Abstract
Lysyl hydroxylase 3 (LH3) is a multifunctional enzyme possessing lysyl hydroxylase, collagen galactosyltransferase, and glucosyltransferase (GGT) activities. We report here an important role for LH3 in the organization of the extracellular matrix (ECM) and cytoskeleton. Deposition of ECM was affected in heterozygous LH3 knock-out mouse embryonic fibroblasts (MEF+/−) and in skin fibroblasts collected from a member of a Finnish epidermolysis bullosa simplex (EBS) family known to be deficient in GGT activity. We show the GGT deficiency to be due to a transcriptional defect in one LH3 allele. The ECM abnormalities also lead to defects in the arrangement of the cytoskeleton in both cell lines. Ultrastructural abnormalities were observed in the skin of heterozygous LH3 knock-out mice indicating that even a moderate decrease in LH3 has deleterious consequences in vivo. The LH3 null allele in the EBS family member and the resulting abnormalities in the organization of the extracellular matrix, similar to those found in MEF+/−, may explain the correlation between the severity of the phenotype and the decrease in GGT activity reported in this family.
Lysyl hydroxylase (LH)2 catalyzes the post-translational formation of hydroxylysines in -X-Lys-Gly- sequences in collagens and other proteins with collagen-like domains (1, 2). Three lysyl hydroxylase isoforms (LH1, LH2, and LH3) have been identified from human, mouse, rat, and zebrafish (3–10). In addition, LH2 has two alternatively spliced forms, LH2a (short) and LH2b (long) (11). Several mutations of the LH1 gene (PLOD1) have been identified in patients with Ehlers-Danlos syndrome (EDS) type VIA, a heritable disorder characterized by kyphoscoliosis, joint laxity, skin fragility, and muscle hypotonia (12, 13). EDS VIA data indicate that LH1 hydroxylates helical cross-linking lysines of type I collagen in bone and type II collagen in cartilage (14, 15). Patients with Bruck syndrome, characterized by joint contractures, fragile bones, and osteoporosis, have mutations in the LH2 gene (PLOD2) that result in the complete absence of telopeptide hydroxylysine residues in bone collagens (16, 17). In addition, overexpression of LH2b has been linked to an increase in the hydroxylysine content of collagen telopeptides seen in fibrotic disorders (18).
LH3 differs from the other lysyl hydroxylase isoforms in that it possesses, in addition to lysyl hydroxylase activity (6, 19), hydroxylysyl galactosyltransferase and galactosylhydroxylysyl glucosyltransferase (GGT) activities (20–22), thus LH3 is able to catalyze the formation of glucosylgalactosylhydroxylysine residues. Recent studies show that LH3 is located not only in the endoplasmic reticulum but also in the extracellular space, and that the GGT activity in serum originates from LH3 (23). Furthermore, LH3 knock-out studies in mice demonstrate that the loss of LH3 leads to embryonic lethality due to disruption of the formation of basement membranes (24, 25). Analyses of LH3 knock-out embryos and embryonic fibroblasts indicate that the loss of hydroxylysine glycosylation prevents the assembly and secretion of type IV and VI collagens (26). Recently, mutations in the human LH3 gene (PLOD3) were shown to cause a severe connective tissue disorder with features that overlap with a number of collagen disorders (27).
The presence of LH3 in the intra- and extracellular space and the drastic consequences to the basement membrane when this enzyme is absent during early development suggest that LH3 plays an important role in the deposition of the ECM. To examine this role of LH3 in more detail, we have investigated the affects of the partial loss of LH3 on cells and tissues. We have examined heterozygous LH3 knock-out mice and mouse embryonic fibroblasts (MEF+/−) at the ultrastructural level as well as cells from a human model of LH3 deficiency, an epidermolysis bullosa simplex (EBS) family member known to exhibit a decrease in GGT activity. The decrease in GGT activity in a Finnish family with dominant EBS, accompanied by a markedly decreased urinary excretion of glucosylgalactosylhydroxylysine, was reported earlier (28). Among the families tested, this was the only family with a deficiency of GGT activity, indicating that this feature is unique to this family and does not represent a typical finding in EBS patients (28). In addition, Savolainen and co-workers (28) reported that the decrease in GGT activity correlated with the severity of the disease. We have re-examined a patient of the EBS family and determined that the decrease in GGT activity is due to a transcriptional defect in one LH3 allele.
Our results indicate that even a moderate decrease in the amount of intracellular LH3 results in a substantial decrease in LH3 secretion and causes changes in the deposition and organization of the ECM as well as the cytoskeletal arrangement of the cell. This was seen in tissues and cells of heterozygous LH3 knock-out mice, as well as in the EBS patient cells, both representing conditions where only one allele produces LH3. The results reveal an important role of LH3 in the organization of ECM.
EXPERIMENTAL PROCEDURES
Fibroblast Cultures
A fibroblast culture was established from a skin biopsy specimen taken from one patient belonging to the Finnish EBS family detailed elsewhere (28). The patient marked 10 in the pedigree was re-examined in this study. One control culture was kindly provided by Dr. Kaisa Tasanen-Määttä and two others have been locally established. All samples were obtained after informed consent, according to the Declaration of Helsinki. Heterozygous LH3 knock-out crosses were used to derive mouse embryonic fibroblasts as described earlier (26). All cells were maintained at 5% CO2 in Dulbecco's modified Eagle's medium with Glutamax (Invitrogen) supplemented with 10% fetal calf serum (Promocell), penicillin/streptomycin, and 50 μg/ml of ascorbic acid. The production of the heterozygous LH3 knock-out mice has been previously described (25). The animal experiments were approved by the Animal Care and Use Committee of the University of Oulu.
