 2), or GFP-MyD88-(27–72) (lane 3). Twenty h after transfection, the cells were collected, and the interaction of GFP-MyD88 and GFP-MyD88-(27–72) with Myc-IRAK1KD was assessed by co-immunoprecipitation. Cell extracts were immunoprecipitated (IP) with anti-GFP antibodies, and immunoprecipitated proteins were analyzed by Western blot with either anti-GFP or anti-Myc antibodies. Densitometric analysis of the interaction of GFP-MyD88 and GFP-MyD88-(27–72) with Myc-IRAK1KD is shown and is expressed as -fold induction with respect to GFP-MyD88.
FIGURE 6.
GFP-MyD88-(27–72) interferes with recruitment of IRAK1 and IRAK4 by MyD88. A, HEK293T cells were transfected with Myc-IRAK1KD alone (lane 1) or in combination with FLAG-MyD88 (lanes 2 and 3) in the presence of GFP (lanes 1 and 2) or GFP-MyD88-(27–72) (lane 3). Twenty h after transfection, the cells were collected, and the effect of either GFP or GFP-MyD88-(27–72) on the interaction of FLAG-MyD88 with Myc-IRAK1KD was evaluated by co-immunoprecipitation. Cell extracts were immunoprecipitated (IP) with anti-FLAG antibodies, and the immunoprecipitated proteins were then analyzed by Western blotting with either anti-FLAG or anti-Myc antibodies to detect association. GFP-MyD88-(27–72) strongly interferes with recruitment of IRAK1 by MyD88 (lane 3). Densitometric analysis of these results is depicted. GFP-MyD88-(27–72) significantly inhibited IRAK1KD recruitment by MyD88 (*, p < 0.01; n = 3). B, HEK293T cells were transfected with Myc-MyD88 alone (lane 1) or in combination with FLAG-MyD88 (lanes 2 and 3) in the presence of GFP (lanes 1 and 2) or GFP-MyD88-(27–72) (lane 3). Twenty h after transfection, the cells were harvested, and the effect of either GFP or GFP-MyD88-(27–72) on MyD88 self-association was assessed by co-immunoprecipitation. Cell extracts were immunoprecipitated with anti-FLAG antibodies, and immunoprecipitated proteins were analyzed by Western blotting with either anti-FLAG or the anti-Myc antibodies to detect MyD88 self-association. GFP-MyD88-(27–72) does not interfere with MyD88 self-association (lane 3). Densitometric analysis of these results is shown. C, HEK293T cells were transfected with Myc-IRAK1KD and Myc-IRAK4KD alone (lane 1) or in combination with FLAG-MyD88 (lanes 2 and 3) in the presence of GFP (lanes 1 and 2) or GFP-MyD88-(27–72) (lane 3). Twenty h after transfection, the cells were collected, and the effect of either GFP or GFP-MyD88-(27–72) on the interaction of FLAG-MyD88 with Myc-IRAK1KD and Myc-IRAK4KD was evaluated by co-immunoprecipitation. Cell extracts were immunoprecipitated with anti-FLAG antibodies, and the immunoprecipitated proteins were then analyzed by Western blotting with either anti-FLAG or anti-Myc antibodies to detect interaction. GFP-MyD88-(27–72) strongly interferes with recruitment of IRAKs by MyD88 (lane 3).
GFP-MyD88-(27–72) Inhibits IRAK1-dependent Activation of NF- κB
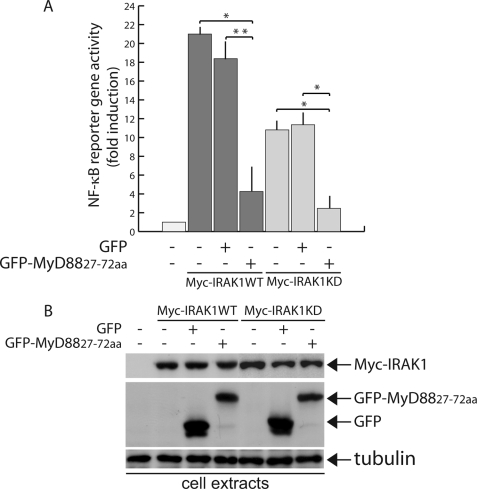
If GFP-MyD88-(27–72) inhibits IL-1 signaling through titration of IRAK1, it should also interfere with the activation of NF-κB elicited when this kinase is overexpressed (8, 31). Using a reporter gene assay, we observed that overexpression of wild type or kinase-dead IRAK1 induces NF-κB activation (Fig. 7A). Strikingly, in both cases, co-expression of GFP-MyD88-(27–72) strongly inhibited IRAK1-induced NF-κB activation, whereas GFP did not exert any effect. Similar amounts of GFP or GFP-MyD88-(27–72) and wild type or kinase-dead Myc-IRAK1 were expressed in the samples (Fig. 7B). In similar experiments, wild type and kinase-dead IRAK4 were unable to induce NF-κB activation in HEK293T cells (data not shown). These results confirm that the region of the MyD88 DD comprised between residues 27 and 72 is required for the recruitment of IRAK1 and for the subsequent activation of the downstream pathway.
