Abstract
In low GC content Gram-positive bacteria, the HPr protein is the master regulator of carbon metabolism. HPr is a key component of the phosphoenolpyruvate (PEP):sugar phosphotransferase system that interacts with and/or phosphorylates proteins relevant to carbon catabolite repression. HPr can be phosphorylated by two distinct kinases as follows: the bifunctional enzyme HPr kinase/Ser(P)-HPr phosphorylase (HprK/P) phosphorylating the serine 46 residue (Ser(P)-HPr) and acting as a phosphorylase on Ser(P)-HPr; and the PEP-requiring enzyme I (EI) generating histidine 15-phosphorylated HPr (His(P)-HPr). The various HPr forms interact with numerous enzymes and modulate their activity. By carrying out a genome-wide yeast two-hybrid screen of a Bacillus subtilis library, we identified a novel HPr-interacting protein, the transcriptional activator YesS, which regulates the expression of pectin/rhamnogalacturonan utilization genes. Remarkably, yeast tri-hybrid assays involving the ATP-dependent HprK/P and the PEP-dependent EI suggested that YesS interacts with HPr and His(P)-HPr but not with Ser(P)-HPr. These findings were confirmed by in vitro interaction assays using the purified HPr-binding domain of the YesS protein. Furthermore, pectin utilization and in vivo YesS-mediated transcriptional activation depended upon the presence of His(P)-HPr, indicating that HPr-mediated YesS regulation serves as a novel type of carbon catabolite repression. In the yeast two-hybrid assays, B. subtilis HprK/P and EI were active and specifically recognized their substrates. Both kinases formed long lived complexes only with the corresponding nonphosphorylatable mutant HPr. These findings suggest that two-hybrid assays can be used for the identification of unknown kinases of phosphorylated bacterial proteins detected in phosphoproteome analyses.
Bacteria developed complex regulatory mechanisms allowing them to adapt to their frequently changing environment and to use preferentially the carbohydrates easily transformable into glycolytic intermediates. This phenomenon is called carbon catabolite repression (CCR).3 Components of the phosphoenolpyruvate:sugar phosphotransferase system (PTS) play a major role in CCR, as they interact with and/or phosphorylate proteins relevant for CCR. Nevertheless, the main function of the PTS is to transport and concomitantly phosphorylate sugars via a PEP-requiring protein phosphorylation cascade, which includes the transient phosphorylation of the PTS component HPr on its His-15 residue providing His(P)-HPr.
In firmicutes (low G + C Gram-positive bacteria), the central regulator of CCR, which affects the expression of about 10% of their genes, is HPr. His(P)-HPr contributes to CCR by phosphorylating various non-PTS proteins involved in the transport and metabolism of secondary carbon sources (for reviews see Refs. 1, 2). HPr is also phosphorylated on Ser-46 by the bifunctional enzyme HPr kinase/Ser(P)-HPr phosphorylase (HprK/P) (3), whose antagonistic activities are controlled by various metabolites. The interaction of Ser(P)-HPr with the catabolite control protein A (CcpA) leads to CCR (4, 5) and with non-PTS transporters to inducer exclusion (6). In B. subtilis, CcpA interacts also with seryl-phosphorylated Crh (catabolite repression HPr), a paralog of HPr (7, 8). However, Ser(P)-Crh plays only a minor role in CCR (for a review, see Ref. 1).
The doubly phosphorylated form of HPr ((His(P),Ser(P))-HPr) is not abundant in Bacillus subtilis (9, 10), and to date no role has been assigned to it in this organism. Nevertheless, it seems that large quantities of (His(P),P∼Ser)-HPr are present in certain glucose-grown streptococci and play a physiological role there (11, 12).
Therefore, numerous fundamental cellular processes in firmicutes are influenced by the ratio of the various intracellular HPr forms. They exert their effects either by interacting with (HPr, Ser(P)-HPr) or by phosphorylating their target proteins (His(P)-HPr, (His(P),Ser(P))-HPr). HPr and Ser(P)-HPr of B. subtilis were reported to bind to glyceraldehyde-3-P dehydrogenase, but only Ser(P)-HPr inhibits GapA activity (13). Moreover, B. subtilis Ser(P)-HPr was shown to interact in vitro with RbsR, a LacI/GalR type regulator that controls the expression of the ribose operon. However, a physiological role has so far not been established for the latter interaction (14).
Here we report that the B. subtilis transcriptional activator YesS is a novel partner of HPr. This interaction was detected in a genome-wide yeast two-hybrid screen and was further studied functionally. We find that the YesS-HPr interaction mediates a novel CCR mechanism.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid and Tri-hybrid Experiments
Saccharomyces cerevisiae strains were grown in rich YEPD medium or in synthetic complete medium (SC) lacking the appropriate amino acids (Leu, Trp, His) or nucleotides (Ade, Ura) (15). B. subtilis wild-type genes were PCR-amplified from genomic DNA of strain 168. The ptsHS46A, ptsHH15A, and crhS46A mutant alleles were PCR-amplified from genomic DNA of the B. subtilis strains RHGR64 (16), GR11,4 and QB7112 (7), respectively. The ΔhprKV267F mutant gene from Lactobacillus casei was PCR-amplified from pΔHPRKLCV267F (17). The various PCR products were cloned into the bait vector pGBDU (URA3), the prey vector pGAD (LEU2) (18), as well as in the p3H vector (TRP1) for tri-hybrid assays (19). The bait and tri-hybrid constructs were introduced in the yeast strain PJ69-4a and the prey constructs in strain PJ69-4α. The DNA sequences of all cloned fragments were verified.
Some baits were used to screen a B. subtilis prey library essentially as described previously (19). For each screen, >30 × 106 diploid cells were plated on selective medium, ensuring maximal coverage of the library. The interaction candidates were identified by PCR amplification and sequencing of the DNA inserts in the prey plasmids. Importantly, false-positive interactions generated by the yeast two-hybrid system were eliminated experimentally as described (19).
For tri-hybrid experiments, the appropriate open reading frames were expressed from the vector p3H (TRP1) (19). Strain PJ69-4a was co-transformed with different combinations of bait and p3H-derived vectors. Ura+ Trp+ colonies were isolated and mated with PJ69-4α strains containing various prey vectors, and diploids were selected on SC-LUW (Synthetic Complete medium lacking leucine, uracil, and tryptophan). Interaction phenotypes were scored by replica plating the diploids onto selective plates SC-LUWH (SC-LUW lacking histidine and containing 0.5 mm 3-aminotriazole) and SC-LUWA (SC-LUW lacking adenine) after 7 and 14 days of incubation at 30 °C.
Bacterial Strains and Growth Conditions
Escherichia coli strains NM522 (Stratagene) and DH10B (Invitrogen) were used for all cloning experiments and for protein overproduction. NM522 harboring pQE30 derivatives (Qiagen) and the derivative of pGEX-2T (GE Healthcare) were grown at 37 °C under agitation in LB medium supplemented with 100 μg/ml ampicillin.
