Abstract
Bacteria/eukaryotes share a common pathway for coenzyme A (CoA) biosynthesis. Although archaeal genomes harbor homologs for most of these enzymes, homologs of bacterial/eukaryotic pantothenate synthetase (PS) and pantothenate kinase (PanK) are missing. PS catalyzes the ATP-dependent condensation of pantoate and β-alanine to produce pantothenate, whereas PanK catalyzes the ATP-dependent phosphorylation of pantothenate to produce 4′-phosphopantothenate. When we examined the cell-free extracts of the hyperthermophilic archaeon Thermococcus kodakaraensis, PanK activity could not be detected. A search for putative kinase-encoding genes widely distributed in Archaea, but not present in bacteria/eukaryotes, led to four candidate genes. Among these genes, TK2141 encoded a protein with relatively low PanK activity. However, higher levels of activity were observed when pantothenate was replaced with pantoate. Vmax values were 7-fold higher toward pantoate, indicating that TK2141 encoded a novel enzyme, pantoate kinase (PoK). A search for genes with a distribution similar to TK2141 led to the identification of TK1686. The protein product catalyzed the ATP-dependent conversion of phosphopantoate and β-alanine to produce 4′-phosphopantothenate and did not exhibit PS activity, indicating that TK1686 also encoded a novel enzyme, phosphopantothenate synthetase (PPS). Although the classic PS/PanK system performs condensation with β-alanine prior to phosphorylation, the PoK/PPS system performs condensation after phosphorylation of pantoate. Gene disruption of TK2141 and TK1686 led to CoA auxotrophy, indicating that both genes are necessary for CoA biosynthesis in T. kodakaraensis. Homologs of both genes are widely distributed among the Archaea, suggesting that the PoK/PPS system represents the pathway for 4′-phosphopantothenate biosynthesis in the Archaea.
Coenzyme A (CoA)2 and its derivative 4′-phosphopantetheine are essential cofactors in numerous metabolic pathways, including the tricarboxylic acid cycle, the β-oxidation pathway, and fatty acid and polyketide biosynthesis pathways. Acyl-CoA derivatives are key intermediates in energy metabolism due to their high energy thioester bonds and have been identified in all three domains of life.
The mechanism of CoA biosynthesis in bacteria and eukaryotes has been well examined and involves common enzymatic conversions (1–3). CoA is synthesized from pantothenate via five enzymatic reactions; pantothenate kinase (PanK), 4′-phosphopantothenoylcysteine synthetase (PPCS), 4′-phosphopantothenoylcysteine decarboxylase (PPCDC), 4′- phosphopantetheine adenylyltransferase (PPAT), and dephospho-CoA kinase (DPCK). Although many animals rely on exogenous pantothenate to initiate CoA biosynthesis, microorganisms and plants can synthesize pantothenate from 2-oxoisovalerate and β-alanine. This is a three-step pathway catalyzed by ketopantoate hydroxymethyltransferase (KPHMT), ketopantoate reductase, and pantothenate synthetase (PS).
In contrast to the wealth of knowledge on CoA biosynthesis in bacteria and eukaryotes, the corresponding pathway in the Archaea remains unclear (4). Sequence data indicate that the bacterial PPCS and PPCDC homologs and eukaryotic PPAT homologs are found on almost all of the archaeal genomes. The archaeal PPCS and PPCDC genes are fused in many cases, and the bifunctional protein from Methanocaldococcus jannaschii has been shown to exhibit both activities (5). The PPAT homolog from Pyrococcus abyssi has also been studied and confirmed to exhibit the expected PPAT activity (6). Bacterial KPHMT and ketopantoate reductase homologs can also be found, to a lesser extent, on the archaeal genomes. They are not found in the methanogens and Thermoplasmatales, and the fact that the structural similarity among archaeal enzymes is not higher than that toward enzymes from hyperthermophilic bacteria suggests that the archaeal KPHMT and ketopantoate reductase are a result of horizontal gene transfer from bacteria (4). In addition, there are candidate genes distantly related to bacterial/eukaryotic DPCK. However, PS homologs are not found in any of the archaeal genomes, and PanK homologs are found only in a few exceptional cases. Recently, Genschel and co-workers have taken a comparative genomics approach to predict the genes corresponding to the archaeal PS and PanK genes, and have also described the identification of a structurally novel PS from Methanosarcina mazei (4, 7).
In this study, we describe the identification of the enzymes responsible for the conversion of pantoate to 4′-phosphopantothenate in Thermococcus kodakaraensis. The organism is a hyperthermophilic archaeon isolated from Kodakara Island, Japan (8, 9). The complete genome sequence is available (10), and gene disruption systems have been developed (11–13). To our surprise, the conversion of pantoate to 4′-phosphopantothenate in T. kodakaraensis is not brought about by the two classic enzyme reactions catalyzed by PS and PanK, but by two novel enzyme reactions; phosphorylation of pantoate (pantoate kinase) followed by the condensation of 4-phosphopantoate and β-alanine (4′-phosphopantothenate synthetase or 4-phosphopantoate:β-alanine ligase). Homologs of these two genes are distributed on almost all of the archaeal genomes, suggesting that the Archaea utilize different chemistry in the conversion from pantoate to 4′-phosphopantothenate.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
The strains and plasmids used in this study are listed in supplemental Table S1. T. kodakaraensis KOD1 and its derivatives were cultivated under strictly anaerobic conditions at 85 °C in a nutrient-rich medium (ASW-YT) or a synthetic medium (ASW-AA). ASW-YT medium (pH 6.6) was composed of 0.8× artificial seawater (0.8× ASW), 5.0 g liter−1 of yeast extract, and 5.0 g liter−1 of Tryptone. Sodium pyruvate (5 g liter−1) or elemental sulfur (2 g liter−1) was supplemented prior to inoculation. ASW-AA medium consisted of 0.8× ASW, a mixture of 20 amino acids, modified Wolfe's trace minerals, a vitamin mixture, and 2.0 g liter−1 of elemental sulfur (11, 14). Solid media used to isolate transformants were based on ASW-YT medium containing 1.0 g liter−1 of gelrite and 0.4 g liter−1 of polysulfide. Specific modifications of the medium to select and investigate the auxotrophy of individual mutant strains are described in the respective sections. When examining the effects of adding 4′-phosphopantothenate to the medium, 4′-phosphopantothenate was enzymatically synthesized (37 °C, 90 min) in a reaction mixture (30 ml) containing 12 mm pantothenate, 13 mm ATP, 10 mm MgCl2, and 3.6 μm Ec-PanK in 50 mm Tris-HCl (pH 8.0). The volume of the reaction mixture added was 10% (v/v) of that of the medium. Escherichia coli strains DH5α and BL21-CodonPlus(DE3)-RIL, used for plasmid construction and heterologous gene expression, respectively, were cultivated at 37 °C in Luria-Bertani (LB) medium containing ampicillin (100 mg liter−1). Unless mentioned otherwise, all chemicals were purchased from Wako Pure Chemicals (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan).