FIGURE 7.
GFP-MyD88-(27–72) interferes with activation of NF-κB induced by overexpression of IRAK1. A, NF-κB activity monitored by luciferase reporter assay. HeLa cells were transfected with GFP or GFP-MyD88-(27–72) together with NF-κB firefly luciferase, Renilla luciferase, and wild type (WT) Myc-IRAK1 (dark gray) or kinase-dead (KD) Myc-IRAK1 (light gray) constructs. Twenty-four h after transfection, the cells were harvested, and luciferase activity was measured in soluble extracts, as described under “Experimental Procedures.” NF-κB-driven firefly luciferase reporter activity was normalized to the control Renilla-luciferase activity, as described above. Activities are plotted as mean -fold induction ± S.D., compared with control only transfected with NF-κB firefly luciferase and Renilla luciferase constructs of at least three experiments. GFP-MyD88-(27–72) significantly reduces NF-κB activity induced by overexpression of Myc-IRAK1WT or Myc-IRAK1KD (*, p < 0.01; **, p < 0.05). B, expression levels of GFP, GFP-MyD88-(27–72) and Myc-IRAK1WT or Myc-IRAK1KD in transfected HeLa as determined by Western blotting with anti-GFP or anti-Myc antibodies. Cell extract loading was normalized with anti-tubulin (lower panel). GFP-MyD88-(27–72) exerts a dominant negative effect on IRAK1-induced activation of NF-κB.
DISCUSSION
MyD88 is an adaptor protein that plays a crucial role in innate immunity (10). MyD88 is required for the intracellular signaling triggered by stimulation of most TLR/IL1R receptors. Although it is generally accepted that MyD88 acts by recruiting the kinases IRAK1 and IRAK4 through interaction between their respective DDs, the specific residues involved in such interactions have not been determined yet. Herein, we identified three residues that are required for the interaction of MyD88 with IRAK1 and/or IRAK4 and whose substitution strongly impairs MyD88-dependent activation of NF-κB. Our results provide strong evidence that the region of the DD of MyD88 composed of residues 27–72 (predicted α1, α2, α3, and NH2-terminal α4 helices) is required for the recruitment of IRAK1. Moreover, we demonstrate that overexpression of this region in cells titrates out IRAK1 and specifically prevents MyD88-dependent signaling. On the other hand, mutation of Lys95 in the predicted α5 helix only affects recruitment of IRAK4. Thus, our results provide the first evidence of single site-specific substitution in the DD of MyD88 that selectively interferes with downstream components of its signaling pathway.
A recent crystallographic study on RAIDD DD and PIDD DD has demonstrated that the DDs can form three types of asymmetric interactions that generate eight different interfaces or complexes (26). Based on previous nomenclature (32), these homomeric and heteromeric surfaces were named type I (a and b), type II (a and b), and type III (a and b) (26). By analogy with these DD-containing proteins, MyD88 might form higher order oligomers in which each monomeric domain can be engaged in three types of interactions, named type I–III. These interfaces can form either homo- or heteroassociation to yield a large multimeric complex competent for signaling.
By following the proposed type of interaction for RAIDD-PIDD complex (26), the residues Glu52 and Tyr58 of MyD88 are comprised in a type Ib interface, which asymmetrically interacts with a type Ia interface; analogously, the corresponding residues Leu136 and Gln142/Tyr814, respectively, in RAIDD-PIDD (Fig. 1A) disrupted in vitro complex assembly following their mutation.
