Abstract
Metallo-β-lactamases (MβLs) constitute an increasingly serious clinical threat by giving rise to β-lactam antibiotic resistance. They accommodate in their catalytic pocket one or two zinc ions, which are responsible for the hydrolysis of β-lactams. Recent x-ray studies on a member of the mono-zinc B2 MβLs, CphA from Aeromonas hydrophila, have paved the way to mechanistic studies of this important subclass, which is selective for carbapenems. Here we have used hybrid quantum mechanical/molecular mechanical methods to investigate the enzymatic hydrolysis by CphA of the antibiotic biapenem. Our calculations describe the entire reaction and point to a new mechanistic description, which is in agreement with the available experimental evidence. Within our proposal, the zinc ion properly orients the antibiotic while directly activating a second catalytic water molecule for the completion of the hydrolytic cycle. This mechanism provides an explanation for a variety of mutagenesis experiments and points to common functional facets across B2 and B1 MβLs.
The most effective β-lactam drug resistance mechanism in bacteria is the expression of β-lactamases (1). These enzymes are able to hydrolyze the β-lactam ring (2), using either a serine residue (as in class A, C, and D β-lactamases) (3–6) or a zinc-bound water/hydroxide group (as in class B or metallo-β-lactamases (MβLs))3 (7). The spreading of MβLs constitutes, however, an ever increasing serious threat for human health. Indeed, in contrast to serine β-lactamases (8), inhibitors targeting MβLs are not yet known (9).
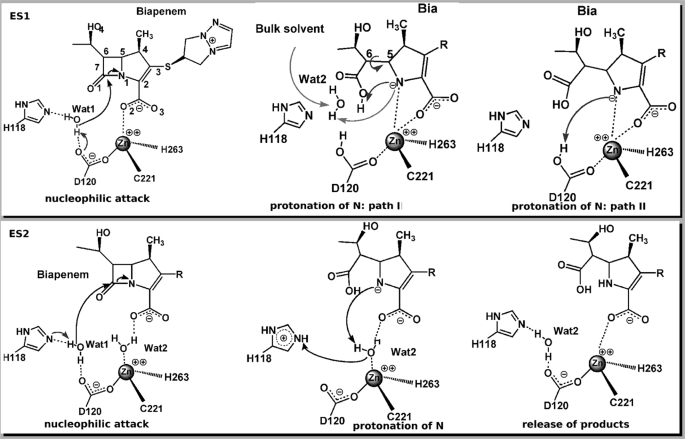
MβLs feature the typical αβ/βα sandwich fold shared with the superfamily of zinc hydrolases (7, 10, 11), and they are classified in three distinct subclasses (B1, B2, and B3) according to their sequence homology (7, 12–14). The active site hosts two potential metal-binding sites (supplemental Fig. S1), in which the zinc ion features a tetrahedral coordination in one site present in B1 and B3 subclasses (called the “Zn1 site” hereafter) and a tetrahedral/trigonal bipyramidal coordination in the other one that is present across the entire MβL family (called the “Zn2 site” hereafter) (7). The B1 and B3 subclasses share similar features, being active in the mono- or di-zinc forms in vitro (7). B2 MβLs are catalytically active only in the mono-zinc form, with the zinc metal accommodated in the Zn2 site (15, 16). In addition, B2 active site cavities are much tighter (17). Consistently their substrate selectivity is confined to carbapenems only, in contrast to B1 and B3 enzymes, which show a broad substrate spectrum (7). In the past years, several groups including ours (18–26) have investigated the reaction of subclass B1 enzymes with quantum chemical methods. Few studies have been performed specifically for B2 MβL carbapenemases yet (17, 27, 28). The mechanistic hypotheses proposed so far are based on the x-ray structure of B2 enzyme CphA from Aeromonas hydrophila in complex with a biapenem (Bia) derivative (15) (Protein Data Bank entry 1X8I; supplemental Figs. S5 and S6). In this structure Asp120, Cys221, His263, and a Bia derivative carboxylate oxygen (O2 in the top panel of Fig. 1, and in supplemental Fig. S5) bind to the metal ion in the Zn2 site. Based on this structure, Garau et al. (15) proposed that Bia acts as the fourth coordinated ligand of the metal ion in the Henry-Michaelis complex (called ES1 in top panel of Fig. 1). Within this proposal, a water molecule (Wat1 in Fig. 1) activated by a general base (His118 or Asp120) (15) would perform a nucleophilic attack to the substrate (C-7 at Bia). The resulting intermediate (Int) would subsequently rearrange by a rotation of the C-5–C-6 bond (path I in top panel of Fig. 1), forming the adduct as found in the x-ray structure (15).
FIGURE 1.
Schematic representation of the proposed reaction mechanisms of B2 MβL CphA. Top panel, the Henry-Michaelis complex ES1 assumes direct binding of the substrate to the zinc ion and one water molecule (Wat1) in the active site. Wat1 is deprotonated by a general base (His118 or Asp120) to perform the nucleophilic attack (15). Nitrogen protonation can take place by a second water molecule (Wat2), involving also an internal rearrangement in the substrate (Path I) or from the general base (in this case, Asp120 is shown as the general base and proton donor) (Path II). Bottom panel, in the Henry-Michaelis complex ES2, the interaction of the substrate with the zinc ion is mediated by a water molecule (Wat2), whereas Wat1 occupies a similar position as in ES1. Wat1 is activated by a general base (His118 in this picture) to perform the nucleophilic attack. The zinc-bound Wat2 acts as a proton donor to the lactam nitrogen.