Overexpression of Genes in E. coli and Partial Purification of the Recombinant Proteins
Plasmids for gene expression of TK0939, TK1473, TK1686, TK2141, and TK2242 in E. coli were constructed as follows. The coding region of each gene was amplified from the genomic DNA of T. kodakaraensis KOD1 by PCR using the primer sets TK0939EF/ER, TK1473EF/ER, TK1686EF/ER, TK2141EF/ER, and TK2242EF/ER for TK0939, TK1473, TK1686, TK2141, and TK2242, respectively (supplemental Table S2). Using the NdeI-EcoRI or NdeI-BamHI restriction enzyme sites incorporated during the PCR, the fragments were inserted into the pET21a(+) expression vector (Novagen). After confirming the absence of unintended mutations, the plasmids were introduced into E. coli strain BL21-CodonPlus(DE3)-RIL. The transformants were cultivated at 37 °C in LB medium with ampicillin until the optical density at 660 nm reached ∼0.5. Isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 0.1 mm to induce overexpression, and cells were incubated for a further 4 h at 37 °C. Cells were harvested by centrifugation, resuspended in 50 mm Tris-HCl buffer (pH 7.5), and disrupted by sonication. After centrifugation (20,000 × g, 15 min), the soluble cell extract was heat treated for 10 or 15 min at 70 or 80 °C, and thermolabile proteins derived from the host were removed by centrifugation (20,000 × g, 15 min). The supernatants were used for initial examination of enzyme activity.
Purification of the Recombinant TK1686 Protein and TK2141 Protein
The recombinant TK1686 protein produced in E. coli and the TK2141 protein produced in T. kodakaraensis (see below) were further purified by anion-exchange chromatography with HiTrap Q HP 5 ml (Amersham Biosciences), and the recombinant proteins were eluted with a linear gradient of KCl (0 to 1.0 m) in 50 mm Tris-HCl (pH 7.5) at a flow rate of 0.4 ml min−1. For TK1686, protein fractions were combined and concentrated using Amicon Ultra-4 10K (Millipore) and were further applied to a Superdex 200 HR 10/30 gel filtration column (Amersham Biosciences) with a mobile phase of 50 mm Tris-HCl (pH 7.5) containing 100 mm KCl at a flow rate of 0.3 ml min−1. The molecular mass of native TK1686 protein and TK2141 protein were determined from a calibration curve constructed with the standard proteins thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67.0 kDa), ovalbumin (43.0 kDa), and chymotrypsinogen A (25.0 kDa). Protein concentration was determined with the Protein Assay System (Bio-Rad), using bovine serum albumin as a standard.
Plasmids for Overexpression of Genes in T. kodakaraensis
Genes whose expression did not lead to soluble proteins in E. coli (TK0939 and TK2141) were expressed in T. kodakaraensis (supplemental Fig. S1). Recombination in T. kodakaraensis was designed so that the individual genes would be fused downstream of the cell surface glycoprotein gene (csg, TK0895) promoter. The csg promoter corresponds to a 200-bp region of the 5′-flanking region of the TK0895 gene. Because TK0939 is the first gene of a putative TK0939-TK0940 operon, we inserted the csg promoter upstream of the original TK0939 gene locus. A His tag sequence was incorporated in the N terminus of TK0939 for use in purification. On the other hand, the TK2141 gene is located in the middle of a putative TK2140-TK2142 operon. We thus constructed a csg promoter/TK2141 gene cassette and inserted the cassette into another region of the genome, which originally harbors the chitinase-encoding chiA gene (TK1765). A His tag was also added at the N terminus of TK2141. pUD3, which contains the pyrF marker gene cassette inserted in the ApaI site of pUC118, was used to construct individual plasmids. For the TK0939 gene, the coding region along with a 1000-bp region of the TK0939 5′-flanking region was amplified from genomic DNA with the primer set TK0939TF1/TR1, digested with BamHI, and inserted into the BamHI site of pUD3. Inverse PCR of this plasmid with the primer set TK0939TF2/TR2 was performed so that a His tag sequence was incorporated after the initiation codon. The primer set PcsgF1/R was used to amplify the csg promoter region from genomic DNA, with the initiation codon of TK0939 and the His tag sequence present at the 3′-terminal region. The fragments amplified by TK0939TF2/TR2 and PcsgF1/R were mixed for fusion PCR. The obtained fragment was self-ligated to complete the promoter exchange plasmid. For TK2141, the coding region was amplified from genomic DNA with the primer set TK2141TF/TR, incorporating a His tag sequence after the initiation codon. chiATF1/TR1 was used to amplify a 1000-bp region of the 3′-flanking region of the chitinase-encoding chiA gene (TK1765). Fusion PCR was performed so that the His tag TK2141 gene was placed upstream of the TK1765 3′-flanking region (fragment 1). Fragments amplified with chiATF2/TR2 (TK1765 5′-flanking region) and PcsgF2/R (csg promoter) were fused (fragment 2). The NdeI-BamHI-digested fragment 1, the EcoRI-NdeI-digested fragment 2, and the EcoRI-BamHI-digested pUD3 were ligated to complete the plasmid.
Transformation of T. kodakaraensis
T. kodakaraensis strain KUW1 (ΔpyrFΔtrpE) (12), was used as the host strain for genetic manipulation. Transformation of KUW1 was performed as follows. Cells at the late log phase were harvested, resuspended in 200 μl of 0.8× ASW, and kept on ice for 30 min. After treatment with 3.0 μg of the plasmid and further incubation on ice for 1 h, the cells were cultivated in ASW-AA medium for 24 h at 85 °C. Cells were consecutively cultivated under the same conditions to enrich the desired transformants that display uracil prototrophy, followed by cultivation in ASW-YT medium for 12 h at 85 °C. The cells grown in ASW-YT medium were harvested, diluted with 0.8× ASW, and spread onto solid medium supplemented with 10 g liter−1 of 5-fluoroorotic acid and 60 mm NaOH. Only cells that have undergone a pop-out recombination that removes the pyrF gene can grow in the presence of 5-fluoroorotic acid. Cells were grown for 2 days at 85 °C for colony formation.
