Abstract
Post-translational stabilization of β-catenin is a key step in Wnt signaling, but the features of β-catenin required for stabilization are incompletely understood. We show that forms of β-catenin lacking the unstructured C-terminal domain (CTD) show faster turnover than full-length or minimally truncated β-catenins. Mutants that exhibit faster turnover show enhanced association with axin in co-transfected cells, and excess CTD polypeptide can compete binding of the β-catenin armadillo (arm) repeat domain to axin in vitro, indicating that the CTD may restrict β-catenin binding to the axin-scaffold complex. Fluorescent resonance energy transmission (FRET) analysis of cyan fluorescent protein (CFP)-arm-CTD-yellow fluorescent protein β-catenin reveals that the CTD of β-catenin can become spatially close to the N-terminal arm repeat region of β-catenin. FRET activity is strongly diminished by the coexpression of β-catenin binding partners, indicating that an unliganded groove is absolutely required for an orientation that allows FRET. Amino acids 733–759 are critical for β-catenin FRET activity and stability. These data indicate that an N-terminal orientation of the CTD is required for β-catenin stabilization and suggest a model where the CTD extends toward the N-terminal arm repeats, shielding these repeats from the β-catenin destruction complex.
The protein β-catenin serves two fundamental roles in the formation and maintenance of tissues. At the cell surface, β-catenin binds the cytoplasmic domain of cadherin-type adhesion receptors, allowing cells to engage their neighbors through robust intercellular adhering junctions. In the cytoplasm and nucleus, a cadherin-independent pool of β-catenin interacts with TCF3-type transcription factors to activate genes that produce cells with distinct identities. The abundance of this cytosolic/nuclear pool of β-catenin is largely determined by the presence of extracellular Wnt factors, which initiate a signaling cascade that prevents the continual destruction of cadherin-free β-catenin.
The destruction of β-catenin is carried out by a highly coordinated series of phosphorylation events. In the absence of Wnt, the N terminus of β-catenin is sequentially phosphorylated by casein kinase 1α and glycogen synthase kinase 3β (GSK3β) (1, 2). Phosphorylation of β-catenin by GSK at residues serine 33 and 37 allows recognition by the E3-ligase component βTrCP, which when part of the SCFβTrCP complex, catalyzes the ubiquitylation and rapid degradation of β-catenin (3). This phosphorylation-dependent degradation of β-catenin depends on the scaffold protein, axin, and the tumor suppressor APC. Axin has binding sites for casein kinase 1α, GSK3β, β-catenin, and APC, so that phosphorylation of axin by casein kinase 1α and GSK3β increases binding to β-catenin (4–6), allowing the N-terminal region of β-catenin to be a more efficient substrate of casein kinase 1α and GSK3β (7). Axin also promotes phosphorylation of APC by casein kinase 1ϵ and GSK3β (8, 9), increasing the affinity of APC for β-catenin (10–12). Because phospho-APC binds to β-catenin with higher affinity than phospho-axin, it is thought that ordered phosphorylations within the axin-APC complex may control the flow of β-catenin through this complex (10, 11). Once β-catenin is bound to APC, APC ensures targeting of N-terminal-phosphorylated β-catenin (Ser(P)-33 and Ser(P)-37) to the proteasome by protecting this βTrCP recognition epitope from the phosphatase PP2A (13).
Understanding how β-catenin moves through the axin-APC-scaffold destruction complex requires an appreciation of β-catenin structure. β-Catenin belongs to the armadillo family of proteins, which are characterized by a central domain consisting of a repeating 42-amino acid motif, termed the “arm repeat” (14). X-ray crystallographic analysis of the β-catenin central armadillo domain shows that its 12 arm repeats form a superhelix of helices that create a long positively charged groove (15). Interestingly, these positive charges are critical for β-catenin binding to many of its negatively charged unstructured ligands such as the cadherin adhesion receptor, axin and APC degradation machinery components, or TCF-DNA binding factors (10, 12, 16–18). Phosphorylation of these binding partners introduces additional negative charges that enhance interactions with the positively charged groove of β-catenin, thereby increasing binding affinity (10–12, 18, 19). Thus, the extent to which β-catenin is used in adhesive or nuclear signaling complexes will critically depend on the phosphorylation state of these major β-catenin ligands (for review, see Ref. 20).