Transmission and Immunoelectron Microscopy
For transmission electron microscopy, skin and muscle samples from newborn and adult mice were fixed in 1% glutaraldehyde, 4% formaldehyde in 0.1 m phosphate buffer (pH 7.4), post-fixed in 1% osmium tetroxide, dehydrated in acetone, and embedded in Epon LX 112 (Ladd Research Industries, Williston, VT). Thin sections were cut with a Leica Ultracut UCT ultramicrotome and examined in a Philips CM100 transmission electron microscope. The diameters of 200–250 collagen fibrils were measured from thin sections of the skin of three newborn and adult heterozygous LH3 knock-out and wild-type mice using ITEM Soft Imaging Solutions (Olympus). Statistical analysis was performed using the Student's t test.
For immunoelectron microscopy, cells on confluent 58-cm2 dishes were fixed in 4% paraformaldehyde in a 0.1 m phosphate buffer (pH 7.3), embedded into gelatin, and immersed in 2.3 m sucrose. Thin cryosections were incubated with a polyclonal LH3 antibody (PLOD3 antibody, ProteinTech Group, Inc.) and then with a protein A-gold complex. The sections were examined as above.
GGT Activity Measurements
Cells on a confluent 58-cm2 dish were homogenized as described earlier (4). Culture medium collected after 72 h was concentrated to about 250 μl using an Amicon Ultra 10k centrifugal filter device (Millipore). The samples were used in the GGT activity assay based on the transfer of a tritium-labeled sugar from UDP-glucose (139 Ci/mol) to galactosyl hydroxylysyl residues in a calf skin gelatin substrate (29, 30).
RNA Isolation, Quantitative PCR, and Sequencing of Genomic DNA and cDNA
Total RNA was isolated from cultured skin fibroblasts with a TRIzol plus RNA purification kit (Invitrogen). cDNA was synthesized from total RNA with a cloned avian myeloblastosis virus first-strand cDNA synthesis kit (Invitrogen) in random hexamer or oligo(dT)20-primed reactions.
The expression level of human LH3 was detected using a TaqMan gene expression assay (Hs00153670_m1, PLOD3) (Applied Biosystems) and TaqMan Universal PCR Master Mix (Applied Biosystems) with a 7500 real time PCR system (Applied Biosystems) by the relative standard curve method. A eukaryotic 18 S rRNA endogenous control (Applied Biosystems) was used to normalize the quantities of LH3 expression.
Genomic DNA was isolated from cultured skin fibroblasts using proteinase K and phenol extraction (31). PCR amplification of the LH3 promoter sequence was performed with promoter and exon 1-specific oligonucleotides covering 1043 nucleotides upstream from the translation start site (32) (AF207069), and the cDNA was amplified with LH3-specific oligonucleotides covering the entire coding region and 303 nucleotides of the 5′- and 259 nucleotides of the 3′-untranslated regions (5, 6) (NM_001084). The overlapping PCR fragments were directly sequenced using the DYEnamic ET terminator cycle sequencing premix kit (GE Healthcare) and an ABI Prism 377 sequencer (PerkinElmer Life Sciences).
Immunofluorescence Staining
Cells grown on glass coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline. 95% Ethanol, 5% acetic acid was used for tubulin and vimentin analyses. After blocking with 1% bovine serum albumin, 0.1% saponin in phosphate-buffered saline, the cells were incubated with monoclonal anti-vimentin (Affinity BioReagents), anti-vimentin (Sigma), or monoclonal anti-α-tubulin (Sigma) primary antibodies diluted in blocking solution. In the collagen and fibronectin experiments, the cells were incubated with polyclonal rabbit anti-collagen type I (Rockland), polyclonal rabbit anti-collagen type VI (Rockland), and anti-human fibronectin (Sigma) in the absence of saponin, and nuclei were stained with Hoechst 33258 (Sigma). Alexa Fluor 488-conjugated anti-rabbit IgG, anti-mouse IgG, or anti-goat IgG (Invitrogen) antibodies were used for secondary detection. For actin microfilament staining, the cells were permeabilized with 1% Triton X-100 and incubated with Alexa Fluor 568 phalloidin (Invitrogen). The cells were embedded with Immu-Mount (Thermo Shandon) and examined using an Olympus epifluorescence microscope or an Olympus Fluoview 1000 confocal microscope.
Immunoblotting and Biosynthetic Labeling
LH3 was partially purified using UDP-hexanolamine-agarose (Sigma) as described earlier (23). Equal amounts of soluble protein from confluent 58-cm2 dishes and equal volumes of the 72-h culture media were analyzed on immunoblots using a mouse LH3 antibody (23) or a polyclonal LH3 antibody (PLOD3 antibody, ProteinTech Group, Inc.).
For collagen analysis, cells on the confluent 58-cm2 dish were washed twice with phosphate-buffered saline, scraped into homogenization buffer (0.2 m NaCl, 0.1 m glycine, 0.1% Triton X-100, 50 μm dithiothreitol, 10 mm EDTA, 20 mm Tris, pH 7.5) and lysed by brief sonication. The cell debris was removed by centrifugation at 15,000 × g for 15 min. The proteins in the 72-h serum-free culture medium were trichloroacetic acid-precipitated and redissolved in SDS sample buffer. The samples, 15 μl of cell lysate and 10 or 20 μl of the solubilized trichloroacetic acid precipitate, were separated by 7.5% SDS-PAGE and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore). Nonspecific binding was blocked with 5% nonfat milk powder in Tris-buffered saline-Tween. The membranes were incubated with polyclonal rabbit anti-collagen type I (Rockland) or polyclonal rabbit anti-collagen type VI (Rockland) and further with a horseradish peroxidase-conjugated anti-rabbit IgG (P.A.R.I.S) secondary antibody. Immunocomplexes were visualized by using ECL+ reagent (GE Healthcare) and Kodak X-AR films. Types I and VI collagen α chain levels were quantified with ImageQuant 5.2 software (GE Healthcare) from the films. The distribution of soluble collagen between cells and medium was compared with control cells and medium. Type VI collagen from EBS and control fibroblasts was biosynthetically labeled with [35S]methionine, immunoprecipitated, and detected as previously described (33).