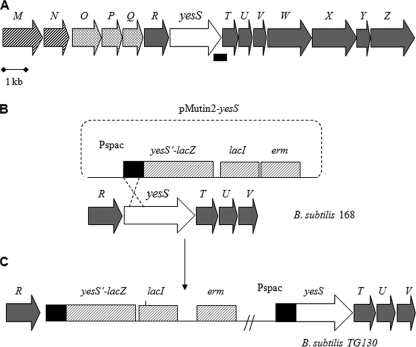
B. subtilis strains used in this study are listed in Table 1. To obtain strain TG130, strain 168 was transformed with plasmid pMUTIN2-yesS to insert a yesS′-lacZ fusion into the chromosome and to newly create a Pspac-yesS fusion (see Fig. 4C). All other B. subtilis mutants were derived from TG130. Insertion of a ptsI disruption marked with spectinomycin, co-transformation of the ptsHH15A allele with a chloramphenicol resistance cassette, and in-frame deletion of codons 7–42 in ptsH (ptsHΔDH allele) in B. subtilis TG130 (Table 1) were confirmed by the inability of the resulting mutants (TG136, TG137, and TG134, respectively) to grow on minimal medium supplemented with 0.2% of the PTS sugar mannitol. Introduction of the ccpA and hprK mutations into TG130 (strains TG131 and TG132, respectively) caused a loss of the repressive effect of glucose on gluconate kinase activity, as was expected from similar previous experiments (20). B. subtilis strains were grown under agitation in LB medium or C mineral medium (21) supplemented with the indicated antibiotics at the following concentrations: erythromycin, 5 μg/ml; chloramphenicol, 6 μg/ml; spectinomycin, 100 μg/ml; kanamycin, 5 μg/ml. When needed, isopropyl β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 0.1 mm (for B. subtilis) or 50 μm (for E. coli).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype | Reference or construction |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| GM1090 | trpC2 pheA1 ccpA ::spc | M. Steinmetz |
| GR11 | trpC2 sacXYΔ3 sacBΔ23 sacTΔ4 bglPΔ3 amyE ::(bglP′-lacZ phl) ptsH(H15A) cat | E. Darbon and J. Deutscher, unpublished results |
| GR14 | trpC2 pheA1 ptsHΔDH cat | 45 |
| GR26 | trpC2 sacXYΔ3 sacBΔ23 sacTΔ4 bglPΔ3 amyE ::(bglP′-lacZ phl) ptsI ::spc | 45 |
| QB1701 | trpC2 crh ::spc | 46 |
| QB7160 | trpC2 hprK ::spc | 47 |
| RHGR64 | trpC2amyE ::phoP′-lacZ ptsH1 spc | 16 |
| TG130 | trpC2 yesS′-lacZ Pspac-yesS | pMUTIN2-yesS → 168a |
| TG131 | trpC2 yesS′-lacZ Pspac-yesS ccpA ::spc | GM1090 → TG130a |
| TG132 | trpC2 yesS′-lacZ Pspac-yesS hprK ::spc | QB7160 → TG130a |
| TG133 | trpC2 yesS′-lacZ Pspac-yesS crh ::spc | QB1701 → TG130a |
| TG134 | trpC2 yesS′-lacZ Pspac-yesS ptsHΔDH cat | GR14 → TG130a |
| TG135 | trpC2 yesS′-lacZ Pspac-yesS ptsH1 spc | RHGR64 → TG130a |
| TG136 | trpC2 yesS′-lacZ Pspac-yesS ptsI ::spc | GR26 → TG130a |
| TG137 | trpC2 yesS′-lacZ Pspac-yesS ptsH(H15A) cat | GR11 → TG130a |
a Arrows point to the recipient strain transformed with the indicated plasmid or chromosomal DNA.
FIGURE 4.
Strain construction to study YesS transcriptional activity. A, gene organization around yesS in B. subtilis. yesM and yesN code for a two-component system, which probably also participates in the regulation of yes operon expression. The other genes were reported to be involved in rhamnogalacturonan utilization (41). B, homologous integration of the pMUTIN2-yesS vector into the chromosome of B. subtilis. pMUTIN2 carries a promoterless lacZ gene followed by the lacI gene, an erythromycin resistance cassette, and the Pspac promoter. Genes of the putative yes operon are indicated as gray arrows except yesS, which is presented as a white arrow with its 5′ part in black. The Shine-Dalgarno box of yesS together with its 5′ part were PCR-amplified and cloned into pMUTIN2 and integrated into strain 168 by a single crossover (C). In the resulting strain the 5′ segment of yesS was duplicated (black boxes), and the incomplete yesS was fused to lacZ (yesS′-lacZ), whereas the resulting entire yesS was placed under control of the Pspac promoter.
Plasmid Constructions
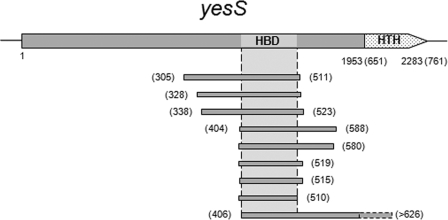
An internal region of yesS coding for residues 403–515 (including the entire HPr binding domain, HBD; see Fig. 2) was amplified by PCR using chromosomal DNA of B. subtilis as a template and primers YesS7 and YesS8. The sequences of primers used in this study will be provided on request. The PCR product was digested with BamHI and KpnI (restriction sites carried in the primers) and inserted into the expression vector pGEX-2T digested with the same enzymes, giving plasmid pGEX2T-HBD, which allows the overproduction of HBD fused to the C-terminal end of glutathione S-transferase (GST-HBD).
FIGURE 2.
HPr interacts with a central domain of the transcription regulator YesS. Schematic representation of the yesS gene (2283 bp) is shown. HTH indicates two conserved helix-turn-helix (HTH) motifs. The gray bars below represent the nine overlapping fragments of YesS isolated from the yeast two-hybrid screens of a fragmented genomic library. They correspond to in-frame fusions to the GAL4 activation domain (AD-YesS) when HPr was used as bait (BD-HPr). The amino acid coordinates are indicated in parentheses. This delineates the minimal domain of YesS (amino acids 406–510) required for HPr binding (HBD, light gray).
By using plasmid pAG2 (7), which allows the synthesis of HPr with a polyhistidine tag fused to the N terminus (His6-HPr), as a template and restriction sites-carrying primers SP31 and SP32, ptsH from B. subtilis was PCR-amplified, digested with KpnI and EcoRI, and cloned into pGEX2T-HBD cleaved with the same enzymes, leading to plasmid pGEX2T-HBD+HPr, which harbors an artificial IPTG-inducible operon allowing the simultaneous synthesis in E. coli of GST-HBD and His6-HPr. Mutagenesis of ptsH was performed by three-step PCR (22) using pGEX2T-HBD+HPr as a template and the primers pairs SP215-SP216 and SP217-SP218 to construct the pGEX2T-HBD+HPrH15D and pGEX2T-HBD+HPrS46D plasmids, respectively.