In the second step of the reaction, a second water molecule (Fig. 1, Wat2), coming from the bulk solvent, would protonate the β-lactam nitrogen with subsequent cleavage of the β-lactam ring at the N-1–C-7 bond. Theoretical calculations performed at the self-consistent charge density functional tight binding level of theory (29) were employed to describe the postulated first step of the reaction, identifying Asp120 as general base (27, 28). The calculated free energy barrier is consistent with experimental data (15) in the hypothesis that such a step is the rate-limiting of the enzymatic cycle. The authors speculated that the β-lactam N-1 would accept a proton directly from Asp120 (path II in top panel of Fig. 1) with consequent breaking of the C–N bond.
However, this hypothesis is not consistent with experimental studies by Crowder and co-workers (30), who have shown that the reaction catalyzed by the B2 MβL ImiS from Aeromonas veronii bv. Sobria is characterized by a rate-determining proton transfer step. In addition, our recent molecular simulation study (17) pointed to the existence of an alternative conformation of the Henry-Michaelis complex, in which a water molecule replaces Bia substrate as the fourth zinc ligand (ES2 complex in the bottom panel of Fig. 1), similar to the reactant state found in B1 MβLs (21, 22).
We perform hybrid Car-Parrinello quantum mechanics/molecular mechanics (QM/MM) calculations of the whole hydrolysis reaction catalyzed by CphA, using as starting reactants both the complex with a single active water (ES1) and the conformation with two water molecules in the catalytic pocket (17).
Our calculations do not support the previously proposed mechanisms by Xu et al. (28) based on ES1 state, because the second step of the reaction turns out to be associated with an activation free energy barrier larger than the experimental data. Instead, the catalytic mechanism derived from the presence of two water molecules at the active pocket in the reactant state is associated with a free energy of activation consistent with experiments (15) and with the findings of Crowder and co-workers (30) on ImiS MβL. Importantly, the reaction occurs via a single-step mechanism, through the activation of a second catalytic water molecule in the active pocket. This result points in turn to functional similarities between B2 and B1 MβL species, in which the conserved zinc metal at the second binding site is positioned to promote a water-mediated reaction mechanism (18–26).
COMPUTATIONAL METHODS
Hybrid Car-Parrinello QM/MM calculations (31) were based on the structures of ES1 and ES2 complexes equilibrated by classical molecular dynamics (MD) simulations of CphA in complex with Bia substrate (17). These initial structures feature one or two water molecules in the zinc-binding pocket, respectively (Fig. 1). In particular, ES1 conformation is the same as that proposed in Refs. 27 and 28, where the Henry-Michaelis complex is characterized by the direct bond between the Bia carboxylate group and zinc metal. As discussed above, ES2 conformation differs from ES1 by the presence of a second water molecule, which links the zinc metal and the carboxylate moiety of Bia β-lactam (Fig. 1 and supplemental Fig. S4).
The atoms treated at the QM level of theory include His118 and His263 imidazole rings (cut at the Cγ), Asp120 and Cys221 (cut at the Cβ), the reactive part of the biapenem that is its backbone, and part of its hydroxyethyl substituent at the 6 position (Bia) (Fig. 1 and supplemental Fig. S3), catalytic water (Wat1), and, for ES2, the additional second water Wat2 (giving a QM box including 59 and 61 atoms, respectively; supplemental Fig. S3). A fictitious electronic mass of 900 atomic units was used. The valence electrons were described by plane wave basis set up to a 70 Rydberg cutoff, and norm-conserving. Martins-Troullier (32) pseudopotentials allowed description of the interaction between valence electron and (pseudo)ionic cores. The Becke-Lee-Yang-Parr (BLYP) exchange correlation functional was employed (33, 34). The QM part was simulated in an isolated orthorhombic box with the dimensions 32.9 × 33.9 × 36.2 Å for the two models. The valence of terminal carbon atoms was terminated by capping hydrogens. The remaining part of the protein frame, along with the solvent, the Cl− counterions, and the characteristic bicyclic side chain of the antibiotic (Fig. 1 and supplemental Fig. S3), was included in the MM part (about 53,000 atoms) and treated with the AMBER PARM99 force field, as in Ref. 35. Our approach combines the Car Parrinello molecular dynamics (CPMD) (36) code for dealing with the QM part and the GROMOS program (37) for treating the MM part. The mechanical and electrostatic interface between these two regions was coupled during the molecular dynamics evolution using a fully Hamiltonian scheme according to Ref. 38.
We equilibrated the systems for 3 ps at 300 K, in the NVT ensemble, by applying the non-Hamiltonian schemes designed by Nosé (39) and Hoover (40); the frequency constant of coupling with external bath was set to a value of 500−1 cm. A time step of 0.09 fs was used. In a second stage, the thermodynamic integration method was used to evaluate free energies of the catalytic reactions, a function of a selected reaction coordinate (41) of the reactive process. The estimated activation free energy in our scheme should be considered, approximated; this is due to several factors: the still limited sampling accessible to current density-functional theory (DFT)/MM MD calculations, the well known limitations of the current GGA exchange-correlation functionals, and the choice of a one-dimensional RC as a first approximation of the real, unknown RC (42, 43).