After incubation, genotypes of the isolated transformants were analyzed by PCR with the primer set PcsgF1/TK0939TR1 for the TK0939 overexpression strain (ETK0939) and PcsgF1/TK2141TR for the TK2141 overexpression strain (ETK2141) (supplemental Table S2). Transformants whose amplified DNA products displayed the expected size were chosen, and relevant sequences were confirmed to have no unintended mutations. The TK0939 and TK2141 overexpression strains were cultivated in ASW-YT medium with 5 g liter−1 sodium pyruvate for 12 h at 85 °C. Cells were harvested by centrifugation, resuspended in 20 mm potassium phosphate buffer (pH 7.4) containing 0.5 m KCl and 40 mm imidazole, and sonicated. After centrifugation (20,000 × g, 15 min), the supernatant was applied onto His GraviTrap (Amersham Biosciences), and the His tag proteins were eluted with elution buffer (20 mm potassium phosphate buffer (pH 7.4), 0.5 m KCl, and 0.5 m imidazole). After exchanging the elution buffer to Tris-HCl (pH 8.0) with Amicon Ultra-4 10K (Millipore), the collected fractions were used for initial examination of activity.
Examination of Pantothenate Kinase and Pantoate Kinase Activity
PanK activity was measured with HPLC or by coupling the PanK reaction with pyruvate kinase/lactate dehydrogenase (PK/LDH). In the HPLC method, reactions were performed at 75 °C. The reaction mixture contained 4 mm d-pantothenic acid (Sigma), 4 mm ATP (Oriental Yeast, Tokyo, Japan), 10 mm MgCl2, in 50 mm Tris-HCl (pH 7.5), with cell-free extract (∼100 μg ml−1) or various amounts of recombinant protein. MgCl2 and Tris-HCl were first incubated at 75 °C for 2 min, and, after adding and mixing the other reagents, the reaction mixture was further incubated at 75 °C. The reaction was started by adding the protein sample and stopped by adding 20 μl of 1 m HCl and cooling on ice. After incubation on ice for 30 min, proteins in the reaction mixture were removed by ultrafiltration using Microcon Ultracel YM-10 (Millipore). An aliquot (10 μl) of this solution was applied to a Synergi 4u Hydro-RP 80A 150 × 4.60 mm (Phenomenex, Torrance, CA) column using an LC-VP system (Shimadzu, Kyoto, Japan). Compounds were eluted with a solution of 20 mm KH2PO4 plus 0.1% hexane sulfonate adjusted to pH 3.0 by phosphoric acid and 1.5% (v/v) acetonitrile at a flow rate of 1.0 ml min−1. Absorbance at 210 nm was measured.
For measurements using PK/LDH, the PanK reaction was either carried out prior to the PK/LDH reaction (80 °C) or simultaneously (42 °C). When the PanK and PK/LDH reactions were carried out separately, the initial reaction mixture was identical to that described for HPLC analysis. After the reaction, samples were put on ice for 30 min, and proteins were removed as described above. An aliquot (100 μl) was added to the second reaction mixture (1 ml) containing 0.2 mm NADH (Oriental Yeast), 5 mm potassium phosphoenolpyruvate, 14.8 units ml−1/18.6 units ml−1 of PK/LDH from rabbit muscle (Sigma), 10 mm MgCl2, and 50 mm Tris-HCl (pH 7.5). MgCl2 and Tris-HCl were first incubated at 37 °C for 2 min, and, after adding the other components, the decrease in absorption at 340 nm was measured by using a spectrophotometer at 37 °C. The millimolar absorbance coefficient (ϵ340 nm = 6.22) of NADH was used to quantify the ADP formed in the first reaction. The amount of NADH that decreased after adding an aliquot of the first reaction without pantothenate was subtracted from each assay result. When the PanK and PK/LDH reactions were carried out simultaneously, the reaction mixture contained 5 mm ATP, 5 mm of pantothenate, 0.2 mm NADH, 5 mm phosphoenolpyruvate, 14.8 units ml−1/18.6 units ml−1 of PK/LDH, 10 mm MgCl2, 50 mm Tris-HCl (pH 8.0), and 6.8 μg ml−1 of recombinant TK2141 protein. After preincubating MgCl2 and Tris-HCl at 42 °C for 2 min, other reagents were added, and the rate of decrease in absorption at 340 nm was measured. The reaction was initiated with the addition of ATP. Pantoate kinase activity was measured with the same methods, except that d-pantothenic acid was replaced with d-pantoic acid. d-Pantoic acid was obtained by hydrolyzing d-pantolactone in 400 mm KOH at 90 °C for 1 h. For kinetic examination of TK2141, kinetic parameters for pantothenate and pantoate were examined in the presence of 5 mm ATP, whereas those for ATP were examined in the presence of 5 mm pantoate.
Examination of Pantothenate Synthetase and Phosphopantothenate Synthetase Activity
The reaction mixture for examining PS activity contained 4 mm d-pantoate, 4 mm β-alanine (Sigma), 4 mm ATP, 10 mm MgCl2 in 50 mm Tris-HCl (pH 8.0), and recombinant TK1686 protein (17 μg ml−1). MgCl2 and Tris-HCl buffer were incubated at 75 °C for 2 min, and, after adding the other reagents, the reaction mixture was incubated at 75 °C. The reaction was started by adding ATP and stopped by adding 20 μl of 1 m HCl and cooling on ice for 30 min. Proteins were removed by ultrafiltration using Microcon Ultracel YM-10, and 10-μl aliquots were applied to a COSMOSIL 5C18-PAQ 250 × 4.60 mm column (Nacalai Tesque). Compounds were separated with 50 mm NaH2PO4 at a flow rate 1.0 ml min−1. Absorbance at 210 nm was measured.
Phosphopantothenate synthetase activity was measured with the same methods, except that the substrate 4-phosphopantoate was supplied by the pantoate kinase activity of the recombinant TK2141 protein. The pantoate kinase reaction was performed either prior to or together with the phosphopantothenate synthetase reaction. In the latter case, recombinant TK2141 (11 μg) was added to the reaction mixture described for the examination of PS activity. When the pantoate kinase reaction was carried out separately, the first reaction mixture was identical to that used for examining pantothenate kinase activity by HPLC, except that pantothenate was replaced by pantoate. After removing proteins, an aliquot (100 μl) was added to the second reaction mixture identical to that described for the examination of PS activity, but without pantoate.