Plasmid pMutin2-yesS was constructed by PCR amplification and cloning of the region from −26 to +496 of the yesS gene using B. subtilis 168 chromosomal DNA as a template and SP150 and SP27 as primers. The PCR product was digested with HindIII and BamHI and cloned into pMutin2 (23) cleaved with the same enzymes. Pfu DNA polymerase was used for all PCR amplifications, and the correct sequence of the PCR products was confirmed by DNA sequencing.
Protein Purification and Phosphorylation Assays
E. coli NM522 harboring either pGEX2T-HBD+HPr, pGEX2T-HBD+HPrH15D, or pGEX2T-HBD+HPrS46D was grown in 1 liter of LB medium containing ampicillin. When the culture reached an A600 = 0.4, the synthesis of the GST-HBD and His6-HPr proteins was induced by the addition of 50 μm IPTG, and incubation was continued for 3 h at 37 °C. Crude cell extracts were prepared, and His6-tagged HPr, HPrS46D, and HPrH15D were purified using a 1-ml Ni-NTA column (Qiagen) as described previously (7).
The purified His6-HPr-GST-HBD complex (∼450 μg of GST-HBD and ∼150 μg of His6-HPr) was incubated in a 1-ml reaction mixture containing 5 mm ATP, 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 10 mm fructose 1,6-biphosphate, and 14 μg of ΔHprK/PV267F for 30 min at 37 °C to allow the formation of Ser(P)-HPr. Similarly, the same amount of purified complex was incubated in a 1-ml reaction mixture containing 5 mm PEP, 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, and 20 μg of EI from B. subtilis for 30 min at 37 °C to allow the formation of His(P)-HPr. After incubation, the reaction mixtures were loaded on Ni-NTA spin columns (Qiagen) and centrifuged for 1 min at 1000 rpm. The columns were subsequently washed with 500 μl of buffer A (50 mm Tris-HCl, pH 7.4; 15% glycerol; 50 mm Na2SO4) and 500 μl of buffer B (buffer A + 30 mm imidazole). The elution of His6-HPr from the column was carried out using 500 μl of buffer C (buffer A + 300 mm imidazole). Fractions were collected from all washing and elution steps, and aliquots were analyzed by SDS-PAGE.
HPr and EI from B. subtilis and ΔHprK/P-V267F from L. casei were purified as His6-tagged proteins from E. coli strain NM522 carrying plasmid pAG2, pAG3, and pQE30-ΔHprK/PV267F, respectively, as described previously (7, 17).
β-Galactosidase Assays
The various B. subtilis strains harboring pMutin2-yesS integrated in the chromosome were grown at 37 °C under agitation in 40 ml of LB medium supplemented with the appropriate antibiotics. IPTG was added at a final concentration of 0.1 mm, and 2-ml samples were withdrawn every 60 min over a period of 2–8 h after the addition of IPTG. Crude extracts were prepared and used for β-galactosidase assays with ortho-nitrophenyl β-d-galactopyranoside as substrate. β-Galactosidase activity is expressed in Miller units/mg of protein (24). Protein concentrations were determined using the Bio-Rad protein assay.
RNA Isolation and RT-PCR Assays
Extraction of B. subtilis total RNA was performed on 4-ml cultures of cells grown to late exponential phase in LB medium supplemented or not with 0.5% pectin from apple (Fluka). Cells from 4-ml cultures were harvested, and total RNA was extracted using the RNeasy mini kit (Qiagen). The remaining contaminating genomic DNA was then digested with the Turbo DNA-free kit (Ambion). Reverse transcription experiments were performed using the Ready-to-Go RT-PCR Beads kit (GE-Healthcare). Beads were resuspended in an appropriate volume of RNase-free water, and 200 ng of DNase-treated total RNA was added to the reaction mixture also containing 2 μl of random primers (pDN6) before it was incubated for 30 min at 42 °C. To check for DNA contamination, a control experiment was performed: the same volume of suspended beads was incubated for 5 min at 95 °C to inactivate the reverse transcriptase before the addition of pDN6 and RNA. Oligonucleotides SP85 and SP86 were added to the reaction mixture, and the PCR amplification of a 450-bp yesS-yesT intergenic fragment was performed as follows: one cycle of 5 min at 95 °C, followed by 39 cycles of 30 s at 95 °C, 30 s at 52 °C, 30 s at 72 °C, and a final step of 10 min at 72 °C.
RESULTS AND DISCUSSION
HprK/P and EI Kinases Interact with and Specifically Modify HPr in Yeast
Because the B. subtilis HPr protein and its partners have been extensively characterized (see Introduction), we used this system as a model to investigate the potentialities of the yeast two- and tri-hybrid assays to monitor phosphorylation-dependent interactions. The effects of the ATP- and PEP-dependent HPr kinases HprK/P and EI, respectively, on the protein interactions of their natural substrate HPr were investigated using the yeast tri-hybrid approach (see “Experimental Procedures”). Because HprK/P is a bifunctional enzyme that catalyzes phosphorylation and dephosphorylation of Ser(P)-HPr (3), we included in one set of our assays the B. subtilis HprK/P and in another the L. casei ΔHprK/PV267F mutant protein, which exhibits normal HPr kinase but drastically reduced Ser(P)-HPr phosphorylase activity (17). Phosphorylation-dependent interactions were revealed when the interaction phenotype appeared in response to the presence of a kinase, whereas binary interactions were detected independently of the presence of a third protein. A binary interaction of AD-ΔHprK/PV267F was observed with BD-HPrS46A but not with BD-HPr, BD-HPrH15A, BD-Crh, and BD-CrhS46A (Fig. 1, A–C, 5th row). In contrast, AD-HprK/P interacted preferentially with BD-CrhS46A, as only this interaction conferred adenine prototrophy to yeast cells (Fig. 1, A–C, 7th row). However, under less stringent selection conditions (histidine prototrophy), AD-HprK/P interacted with all HPr and Crh baits (Fig. 1, A and B, 8th row). This is in agreement with the previously observed co-crystallization of truncated L. casei HprK/P and B. subtilis HPr (25). Finally, AD-EI interacted solely with BD-HPrH15A (Fig. 1, B and C, 6th row). Strikingly, both AD-HprK/P and AD-EI apparently form more stable complexes with the nonphosphorylatable protein than with their wild-type substrate. Furthermore, BD-HPr interacted with AD-HprK/P (Fig. 1, A–C, 8th row) but not with AD-HprK/PV267F (Fig. 1, A–C, row 5) even under the less stringent conditions (data not shown), suggesting that the bifunctional enzyme catalyzing both the phosphorylation of HPr and dephosphorylation of Ser(P)-HPr (3) yields a detectable complex with HPr, whereas a mutant form with only the kinase function does not.
FIGURE 1.