During Wnt activation, GSK3β activity is locally inhibited within the axin complex (21–23), allowing β-catenin to escape this phosphorylation-dependent degradation mechanism and accumulate in both cytoplasmic and nuclear compartments (24). Although the accumulation of β-catenin is not sufficient for signaling (25, 26), it is a key feature of Wnt activation. Given the number and variety of components that can impact β-catenin signaling at the level of the axin-scaffold phospho-destruction complex (for review, see Ref. 20), knowing the intrinsic stabilizing features of β-catenin will be important for understanding how these various factors cross-regulate β-catenin signaling.
A previous study found that forms of β-catenin lacking the C-terminal domain (CTD) failed to accumulate in flies, even during Wnt signaling (27). Recent evidence indicates that the CTD negatively impacts β-catenin binding to axin and APC using the isothermal titration calorimetry method (19), but how the CTD restricts β-catenin binding to axin has remained unresolved. Although the CTD plays an established role in recruiting factors required for gene expression, most interaction partners depend on the more proximal, structured elements of this region, such as helix C, which directly follows the 12th arm repeat (28, 29). Functions for the more distal, unstructured region of the CTD remain poorly defined despite its contribution to β-catenin nuclear signaling (27). The following study demonstrates that the CTD is required for the accumulation of β-catenin through shielding β-catenin from the axin-scaffold phospho-destruction complex.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Recombinant Protein Purification
YFP cDNA was amplified by PCR from pEYFP vector (Clontech) and subcloned into BamHI and XbaI restriction sites of pECFP-C1 (Clontech) vector to generate pECFP-EYFP plasmid. β-Catenin fragments were amplified by PCR and cloned into XhoI and BamHI restriction sites of the pECFP-EYFP plasmid. Coding sequences of β-catenin amino acids 140–663 and ICAT were fused using PCR and also cloned into the XhoI and BamHI restriction sites of pECFP-EYFP. The coding sequence of peptide linker GGGGSGGGGS or PPPPPPPPPP was introduced into overlapping primer regions between β-catenin and ICAT. N-terminal FLAG-tagged full-length β-catenin and S33YD695 were kind gifts of Dr. Eric Fearon (University of Michigan). FLAG-tagged β-catenin truncations were amplified from FLAG-tagged full-length β-catenin constructs and cloned into pCDNA 3 vector (Invitrogen). β-Catenin amino acids 663–666 were changed to 4 prolines using the QuikChange site-directed mutagenesis kit (Stratagene). To introduce alanine mutations into β-catenin residues 733–759 and 760–773, corresponding DNA sequences for the alanine stretches were synthesized (IDT), annealed in vitro, and PCR-fused into β-catenin fragments. Coding sequence of β-catenin armadillo repeats (amino acids 140–663) was amplified by PCR and inserted into BamHI and XbaI restriction sites of pm vector (Clontech). Coding sequences of β-catenin CTD (amino acids 664–781) and ICAT were also amplified using PCR and cloned into BamHI and XbaI restriction sites of pVP16 vector (Clontech). The coding sequence of β-catenin amino acids 520–781 was PCR-amplified and inserted into HindIII and XbaI restriction sites of p3xFLAG-CMV-10 vector (Sigma). pCS2+ Myc-Zebrafish β-catenin 1 (zβ-cat-1) and pCS2+ MycZebrafish β-catenin 2 (zβ-cat-2) plasmids are described in Bellipanni et al. (30). GST-ICAT, FLAG-ICAT, and Myc-Xenopus C-cadherin cyto-domain are described in Gottardi et al. (31). GST-axin-catenin binding domain (CBD) (amino acids 436–498) and His-β59 (β-catenin arm repeats 1∼12) were kindly provided by William Weis (Stanford University), and GST-β-catenin CTD (amino acids 695∼781) was provided by García de Herreros (Universitat Autònoma de Barcelona). GST fusion proteins were expressed in BL-21 cells (GE Healthcare) and purified using glutathione-Sepharose beads 4B (GE Healthcare) according to standard methods.
Antibodies, Immunoblots, Immunoprecipitation, and Affinity Precipitation
The following antibodies were used for Western blots: mouse monoclonal anti-FLAG (Sigma, F3165), mouse monoclonal anti-β-catenin clone 14 (BD Transduction Laboratories), rabbit polyclonal anti-GAPDH (Santa Cruz Biotechnology, SC-25778), mouse monoclonal anti-c-Myc clone 9E10 (Sigma), anti-RGS-His (Qiagen), rabbit polyclonal anti-ICAT (31), and goat anti-mouse, anti-rabbit, and rabbit anti-goat IgG-horseradish peroxidase secondary antibodies (Bio-Rad).