As in the B1 MβLs enzymes (21, 22), the first step of the reaction was studied by pulling the catalytic Wat1 oxygen toward the β-lactam carbonyl carbon (C-7). dO–C-7 (the distance between O at Wat1 and C-7 at Bia) (Figs. 2 and 3) was selected as the RC during the first step of the reaction (∼3.1 Å for ES1 and ES2 complexes).
FIGURE 2.
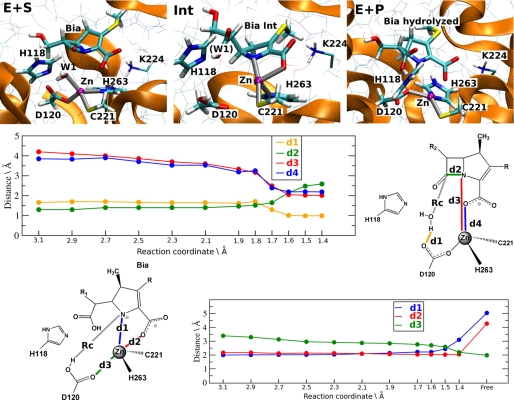
QM/MM calculations of ES1. Top panel, representative structures of the Henry-Michaelis complex (E+S), of the Int and of the products (E+P). QM atoms along with the key Lys224 are shown in bold and thin licorice, respectively. Middle panel, key distances plotted as a function of our chosen reaction coordinates (RC = distance between O at Wat1 and C-7 at Bia)), during the first step of the reaction (the nucleophilic attack). d1, distance between H at Wat1 and O-2 at Asp120; d2, distance to N-1 at Bia-zinc; d3, distance between N-1 at Bia and C-7 at Bia; d4, distance to O-2 at Bia-zinc. Bottom panel, key distances plotted as a function of our chosen reaction coordinates (RC = H at Asp120-N-1 at Bia) during the second step of the reaction (the protonation of the nitrogen). d1, distance to N-1 at Bia-zinc; d2, distance to O-2 at Bia-zinc; d3, distance to O-1 at Asp120-zinc.
FIGURE 3.
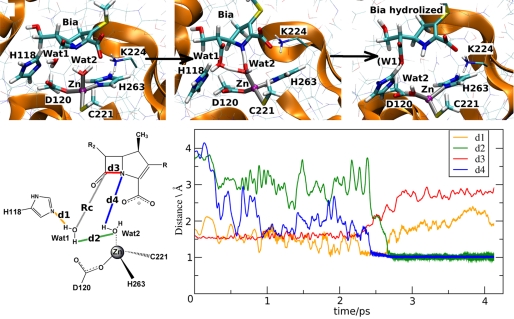
QM/MM calculations of ES2. The top panel shows the rearrangement of the QM atoms (in bold licorice) and those of Lys224 (thin licorice) at TS2. This is characterized by dO–C-7 = 1.7 Å. The bottom panel plots key distances as a function of simulated time at the transition state. d1, distance between Nϵ at His118 and H-1 at Wat1; d2, distance between H-2 at Wat1 and O at Wat2; d3, distance between C-7 at Bia and N-1 at Bia; d4, distance between H at Wat2 and N-1 at Bia.
The RC distance was shortened in steps of 0.2 Å (0.1 Å near transition state region), until the constrained force changed sign, indicating the formation of a transition state in the free energy surface. The free energy surface describing the reaction starting from the ES1 state was completed by the characterization of the Int state and the second transition state calculated as a function of dH–N (the distance between H at Asp120 and N-1 at Bia RC) (Fig. 2). No intermediate was instead formed for the reaction path involving ES2. In each step ∼3 ps of QM/MM trajectory were collected, and the averaged force on constraint (Fc) was obtained over the last 2 ps of trajectory or when the constrained forces had reached convergence (running averages over 1 ps differed less then 5%).
The structural analyses of trajectories have been performed using the Visual Molecular Dynamics (VMD) visualization software (44). The root mean square displacement was calculated considering the heavy backbone atoms only. Hydrogen bonds were assumed to be present if donor and acceptor were located within 3.4 Å and the angle between donor, hydrogen, and acceptor was between 150 and 210 degrees.
RESULTS AND DISCUSSION
Based on classical MD simulations, we suggested that both ES1 (17, 28, 15) and ES2 are plausible Henry-Michaelis complexes (17) (Fig. 1). They differ essentially by the identity of one nonprotein ligand coordinated to the zinc ion, which is either the substrate (in ES1) (15, 28) or a water molecule (in ES2), as already proposed for B1 enzymes (18, 20–23, 25, 26). The nucleophilic water is activated in both the cases by either His118 (15) or Asp120 (28), which are proposed to act as H-bond acceptors to the nucleophile. Here, we used QM/MM simulations (31, 33, 34, 36, 46) to investigate the reaction mechanism of CphA considering both the ES1 and ES2 complex as initial reactants.