Plasmids for Gene Disruption of TK1686 and TK2141 in T. kodakaraensis
Gene disruption plasmids for TK1686 and TK2141 were constructed by first amplifying the genes along with 1000 bp of their 5′- and 3′-flanking regions using the primer sets TK1686DF1/DR1 and TK2141DF1/DR1. After digesting with BamHI, the fragments were inserted into the BamHI site of pUD3. Inverse PCR was performed with TK1686DF2/DR2 and TK2141DF2/DR2 to remove the respective coding regions, and the amplified fragment was self-ligated. The proper sequences of the 5′- and 3′-flanking regions of each gene were confirmed. The plasmids were introduced into T. kodakaraensis KUW1 as described above. PCR analyses of transformants were performed with the primer sets TK1686CF/CR (TK1686 gene disruptant) and TK2141CF/CR (TK2141 gene disruptant). The loci of candidate strains were sequenced to confirm gene disruption.
RESULTS
Growth of T. kodakaraensis in the Absence of CoA and Its Immediate Precursors
We first examined whether T. kodakaraensis was able to synthesize CoA de novo. Cells were grown in a synthetic ASW-AA medium, including salts, trace minerals, amino acids, elemental sulfur, and trace vitamins but excluding pantothenate. Cell growth was consistently observed in the synthetic medium during three rounds of cultivation, indicating that T. kodakaraensis cells harbor the ability to synthesize CoA.
Examination of Pantothenate Kinase Activity in the Cell-free Extracts of T. kodakaraensis
In terms of T. kodakaraensis, homologs with high similarity to the bacterial KPHMT, ketopantoate reductase, PPCS, and PPCDC and the eukaryotic PPAT are present on the genome. In addition, two candidate genes that are distantly related to the bacterial/eukaryotic DPCK can also be identified. As in the case of the majority of the archaea, genes structurally related to PS and PanK genes are not present on the T. kodakaraensis genome.
We have previously encountered numerous cases in which structurally novel, archaea-specific proteins replace the functions of proteins homologous to previously characterized enzymes from bacteria and eukaryotes (15–18). We thus examined the presence of ATP-dependent PanK activity in the cell-free extracts of T. kodakaraensis cells. Activity was first measured by coupling the generation of ADP with the PK/LDH enzyme system. No notable levels of activity could be detected. We next examined activity by monitoring the generation of 4′-phosphopantothenate with HPLC. For use as a standard compound, 4′-phosphopantothenate was synthesized enzymatically with PanK from E. coli (Ec-PanK). The Ec-PanK gene was amplified from E. coli genomic DNA and expressed with the pET21a(+)/E. coli BL21(DE3)CodonPlus RIL system. A His tag sequence was introduced in the N terminus region, and the enzyme was purified with a nickel affinity column. Incubation of Ec-PanK with ATP and pantothenate led to increases in a peak corresponding to ADP (retention time: 1.4–1.5 min) and another peak corresponding to 4′-phosphopantothenate with a retention time of 1.9–2.0 min using a Synergi 4u Hydro-RP 80A column. When we examined the cell-free extract of T. kodakaraensis for PanK activity, a peak corresponding to 4′-phosphopantothenate could not be detected. In addition, pantothenate consumption was not observed, suggesting that activity levels, if any, were trace.
Selection of Candidate Genes Encoding a Structurally Novel PanK
As we could not detect sufficient levels of ATP-dependent pantothenate kinase (PanK) activity in the cell-free extracts of T. kodakaraensis, we took a comparative genomics approach to identify candidate genes that might encode a structurally novel PanK. Among the 51 genes annotated as putative kinases on the T. kodakaraensis genome (10), we were able to identify ten kinase genes that were widely distributed in the archaeal genomes and whose specific functions could not be predicted. Among these, only four were specific to the Archaea, and thus selected for further examination; TK0939 (predicted archaeal sugar kinase), TK1473 (predicted archaeal kinase), TK2141 (predicted archaeal sugar kinase), and TK2242 (predicted archaeal sugar kinase). TK0939, TK2141, and TK2242 are members of the galacto-/homoserine/mevalonate/phosphomevalonate kinase family (19).
Expression of the Four Candidate Genes and Activity Measurements of the Recombinant Proteins
The TK0939, TK1473, TK2141, and TK2242 genes were individually inserted into pET21a(+) and expressed in E. coli BL21(DE3)CodonPlus RIL. Soluble proteins were obtained for TK1473 and TK2242. The two proteins were partially purified by heat treatment and examined for PanK activity by HPLC. When recombinant TK1473 and TK2242 proteins were incubated in the presence of ATP and pantothenate at 75 °C, we observed neither the formation of 4′-phosphopantothenate nor a decrease in pantothenate, indicating that the two proteins did not harbor PanK activity.
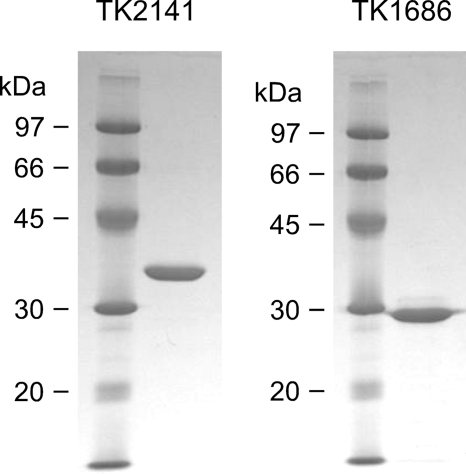
We then examined the two remaining gene products. Because a gene manipulation system has been developed for T. kodakaraensis (11, 12), the genes were individually introduced into the T. kodakaraensis genome under the control of a strong constitutive promoter from the cell surface glycoprotein (csg) gene. A His tag sequence was introduced directly downstream of the initiation codon. Plasmids for gene exchange or gene insertion were designed so that the plasmids were able to undergo an initial single crossover insertion, followed by a pop-out recombination event (see “Experimental Procedures”). The recombinant strains were isolated and grown in ASW-YT medium supplemented with sodium pyruvate. Cell-free extracts were subjected to nickel affinity columns, and partially purified proteins with the expected molecular mass could be observed with SDS-PAGE (TK0939, 36 kDa; TK2141, 34 kDa). The two proteins were thus examined for PanK activity at 75 °C. No activity could be detected with the TK0939 protein. In the case of the TK2141 protein, we observed a notable decrease in the substrate pantothenate as well as the formation of the reaction product 4′-phosphopantothenate. We further purified the TK2141 protein to apparent homogeneity by anion-exchange chromatography (Fig. 1). We examined the TK2141 activity by monitoring the formation of ADP using the PK/LDH system at 42 °C. We found that the protein harbored an ATP-dependent PanK activity of 157 nmol min−1 mg protein−1. Activity was not observed in the absence of pantothenate.
FIGURE 1.
SDS-PAGE analysis of the recombinant TK2141 (3 μg, left panel) and TK1686 (4 μg, right panel) proteins after purification. After electrophoresis, the gel was stained with Coomassie Brilliant Blue.