Protein interactions with HPr are specifically affected by the presence of its two cognate kinases in a yeast tri-hybrid assay. Protein interactions of HPr and Crh with YesS, CcpA, HprK/P, and EI were assayed in the presence or absence of the ATP-dependent, truncated, constitutively active L. casei V267F mutant HprK/P (A), full-length wild-type B. subtilis HprK/P (B), and PEP-dependent B. subtilis EI (C) in a yeast tri-hybrid approach. Full-length wild-type and mutant HPr and Crh proteins were fused with the Gal4 DNA-binding domain (BD, top row). Two fragments of YesS, the full-length CcpA, L. casei mutant HprK/PV267F, a fragment of BioI (only in A), B. subtilis wild-type HprK/P, and B. subtilis wild-type EI were fused to the Gal4 activation domain (AD, left column). In the line above the figure, + indicates that truncated L. casei mutant HprK/PV267F (A), B. subtilis wild-type HprK/P (B), and B. subtilis wild-type EI (C) were also synthesized without fusion to a Gal4 domain (3HB, right column). The relevant amino acid substitutions are indicated for mutant proteins. Similarly, amino acid coordinates are indicated for protein fragments. Independent yeast colonies containing the various binding domain/3HB combinations were arrayed and mated with the strains expressing the different activation domain fusion proteins, and diploid cells were subjected to selection for the expression of the HIS3 and more stringent ADE2 interaction reporters. Interaction phenotypes on SC-LUWA selective plates after incubation at 30 °C for 7 days are shown for the complete set of experiments, whereas interaction phenotypes on SC-LUWH selective plates after incubation at 30 °C for 14 days are shown only for the experiments with AD-HprK/P (bottom rows in A–C). For all other activation domain fusions, colonies that grew only on SC-LUWH- but not SC-LUWA-selective plates are indicated with blue arrowheads. Binary interactions, which are independent of a 3HB partner, appeared as pairs of colonies in a row. Ternary interactions were revealed as a positive or negative effect of the 3HB partner on the growth of the yeast colonies. Each experiment was at least twice independently reproduced. Row numbers are indicated on the right side of each image.
To investigate whether the specific kinase-substrate interactions in yeast also entailed phosphorylation of the protein substrate, we used the known interaction of Ser(P)-HPr with CcpA. Full-length CcpA was synthesized as prey (AD-CcpA) and bait (BD-CcpA). In agreement with the finding that CcpA forms dimers already in the absence of its co-repressor Ser(P)-HPr (26), AD-CcpA specifically interacted with BD-CcpA (data not shown). Interestingly, AD-CcpA only interacted with BD-HPr and BD-Crh when either ΔHprK/PV267F or HprK/P was co-expressed in the yeast cells (Fig. 1, A and B, 4th row). However, when the phosphorylation of BD-HPr and BD-Crh was prevented by the S46A replacement, no interaction with AD-CcpA occurred (Fig. 1, A and B, 4th row). These findings suggest that HprK/P phosphorylates BD-HPr and BD-Crh on their Ser-46 residue, thus allowing their interaction with AD-CcpA. This is in agreement with previous studies demonstrating that Ser(P)-HPr but not HPr and His(P)-HPr can bind to CcpA in vitro and function in CCR in vivo (9, 27–29). Therefore, this tri-hybrid assay reveals the phosphorylation-dependent formation of a binary complex (BD-Ser(P)-HPr and AD-CcpA) rather than formation of a ternary complex. It also provided evidence for HPr phosphorylation by EI, because the presence of 3HB-EI specifically prevented the interaction of AD-HprK/P with wild-type but not with H15A mutant BD-HPr (Fig. 1C, 8th row). These results are consistent with the finding that His-15 is part of the interface in the HPr-HprK/P complex (25). EI-catalyzed phosphorylation at His-15 probably weakens the HPr-HprK/P complex, which probably explains why in vitro HprK/P-mediated phosphorylation at Ser-46 is much slower with His(P)-HPr than with HPr (30, 31). Altogether, our results indicate that the ATP-dependent HprK/P and the PEP-dependent EI specifically phosphorylate HPr in yeast cells and establish that the yeast tri-hybrid assay is a sensitive approach to study the effect of phosphorylation on protein-protein interactions.
The expression of bacterial kinases exhibiting substrate-specific activities in yeast might be a useful methodology to identify yet unknown kinases. Recent studies of bacterial phosphoproteomes revealed a certain evolutionary conservation of the phosphorylated proteins and allowed the identification of the serine/threonine/tyrosine phosphorylation sites in a large number of proteins (32–36). However, in most cases the kinases that phosphorylate these proteins remain unknown. Our findings that kinases can form a long lived complex with a mutant substrate that cannot be phosphorylated open the door for a proteome-wide yeast two-hybrid strategy for the discovery of novel bacterial protein kinases. In a protein detected by phosphoproteome analyses, the phosphorylated residue(s) can be changed to prevent its phosphorylation, and the mutant protein can be used in a proteome-wide yeast two-hybrid screen to identify its cognate protein kinase, provided a long lived substrate-kinase complex will be formed.
HPr Interacts with the Transcriptional Regulator YesS
B. subtilis HPr was fused to the Gal4 DNA-binding domain (BD-HPr) and used in a yeast two-hybrid screen of a library of fragmented B. subtilis genomic DNAs to search for potential interacting partners (see under “Experimental Procedures”). Over 400 candidate colonies showing an interaction phenotype (Ade+, His+) were isolated, and the DNA inserts of prey plasmids from 120 positive candidates were sequenced. Remarkably, the vast majority of the prey proteins were in-frame fusions of the Gal4 activation domain to at least nine different overlapping fragments of the YesS protein, thus defining the core HPr binding domain of YesS (HBD, amino acids 406–510) (Fig. 2). The AD-YesS protein fragments interacted specifically with BD-HPr, suggesting that the HPr-YesS interaction likely has a biological significance.
YesS Interacts with Unphosphorylated HPr and Likely with His(P)-HPr
The bait protein BD-HPr synthesized in yeast is probably present in unphosphorylated form (as the cognate kinases are absent). We tried to determine the various forms of HPr interacting with YesS by using the tri-hybrid assay and also tested whether the HPr paralog Crh can bind to YesS. Two fragments of YesS (amino acids 404–510 and 305–511) carrying the HBD were used for the yeast tri-hybrid assays (Fig. 2). The AD-YesS fragments interacted with wild-type and the two mutant BD-HPr proteins and with BD-Crh (Fig. 1, A–C, 2nd and 3rd rows). A weak interaction was detected with BD-CrhS46A as yeast cells grew only under the less stringent screening conditions. Co-synthesis of the “constitutively” active kinase 3HB-ΔHprK/PV267F completely abolished the interaction of AD-YesS with all variants of BD-HPr and with BD-CrhS46A, whereas it only weakened the interaction with BD-Crh (Fig. 1A, 2nd and 3rd rows). The co-synthesis of wild-type 3HB-HprK/P also abolished the interaction of AD-YesS with BD-CrhS46A, but it did not affect the other binary interactions except that it weakened the interaction with BD-HPrS46A (Fig. 1B, 2nd and 3rd rows). These results indicate that AD-YesS interacts with the unphosphorylated forms of BD-HPr and BD-Crh and that phosphorylation of Ser-46 strongly weakens this interaction. The lack of interaction of the AD-YesS fragments with the nonphosphorylatable BD-HPrS46A in the presence of 3HB-ΔHprK/PV267F and with BD-CrhS46A in the presence of both 3HB-ΔHprK/PV267F and 3HB-HprK/P might result from trapping the binding domain proteins in a stable complex with the kinase, thus preventing their interaction with AD-YesS. This assumption is supported by the finding that AD-YesS interacts with BD-CrhS46A when 3HB-EI is co-synthesized in the cells (Fig. 1C, 2nd and 3rd rows). Crh cannot become phosphorylated at position 15 because of the natural His → Gln exchange. Finally, co-synthesis of 3HB-EI did not affect the binary interactions of AD-YesS with BD-HPr and BD-HPrH15A but weakened the interaction with BD-HPrS46A (Fig. 1C, 2nd and 3rd rows). This weaker interaction cannot simply be explained from the binary interaction data; it is likely that either phosphorylation of the mutant HPr by 3HB-EI or interaction of these two proteins specifically reduces the stability of the AD-YesS-BD-HPrS46A complex. As already discussed, phosphorylation of BD-HPr at His-15 prevented the interaction of wild-type BD-HPr with AD-HprK/P (Fig. 1C, 8th row). These results suggest that AD-YesS can also bind to BD-His(P)-HPr. However, because the activity of 3HB-EI in yeast might not be sufficient to convert all BD-HPr into BD-His(P)-HPr, it cannot be ruled out that the observed interaction in Fig. 1C stems partly from the remaining unphosphorylated BD-HPr.