Reaction Mechanism Based on ES1 Complex
The Henry-Michaelis Complex
All of the substrate-enzyme interactions were maintained during the QM/MM simulations. In particular, the substrate interacts with Val67 and Lys224 (supplemental Table S1), in agreement with mutagenesis studies (15, 47), which suggest an important role for these two residues in the carbapenem binding. However, the carboxylate-zinc bond assumed in ES1, which is a key facet of this conformation (15, 28), was readily disrupted during simulations (see O-2 at Bia-zinc distance evolution in supplemental Table S1). This might be a consequence of the salt bridge formed between the substrate and Lys224 (supplemental Fig. S3). As a result, Wat1 enters the coordination sphere of the zinc ion, resulting in a distorted tetrahedral geometry of the metal center (supplemental Table S1). Minimization calculations of the atom described at the QM level of theory (i.e. gas phase calculations) turned out to describe a direct binding of the Bia carboxylate group to the metal, a result similar to those reported in (28), in which a different QM/MM set-up was used. Thus, several effects, including the mobility of Lys224, the presence of the protein frame, and temperature effects are for a proper modeling of the correct conformation of the Henri-Michaelis complex.
The Enzymatic Reaction
The distance between the oxygen of the nucleophilic water molecule, Wat1, and the C-7 β-lactam carbon (dO–C-7) was selected as RC (see under “Computational Methods”). This choice is similar to those used in other studies (21, 22, 28) and, to a first approximation, is the natural one to describe the direct nucleophilic attack. dO–C-7 was progressively shortened until the formation of the transition state (TS1) (at 1.7 Å). The coordination sphere of the zinc ion in ES1 was maintained until TS1 was reached; then the polyhedron was subsequently modified because of the simultaneous occurrence of several events (Fig. 2): (i) Bia carboxylate and the β-lactam nitrogen N-1 entered the ligand shell of zinc; (ii) Asp120 and Wat1 detached from the metal ion; (iii) the catalytic Wat1 was deprotonated by Asp120, becoming an activated hydroxide; and (iv) the β-lactam C–N bond weakened, as evidenced by the lengthening of the C–N bond from 1.5 to 1.7 Å (Fig. 2, middle panel).
The calculated activation free energy for this step is ΔF‡ = ∼15 ± 3 kcal/mol, which is close both to the experimentally measured value (ΔG‡ = 14 kcal/mol) (15) and to the computational estimation by Xu et al. (28) (ΔF‡ = ∼14).
By further decreasing RC from TS1 (supplemental Table S1), we identified a stable Int state characterized by a free energy of ΔFInt = ∼9 ± 2 kcal/mol. In this intermediate state, the coordination sphere of the zinc ion was characterized as a distorted tetrahedral geometry. Detachment of Asp120 leaves Cys221 and His263 as metal ligands.
The coordination sphere is completed by the N-1 and O-2 atoms from the substrate (Fig. 2 and supplemental Table S1). The remaining active site residues maintain the H-bond network described for the ES1 state, apart from the interactions subtended by the nucleophile. The N-1 and O-2 atoms from biapenem in this intermediate were located in a position similar to that occupied in the crystal structure of the complex Bia-derivative/CphA (15).
After formation of this intermediate, two different scenarios have been proposed for completion of the reaction: either a proton is transferred from an incoming water molecule in the catalytic pocket, accompanied by rotation of C(5)-C(6) substrate bond (path I in Fig. 1) (15), or a proton transfers directly from Asp120 to β-lactam nitrogen N-1 (path II in top panel of Fig. 1) (28). We explored both pathways using the Int state as a starting configuration for molecular simulations.
Given that large internal structural rearrangement and/or possible resolvation of the active site require time scales well beyond those accessible to first principle QM/MM calculations, we resorted to classical MD, which allows us to extend the time scale investigated. MD calculations performed for 5 ns did not show any relevant rearrangement or resolvation events within the active site pocket. Thus, our calculations provide no support for this possible second step (15). However, because these rearrangements may occur in longer time scales, the mechanism proposed by Garau et al. (15) is not ruled out by the MD calculations.
As for the second possible mechanism, we performed constrained QM/MM simulations, in which the distance between the hydrogen atom on Asp120 and N-1 atom of the substrate was selected as reaction coordinate for this step and was progressively decreased (Fig. 2 and supplemental Table S2). The resulting free energy barrier needed to reach TS2 from the Int state is ΔF‡ = ∼15 ± 2 kcal/mol, giving a total free energy barrier of ∼ 24 ± 3 kcal/mol for the overall reaction with respect to the reactant state ES1. This value largely exceeds the experimental findings, even more considering the well known deficiencies of DFT-BLYP scheme in calculating the energy of transition states and intermediates (42, 43). Thus, the two-step mechanism proposed by Xu et al. (28) appears to be inconsistent with the total activation free energy of the reaction.
Reaction Mechanism Based on ES2 Complex
The Henry-Michaelis Complex
In this case, the geometry in the reactant state ES2 involves an indirect interaction of the substrate carboxylate with the zinc ion, mediated by a water molecule that is the fourth metal ligand (Fig. 1). In contrast to the reaction calculated starting from the ES1 state, the zinc coordination sphere was maintained during the whole QM/MM simulation, preserving a regular tetrahedral geometry (supplemental Table S3). Remarkably, the ligand-zinc distances are within the characteristic values reported for a variety of zinc proteins (49). In particular, Wat2 remained within a distance of 2.1 Å from the zinc ion, keeping also its H-bond to the carboxylate of Bia. The ligand-protein interactions were also maintained (supplemental Table S3) and were similar to those observed in ES1, except that here His196 H-bonds either to carbonyl and to the carboxylate group (O-1 and O-2, respectively) of the antibiotic. The interactions of the substrate with residues Val67 and Lys224 are also conserved, in agreement with mutagenesis data (47), which showed the involvement of these two residues in substrate binding.