RESULTS
Lack of One LH3 Allele Causes Ultrastructural Changes in the Skin of Heterozygous LH3 Knock-out Mice
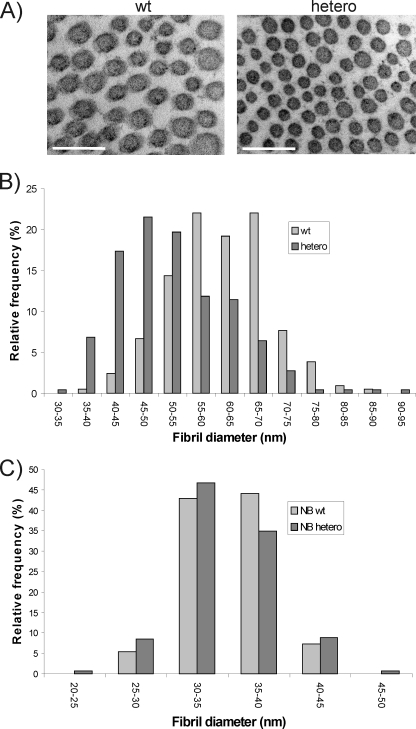
LH mutant mice showed ultrastructural alterations in their skin and muscle (25, 26). Therefore we examined these tissues in newborn and adult heterozygous LH3 knock-out mice by transmission electron microscopy. The muscle tissue had no obvious abnormalities (data not shown). However, the dermis appeared looser and more edematous than the wild-type dermis. Analysis of collagen cross-sections in adult skin showed that the mean fibril diameter was significantly (p < 0.001) decreased in heterozygous LH3 knock-out mice (Fig. 1, A and B) compared with wild-type, being 52 ± 10 and 61 ± 8 nm, respectively. In addition, the relative frequency of collagen fibrils with variable diameters in the skin of adult heterozygous LH3 knock-out mice differed from that in wild-type mice (Fig. 1B). In newborn skin, no variation in mean fibril diameter was observed between the genotypes. However, the skin of newborn heterozygous LH3 knock-out mice contained small diameter fibrils with higher frequency and also the range of the fibril diameter was wider than in wild-type skin, although the changes were not statistically significant (Fig. 1C). These data from the skin of heterozygous LH3 knock-out mice demonstrate that a moderate decrease in the amount of LH3 affects collagen fibrillogenesis in vivo.
FIGURE 1.
A, transmission electron microscopy analysis of collagen fibrils in skin of adult heterozygous LH3 knock-out (hetero) and wild-type (wt) mice. Scale bar, 200 nm. B and C, relative frequencies (%) of fibril diameters in the skin of adult (hetero) and newborn (NB hetero) heterozygous LH3 knock-out mice that differed from wild-type mice (wt or NB wt). The diameters of the collagen fibril cross-sections were measured from digital images using image analysis software.
Finnish EBS Patient Cell Line Represents One LH3 Allele Model in Human
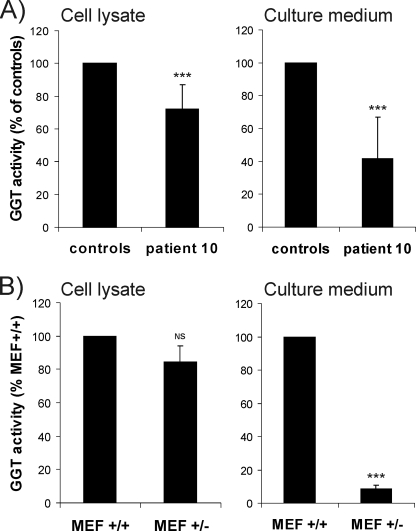
A Finnish family with EBS (28) was reported earlier to have reduced GGT activity and a markedly decreased urinary excretion of glucosylgalactosylhydroxylysine. We re-examined this family to determine whether a defect in LH3 could explain these observations. Our data (Fig. 2A) are consistent with previous results (28), showing a decrease in the GGT activity of skin fibroblasts (about 70% of control) for patient 10. The GGT activity measured in the 72-h culture medium from confluent cells was 42% of controls. A similar decrease in the GGT level (80% of normal mouse embryonic fibroblast (MEF+/+), Fig. 2B) was also found in our heterozygous LH3 knock-out mouse embryonic fibroblasts (MEF+/−). The GGT activity detected in the culture medium of LH3 knock-out MEF+/− (Fig. 2B) was only about 10% of that found in medium from control MEF+/+, indicating that a reduction of GGT/LH3 inside the cells markedly affected the secretion of activity from the cells. This finding is thus consistent with results obtained with EBS patient fibroblasts.
FIGURE 2.
GGT activity obtained from cell lysate and cell culture media of a (A) EBS patient and normal human skin fibroblasts, and (B) heterozygous LH3 knock-out (MEF+/−) and normal (MEF+/+) mouse embryonic fibroblasts. The value of control skin fibroblasts is the mean of three healthy individuals. Values represent the average ± S.D. of five measurements, calculated per micrograms of soluble protein and expressed as a percentage of control fibroblasts (A) or MEF+/+ (B). NS, not significant; ***, p < 0.001.