Purified HBD of YesS Interacts with HPr and His(P)-HPr
To confirm the above preliminary findings that YesS likely interacts with HPr and His(P)-HPr but not Ser(P)-HPr by independent experimental evidence, we tried to overproduce the entire YesS and various fragments in E. coli. However, all formed insoluble inclusion bodies. To overcome this problem, a yesS fragment encoding the entire HBD (amino acids 403–515) fused to the C terminus of glutathione S-transferase (GST-HBD) was placed in an artificial operon with ptsH encoding HPr with a hexahistidine tag at its N terminus (His6-HPr, see “Experimental Procedures”). Upon induction with IPTG, GST-HBD and His6-HPr were simultaneously overproduced (Fig. 3A). When the soluble protein fractions were loaded on a Ni-NTA column, most of GST-HBD co-eluted with His6-HPr (Fig. 3, A and B). Conversely, His6-HPr co-purified with GST-HBD on a glutathione-Sepharose 4B column (data not shown), indicating that the two proteins form a stable complex. When aliquots from both purification procedures were separated by electrophoresis, coloration of GST-HBD was about three times stronger than that of His6-HPr (Fig. 3B, lane 1). As GST-HBD (39 kDa) is about three times bigger than His6-HPr (11 kDa), it is likely that one molecule of GST-HBD binds one molecule of His6-HPr. The mutated HPr alleles HPrH15D and HPrS46D, which mimic in vivo phosphorylation of HPr at His-15 and Ser-46, respectively (28), were co-expressed with GST-HBD. Interestingly, GST-HBD co-eluted with HPrH15D, although it did not with HPrS46D (Fig. 3A). In the latter case, GST-HBD was detected in insoluble inclusion bodies (data not shown). These findings support the yeast two-hybrid data that YesS interacts with HPr and His(P)-HPr but not with Ser(P)-HPr.
FIGURE 3.
Phosphorylation of HPr on Ser-46, but not on His-15, dissociates the HPr-YesS-HBD complex. A, His-tagged wild-type (WT), S46D, and H15D HPr proteins were co-expressed with GST-HBD in E. coli NM522. In each strain, the two proteins were expressed to similar extents (lanes 2–4). When the soluble extracts were loaded on a Ni-NTA column and HPr was eluted with 300 mm imidazole, co-elution of GST-HBD was observed only for wild-type and H15D HPr but not for S46D HPr (lanes 5–7). B–D, complex between GST-HBD and His6-HPr was purified from extracts of E. coli strain NM522 carrying the pGEX2T-HBD+HPr plasmid, using a Ni-NTA column. Aliquots of the GST-HBD-His6-HPr complex (∼450 μg of GST-HBD and ∼150 μg of His6-HPr) were incubated for 30 min at 37 °C in a 400-μl reaction mixture containing either control buffer (B), 5 mm PEP, and 20 μg of purified B. subtilis EI (C), and 2 mm ATP, 10 mm fructose 1,6-biphosphate, and 14 μg of purified ΔHprK/PV267F (D). The reaction mixtures were passed over Ni-NTA columns, and proteins retained on the column were washed twice to eliminate nonspecific interactions with the resin and finally eluted with buffer containing 300 mm imidazole. Aliquots of 50 μl (B and D) or 20 μl (C) of the untreated original sample (lane 1), the flow-through (lane 2), the wash (lanes 3 and 4), and elution fractions (lane 5) were analyzed by SDS-PAGE, and proteins were stained with Coomassie Blue. Each experiment was repeated twice independently, giving similar results. Gels with typical results are shown.
To investigate further the effect of phosphorylation of HPr on the interaction with GST-HBD, the purified His6-HPr-GST-HBD protein complex was incubated with either purified L. casei ΔHprK/PV267F to allow the formation of Ser(P)-HPr or with purified B. subtilis EI to allow the formation of His(P)-HPr (see under “Experimental Procedures”). To verify that both ΔHprK/PV267F and EI were active, aliquots of the reaction mixtures were separated on a 12.5% polyacrylamide gel under nondenaturing conditions. Under these conditions, Ser(P)-HPr and His(P)-HPr migrate faster than HPr, whereas the His6-HPr-GST-HBD complex migrated significantly slower. Coomassie Blue staining revealed that in the presence of ΔHprK/PV267F most of the His6-HPr was converted into His6-Ser(P)-HPr and released from the complex with GST-HBD. In contrast, in the presence of EI, most of the His6-HPr remained in complex with GST-HBD (data not shown). To confirm that in the latter case HPr complexed to GST-HBD was indeed phosphorylated by EI, a phosphorylation experiment with radioactive [32P]PEP was carried out. After separation on an SDS-polyacrylamide gel, a strong radioactive band migrating to the position of HPr was observed, suggesting that HPr in the His6-HPr-GST-HBD complex becomes indeed phosphorylated by EI and PEP (data not shown).