TK2141 Displays a Novel Pantoate Kinase Activity
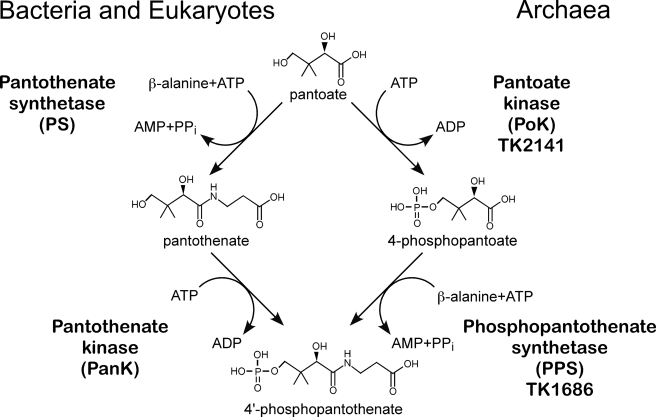
Although the TK2141 protein exhibited PanK activity, its relatively low specific activity led us to examine the possibilities of the protein to be a pantoate kinase. The conversion of pantoate to 4′-phosphopantothenate in the classic CoA biosynthesis pathway is brought about by an initial condensation reaction with β-alanine (pantothenate synthetase) followed by phosphorylation (PanK). Because the two reactions involve independent functional groups of pantoate, we realized that the formation of 4′-phosphopantothenate from pantoate is also possible by a phosphorylation step occurring before the condensation with β-alanine (Fig. 2). In this case, the phosphorylation reaction would be a pantoate kinase reaction. When we replaced the pantothenate in our reaction mixture with pantoate, we observed significantly higher kinase activity. The specific activity of TK2141 for the pantoate kinase reaction was 1650 nmol min−1 mg protein−1 at 42 °C, over 10-fold higher than the PanK activity of the protein.
FIGURE 2.
The classic bacterial/eukaryotic pathway and the novel archaeal pathway for the formation of 4′-phosphopantothenate from pantoate.
We examined whether the pantoate kinase activity of TK2141 was inhibited in the presence of CoA. The PanK of E. coli is significantly inhibited in the presence of CoA (20). We measured pantoate kinase activity in the presence of 0.1 mm CoA, acetyl-CoA, dephospho-CoA, or 4′-phosphopantothenate. In each case, no notable decreases in activity could be detected, suggesting that the enzyme is not inhibited by these compounds. When CoA was present at higher concentrations of 1 or 2 mm, enzyme activity decreased by 20 and 25%, respectively. The effects, however, were much lower than those observed for Ec-PanK, where a 50% decrease in activity is observed at 165 μm (20).
Kinetic Examinations of the Pantoate- and Pantothenate Kinase Activities of TK2141
We performed kinetic analyses on the pantoate- and pantothenate kinase activities of TK2141. Activity measurements were performed at 42 °C with the PK/LDH system. The kinetic parameters for pantoate and pantothenate were determined in reactions mixtures containing 5 mm ATP, whereas those for ATP were determined in the presence of 5 mm pantoate. All reactions followed Michaelis-Menten kinetics, and the kinetic parameters are shown in Table 1. The kcat/Km value with pantoate as a substrate was over 7-fold higher than that observed with pantothenate. The results indicate that the TK2141 protein catalyzes a novel enzyme reaction, the ATP-dependent phosphorylation of pantoate, and was thus designated as pantoate kinase (PoK).
TABLE 1.
Kinetic parameters of the pantoate and pantothenate kinase activities of TK2141 protein
| Substrate | Km | Vmax | Kcat | Kcat/km |
|---|---|---|---|---|
| mm | nmol min−1mg−1 | s−1 | s−1mm−1 | |
| Pantothenate | 1.3 ± 0.1 | 390 ± 10 | 0.21 ± 0.01 | 0.17 |
| Pantoate | 1.2 ± 0.1 | 2,870 ± 60 | 1.56 ± 0.03 | 1.3 |
| ATP | 0.47 ± 0.01 | 2,710 ± 20 | 1.48 ± 0.01 | 3.1 |
The PoK from T. kodakaraensis, the product of TK2141 (accession number BAD86330), consists of 300 amino acid residues with a deduced molecular mass of 32,748 Da. It is classified in arCOG04263 and COG01829 and is annotated as a member of the galacto-/homoserine/mevalonate/phosphomevalonate kinase family (19). PoK harbors the well conserved PX3GL(G/S)SSA motif (96PNGYGFGNSA105), whose residues are involved in phosphate binding in galacto-/homoserine/mevalonate/phosphomevalonate kinases (21, 22). The purified recombinant protein, judged by gel-filtration chromatography, was a dimer.
Search for a Gene Encoding a Phosphopantothenate Synthetase (4-Phosphopantoate:β-alanine Ligase)
The high level of pantoate kinase activity in TK2141 implies the presence of a novel enzyme that can convert 4-phosphopantoate to 4′-phosphopantothenate. This activity corresponds to a 4-phosphopantoate:β-alanine ligase activity or phosphopantothenate synthetase. We again performed a search in the archaeal genomes for genes that displayed a distribution similar to that of TK2141. We utilized the arCOG database (New Archaeal Clusters of Orthologous Genes, ftp.ncbi.nih.gov/pub/wolf/COGs/arCOG), which includes sequence data of 41 archaeal genomes (23). TK2141 homologs are present on all of the sequenced archaeal genomes with the exception of Nanoarchaeum equitans and the three organisms of the Thermoplasmatales, Thermoplasma volcanium, Thermoplasma acidophilum, and Picrophilus torridus. Among the 5961 arCOGS that do not have members in any of these four organisms, only two are present in all the 37 remaining genomes; arCOG01028, which corresponds to mevalonate kinase, and arCOG04263, which corresponds to PoK. Only three arCOGs are present in 36 genomes (arCOG01008, arCOG01723, and arCOG04262). Because we have previously determined the functions of the members of arCOG01008 (TK1439, transcriptional regulator)3 and arCOG01723 (TK0507 and TK1179, phosphatases),4 we thus expressed and examined TK1686, the arCOG04262 member in T. kodakaraensis. It should be noted that TK1686 corresponds to the novel archaeal PS proposed by the group of Genschel (7).