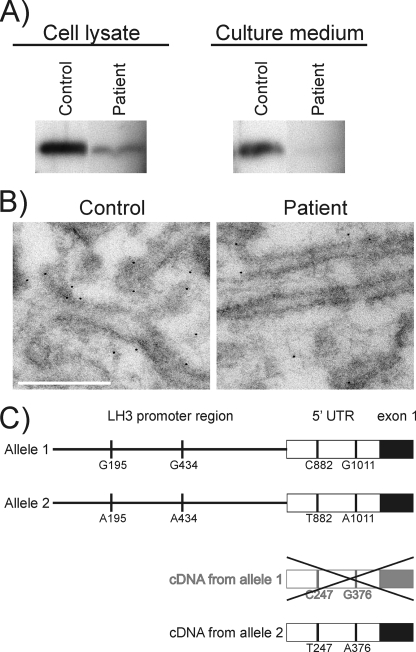
We have previously shown that LH3 is the main molecule responsible for GGT activity in mouse embryos and the LH3 protein level correlates with the GGT activity of adult mouse tissues (23, 25). Therefore the LH3 protein levels in EBS patient fibroblasts and cell culture media were assayed by immunoblotting and immunoelectron microscopy. On immunoblots, the amount of LH3 protein was reduced in EBS patient fibroblasts and also in the cell culture media (Fig. 3A). The molecular weight of LH3 did not differ between the control and EBS patient suggesting that glycosylation and processing of LH3 is normal in the EBS patient. In immunoelectron microscopy (Fig. 3B), LH3 was observed to line the endoplasmic reticulum membranes in both EBS patient and control cells, confirming earlier results from mouse tissues (23). When the gold particles in a 4.4-μm2 area of the endoplasmic reticulum were counted in 30 cells, fewer particles, about 57% of controls, were observed in the EBS fibroblasts, revealing a reduction in the amount of LH3 in these cells. Taken together, these results indicate that although there are no molecular weight differences between the LH3 proteins produced by the EBS patient and control cells, the quantity of LH3, measured either as GGT activity or as LH3 protein, was decreased in the EBS patient and the decrease was more pronounced in the cell culture medium.
FIGURE 3.
A, immunoblot analysis of LH3 in skin fibroblasts and culture medium of the EBS patient and a control. LH3 was partially purified from cell lysates that contained equal amounts of soluble protein. The cell culture media corresponding to the lysates were also analyzed by immunoblot. Similar results were obtained in three different experiments. B, immunoelectron microscopy analysis of LH3 in control and EBS patient fibroblasts. LH3 lines the endoplasmic reticulum membranes in both cell lines. Less LH3 is observed in EBS patient fibroblasts than in control fibroblasts. Scale bar, 300 nm. C, schematic presentation of LH3 alleles and cDNA in the EBS patient. Four polymorphic nucleotides, A195G, A434G, C882T, and A1011G, were observed in the promoter and 5′-untranslated (5′UTR) region of the LH3 gene (PLOD3) (AF207096) in the EBS patient. The figure shows the possible allelic distribution of the polymorphic nucleotides. The C882T change was present as T247 and the A1011G as A376 in the LH3 cDNA of the EBS patient suggesting that only one of the LH3 alleles (allele 2) is transcribed in the EBS patient.
To study the reason for the reduced amount of LH3 in EBS patient fibroblasts, the expression level of the LH3 gene (PLOD3) was analyzed by quantitative PCR, and the LH3 cDNA and promoter and exon 1 regions of the LH3 gene (PLOD3) were sequenced. Our data indicate that the LH3 mRNA level in the EBS patient was 67% (S.D. ± 11) of the control level. Sequencing of the LH3 cDNA did not reveal any mutations in the LH3 coding region in the EBS patient. In the genomic DNA (Fig. 3C), heterozygous A195G and A434G changes were observed upstream, and heterozygous C882T and A1011G changes were found downstream of the main transcription start site of the PLOD3 (AF207096) in the EBS patient. Changes at positions A195G, A434G, and A1011G were also found in the genomic DNA of healthy controls, which reveals them as polymorphic changes. The heterozygous A1011G change in the genomic DNA was present as A376 in the cDNA (NM_001084) of the EBS patient (Fig. 3C). Furthermore, LH3 cDNA of the EBS patient contained only T at position 247 corresponding to a heterozygous C882T in the genomic DNA (Fig. 3C). Our sequencing data thus indicate that only one of the LH3 alleles is transcribed in the EBS patient. This is in agreement with the reduced amount of LH3 mRNA seen in our quantitative PCR analysis. These findings explain the low level of GGT activity and LH3 protein in EBS patient fibroblasts, and thus represent a human heterozygous null model of LH3 gene (PLOD3).
Abnormalities in the Deposition and Organization of the ECM in Cells with a Decreased Amount of LH3
Our recent analysis of LH3 knock-out mouse embryonic fibroblasts have indicated that a loss of glycosylated hydroxylysines prevents intracellular tetramerization of type VI collagen and leads to impaired secretion of this collagen (26). Furthermore, underglycosylation of the type VI collagen causes aggregation and changes in collagen distribution in tissues (26). As indicated earlier, type VI collagen controls the organization of fibronectin (34), and furthermore, type VI collagen and fibronectin fibrils play a role in the fibrillogenesis of type I collagen (35, 36). Therefore these proteins were also analyzed in the heterozygous LH3 knock-out mouse embryonic fibroblasts (MEF+/−) and in EBS patient fibroblast cultures. Immunofluorescence staining was carried out with non-permeabilized cells cultured for 3 days after confluence to observe the ECM of the cells. In LH3 knock-out MEF+/−, the arrangement of the ECM components, type VI collagen, fibronectin, and type I collagen appeared abnormal when compared with the normal mouse embryonic fibroblasts (MEF+/+) (Fig. 4). Type VI collagen formed a fine and slightly interconnected three-dimensional network in LH3 knock-out MEF+/− (Fig. 4B), whereas a thick and heavily cross-linked network was observed in control MEF+/+ (Fig. 4A). Subsequently, an abnormal arrangement of fibronectin was also observed in the LH3 knock-out MEF+/− (Fig. 4F). In control MEF+/+, fibronectin formed a three-dimensional network of thick interconnected fibrils arranged in a nest-like pattern (Fig. 4E), whereas, in the LH3 knock-out MEF+/−, thin fibrils with few interconnections in the three-dimensional network were observed (Fig. 4F). Type I collagen was also abnormal in the LH3 knock-out MEF+/− (Fig. 4J); the fibrils were short and faint, differing from the type I collagen fibrils in normal MEF+/+ (Fig. 4I). The findings with the EBS patient fibroblasts (Fig. 4, D, H, and L) were similar to those of the LH3 knock-out MEF+/− that revealed abnormalities in the arrangements of fibronectin, and type VI and I collagens. These data suggest that even a moderate decrease in the amount of LH3 affects the deposition and organization of the ECM.