The Enzymatic Reaction
The same reaction coordinate as in ES1 complex was used, i.e. the distance between the Wat1 nucleophile and C-7 of Bia (starting value dO–C-7 = 3.1 Å). The chemical bonds present in the Henry-Michaelis complex were maintained until formation of the transition state. The TS was reached at dO–C-7 = 1.7 Å, i.e. at the same distance observed for the reaction starting from ES1. However, in this case the complete cleavage of the β-lactam substrate took place in a single step. Upon formation of the TS, the following events occurred in the sub-ps timescale leading to the enzyme-product adduct (Fig. 3): (i) deprotonation of the nucleophile by His118, which became positively charged; (ii) protonation of the nitrogen N-1 by Wat2, which transiently became a metal-bound hydroxide group; (iii) proton transfer from Wat1 to the metal bound hydroxide, restoring Wat2 as a water molecule; and (iv) proton transfer from His118 to the transient form of Bia. These synchronous events led to the enzyme product state, where the zinc coordination sphere reported for the reactant state is recovered, in which Wat2 has occupied the position of Wat1 (Fig. 3).
The calculated activation free energy for the entire concerted process was ΔF‡ = ∼15 ± 3 kcal/mol, in good agreement with experimental findings (15). We conclude that this pathway is in turn consistent with the finding that the rate-limiting step in this case is the C–N bond cleavage, simultaneous with a proton transfer (47). The present mechanism supports the role of His118 as a general base, and it is therefore crucial for the function of CphA. His118 has been shown to be an important residue for the reaction, because the kcat of the H118A mutant is 3 orders of magnitude lower than the wild type (47). A role for His196 has been more recently proposed studying the kinetics of the H196A mutant, which produces a decrease of kcat by 2 order of magnitudes compared with the wild type (47). Moreover, the role of these two His residues has been recently further confirmed by another experimental study: a B1 MβL, BCII from Bacillus cereus (50) has been engineered so as to be similar to a mononuclear MβL, in which the metal ion binds to the Zn2 site (Zn2 MβL), as usually found in the B2 subclass (supplemental Fig. S1). In the presence of only one equivalent of zinc, two point mutations, H118S and H196S, completely impaired the catalytic activity, suggesting that residues His118 and His196 located at the Zn1 site play a key role for catalysis of mono Zn2 MβLs.
Other residues have been shown to play important roles for the binding affinity of the substrate. The mutation of Lys224 (K224Q) increases the Km measured for the carbapenem by 10-fold compared with the wild type enzyme (47). A structural basis for the role of this residue is obviously provided by our model of the Henry-Michaelis complex. Indeed, Lys224 forms a strong salt bridge with the substrate (supplemental Fig. S3), as also found for the B1 subclass (51). In addition, the mutation of the Val67 residue with longer or more polar amino acids (i.e. V67I and V67D, respectively) invariantly led to a major increase of Km values (47).
Our simulations of ES2 complex offer a rationale for this effect: Val67 forms in fact conserved hydrophobic interactions with the methyl group at position 4 of the substrate. Thus, we conclude that our proposed mechanism provides a molecular basis for the role of these key residues, which have been shown to affect both kcat or Km values.
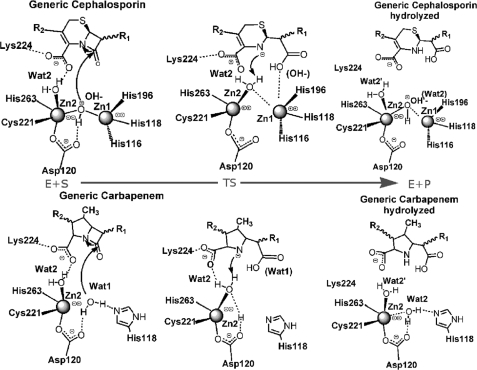
Mechanistic Comparison between B1 and B2 MβLs
Based on the herein proposed mechanism, we can suggest that the zinc ion in the Zn2 site contributes: (i) to substrate binding, by using a water molecule (Wat2), which H-bonds to the carbapenem carboxylate group, thus positioning the β-lactam ring in a favorable orientation for the nucleophilic attack; and (ii) to substrate catalysis, activating a water molecule (Wat2) as a proton donor for the completion of β-lactam hydrolysis. These features closely resemble those already found for the dizinc B1 MβL CcrA complexed with cefotaxime (22) (Fig. 4). Indeed, in both the enzymes the metal ion at the Zn2 site coordinates an additional water molecule present in the active pocket (Wat2), which has the role to anchor the β-lactam carboxylate (a substituent motif present in all β-lactam substrates), allowing an optimal binding orientation of the antibiotic with respect to the nucleophile. Moreover, upon nucleophilic attack this zinc-bound water molecule can readily protonate the β-lactam nitrogen for efficient cleavage of the substrate in a single-step mechanism.