Theoretically, the region encompassing the residues Glu52–Tyr58 could interact with a type Ia surface of MyD88 itself, IRAK1, or IRAK4. However, our results suggest that in the case of MyD88, this type Ib surface recruits IRAK1. This is supported by the evidence that mutations in these residues strongly reduce the recruitment of IRAKs but do not impair MyD88 self-association. Moreover, a mini-MyD88 protein encompassing these residues associates with IRAK1 but not with IRAK4. Although GFP-MyD88-(27–72) is an artificial chimeric protein, it appeared to fold properly, because its ability to associate with IRAK1 was comparable with that of full-length MyD88.
Our co-immunoprecipitation experiments indicate that residue Lys95 of MyD88 is required for the association between MyD88 and IRAK4. By following the alignment depicted in Fig. 1A, the residues of RAIDD and PIDD corresponding to Lys95 are Ala186 and Gln859, respectively, and lie in the α5-α6 loop. Both Ala186 of RAIDD and Gln859 of PIDD are exposed to solvent, and Gln859 results in participation in a type I heteromeric interaction with the Gln125 residue of RAIDD, which is located at the end of the α1 helix (26). Thus, based on these findings and our new experimental results, we might speculate that MyD88-IRAK4 heteromeric association also occurs via an extended type I interface comprising the α5-α6 loop.
A previous study reported that the first 172 residues of MyD88 are required for the interaction with IRAK4. This region contains the whole DD and ID and a small portion of the TIR domain (6). However, neither the isolated DD nor the ID was sufficient for this interaction, suggesting that the binding region is composed of residues spanning both the DD and the ID or that the ID induces a change in the conformation of the DD that is required for association with IRAK4 (6). Our results are compatible with both models proposed in the previous study and suggest that Lys95 is one of the residues in the DD that are crucial for the recruitment of IRAK4. Since Lys95 is replaced by a leucine in mouse MyD88, it is possible that the hydrophobic contribution of the lysine, rather than its charge, is important for this interaction. Crystal structure analyses of MyD88-IRAK complexes will be required to fully elucidate the mode(s) of interaction between these proteins.
While this manuscript was in preparation, it was reported that children with recurrent pyogenic infections display MyD88 mutations in the DD or TIR domain of the protein (33). Interestingly, one of the TIR mutations (the heterozygous missense mutation R196C) was within the heptapeptide MyD88-(196–202) (i.e. Ac-RDVLPGT-NH2) that we previously showed to be critically involved in promoting dimerization of the TIR domains of MyD88 (34). Intriguingly, von Bernuth and co-workers (33) have also found that a homozygous in-frame MyD88 deletion (i.e. E52 del) provokes functional MyD88 deficiency due to defective IRAK4 recruitment. Although they did not report whether IRAK1 is similarly affected, our present data indicate that MyD88E52A is defective in the recruitment of both IRAK1 and IRAK4. Moreover, the region composed of residues 27–72 preferentially associates with IRAK1 and not IRAK4. It is possible that mutation of Glu52 also affects the region of interaction with IRAK4, possibly due to their common presence in an extended heteromeric type Ib interface similar to that described in the RAIDD-PIDD model (see Fig. 3D in Ref. 26). Since our mutagenesis study has identified a MyD88 loss-of-function mutation found in patients, it is possible that Tyr58 and Lys95 are additional residues that, if mutated, cause defective MyD88 signaling and might pave the way for selected genetic analyses of these residues in patients affected by other immunological disorders.
The work presented herein is based on structural/functional analyses of MyD88 and on site-directed mutagenesis of residues potentially involved in protein-protein interactions. Although most of our experiments are performed in transfected cells overexpressing recombinant proteins, several observations indicate that the conclusions that can be reached with this approach are physiologically relevant. First, a previous study using the same approach with the TIR domain of MyD88 has allowed us to identify a region composed of residues 196–202 that are required for MyD88 function (34). These results were then independently confirmed by other investigators (35). Remarkably, a small cell-permeable peptide encompassing these amino acids specifically inhibits MyD88 function also in live cells (34). To our knowledge, this peptide is currently the only commercially available MyD88-specific inhibitor and has contributed to defining the role of TLR/MyD88 function in several cellular and physiological systems (36–39). Second, a synthetic molecule mimicking the structure of this peptide reproduced the effect on MyD88 dimerization and specific inhibition of IL1-R-mediated NF-κB activation both in live cells and animals (28). Third, two of the three mutations (Glu52 and Arg196) identified in immunologically deficient children by von Bernuth and co-workers (33) have been anticipated by our structural/functional and mutagenesis analyses (this work) (28), strongly indicating the validity of this approach. Thus, the results presented herein, which point to Glu52, Tyr58, and Lys95 as crucial residues for MyD88 signaling, might provide the rational basis for the design of new cell-permeable peptides or synthetic molecules that specifically suppress TLR/MyD88-mediated responses. Since aberrant regulation of this pathway is relevant for several inflammatory and autoimmune diseases (40, 41), it may be predicted that the development of specific inhibitors of this pathway will be valuable for a novel therapeutic approach of such diseases (14, 27).