Expression of TK1686 in E. coli and Characterization of the Recombinant Protein
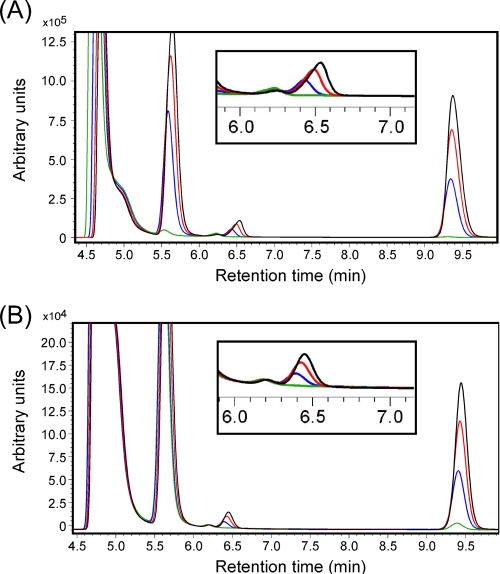
The TK1686 gene was inserted into pET21a(+) and expressed in E. coli BL21(DE3)CodonPlus RIL. The protein was purified by heat treatment, followed by anion-exchange chromatography and gel-filtration chromatography (Fig. 1). Because phosphopantoate is not commercially available, we first examined whether the TK2141 and TK1686 proteins together could convert pantoate and β-alanine to 4′-phosphopantothenate in the presence of ATP. Here we used a COSMOSIL 5C18-PAQ column, which displayed improved separation of the substrates and products. The retention time of 4′-phosphopantothenate produced with Ec-PanK was 6.4–6.6 min. After the TK2141 and TK1686 reactions, we clearly observed the formation of a peak with a retention time corresponding to 4′-phosphopantothenate (Fig. 3). The generation of ADP and AMP was also observed. The fractions presumed to include 4′-phosphopantothenate were collected and analyzed by time-of-flight secondary ion mass spectrometry using Ga+ as the primary ion. Among the resulting secondary anions, we detected clear m/z signals of 297.12, 298.15, and 299.10 (data not shown), corresponding to the molecular masses of 4′-phosphopantothenate with various degrees of protonation. Because we could not provide saturating concentrations of phosphopantoate, we could not determine the specific activity for the phosphopantothenate synthetase activity of TK1686. However, the generation rate of AMP in a coupled reaction with TK2141 indicates that the specific activity is at least 2150 nmol min−1 mg protein−1 at 75 °C.
FIGURE 3.
Chromatograms of the reaction products after the PoK and PPS reactions. A COSMOSIL 5C18-PAQ column was used for separation. The peaks with approximate retention times of 4.7, 5.6, 6.5, and 9.4 min correspond to ATP, ADP, 4′-phosphopantothenate, and AMP, respectively. The peak corresponding to 4′-phosphopantothenate is enlarged in the insets. Line colors indicate reaction times of 0 (green), 5 (blue), 10 (red), and 15 (black) min. All reactions were performed at 75 °C. A, the reaction mixture included both PoK and PPS along with ATP, pantoate, and β-alanine (see “Experimental Procedures”) and was applied to HPLC after various periods of time. B, an initial reaction was performed with PoK, ATP, and pantoate (1 h). After stopping the first reaction and removing PoK, an aliquot was added to a second reaction mixture, including PPS, ATP, and β-alanine. The second reaction mixture was applied to HPLC after various periods of time.
Because TK2141 also exhibits low levels of PanK activity in addition to its PoK activity, there was a remote possibility that the formation of 4′-phosphopantothenate described above was the sum of a PS activity of TK1686 and the PanK activity of TK2141. To rule out this possibility, we first incubated the TK2141 protein in the presence of ATP and pantoate to produce 4-phosphopantoate, and then removed the protein from the reaction mixture. We then mixed an aliquot of this solution with TK1686, β-alanine, and ATP. This also resulted in the formation of 4′-phosphopantothenate, indicating that the conversion of pantoate to 4′-phosphopantothenate was due to PoK and phosphopantothenate synthetase activities, and not PS and PanK activities. Furthermore, we examined whether TK1686 harbored PS activity. No formation of pantothenate was observed when the TK1686 protein was incubated with pantoate, β-alanine, and ATP. The results indicate that TK1686 is not a PS and should be designated as a phosphopantothenate synthetase (PPS) or 4-phosphopantoate:β-alanine ligase.
The PPS from T. kodakaraensis, the product of TK1686 (accession number BAD85875), consists of 261 amino acid residues, corresponding to a molecular mass of 29,843 Da. It is a member of arCOG04262 and COG01701 and is annotated as a hypothetical protein. The protein harbors a putative Walker A-like motif (227GEYDNGKT234) (24, 25) in its C-terminal domain. The purified recombinant protein displayed a molecular mass corresponding to a dimer.
Our biochemical analyses indicate that the TK2141 and TK1686 proteins exhibit two novel enzyme activities, PoK and PPS, respectively, and together are able to convert pantoate, β-alanine, and two molecules of ATP to 4′-phosphopantothenate, ADP, AMP, and pyrophosphate.
Gene Disruption of TK2141 and TK1686
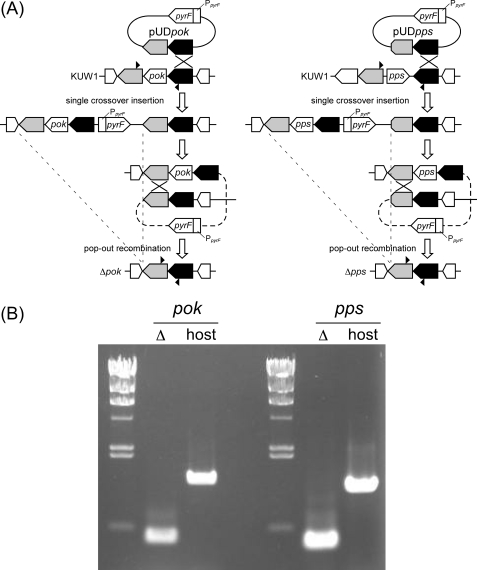
To confirm that TK2141 and TK1686 were involved in CoA biosynthesis in T. kodakaraensis, we set out to individually disrupt the genes and compare their phenotypes with the host strain T. kodakaraensis KUW1. Although there was the possibility that the gene disruptions would lead to lethal phenotypes, we had observed that the cell yield of T. kodakaraensis increased when 1 mm CoA was supplemented to growth medium, suggesting the uptake of CoA by T. kodakaraensis cells. Plasmids were designed so that gene disruption would occur via single crossover insertion of the plasmid, followed by pop-out recombination (Fig. 4A). Cells that had undergone single crossover insertion were enriched by growing the transformants in a uracil-free medium, and cells that had further undergone pop-out recombination were selected on solid media that included 5-fluoroorotic acid. In principle, if the gene disruption does not affect cell growth, half of the transformants isolated after the second recombination event should correspond to gene disruption strains. In this case, CoA (1 mm) was supplemented to the solid medium used for the isolation of gene disruption strains. By examining 16 colonies for each gene disruption, we were able to isolate gene disruption strains for both TK1686 (13/16 colonies) and TK2141 (6/16 colonies). PCR analysis (Fig. 4B) and DNA sequencing confirmed the complete deletion of each gene from the genome of T. kodakaraensis KUW1.