FIGURE 4.
Comparison between MβL subclasses. Proposed reaction mechanism for B1 (in the top panel) and our proposed for B2 MβL (in the bottom panel), in which the key role of the Zn2-binding site in both subclasses emerges clearly. The metal loaded in this site is involved, almost simultaneously, either in the nucleophilic attack of the β-lactam ring and protonation of the β-lactam nitrogen.
The present mechanism discards the hypothesis of Asp120 behaving as a general base. Instead Asp120 plays a role in positioning the zinc ion, as also shown for B1 enzymes (52).
The B1 mechanistic similarity is paralleled by mutagenesis data of residues in the active pocket, namely Val67, His118, His196, and Lys224 (47, 53). The role of Lys224, for instance, is identical to that already found in the B1 subclass (51), further pointing to a structural similarity in the substrate binding across the two subclasses.
We conclude that the zinc metal at the second binding site has a similar role for the reaction mechanism in B1 and B2 subclasses (and likely in B3 species, which is functionally similar to B1). Indeed, the metal at the Zn2 site promotes the hydrolysis of the substrate, using its conserved coordination sphere for binding recognition and a conserved hydroxide/water switch (Wat1 and Wat2 in CphA) for the nucleophilic attack and β-lactam ring cleavage. Thus, regardless of the metal content in B1 and B2 MβLs, such a specific site emerges as a key player during catalysis. This hypothesis is supported by recent mechanistic studies. Despite steady-state kinetic studies at different pH values (54) suggesting the requirement of two bound metal ions for the hydrolytic activity of the B1 MβL BcII, it has been recently shown that BcII can be active as a mono-zinc species with the metal ion accommodated at the Zn2 site, i.e. resembling the conformation of the B2 active site (55, 56). Recent studies have revealed that the B3 lactamase GOB (57) is able to act as a mononuclear enzyme with the metal ion in the putative Zn2 site, i.e. without a metal-activated nucleophile. The importance of the Zn2 site emerges indirectly also from another experimental study. Adaptive protein evolution of B1 MβL has shown that modulating the ionic strength of the metal in the Zn2 site resulted in improved catalytic efficiency (45).
Conclusions
We have presented a computational study of the complete reaction mechanism of B2 CphA MβL from A. hydrophila. Our calculations allow us to propose a new, single-step mechanism, in which a water molecule (Wat1) performs the nucleophilic attack to the β-lactam carbonyl carbon, and another Zn-bound water (Wat2) protonates at the same time the β-lactam nitrogen.
The calculated activation free energy for the reaction is in good agreement with experimental findings, and the proposed mechanism is consistent with the studies by Crowder and co-workers (30), who showed that the rate-limiting step is the C–N β-lactam cleavage and involves a proton transfer event. The catalytic mechanism is also consistent with previous experimental (52) and theoretical (22) studies, while pointing to the presence of a second water molecule in the active site as crucial element for an efficient β-lactam hydrolysis. In addition, our calculations provide structural explanations for the role of the residues that have been shown to be important for the function of B2 MβLs. These include His118, which orients the catalytic water and acts as generalized base; His196, which creates an oxyanion hole, as proposed by Garau et al. (15); and Val67 and Lys224, which affect substrate binding.
Finally, our study, together with recent experimental (45, 48, 52, 54, 55, 57) and theoretical (22) findings, further remarks the central structural and chemical role of the zinc metal at the Zn2 site in MβL family. Thus, future directions in development of MβL inhibitors should specifically target this metal motif, which appears to be relevant to both binding affinity and hydrolysis of β-lactam antibiotics.
In particular, two possible ways of exploiting the outcomes of the present study for drug design purposes can be hypothesized: (i) the TS electronic properties can be used to screen virtual compound libraries aimed at identifying small organic molecules resembling the physicochemical features of the TSs here found and (ii) the TS conformation of the enzyme can be exploited to perform structure-based docking experiments aimed at discovering small organic molecules able to bind the enzyme in the TS configuration rather than in the Henry-Michaelis complex. In the latter case, refinement calculations in the QM (or QM/MM) framework should also be carried out to improve reliability of docking studies of ligands to metal-carrying proteins. This kind of application can open up new ways for an actual exploitation of first principles quantum chemical approaches to drug design and discovery projects.
Supplementary Material
Acknowledgments
We thank Distributed European Infrastructure for Supercomputing Applications (DEISA) and Consorzio Interuniversitario (CINECA) and its staff for CPU time and support.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, references, Tables S1–S4, and Figs. S1–S6.
- MβL
- metallo-β-lactamase
- Bia
- biapenem
- Int
- intermediate
- QM
- quantum mechanics
- MM
- molecular mechanics
- MD
- molecular dynamics
- RC
- reaction coordinate
- TS
- transition state.