FIGURE 4.
Immunofluorescence analysis of type VI collagen, fibronectin, and type I collagen deposited in heterozygous LH3 MEF+/− and EBS patient fibroblasts. The deposition and organization of type VI collagen (B), fibronectin (F), and type I collagen (J) are abnormal in the LH3 knock-out MEF+/− compared with normal MEF+/+ (A, E, and I). Similar abnormalities were seen in the EBS patient fibroblasts (D, H, and L) compared with the control cells (C, G, and K). The findings were evident both in LH3 knock-out MEF+/− and EBS patient fibroblasts in two sets of experiments. Scale bar, 20 μm.
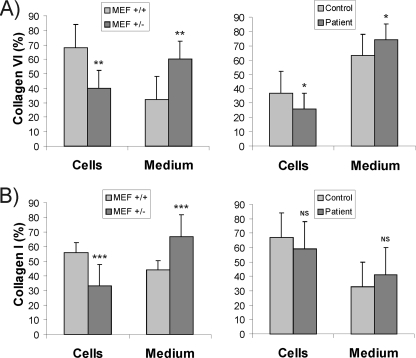
Immunoblot analysis was used to further examine the distribution of soluble type VI and I collagen α-chains between the cell lysate and the 72-h culture medium. Differences were seen in collagen distribution in both the LH3 knock-out MEF+/− and the EBS patient fibroblast cultures (Fig. 5). In the LH3 knock-out MEF+/− culture, quantification of the collagen band intensities on immunoblots revealed that, on average, 60% of the α(VI) chains (Fig. 5A) and 67% of the α(I) chains (Fig. 5B) were soluble in the medium, whereas in normal MEF+/+ cultures, on average, 32 and 44% were soluble, respectively. A similar tendency in the distribution of these collagens was seen in the EBS patient fibroblast culture (Fig. 5, A and B). However, type VI and I collagen α-chains of the LH3 knock-out MEF+/− and the EBS fibroblasts migrated normally on SDS-PAGE (not shown). In addition, type VI collagen formed tetramers that were secreted from the EBS fibroblasts with no obvious abnormalities when compared with the control (not shown).
FIGURE 5.
The distribution (%) of soluble (A) type VI and (B) type I collagen α-chains between the cell lysate and culture medium of heterozygous LH3 MEF+/− and EBS patient fibroblast cultures. The amount of the collagen α-chains was quantified from immunoblots and the values represent the average ± S.D. of three experiments. More soluble collagen was observed in the culture media of the LH3 knock-out (MEF+/−) and the EBS patient fibroblast cultures than in media from normal MEF+/+ and control fibroblast cultures, respectively. NS, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
In conclusion, our results revealed no marked differences in molecular weight, i.e. in post-translational modifications of lysine residues, in type VI and I collagens. Nevertheless, these collagens were found in abnormally large amounts in the medium as a soluble form. Furthermore, immunofluorescence data revealed that collagen deposition was clearly reduced compared with the deposition in the control cultures. Our data thereby indicate that in LH3 knock-out MEF+/− and EBS patient fibroblast cultures types VI and I collagens are not deposited normally into the extracellular matrix.
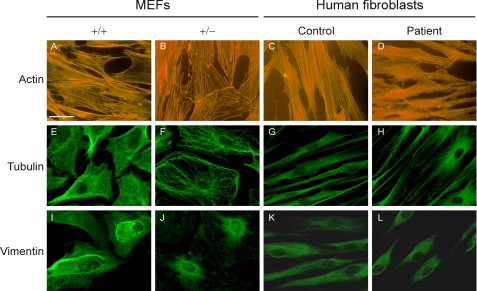
Decreased LH3 in Cultured Cells Causes Abnormalities in the Organization of Cytoskeleton Components
When fibroblasts with decreased LH3 (LH3 knock-out MEF+/− and EBS patient fibroblasts) were cultured in vitro, the cell shape differed from the corresponding control fibroblasts under light microscopy. LH3 knock-out MEF+/− and EBS patient fibroblasts appeared smaller and rounder at confluence, and were not in such close contact with each other compared with the control cells. To visualize the differences, the major cytoskeletal components of fibroblasts, actin microfilaments, tubulin microtubules, and vimentin intermediate filaments were examined by immunofluorescence. As seen in Fig. 6, the cytoskeleton of LH3 knock-out MEF+/− and EBS patient fibroblasts differed from that of the corresponding controls. In control cells, the actin stress fibers extended from one end to the other along the long axis of the cell (Fig. 6, A and C). In fibroblasts with decreased LH3 (Fig. 6, B and D), they appeared tangled and thinner and did not always extend the whole long axis of the cell. Microtubules were also faint in fibroblasts with decreased LH3 (Fig. 6, F and H), and appeared curlier than in controls (Fig. 6, E and G). The number of vimentin intermediate filaments appeared lower in fibroblasts with decreased LH3 (Fig. 6, J and L), and the filaments were shorter than those in control cells (Fig. 6, I and K), covering only the area surrounding the nucleus. Our data thus indicate that a decrease in LH3 is reflected in changes in cytoskeletal organization.
FIGURE 6.