These functional kinase assays allowed us to study the effect of HPr phosphorylation on the stability of the His6-HPr-GST-HBD complex. The complex was treated with EI-PEP, HprK/P-ATP, and left untreated as a control. Samples were loaded on Ni-NTA columns, washed twice, and eluted with imidazole-containing buffer. The proteins in each fraction were analyzed by SDS-PAGE (Fig. 3, B–D). In the absence of any kinase as well as in the presence of EI, small amounts of GST-HBD and His6-HPr were detected in the flow-through (Fig. 3, B and C, lane 2). Whereas virtually no proteins were detected in the wash fractions (Fig. 3, B and C, lanes 3 and 4), both proteins were eluted with imidazole and were present in a similar proportion as in the sample loaded on the Ni-NTA column (compare lanes 1 and 5 in Fig. 3, B and C). Because under the conditions used EI converts most of the His6-HPr into His6-His(P)-HPr, these results suggest that phosphorylation of HPr on His-15 does not destabilize the interaction with the HBD of YesS. In contrast, upon phosphorylation of His6-HPr with ΔHprK/PV267F, the majority of GST-HBD eluted in the flow-through and wash fractions (Fig. 3D, lanes 2–4). Only a small part of GST-HBD co-eluted with His6-HPr (Fig. 3D, lane 5), which is likely due to an incomplete phosphorylation of HPr. These results indicate that phosphorylation of HPr on Ser-46 weakens or even prevents the interaction with the HBD of YesS. Altogether, the data obtained with the phosphomimetic mutants and the in vitro phosphorylation assays are qualitatively consistent with the yeast two- and tri-hybrid data, confirming that YesS interacts with both HPr and His(P)-HPr, whereas phosphorylation of HPr on its Ser-46 promotes dissociation of the YesS-HPr complex.
YesS Controls Expression of the Yes Operon
The yesS gene encodes a protein of unknown function composed of 761 amino acids, which is a putative transcriptional regulator of the AraC/XylS family. Most of the AraC/XylS family members are transcription activators, and some of them control carbon catabolic operons (AraC, regulator of the l-arabinose operon in E. coli) (37). Members of the huge AraC/XylS family (38) usually contain a well conserved ∼100-amino acid-long C-terminal region containing two helix-turn-helix motifs for DNA binding. The N-terminal and central regions of AraC/XylS regulators are not conserved and are presumed to bind small effector molecules, which in turn enable the regulator to interact with specific DNA sequences. In E. coli, AraC is activated by arabinose, and RhaS and RhaR, which regulate transcription of the rhaBAD and rhaSR operons, respectively, bind rhamnose (39). TxtR, which controls virulence in Streptomyces scabies, binds cellobiose (40). Interestingly, our results show that the central region of YesS binds HPr. To the best of our knowledge, this is the first report that an AraC/XylS member interacts with a regulatory protein.
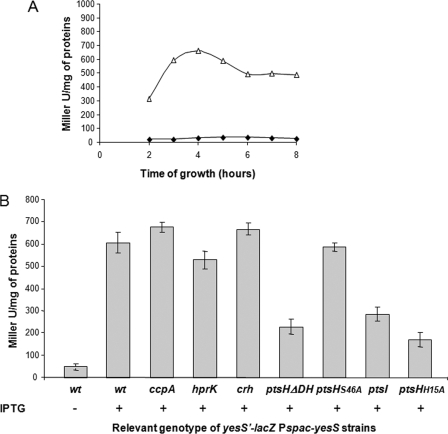
The chromosomal gene arrangement suggests that yesS belongs to a large operon (Fig. 4A). yesS is preceded by genes encoding a protein homologous to a glycosyltransferase from Enterococcus faecalis (yesR), proteins exhibiting similarity to a lactose-ABC transporter (yesO, yesP, and yesQ), and the sensor histidine kinase and response regulator of a two-component system (yesM-yesN, respectively) (Fig. 4A). yesS is followed by genes (yesT to yesZ) encoding proteins with similarity to degradative enzymes involved in utilization of rhamnogalacturonan, the backbone component of rhamnose-containing pectins (41). These genes seem to be organized in an operon, whose expression is induced by rhamnogalacturonan (41). To test whether YesS is a transcriptional regulator involved in the control of the large yes operon, including its own expression, the plasmid pMUTIN2-yesS was integrated in the B. subtilis 168 chromosome (Fig. 4B) to generate a yesS′-lacZ fusion and an entire yesS gene placed under control of the IPTG-inducible Pspac promoter (Fig. 4C). When this strain (TG130) was grown in LB medium supplemented with erythromycin, a very low background expression of lacZ was detected (Fig. 5A, filled diamonds). In contrast, upon addition of 0.1 mm IPTG, a significant increase of lacZ expression was detected during exponential growth, which reached a maximum at A ∼0.7, 4 h after IPTG addition (Fig. 5A, open triangles). This result demonstrates that yesS encodes a positive regulator that controls its own expression.
FIGURE 5.
YesS transcriptional activity is stimulated by His(P)-HPr. A, expression of the yesS′-lacZ fusion during growth of B. subtilis 168 in the absence (filled diamonds) or presence (open triangles) of 0.1 mm IPTG. β-Galactosidase activities were measured as described under “Experimental Procedures.” B, level of yesS′-lacZ expression was measured in the wild-type (wt) 168 strain and in isogenic mutant strains grown to A = 0.7 after 4 h of induction with IPTG. Complete strain genotypes are indicated in Table 1. Mean values and standard deviations were calculated from at least three independent experiments.
To test whether yesS expression is under CcpA/Ser(P)-HPr-mediated CCR control, we compared the expression of the yesS′-lacZ fusion in isogenic ccpA− and ccpA+ strains (strains TG130 and TG131, respectively, see Table 1). No significant difference was observed independently of whether the strains were grown in the presence or absence of glucose (Fig. 5B and data not shown). A similar observation was made with an hprK-inactivated strain (Fig. 5B). These results indicate that CcpA/Ser(P)-HPr-mediated CCR plays no major role in yesS expression.
His(P)-HPr Stimulates YesS Activity
The roles of HPr and Crh in the regulation of yesS expression were also investigated. Disruption of crh (strain TG133) had no effect on yesS′-lacZ expression compared with the wild-type strain (Fig. 5B). In contrast, the ptsHΔDH mutation corresponding to an in-frame deletion of codons 7–42 (strain TG134) led to a 2.5-fold decrease of yesS′-lacZ expression. These results suggest that HPr or one of its phospho-forms, but not Crh, plays a positive role in yesS expression. To determine which form of HPr was implicated in the activation of yesS expression, a strain harboring the ptsH1 allele encoding a mutant HPr, in which Ser-46 is replaced with Ala, was constructed (strain TG135). The absence of a phosphorylatable Ser-46 residue did not affect β-galactosidase activity (Fig. 5B), indicating that Ser(P)-HPr does not stimulate the expression of yesS. To determine the role of His(P)-HPr in yesS expression, the effects of a ptsI gene disruption and of the ptsHH15A mutation were tested (strains TG136 and TG137, respectively). Both mutations reduced the β-galactosidase activity to a level similar to that measured in the ptsHΔDH deletion mutant (Fig. 5B). Thus, the presence of HPr molecules unphosphorylated at His-15 because of the absence of EI or because of the replacement of the His-15 residue with an alanine lowers the transcriptional activity of YesS. These findings suggest that His(P)-HPr stimulates the transcriptional activation mediated by YesS. They are in agreement with the observation that the absence of Crh, in which His-15 is naturally replaced with a glutamine, had no effect on YesS activity.