REFERENCES
- 1.Babic M., Hujer A. M., Bonomo R. A. (2006) Drug Resist. Updat. 9, 142–156 [DOI] [PubMed] [Google Scholar]
- 2.Fisher J. F., Meroueh S. O., Mobashery S. (2005) Chem. Rev. 105, 395–424 [DOI] [PubMed] [Google Scholar]
- 3.Hata M., Fujii Y., Tanaka Y., Ishikawa H., Ishii M., Neya S., Tsuda M., Hoshino T. (2006) Biol. Pharm. Bull. 29, 2151–2159 [DOI] [PubMed] [Google Scholar]
- 4.Strynadka N. C., Adachi H., Jensen S. E., Johns K., Sielecki A., Betzel C., Sutoh K., James M. N. (1992) Nature 359, 700–705 [DOI] [PubMed] [Google Scholar]
- 5.Sulton D., Pagan-Rodriguez D., Zhou X., Liu Y., Hujer A. M., Bethel C. R., Helfand M. S., Thomson J. M., Anderson V. E., Buynak J. D., Ng L. M., Bonomo R. A. (2005) J. Biol. Chem. 280, 35528–35536 [DOI] [PubMed] [Google Scholar]
- 6.Pagan-Rodriguez D., Zhou X., Simmons R., Bethel C. R., Hujer A. M., Helfand M. S., Jin Z., Guo B., Anderson V. E., Ng L. M., Bonomo R. A. (2004) J. Biol. Chem. 279, 19494–19501 [DOI] [PubMed] [Google Scholar]
- 7.Bebrone C. (2007) Biochem. Pharmacol. 74, 1686–1701 [DOI] [PubMed] [Google Scholar]
- 8.Georgopapadakou N. H. (2004) Expert Opin. Investig. Drugs 13, 1307–1318 [DOI] [PubMed] [Google Scholar]
- 9.Toney J. H., Moloughney J. G. (2004) Curr. Opin. Investig. Drugs 5, 823–826 [PubMed] [Google Scholar]
- 10.Carfi A., Pares S., Duée E., Galleni M., Duez C., Frère J. M., Dideberg O. (1995) EMBO J. 14, 4914–4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowder M. W., Spencer J., Vila A. J. (2006) Acc. Chem. Res. 39, 721–728 [DOI] [PubMed] [Google Scholar]
- 12.Galleni M., Lamotte-Brasseur J., Rossolini G. M., Spencer J., Dideberg O., Frère J. M. (2001) Antimicrob. Agents Chemother. 45, 660–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garau G., García-Sáez I., Bebrone C., Anne C., Mercuri P., Galleni M., Frère J. M., Dideberg O. (2004) Antimicrob. Agents Chemother. 48, 2347–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garau G., Di Guilmi A. M., Hall B. G. (2005) Antimicrob. Agents Chemother. 49, 2778–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garau G., Bebrone C., Anne C., Galleni M., Frère J. M., Dideberg O. (2005) J. Mol. Biol. 345, 785–795 [DOI] [PubMed] [Google Scholar]
- 16.Costello A. L., Sharma N. P., Yang K. W., Crowder M. W., Tierney D. L. (2006) Biochemistry 45, 13650–13658 [DOI] [PubMed] [Google Scholar]
- 17.Simona F., Magistrato A., Vera D. M., Garau G., Vila A. J., Carloni P. (2007) Proteins 69, 595–605 [DOI] [PubMed] [Google Scholar]
- 18.Suárez D., Merz K. M., Jr. (2001) J. Am. Chem. Soc. 123, 3759–3770 [DOI] [PubMed] [Google Scholar]
- 19.Díaz N., Suárez D., Merz K. M., Jr. (2001) J. Am. Chem. Soc. 123, 9867–9879 [DOI] [PubMed] [Google Scholar]
- 20.Suárez D., Díaz N., Merz K. M., Jr. (2002) J. Comput. Chem. 23, 1587–1600 [DOI] [PubMed] [Google Scholar]
- 21.Dal Peraro M., Llarrull L. I., Rothlisberger U., Vila A. J., Carloni P. (2004) J. Am. Chem. Soc. 126, 12661–12668 [DOI] [PubMed] [Google Scholar]
- 22.Dal Peraro M., Vila A. J., Carloni P., Klein M. L. (2007) J. Am. Chem. Soc. 129, 2808–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park H., Brothers E. N., Merz K. M., Jr. (2005) J. Am. Chem. Soc. 127, 4232–4241 [DOI] [PubMed] [Google Scholar]
- 24.Oelschlaeger P., Schmid R. D., Pleiss J. (2003) Prot. Eng. 16, 341–350 [DOI] [PubMed] [Google Scholar]
- 25.Krauss M., Gresh N., Antony J. (2003) J. Phys. Chem. B 107, 1215–1229 [Google Scholar]
- 26.Olsen L., Rasmussen T., Hemmingsen L., Ryde U. (2004) J. Phys. Chem. B. 108, 17639–17648 [Google Scholar]
- 27.Xu D., Zhou Y., Xie D., Guo H. (2005) J. Med. Chem. 48, 6679–6689 [DOI] [PubMed] [Google Scholar]
- 28.Xu D., Xie D., Guo H. (2006) J. Biol. Chem. 281, 8740–8747 [DOI] [PubMed] [Google Scholar]
- 29.Elstner M., Porezag D., Jungnickel G., Elsner J., Haugk M., Frauenheim T., Suhai S., Seifert G. (1998) Phys. Rev. B. 58, 7260–7268 [Google Scholar]