Acknowledgments
We acknowledge Dr. M. Muzio for providing the AU1-MyD88 cDNA and are very grateful for the help from Dr. S. Vincenti in sequence alignment.
This work was supported by grants from the Istituto Superiore della Sanità (ISS Project 527/B/3A/5), the Association for International Cancer Research, and the Fondazione Italiana Sclerosi Multipla.
- DD
- death domain
- TIR
- Toll-interleukin-1 receptor
- TLR
- Toll-like receptor
- IL
- interleukin
- IL-1R
- interleukin-1 receptor
- ID
- intermediate domain
- GFP
- green fluorescent protein
- HEK
- human embryonic kidney
- PBS
- phosphate-buffered saline
- IRAK
- interleukin-1 receptor-associated kinase.
REFERENCES
- 1.Lord K. A., Hoffman-Liebermann B., Liebermann D. A. (1990) Oncogene 5, 387–396 [PubMed] [Google Scholar]
- 2.Hardiman G., Rock F. L., Balasubramanian S., Kastelein R. A., Bazan J. F. (1996) Oncogene 13, 2467–2475 [PubMed] [Google Scholar]
- 3.Hofmann K., Tschopp J. (1995) FEBS Lett. 371, 321–323 [DOI] [PubMed] [Google Scholar]
- 4.Park H. H., Lo Y. C., Lin S. C., Wang L., Yang J. K., Wu H. (2007) Annu. Rev. Immunol. 25, 561–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hultmark D. (1994) Biochem. Biophys. Res. Commun. 199, 144–146 [DOI] [PubMed] [Google Scholar]
- 6.Burns K., Janssens S., Brissoni B., Olivos N., Beyaert R., Tschopp J. (2003) J. Exp. Med. 197, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muzio M., Ni J., Feng P., Dixit V. M. (1997) Science 278, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 8.Wesche H., Henzel W. J., Shillinglaw W., Li S., Cao Z. (1997) Immunity 7, 837–847 [DOI] [PubMed] [Google Scholar]
- 9.Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway C. A., Jr. (1998) Mol. Cell. 2, 253–258 [DOI] [PubMed] [Google Scholar]
- 10.Janssens S., Beyaert R. (2002) Trends Biochem. Sci. 27, 474–482 [DOI] [PubMed] [Google Scholar]
- 11.Ulrichts P., Peelman F., Beyaert R., Tavernier J. (2007) FEBS Lett. 581, 629–636 [DOI] [PubMed] [Google Scholar]
- 12.Brikos C., Wait R., Begum S., O'Neill L. A., Saklatvala J. (2007) Mol. Cell. Proteomics 6, 1551–1559 [DOI] [PubMed] [Google Scholar]
- 13.Cao Z., Henzel W. J., Gao X. (1996) Science 271, 1128–1131 [DOI] [PubMed] [Google Scholar]
- 14.Hong-Geller E., Chaudhary A., Lauer S. (2008) Curr. Drug Discov. Technol. 5, 29–38 [DOI] [PubMed] [Google Scholar]
- 15.Wesche H., Gao X., Li X., Kirschning C. J., Stark G. R., Cao Z. (1999) J. Biol. Chem. 274, 19403–19410 [DOI] [PubMed] [Google Scholar]
- 16.Li S., Strelow A., Fontana E. J., Wesche H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5567–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann D., Kollewe C., Resch K., Martin M. U. (2007) Biochem. Biophys. Res. Commun. 354, 1089–1094 [DOI] [PubMed] [Google Scholar]
- 18.Tartaglia L. A., Ayres T. M., Wong G. H., Goeddel D. V. (1993) Cell 74, 845–853 [DOI] [PubMed] [Google Scholar]
- 19.Feinstein E., Kimchi A., Wallach D., Boldin M., Varfolomeev E. (1995) Trends Biochem. Sci. 20, 342–344 [DOI] [PubMed] [Google Scholar]
- 20.Jeong E. J., Bang S., Lee T. H., Park Y. I., Sim W. S., Kim K. S. (1999) J. Biol. Chem. 274, 16337–16342 [DOI] [PubMed] [Google Scholar]