Immunofluorescence analysis of actin microfilaments, tubulin microtubules, and vimentin intermediate filaments in heterozygous LH3 MEF+/− and EBS patient fibroblast. A divergent organization of actin (B and D), tubulin (F and H), and vimentin (J and L) were seen in cells with decreased LH3 compared with the normal cells (A and C, E and G, and I and K), respectively. The finding was evident both in LH3 knock-out MEF+/− and EBS patient fibroblasts in two sets of experiments. Scale bar, 20 μm.
DISCUSSION
LH3 is a multifunctional enzyme, possessing lysyl hydroxylase (5), hydroxylysyl galactosyltransferase (21), and galactosylhydroxylysyl glucosyltransferase (20) activities both in vitro and in vivo (25). Knocking out the LH3 gene (Plod3) from the mouse genome eliminates GGT activity in mouse embryos and causes embryonal lethality (25). As discussed earlier (26), it is probable that LH1 and LH2 to some extent compensate for the lack of LH activity of LH3. However, there are apparently some lysine residues that are specific targets for LH3, because underglycosylation and abnormalities in collagen distribution were observed in our LH mutant mice (25, 26). These findings suggested that the moderate decrease of LH3 observed in heterozygous LH3 knock-out mouse embryonic fibroblasts (MEF+/−) and mice might be sufficient to cause abnormal deposition of the extracellular matrix. In addition, we re-examined a member of the Finnish EBS family with decreased GGT activity (28), to determine whether a defect in LH3 is responsible for the GGT deficiency. If so, then the cells would serve as a human model for moderately decreased LH3.
In the current study, we have shown that the LH3 protein level is decreased in the EBS patient with decreased GGT activity. The data confirm our earlier findings that LH3 is responsible for GGT activity in vivo (25). The reduction of LH3 was more evident in the cell media from both in EBS patient fibroblast and LH3 knock-out MEF+/− cultures, indicating that whereas the amount of LH3 secreted into the medium is dependent on the amount of LH3 present in the cells, the association is not direct. A small decrease in the amount of intracellular LH3 causes a much larger reduction in LH3 secretion. Our results reveal that only one LH3 allele produces a transcript in the EBS patient, which explains the low level of LH3 mRNA, GGT activity, and LH3 protein. This is the first heterozygous LH3 null case found in humans, although the reason for the LH3 allele silencing remains unknown.
Our results from the LH3 knock-out MEF+/−, as well as from the EBS patient fibroblast cultures indicate that the reduced amount of LH3 alters extracellular matrix organization. Abnormalities were observed in the organization of type VI collagen, fibronectin, and type I collagen. Type VI collagen normally forms a beaded microfibrillar network and is widely expressed in tissues (37). It is a unique collagen, because it forms tetramers inside cells prior to secretion (38) and nearly all the lysine residues in the α1(VI), α2(VI), and α3(VI) chains are hydroxylated and further glycosylated (39). As shown here, the moderate reduction of LH3 does not have a dramatic effect on lysine hydroxylation and glycosylation in type VI collagen, because size differences were not observed by SDS-PAGE compared with controls. Furthermore, the type VI collagen, studied in more detail in the EBS patient fibroblasts, formed tetramers, which were secreted into the cell medium. Nevertheless, we observed abnormal type VI collagen deposition into filament networks in the ECM in both cell lines with moderately decreased LH3, suggesting that some alterations in hydroxylation and/or glycosylation may change the normal deposition of type VI collagen. Therefore, our results further establish the crucial role of the LH3-dependent modification of type VI collagen in its normal assembly to the beaded microfibrillar network in the ECM. It has been shown elsewhere that a total lack or dramatic reduction in the expression of type VI collagen alters the organization of fibronectin fibrils in fibroblast cultures (34, 40). It was noteworthy that fibronectin organization was also impaired both in LH3 knock-out MEF+/− and EBS patient fibroblasts. Our data thus further establish the link between the organization of type VI collagen and fibronectin networks. Furthermore, type VI collagen has been shown to bind directly to type I collagen, thus affecting collagen fibril formation (36). In addition, interactions of type I collagen molecules with fibronectin and specific integrins are also required in fibrillogenesis as demonstrated in cell cultures by others (35, 41, 42). Our results suggest that even small changes in the type VI collagen network, probably due to small alterations in glycosylation of hydroxylysine residues, are enough to affect the organization of fibronectin fibrils and, directly or via the fibronectin network, type I collagen fibrillogenesis in the ECM.
We observed more round cell shape both in the LH3 knock-out MEF+/− and the EBS patient fibroblasts in culture, and analysis of cytoskeleton components revealed changes in the organization of actin microfilaments, tubulin microtubules, and vimentin intermediate filaments. The polymerization of ECM molecules has been shown to regulate the organization of the cytoskeleton (43, 44), therefore the impaired extracellular protein organization seen in LH3 knock-out MEF+/− and EBS patient fibroblasts may cause the changes seen in the cytoskeleton. In addition, the ECM transmits environmental signals via integrins and other cell receptors to the cytoskeleton and these signals affect, for example, cell proliferation, differentiation, and death (45, 46). Our data suggest that LH3 is a key enzyme in regulation of the deposition of ECM components, and thereby also transmits signals to the cytoskeleton. It is probable that glycosylation by LH3 is an important extracellular factor in the regulation of the extracellular matrix and cytoskeletal arrangements as seen in LH3 knock-out MEF+/− and EBS patient fibroblasts. This is supported by our recent data with HT-1080 cells (47) indicating that a deficiency of LH3 glycosyltransferase activities, especially in the extracellular space, results in abnormal cell morphology, abnormalities in the organization of cytoskeletal proteins, and arrest of cell growth followed by cell death.