Utilization of Pectin and Expression of the yesS Operon
B. subtilis 168 grows in liquid C mineral medium supplemented with either 0.5% pectin or rhamnogalacturonan or 0.2% polygalacturonic acid, whereas the growth of the ptsHΔDH strain GR14 is completely abolished and that of the ptsI null strain GR26 is impaired (Fig. 6A). These results indicate that efficient pectin utilization by B. subtilis requires the presence of His(P)-HPr. To test the role of His(P)-HPr on the transcription of the pectin-inducible yesS operon, RT-PCR experiments were performed on total RNA extracted from wild-type and mutant strains grown in LB with or without pectin 0.5% (Fig. 6B). RT-PCR amplification of a 450-bp DNA fragment containing the yesS-yesT intergenic region was detected only with the wild-type strain grown in the presence of pectin (compare Fig. 6B, lanes 4 and 10). This confirmed that yesS transcription is induced by pectin and that yesS and yesT are co-transcribed, further supporting the organization of the yesS to yesZ genes in a single transcription unit. Interestingly, yesS expression could not be detected from pectin-grown ptsH and ptsI mutant strains (Fig. 6B, lanes 6 and 8), suggesting that the presence of His(P)-HPr is required for the transcription of the yes operon. However, we do not know whether the other operons involved in pectin utilization (41) are subjected to the same control.
FIGURE 6.
Pectin utilization and pectin-induced His(P)-HPr-mediated transcription of the yes operon. A, growth of wild-type (WT, squares), ptsHΔDH (GR14, triangles), and ptsI::spc (GR26, circles) strains in C mineral medium supplemented with pectin 0.5% (w/v). B, RT-PCR performed on RNA samples isolated from wild-type (WT), ptsHΔDH (GR14) and ptsI::spc (GR26) strains grown in LB medium supplemented with pectin as indicated. The amplified region covers parts of yesS and yesT and is indicated with a black bar in Fig. 4A. The RT-PCR products were separated on an agarose gel, stained with ethidium bromide, and photographed. For each strain, two experiments are shown as follows: −, a negative control using RNA as template (without RT); and +, RT-PCR. t+ is a positive control using B. subtilis 168 genomic DNA as a template.
Conclusions
Taken together, our results suggest that YesS is a transcriptional activator involved in pectin/rhamnogalacturonan utilization. Moreover, although both HPr and His(P)-HPr interact with YesS, we found that only His(P)-HPr efficiently stimulates the transcriptional activation mediated by YesS and allows the cell to grow on rhamnogalacturonan precursors. YesS is therefore a new member of proteins that interact with both the unphosphorylated and phosphorylated forms of a regulatory protein partner. Known examples include E. coli glycogen phosphorylase, which interacts with HPr as well as His(P)-HPr (42). Interaction with HPr enhances the activity of this enzyme about 2.5-fold. However, although His(P)-HPr exhibits a 4-fold higher affinity for glycogen phosphorylase than HPr, it does not stimulate the activity of this enzyme. In E. coli, the two forms of HPr are much more abundant than glycogen phosphorylase, and it is therefore likely that this enzyme is always complexed with either HPr or His(P)-HPr. Physiological changes leading to an alteration of the HPr/His(P)-HPr ratio are therefore expected to also change the activity of glycogen phosphorylase, thus linking glycogen metabolism to the central regulation of carbon metabolism (42). Another example is the interaction between adenylyl cyclase and EIIAGlc. Genetic experiments suggested that in E. coli and other enterobacteriaceae P∼EIIAGlc stimulate the activity of the cAMP-synthesizing adenylyl cyclase. CRP complexed with cAMP regulates the expression of numerous catabolic operons (1). In vitro experiments with membrane-tethered adenylyl cyclase had revealed that EIIAGlc as well as P∼EIIAGlc interact with the cyclase, but only P∼EIIAGlc in the presence of a not yet identified cellular component seems to be able to stimulate its activity (43). Finally, B. subtilis HPr and Ser(P)-HPr were recently reported to interact with the glyceraldehyde-3-P dehydrogenase GapA (13). However, only Ser(P)-HPr was found to inhibit the activity of the enzyme. A potential interaction of His(P)-HPr with GapA has not been studied.
All these regulatory systems, including the control of the transcription activator YesS described here, are based on the interaction of the target protein with both the dephospho- and phospho-form of their regulator protein and on the sensitivity of the target protein to only one form of the regulator. With HPr or EIIAGlc probably being more abundant than any of their target proteins, the latter will be present in the cell mainly in complex with their dephospho- and phospho-regulators. The ratio of activated to nonactivated (or inhibited to noninhibited) target protein will therefore be strictly related to the ratio of dephospho- to phospho-regulator and their affinities for the target protein, which can differ, as has been shown for glycogen phosphorylase (42). This type of regulation probably allows a very subtle fine-tuning of protein/enzyme functions. The phosphorylation state of the PTS proteins primarily responds to the presence of a PTS substrate in the medium and to the PEP to pyruvate ratio in the cell (44).
HPr-mediated YesS regulation probably serves as a novel type of CCR. If glucose is taken up, the fraction of HPr present in the dephospho form is much larger than the fraction present as His(P)-HPr (9). Under these conditions, YesS will mainly be complexed with HPr and therefore be present in the less active form. Glucose and probably other rapidly metabolizable carbon sources will therefore inhibit the expression of YesS-controlled genes, which do not appear to be submitted to CCR via CcpA/Ser(P)-HPr.
Acknowledgments
We are very grateful to Marie-Françoise Noirot-Gros for invaluable help with the tri-hybrid assays and to Etienne Dervyn, Sophie Brinster, Carine Ganem, and Berengère Dalmais for stimulating discussions.
This work was supported in part by European BaSysBio Project LSHG-CT-2006-037469 (to P. N.) and in part by funding from the INRA Programme Transversalité-AGMIAL (to P. N. and J. D.).