- 21.Xiao T., Towb P., Wasserman S. A., Sprang S. R. (1999) Cell 99, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 24.Burns K., Martinon F., Esslinger C., Pahl H., Schneider P., Bodmer J. L., Di Marco F., French L., Tschopp J. (1998) J. Biol. Chem. 273, 12203–12209 [DOI] [PubMed] [Google Scholar]
- 25.Berglund H., Olerenshaw D., Sankar A., Federwisch M., McDonald N. Q., Driscoll P. C. (2000) J. Mol. Biol. 302, 171–188 [DOI] [PubMed] [Google Scholar]
- 26.Park H. H., Logette E., Raunser S., Cuenin S., Walz T., Tschopp J., Wu H. (2007) Cell 128, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gearing A. J. (2007) Immunol. Cell Biol. 85, 490–494 [DOI] [PubMed] [Google Scholar]
- 28.Loiarro M., Capolunghi F., Fantò N., Gallo G., Campo S., Arseni B., Carsetti R., Carminati P., De Santis R., Ruggiero V., Sette C. (2007) J. Leukocyte Biol. 82, 801–810 [DOI] [PubMed] [Google Scholar]
- 29.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 30.Ding G. J., Fischer P. A., Boltz R. C., Schmidt J. A., Colaianne J. J., Gough A., Rubin R. A., Miller D. K. (1998) J. Biol. Chem. 273, 28897–28905 [DOI] [PubMed] [Google Scholar]
- 31.Li X., Commane M., Jiang Z., Stark G. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4461–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber C. H., Vincenz C. (2001) Trends Biochem. Sci. 26, 475–481 [DOI] [PubMed] [Google Scholar]
- 33.von Bernuth H., Picard C., Jin Z., Pankla R., Xiao H., Ku C. L., Chrabieh M., Mustapha I. B., Ghandil P., Camcioglu Y., Vasconcelos J., Sirvent N., Guedes M., Vitor A. B., Herrero-Mata M. J., Aróstegui J. I., Rodrigo C., Alsina L., Ruiz-Ortiz E., Juan M., Fortuny C., Yagüe J., Antón J., Pascal M., Chang H. H., Janniere L., Rose Y., Garty B. Z., Chapel H., Issekutz A., Maródi L., Rodriguez-Gallego C., Banchereau J., Abel L., Li X., Chaussabel D., Puel A., Casanova J. L. (2008) Science 321, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loiarro M., Sette C., Gallo G., Ciacci A., Fantò N., Mastroianni D., Carminati P., Ruggiero V. (2005) J. Biol. Chem. 280, 15809–15814 [DOI] [PubMed] [Google Scholar]
- 35.Bartfai T., Behrens M. M., Gaidarova S., Pemberton J., Shivanyuk A., Rebek J., Jr. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7971–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan J., Li Y., Levy R. M., Fan J. J., Hackam D. J., Vodovotz Y., Yang H., Tracey K. J., Billiar T. R., Wilson M. A. (2007) J. Immunol. 178, 6573–6580 [DOI] [PubMed] [Google Scholar]
- 37.Funderburg N., Lederman M. M., Feng Z., Drage M. G., Jadlowsky J., Harding C. V., Weinberg A., Sieg S. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18631–18635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rolls A., Shechter R., London A., Ziv Y., Ronen A., Levy R., Schwartz M. (2007) Nat. Cell Biol. 9, 1081–1088 [DOI] [PubMed] [Google Scholar]
- 39.Scott M. J., Billiar T. R. (2008) J. Biol. Chem. 283, 29433–29446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Neill L. A. (2003) Curr. Opin. Pharmacol. 3, 396–403 [DOI] [PubMed] [Google Scholar]
- 41.Fischer M., Ehlers M. (2008) Ann. N.Y. Acad. Sci. 1143, 21–34 [DOI] [PubMed] [Google Scholar]