The crucial role of glycosyltransferase activities of the multifunctional LH3 is also clearly seen in vivo in our LH3 knock-out mice, which die at an early developmental stage primarily due to lack of GGT activity (25), as well as in zebrafish, where motor growth cone migration is critically dependent on GGT activity of LH3 (9). It is important to note, that although heterozygous LH3 knock-out mice appear externally normal, they demonstrated ultrastructural changes in the skin, indicating that a moderate decrease in LH3 affects organization of the ECM in tissues. Recent studies have indicated that LH3 mutations are linked to skin abnormalities. Skin blisters that resemble epidermolysis bullosa were one of the unique features seen in a patient with compound heterozygous mutations of LH3, which caused a decrease of GGT activity to less than 20% of normal (27). In addition, hypomorphic LH3 knock-out mouse embryos, having 11–20% of normal GGT activity, developed blisters at E12.5 and thereafter (25). Our findings, together with observations that LH3 is associated with skin blisters, suggest that the moderately decreased LH3 activity in the EBS patient could also cause changes in the extracellular matrix of skin and thereby be a reason for the more severe phenotype of EBS seen in patients with decreased GGT activity (28).
Acknowledgments
We gratefully acknowledge Piia Mäkelä, Mervi Matero, the staff of the dermatology clinic of Kainuu Central Hospital, and the staff of the EM core facility of Biocenter Oulu for expert technical assistance. We thank docent Sinikka Eskelinen for help with the confocal microscope.
This work was supported by grants from the Research Council for Biosciences and Environment within the Academy of Finland, the Sigrid Juselius Foundation, Biocenter Oulu, Glycoscience Graduate School, Research and Science Foundation of Farmos, Societas Scientiarum Fennica, and the Maud Kuistila Memorial Foundation.
- LH
- lysyl hydroxylase
- GGT
- glucosyltransferase
- MEF
- mouse embryonic fibroblasts
- EBS
- epidermolysis bullosa simplex
- ECM
- extracellular matrix.
REFERENCES
- 1.Kivirikko K. I., Myllylä R., Pihlajaniemi T. (1992) in Post-translational Modifications of Proteins (Harding J. J., Crabbe J. C. eds) pp. 1–51, CRC Press, Boca Raton, FL [Google Scholar]
- 2.Kivirikko K. I., Pihlajaniemi T. (1998) Adv. Enzymol. Relat. Areas Mol. Biol. 72, 325–398 [DOI] [PubMed] [Google Scholar]
- 3.Hautala T., Byers M. G., Eddy R. L., Shows T. B., Kivirikko K. I., Myllylä R. (1992) Genomics 13, 62–69 [DOI] [PubMed] [Google Scholar]
- 4.Valtavaara M., Papponen H., Pirttilä A. M., Hiltunen K., Helander H., Myllylä R. (1997) J. Biol. Chem. 272, 6831–6834 [DOI] [PubMed] [Google Scholar]
- 5.Valtavaara M., Szpirer C., Szpirer J., Myllylä R. (1998) J. Biol. Chem. 273, 12881–12886 [DOI] [PubMed] [Google Scholar]
- 6.Passoja K., Rautavuoma K., Ala-Kokko L., Kosonen T., Kivirikko K. I. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10482–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruotsalainen H., Sipilä L., Kerkelä E., Pospiech H., Myllylä R. (1999) Matrix Biol. 18, 325–329 [DOI] [PubMed] [Google Scholar]
- 8.Mercer D. K., Nicol P. F., Kimbembe C., Robins S. P. (2003) Biochem. Biophys. Res. Commun. 307, 803–809 [DOI] [PubMed] [Google Scholar]
- 9.Schneider V. A., Granato M. (2006) Neuron 50, 683–695 [DOI] [PubMed] [Google Scholar]
- 10.Schneider V. A., Granato M. (2007) Matrix Biol. 26, 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeowell H. N., Walker L. C. (1999) Matrix Biol. 18, 179–187 [DOI] [PubMed] [Google Scholar]
- 12.Yeowell H. N., Walker L. C. (2000) Mol. Genet. Metab. 71, 212–224 [DOI] [PubMed] [Google Scholar]
- 13.Steinmann B., Eyre D. R., Superti-Furga A. (2002) in Connective Tissue and Its Heritable Disorders (Royce P. M., Steinmann B. eds) 2nd Ed., pp. 431–523, Wiley-Liss Inc., New York [Google Scholar]
- 14.Eyre D., Shao P., Weis M. A., Steinmann B. (2002) Mol. Genet. Metab. 76, 211–216 [DOI] [PubMed] [Google Scholar]
- 15.Uzawa K., Yeowell H. N., Yamamoto K., Mochida Y., Tanzawa H., Yamauchi M. (2003) Biochem. Biophys. Res. Commun. 305, 484–487 [DOI] [PubMed] [Google Scholar]
- 16.van der Slot A. J., Zuurmond A. M., Bardoel A. F., Wijmenga C., Pruijs H. E., Sillence D. O., Brinckmann J., Abraham D. J., Black C. M., Verzijl N., DeGroot J., Hanemaaijer R., TeKoppele J. M., Huizinga T. W., Bank R. A. (2003) J. Biol. Chem. 278, 40967–40972 [DOI] [PubMed] [Google Scholar]
- 17.Ha-Vinh R., Alanay Y., Bank R. A., Campos-Xavier A. B., Zankl A., Superti-Furga A., Bonafé L. (2004) Am. J. Med. Genet. A 131, 115–120 [DOI] [PubMed] [Google Scholar]