FIGURE 4.
A, the plasmid and recombination strategies to construct the gene disruption strains of TK2141 (left) and TK1686 (right). T. kodakaraensis KUW1 (ΔpyrF and ΔtrpE) was used as the host strain. Cells that had undergone the initial single crossover insertion were enriched by growing cells in medium without uracil. Cells that had undergone the second, pop-out recombination were selected with resistance toward 5-fluoroorotic acid. B, PCR analyses of the TK2141 (left) and TK1686 (right) loci. The positions of the primers used are indicated with arrowheads in A.
Growth Characterization of the TK2141 and TK1686 Gene Disruption Strains
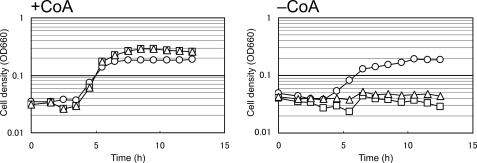
The TK2141 and TK1686 gene disruption strains (ΔTK2141 and ΔTK1686, respectively) were grown in ASW-YT medium with elemental sulfur in the presence or absence of 1 mm CoA (Fig. 5). We found that both ΔTK2141 and ΔTK1686 grew well in media supplemented with 1 mm CoA. However, we found that both strains could not grow at all in the absence of CoA, clearly indicating that the TK2141 and TK1686 genes are involved in CoA biosynthesis in T. kodakaraensis, and that no other pathway can complement the function of these two enzymes in this organism. We also observed that the addition of a crude preparation of 4′-phosphopantothenate, synthesized enzymatically from pantothenate and ATP with Ec-PanK, resulted in growth of ΔTK2141 and ΔTK1686, whereas the addition of 1 mm pantothenate did not (data not shown). These observations further support that the PoK/PPS reactions, which do not proceed via pantothenate, are responsible for the conversion from pantoate to 4′-phosphopantothenate in T. kodakaraensis.
FIGURE 5.
Growth characteristics of T. kodakaraensis KUW1, ΔTK2141 and ΔTK1686 in the presence (left) or absence (right) of 1 ml CoA. The cells were cultured in ASW-YT medium with 0.2% elemental sulfur at 85 °C. Symbols: circles, KUW1; squares, ΔTK2141; and triangles, ΔTK1686.
DISCUSSION
In this study, we have identified the proteins responsible for the conversion of pantoate to 4′-phosphopantothenate in the CoA biosynthesis pathway of T. kodakaraensis. The two proteins, encoded by TK2141 and TK1686, both catalyze novel reactions, the ATP-dependent phosphorylation of pantoate (PoK) to produce 4-phosphopantoate and the ATP-dependent condensation of 4-phosphopantoate and β-alanine to form 4′-phosphopantothenate (PPS), respectively. Gene disruption experiments have clarified that these two enzymes are necessary for CoA biosynthesis in this organism.
PoK (TK2141) and PPS (TK1686) homologs are present on almost all archaeal genomes with the exception of three organisms of the Thermoplasmatales and the parasitic N. equitans. 4′-Phosphopantothenate may be required as a vitamin in the Thermoplasmatales, whereas in the case of N. equitans, because its genome lacks all CoA biosynthesis genes, CoA may be supplied from its host, Ignicoccus. The extensive distribution of PoK and PPS homologs among the archaeal genomes suggests that the two enzymes may represent the main route from pantoate to 4′-phosphopantothenate in the Archaea. Because homologs of these genes cannot be found in bacterial and eukaryotic genomes, the PoK/PPS pathway for CoA biosynthesis can be considered to be specific to the Archaea.
The homologs of TK2141 and TK1686 in Methanosarcina mazei are MM2282 and MM2281, respectively. The two genes precisely correspond to those predicted by Genschel and co-workers to be a novel PS (MM2281) and PanK (MM2282) (4, 7). The MM2281 protein was shown to complement an E. coli strain deficient in PS activity, and also, in cooperation with Ec-PanK, synthesize 4′-phosphopantothenate from pantoate, β-alanine, and ATP (7). It may well be that the reaction mechanisms in M. mazei differ to those reported in this study on T. kodakaraensis. However, there is a possibility that MM2281 also exhibits PPS activity. If Ec-PanK were to have a low level of kinase activity toward pantoate, this activity together with a PPS activity would be able to convert pantoate to 4′-phosphopantothenate. Further studies will be necessary to clarify the differences between the T. kodakaraensis and M. mazei enzymes. In any case, their prediction, based on genomic context and phylogenetic pattern analysis, that MM2281/MM2282 were involved in CoA biosynthesis was correct (4, 7).
In general, CoA biosynthesis in bacteria and eukaryotes is regulated by a feedback-inhibition mechanism that targets PanK (2, 20, 26–28). Ec-PanK is inhibited most by CoA itself (20, 26, 27), whereas other enzymes have been found to be inhibited by CoA and/or acyl-CoA compounds such as acetyl-CoA and malonyl-CoA (28–32). Inhibition was not observed in the PoK of T. kodakaraensis in the presence of CoA, acetyl-CoA, dephospho-CoA, or 4′-phosphopantothenate. This suggests that CoA biosynthesis is regulated via a different enzyme of the pathway or at the transcriptional level. There are organisms whose CoA biosynthesis is not regulated (33–35) and in Staphylococcus aureus results in the accumulation of high concentrations of intracellular CoA. Further studies will be necessary to determine whether regulation of CoA biosynthesis is present in the Archaea.
At present, we cannot estimate what advantages are brought about to the Archaea by utilizing the PoK/PPS pathway, or to the bacteria/eukaryotes by utilizing the PS/PanK pathway. In terms of evolution, the fact that the PoK/PPS system is confined to, but widely distributed among the Archaea, suggests that the pathway evolved at an early stage in archaeal evolution, but after the divergence of the Archaea from the bacteria and eukaryotes.
Supplementary Material
This work was supported in part by the Japan Society for the Promotion of Science under a grant-in-aid for Creative Scientific Research (project 18GS0421 to T. I.) and by a grant-in-aid for Scientific Research (B) (19310126 to H. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1 and S2.