E. Darbon and J. Deutscher, unpublished results.
- CCR
- carbon catabolite repression
- PEP
- phosphoenolpyruvate
- EI
- enzyme I
- PTS
- phosphotransferase system
- HBD
- HPr-binding domain
- RT
- reverse transcription
- IPTG
- isopropyl β-d-thiogalactopyranoside
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1.Deutscher J., Francke C., Postma P. W. (2006) Microbiol. Mol. Biol. Rev. 70, 939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Görke B., Stülke J. (2008) Nat. Rev. Microbiol. 6, 613–624 [DOI] [PubMed] [Google Scholar]
- 3.Kravanja M., Engelmann R., Dossonnet V., Blüggel M., Meyer H. E., Frank R., Galinier A., Deutscher J., Schnell N., Hengstenberg W. (1999) Mol. Microbiol. 31, 59–66 [DOI] [PubMed] [Google Scholar]
- 4.Yoshida K., Kobayashi K., Miwa Y., Kang C. M., Matsunaga M., Yamaguchi H., Tojo S., Yamamoto M., Nishi R., Ogasawara N., Nakayama T., Fujita Y. (2001) Nucleic Acids Res. 29, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorca G. L., Chung Y. J., Barabote R. D., Weyler W., Schilling C. H., Saier M. H., Jr. (2005) J. Bacteriol. 187, 7826–7839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monedero V., Yebra M. J., Poncet S., Deutscher J. (2008) Res. Microbiol. 159, 94–102 [DOI] [PubMed] [Google Scholar]
- 7.Galinier A., Haiech J., Kilhoffer M. C., Jaquinod M., Stülke J., Deutscher J., Martin-Verstraete I. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8439–8444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher M. A., Seidel G., Hillen W., Brennan R. G. (2006) J. Biol. Chem. 281, 6793–6800 [DOI] [PubMed] [Google Scholar]
- 9.Monedero V., Poncet S., Mijakovic I., Fieulaine S., Dossonnet V., Martin-Verstraete I., Nessler S., Deutscher J. (2001) EMBO J. 20, 3928–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig H., Rebhan N., Blencke H. M., Merzbacher M., Stülke J. (2002) Mol. Microbiol. 45, 543–553 [DOI] [PubMed] [Google Scholar]
- 11.Plamondon P., Brochu D., Thomas S., Fradette J., Gauthier L., Vaillancourt K., Buckley N., Frenette M., Vadeboncoeur C. (1999) J. Bacteriol. 181, 6914–6921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cochu A., Roy D., Vaillancourt K., Lemay J. D., Casabon I., Frenette M., Moineau S., Vadeboncoeur C. (2005) Appl. Environ. Microbiol. 71, 1364–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pompeo F., Luciano J., Galinier A. (2007) J. Bacteriol. 189, 1154–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller W., Horstmann N., Hillen W., Sticht H. (2006) FEBS J. 273, 1251–1261 [DOI] [PubMed] [Google Scholar]
- 15.Guthrie C., Fink G. R. (1991) Guide to Yeast Genetics and Molecular Biology, (Guthrie C., Fink G. R. eds) pp. 3–21, Academic Press, Inc., San Diego, CA [Google Scholar]
- 16.Herro R., Poncet S., Cossart P., Buchrieser C., Gouin E., Glaser P., Deutscher J. (2005) J. Mol. Microbiol. Biotechnol. 9, 224–234 [DOI] [PubMed] [Google Scholar]
- 17.Chaptal V., Vincent F., Gueguen-Chaignon V., Monedero V., Poncet S., Deutscher J., Nessler S., Morera S. (2007) J. Biol. Chem. 282, 34952–34957 [DOI] [PubMed] [Google Scholar]
- 18.James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noirot-Gros M. F., Dervyn E., Wu L. J., Mervelet P., Errington J., Ehrlich S. D., Noirot P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deutscher J., Reizer J., Fischer C., Galinier A., Saier M. H., Jr., Steinmetz M. (1994) J. Bacteriol. 176, 3336–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aymerich S., Gonzy-Tréboul G., Steinmetz M. (1986) J. Bacteriol. 166, 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darbon E., Servant P., Poncet S., Deutscher J. (2002) Mol. Microbiol. 43, 1039–1052 [DOI] [PubMed] [Google Scholar]
- 23.Vagner V., Dervyn E., Ehrlich S. D. (1998) Microbiology 144, 3097–3104 [DOI] [PubMed] [Google Scholar]
- 24.Platt T., Müller-Hill B., Miller J. H. (1972) in Experiments in Molecular Genetics (Miller J. H. ed) pp. 352–374, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25.Fieulaine S., Morera S., Poncet S., Monedero V., Gueguen-Chaignon V., Galinier A., Janin J., Deutscher J., Nessler S. (2001) EMBO J. 20, 3917–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miwa Y., Saikawa M., Fujita Y. (1994) Microbiology 140, 2567–2575 [DOI] [PubMed] [Google Scholar]
- 27.Deutscher J., Küster E., Bergstedt U., Charrier V., Hillen W. (1995) Mol. Microbiol. 15, 1049–1053 [DOI] [PubMed] [Google Scholar]
- 28.Reizer J., Bergstedt U., Galinier A., Küster E., Saier M. H., Jr., Hillen W., Steinmetz M., Deutscher J. (1996) J. Bacteriol. 178, 5480–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones B. E., Dossonnet V., Küster E., Hillen W., Deutscher J., Klevit R. E. (1997) J. Biol. Chem. 272, 26530–26535 [DOI] [PubMed] [Google Scholar]
- 30.Deutscher J., Engelmann R. (1984) FEMS Microbiol. Lett. 23, 157–162 [Google Scholar]
- 31.Reizer J., Sutrina S. L., Wu L. F., Deutscher J., Reddy P., Saier M. H., Jr. (1992) J. Biol. Chem. 267, 9158–9169 [PubMed] [Google Scholar]
- 32.Eymann C., Becher D., Bernhardt J., Gronau K., Klutzny A., Hecker M. (2007) Proteomics 7, 3509–3526 [DOI] [PubMed] [Google Scholar]
- 33.Lévine A., Vannier F., Absalon C., Kuhn L., Jackson P., Scrivener E., Labas V., Vinh J., Courtney P., Garin J., Séror S. J. (2006) Proteomics 6, 2157–2173 [DOI] [PubMed] [Google Scholar]
- 34.Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., Mann M. (2007) Mol. Cell. Proteomics 6, 697–707 [DOI] [PubMed] [Google Scholar]
- 35.Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 36.Soufi B., Gnad F., Jensen P. R., Petranovic D., Mann M., Mijakovic I., Macek B. (2008) Proteomics 8, 3486–3493 [DOI] [PubMed] [Google Scholar]
- 37.Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 3346–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egan S. M. (2002) J. Bacteriol. 184, 5529–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolin A., Balasubramaniam V., Skredenske J. M., Wickstrum J. R., Egan S. M. (2008) Mol. Microbiol. 68, 448–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joshi M. V., Bignell D. R., Johnson E. G., Sparks J. P., Gibson D. M., Loria R. (2007) Mol. Microbiol. 66, 633–642 [DOI] [PubMed] [Google Scholar]
- 41.Ochiai A., Itoh T., Kawamata A., Hashimoto W., Murata K. (2007) Appl. Environ. Microbiol. 73, 3803–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seok Y. J., Sondej M., Badawi P., Lewis M. S., Briggs M. C., Jaffe H., Peterkofsky A. (1997) J. Biol. Chem. 272, 26511–26521 [DOI] [PubMed] [Google Scholar]
- 43.Park Y. H., Lee B. R., Seok Y. J., Peterkofsky A. (2006) J. Biol. Chem. 281, 6448–6454 [DOI] [PubMed] [Google Scholar]
- 44.Hogema B. M., Arents J. C., Bader R., Eijkemans K., Yoshida H., Takahashi H., Aiba H., Postma P. W. (1998) Mol. Microbiol. 30, 487–498 [DOI] [PubMed] [Google Scholar]
- 45.Darbon E., Galinier A., Le Coq D., Deutscher J. (2001) J. Mol. Microbiol. Biotechnol. 3, 439–444 [PubMed] [Google Scholar]
- 46.Presecan-Siedel E., Galinier A., Longin R., Deutscher J., Danchin A., Glaser P., Martin-Verstraete I. (1999) J. Bacteriol. 181, 6889–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin-Verstraete I., Deutscher J., Galinier A. (1999) J. Bacteriol. 181, 2966–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]