- 18.van der Slot A. J., Zuurmond A. M., van den Bogaerdt A. J., Ulrich M. M., Middelkoop E., Boers W., Karel Ronday H., DeGroot J., Huizinga T. W., Bank R. A. (2004) Matrix Biol. 23, 251–257 [DOI] [PubMed] [Google Scholar]
- 19.Valtavaara M., Szpirer C., Szpirer J., Myllylä R. (1998) J. Biol. Chem. 273, 12881–12886 [DOI] [PubMed] [Google Scholar]
- 20.Heikkinen J., Risteli M., Wang C., Latvala J., Rossi M., Valtavaara M., Myllylä R. (2000) J. Biol. Chem. 275, 36158–36163 [DOI] [PubMed] [Google Scholar]
- 21.Wang C., Luosujärvi H., Heikkinen J., Risteli M., Uitto L., Myllylä R. (2002) Matrix Biol. 21, 559–566 [DOI] [PubMed] [Google Scholar]
- 22.Myllylä R., Wang C., Heikkinen J., Juffer A., Lampela O., Risteli M., Ruotsalainen H., Salo A., Sipilä L. (2007) J. Cell. Physiol. 212, 323–329 [DOI] [PubMed] [Google Scholar]
- 23.Salo A. M., Wang C., Sipilä L., Sormunen R., Vapola M., Kervinen P., Ruotsalainen H., Heikkinen J., Myllylä R. (2006) J. Cell. Physiol. 207, 644–653 [DOI] [PubMed] [Google Scholar]
- 24.Rautavuoma K., Takaluoma K., Sormunen R., Myllyharju J., Kivirikko K. I., Soininen R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14120–14125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruotsalainen H., Sipilä L., Vapola M., Sormunen R., Salo A. M., Uitto L., Mercer D. K., Robins S. P., Risteli M., Aszodi A., Fässler R., Myllylä R. (2006) J. Cell Sci. 119, 625–635 [DOI] [PubMed] [Google Scholar]
- 26.Sipilä L., Ruotsalainen H., Sormunen R., Baker N. L., Lamandé S. R., Vapola M., Wang C., Sado Y., Aszodi A., Myllylä R. (2007) J. Biol. Chem. 282, 33381–33388 [DOI] [PubMed] [Google Scholar]
- 27.Salo A. M., Cox H., Farndon P., Moss C., Grindulis H., Risteli M., Robins S. P., Myllylä R. (2008) Am. J. Hum. Genet. 83, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savolainen E. R., Kero M., Pihlajaniemi T., Kivirikko K. I. (1981) N. Engl. J. Med. 304, 197–204 [DOI] [PubMed] [Google Scholar]
- 29.Kivirikko K. I., Myllylä R. (1982) Methods Enzymol. 82, 245–304 [DOI] [PubMed] [Google Scholar]
- 30.Myllylä R., Risteli L., Kivirikko K. I. (1975) Eur. J. Biochem. 52, 401–410 [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J., Russel D. W. (2001) Molecular Cloning, A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32.Rautavuoma K., Passoja K., Helaakoski T., Kivirikko K. I. (2000) Matrix Biol. 19, 73–79 [DOI] [PubMed] [Google Scholar]
- 33.Lamandé S. R., Mörgelin M., Adams N. E., Selan C., Allen J. M. (2006) J. Biol. Chem. 281, 16607–16614 [DOI] [PubMed] [Google Scholar]
- 34.Sabatelli P., Bonaldo P., Lattanzi G., Braghetta P., Bergamin N., Capanni C., Mattioli E., Columbaro M., Ognibene A., Pepe G., Bertini E., Merlini L., Maraldi N. M., Squarzoni S. (2001) Matrix Biol. 20, 475–486 [DOI] [PubMed] [Google Scholar]
- 35.Li S., Van Den Diepstraten C., D'Souza S. J., Chan B. M., Pickering J. G. (2003) Am. J. Pathol. 163, 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minamitani T., Ikuta T., Saito Y., Takebe G., Sato M., Sawa H., Nishimura T., Nakamura F., Takahashi K., Ariga H., Matsumoto K. (2004) Exp. Cell Res. 298, 305–315 [DOI] [PubMed] [Google Scholar]
- 37.Timpl R., Chu M. L. (1994) in Extracellular Matrix Assembly and Structure (Yurchenco P. D., Birk D., Mecham R. P. eds) pp. 207–242, Academic Press, Inc., Orlando, FL [Google Scholar]
- 38.Engvall E., Hessle H., Klier G. (1986) J. Cell Biol. 102, 703–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu M. L., Conway D., Pan T. C., Baldwin C., Mann K., Deutzmann R., Timpl R. (1988) J. Biol. Chem. 263, 18601–18606 [PubMed] [Google Scholar]
- 40.Squarzoni S., Sabatelli P., Bergamin N., Guicheney P., Demir E., Merlini L., Lattanzi G., Ognibene A., Capanni C., Mattioli E., Columbaro M., Bonaldo P., Maraldi N. M. (2006) J. Cell. Physiol. 206, 160–166 [DOI] [PubMed] [Google Scholar]
- 41.McDonald J. A., Kelley D. G., Broekelmann T. J. (1982) J. Cell Biol. 92, 485–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Velling T., Risteli J., Wennerberg K., Mosher D. F., Johansson S. (2002) J. Biol. Chem. 277, 37377–37381 [DOI] [PubMed] [Google Scholar]
- 43.Hocking D. C., Sottile J., Langenbach K. J. (2000) J. Biol. Chem. 275, 10673–10682 [DOI] [PubMed] [Google Scholar]
- 44.Putnam A. J., Schultz K., Mooney D. J. (2001) Am. J. Physiol. Cell. Physiol. 280, C556–C564 [DOI] [PubMed] [Google Scholar]
- 45.Geiger B., Bershadsky A., Pankov R., Yamada K. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 793–805 [DOI] [PubMed] [Google Scholar]
- 46.Aszódi A., Legate K. R., Nakchbandi I., Fässler R. (2006) Annu. Rev. Cell Dev. Biol. 22, 591–621 [DOI] [PubMed] [Google Scholar]
- 47.Wang C., Kovanen V., Raudasoja P., Eskelinen S., Pospiech H., Myllylä R. (2009) J. Cell. Mol. Med. 13, 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]