S. Takedomi, T. Kanai, H. Atomi, and T. Imanaka, unpublished data.
A. Iwata, T. Fukui, H. Atomi, and T. Imanaka, unpublished data.
- CoA
- coenzyme A
- PanK
- pantothenate kinase
- PPCS
- 4′-phosphopantothenoylcysteine synthetase
- PPCDC
- 4′-phosphopantothenoylcysteine decarboxylase
- PPAT
- 4′-phosphopantetheine adenylyltransferase
- DPCK
- dephospho-CoA kinase
- KPHMT
- ketopantoate hydroxymethyltransferase
- PS
- pantothenate synthetase
- Ec-PanK
- PanK from E. coli
- HPLC
- high-performance liquid chromatography
- PK
- pyruvate kinase
- LDH
- lactate dehydrogenase
- PoK
- pantoate kinase
- arCOG
- Archaeal clusters of orthologous genes
- ASW
- artificial seawater.
REFERENCES
- 1.Kupke T., Hernández-Acosta P., Culiáñez-Macià F. A. (2003) J. Biol. Chem. 278, 38229–38237 [DOI] [PubMed] [Google Scholar]
- 2.Leonardi R., Zhang Y. M., Rock C. O., Jackowski S. (2005) Prog. Lipid Res. 44, 125–153 [DOI] [PubMed] [Google Scholar]
- 3.Spry C., Kirk K., Saliba K. J. (2008) FEMS Microbiol. Rev. 32, 56–106 [DOI] [PubMed] [Google Scholar]
- 4.Genschel U. (2004) Mol. Biol. Evol. 21, 1242–1251 [DOI] [PubMed] [Google Scholar]
- 5.Kupke T., Schwarz W. (2006) J. Biol. Chem. 281, 5435–5444 [DOI] [PubMed] [Google Scholar]
- 6.Armengaud J., Fernandez B., Chaumont V., Rollin-Genetet F., Finet S., Marchetti C., Myllykallio H., Vidaud C., Pellequer J. L., Gribaldo S., Forterre P., Gans P. (2003) J. Biol. Chem. 278, 31078–31087 [DOI] [PubMed] [Google Scholar]
- 7.Ronconi S., Jonczyk R., Genschel U. (2008) FEBS J. 275, 2754–2764 [DOI] [PubMed] [Google Scholar]
- 8.Morikawa M., Izawa Y., Rashid N., Hoaki T., Imanaka T. (1994) Appl. Environ. Microbiol. 60, 4559–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atomi H., Fukui T., Kanai T., Morikawa M., Imanaka T. (2004) Archaea 1, 263–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukui T., Atomi H., Kanai T., Matsumi R., Fujiwara S., Imanaka T. (2005) Genome Res. 15, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T., Fukui T., Atomi H., Imanaka T. (2003) J. Bacteriol. 185, 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato T., Fukui T., Atomi H., Imanaka T. (2005) Appl. Environ. Microbiol. 71, 3889–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumi R., Manabe K., Fukui T., Atomi H., Imanaka T. (2007) J. Bacteriol. 189, 2683–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robb F. T., Place A. R. (1995) in Archaea: A Laboratory Manual, thermophiles (Robb F. T., Place A. R. eds) pp. 167–168, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 15.Rashid N., Imanaka H., Kanai T., Fukui T., Atomi H., Imanaka T. (2002) J. Biol. Chem. 277, 30649–30655 [DOI] [PubMed] [Google Scholar]
- 16.Sato T., Imanaka H., Rashid N., Fukui T., Atomi H., Imanaka T. (2004) J. Bacteriol. 186, 5799–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orita I., Sato T., Yurimoto H., Kato N., Atomi H., Imanaka T., Sakai Y. (2006) J. Bacteriol. 188, 4698–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato T., Atomi H., Imanaka T. (2007) Science 315, 1003–1006 [DOI] [PubMed] [Google Scholar]
- 19.Bork P., Sander C., Valencia A. (1993) Protein Sci. 2, 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vallari D. S., Jackowski S., Rock C. O. (1987) J. Biol. Chem. 262, 2468–2471 [PubMed] [Google Scholar]
- 21.Yang D., Shipman L. W., Roessner C. A., Scott A. I., Sacchettini J. C. (2002) J. Biol. Chem. 277, 9462–9467 [DOI] [PubMed] [Google Scholar]
- 22.Hartley A., Glynn S. E., Barynin V., Baker P. J., Sedelnikova S. E., Verhees C., de Geus D., van der Oost J., Timson D. J., Reece R. J., Rice D. W. (2004) J. Mol. Biol. 337, 387–398 [DOI] [PubMed] [Google Scholar]
- 23.Makarova K. S., Sorokin A. V., Novichkov P. S., Wolf Y. I., Koonin E. V. (2007) Biol. Direct 2, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. (1999) Genome Res. 9, 27–43 [PubMed] [Google Scholar]
- 26.Song W. J., Jackowski S. (1994) J. Biol. Chem. 269, 27051–27058 [PubMed] [Google Scholar]
- 27.Rock C. O., Park H. W., Jackowski S. (2003) J. Bacteriol. 185, 3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong B. S., Senisterra G., Rabeh W. M., Vedadi M., Leonardi R., Zhang Y. M., Rock C. O., Jackowski S., Park H. W. (2007) J. Biol. Chem. 282, 27984–27993 [DOI] [PubMed] [Google Scholar]
- 29.Kotzbauer P. T., Truax A. C., Trojanowski J. Q., Lee V. M. (2005) J. Neurosci. 25, 689–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rock C. O., Karim M. A., Zhang Y. M., Jackowski S. (2002) Gene 291, 35–43 [DOI] [PubMed] [Google Scholar]
- 31.Rock C. O., Calder R. B., Karim M. A., Jackowski S. (2000) J. Biol. Chem. 275, 1377–1383 [DOI] [PubMed] [Google Scholar]
- 32.Calder R. B., Williams R. S., Ramaswamy G., Rock C. O., Campbell E., Unkles S. E., Kinghorn J. R., Jackowski S. (1999) J. Biol. Chem. 274, 2014–2020 [DOI] [PubMed] [Google Scholar]
- 33.Leonardi R., Chohnan S., Zhang Y. M., Virga K. G., Lee R. E., Rock C. O., Jackowski S. (2005) J. Biol. Chem. 280, 3314–3322 [DOI] [PubMed] [Google Scholar]
- 34.Brand L. A., Strauss E. (2005) J. Biol. Chem. 280, 20185–20188 [DOI] [PubMed] [Google Scholar]
- 35.Yang K., Eyobo Y., Brand L. A., Martynowski D., Tomchick D., Strauss E., Zhang H. (2006) J. Bacteriol. 188, 5532–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.