- 30.Sharma N. P., Hajdin C., Chandrasekar S., Bennett B., Yang K. W., Crowder M. W. (2006) Biochemistry 45, 10729–10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carloni P., Rothlisberger U., Parrinello M. (2002) Acc. Chem. Res. 35, 455–464 [DOI] [PubMed] [Google Scholar]
- 32.Troullier N., Martins J. L. (1991) Phys. Rev. B Condens. Matter 43, 1993–2006 [DOI] [PubMed] [Google Scholar]
- 33.Becke A. D. (1988) Phys. Rev. A 38, 3098–3100 [DOI] [PubMed] [Google Scholar]
- 34.Lee C., Yang W., Parr R. G. (1988) Phys. Rev. B Condens. Matter 37, 785–789 [DOI] [PubMed] [Google Scholar]
- 35.Case D. A., Pearlman D. A., Caldwell J. W., S., T. C. I., Wang J., Ross , Simmerling C., Darden T., Merz K., Stanton R. V., Cheng A., Vincent J. J., Crowley M., Tsui V., Gohlke H., Radmer R. J., Duan Y., Pittera J., Massova I., Seibel G. L., Singh U. C., Weiner P., Kollman P. A. (2002) AMBER version 9.0, University of California, San Francisco, CA [Google Scholar]
- 36.Car R., Parrinello M. (1985) Phys. Rev. Lett. 55, 2471–2474 [DOI] [PubMed] [Google Scholar]
- 37.Scott W., Hunenberger P., Tironi I., Mark A., Billeter S., Fennen J., Torda A., Huber T., Kruger P., van Gunsteren W. (1999) J. Phys. Chem. A 103, 3596–3607 [Google Scholar]
- 38.Laio A., VandeVondele J., Rothlisberger U. (2002) J. Chem. Phys. 116, 6941–6947 [Google Scholar]
- 39.Nosé S. (1984) Mol. Phys. 52, 255–268 [Google Scholar]
- 40.Hoover W. G. (1985) Phys. Rev. A. 31, 1695–1697 [DOI] [PubMed] [Google Scholar]
- 41.Ciccotti G., Ferrario M., Hynes J. T., Karpal R. (1989) Chem. Phys. 129, 241–251 [Google Scholar]
- 42.Piana S., Bucher D., Carloni P., Rothlisberger U. (2004) J. Phys. Chem. B. 108, 11139–11149 [Google Scholar]
- 43.Magistrato A., Vondele J. V., Rothlisberger U. (2000) Inorg. Chem. 39, 5553–5560 [DOI] [PubMed] [Google Scholar]
- 44.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graphics 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 45.Tomatis P. E., Fabiane S. M., Simona F., Carloni P., Sutton B. J., Vila A. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20605–20610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dal Peraro M., Ruggerone P., Raugei S., Gervasio F. L., Carloni P. (2007) Curr. Opin. Struct. Biol. 17, 149–156 [DOI] [PubMed] [Google Scholar]
- 47.Bebrone C., Anne C., Kerff F., Garau G., De Vriendt K., Lantin R., Devreese B., Van Beeumen J., Dideberg O., Frère J. M., Galleni M. (2008) Biochem. J. 414, 151–159 [DOI] [PubMed] [Google Scholar]
- 48.González J. M., Medrano Martín F. J., Costello A. L., Tierney D. L., Vila A. J. (2007) J. Mol. Biol. 373, 1141–1156 [DOI] [PubMed] [Google Scholar]
- 49.Tamames B., Sousa S. F., Tamames J., Fernandes P. A., Ramos M. J. (2007) Proteins 69, 466–475 [DOI] [PubMed] [Google Scholar]
- 50.Abriata L. A., González L. J., Llarrull L. I., Tomatis P. E., Myers W. K., Costello A. L., Tierney D. L., Vila A. J. (2008) Biochemistry 47, 8590–8599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dal Peraro M., Vila A. J., Carloni P. (2004) Proteins 54, 412–423 [DOI] [PubMed] [Google Scholar]
- 52.Llarrull L. I., Fabiane S. M., Kowalski J. M., Bennett B., Sutton B. J., Vila A. J. (2007) J. Biol. Chem. 282, 18276–18285 [DOI] [PubMed] [Google Scholar]
- 53.Bebrone C., Anne C., De Vriendt K., Devreese B., Rossolini G. M., Van Beeumen J., Frère J. M., Galleni M. (2005) J. Biol. Chem. 280, 28195–28202 [DOI] [PubMed] [Google Scholar]
- 54.Badarau A., Page M. I. (2008) J. Biol. Inorg. Chem. 13, 919–928 [DOI] [PubMed] [Google Scholar]
- 55.Llarrull L. I., Tioni M. F., Vila A. J. (2008) J. Am. Chem. Soc. 130, 15842–15851 [DOI] [PubMed] [Google Scholar]
- 56.Tioni M. F., Llarrull L. I., Poeylaut-Palena A. A., Martí M. A., Saggu M., Periyannan G. R., Mata E. G., Bennett B., Murgida D. H., Vila A. J. (2008) J. Am. Chem. Soc. 130, 15852–15863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morán-Barrio J., González J. M., Lisa M. N., Costello A. L., Peraro M. D., Carloni P., Bennett B., Tierney D. L., Limansky A. S., Viale A. M., Vila A. J. (2007) J. Biol. Chem. 282, 18286–18293 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.