Cells were lysed in standard 1% Triton X-100 lysis buffer (20 mm Tris pH 7.5, 1%Triton X-100, 150 mm NaCl, 5 mm EDTA, 10% glycerol) containing protease inhibitors (Roche Applied Science). Protein concentrations were measured using the Bradford reagent (Bio-Rad). For immunoprecipitation and affinity precipitations, protein samples were incubated with a 1:200 ratio of specific antibody or GST fusion proteins for 2 h at 4 °C followed by 4 washes in 1% Triton X-100 lysis buffer, one wash in 0.1% Triton X-100 lysis buffer (same composition as 1% Triton X-100 lysis buffer except with 0.1% Triton X-100), and denatured by heating in SDS protein loading buffer. Proteins were separated on SDS-PAGE, transferred onto nitrocellulose membrane, and immunoblotted. Immunoblots were developed in ECL solution (GE Healthcare) and exposed to Hyperfilm-ECL (GE Healthcare).
Cell Culture, Transfections, and Pulse-Chase
1 × 106 cells per well of HEK293T cells were seeded into 6-well dishes and transiently transfected with 1.0 μg of various FLAG-tagged β-catenin constructs. 24 h after transfection, cells were split into equal parts and cultured another 24 h before treatment with 20 μg/ml cycloheximide. Cells were washed with ice-cold phosphate-buffered saline at 0-, 1-, 2-, 4- and 6-h time points and lysed in 1% Triton X-100 lysis buffer. Protein concentrations were measured by the Bradford assay. Equivalent protein amounts were separated on 8% SDS-PAGE. Western blots were performed using anti-FLAG and anti-GAPDH antibody. ECL Western blot films were scanned, and ImageJ software was used for quantification. Thresholds were set to eliminate the background, and the integrated densities were calculated. Ectopic expressed β-catenin levels obtained from anti-FLAG immunoblots were normalized to GAPDH levels obtained from the same blot re-probed with anti-GAPDH antibody. Protein levels at different time points were normalized to the 0-h time points, and protein turnover rates were shown in percentage remaining relative to the 0-h time points. Experiments were performed at least three times, and the final results were shown as the mean ± S.D.
For [35S]methionine/cysteine metabolic labeling and pulse-chase experiments, transiently transfected Cos-7 cells were incubated in methionine/cysteine-free Dulbecco's modified Eagle's medium for 30 min at 37 °C and subsequently labeled with 0.1mCi/ml PerkinElmer Life Sciences protein labeling mix (NEG772007MC) for 20 min at 37 °C. Cells were lysed in 1% Triton X-100 lysis buffer at various time points, and equal amounts of proteins were incubated with 10 μg of GST-ICAT for 2 h at 4 °C. Sepharose beads were washed 4 times with 1% Triton X-100 lysis buffer and once with 0.1% Triton X-100 buffer and boiled in SDS protein loading buffer. Protein samples were separated on SDS-PAGE, dried, and subjected to autoradiography and phosphorimage analysis (FujiFilm FLA-5100 Imager).
[32P]Orthophosphate Labeling in Cells
Cos7 cells were plated at 1 × 106 cells per well and transfected accordingly. After 36 h, cells were washed twice with phosphate-buffered saline and incubated in phosphate-free labeling media for 30 min. Cells were then labeled with 120 μCi of [32P]orthophosphate (PerkinElmer Life Sciences) in 2 ml of labeling media for 3 h, washed with phosphate-buffered saline twice, and lysed in 1% Triton X-100 lysis buffer. Immunoprecipitations were performed using anti-FLAG antibody, separated on Criterion pre-cast gels (Bio-Rad), and subjected to autoradiography. Western blot was performed on ⅓ of each sample to confirm immunoprecipitation efficiency.
Mammalian Two-hybrid Assay
A mammalian two-hybrid assay was performed in HEK293T cells according to Mammalian Matchmaker Two-hybrid Assay kit (Clontech) manual. Briefly, 0.1 μg of pM-arm (amino acid 140–663) and 0.1 μg of pVP16-CTD were co-transfected together with 0.3 μg GAL4-Luc reporter and 0.1 μg of Renilla luciferase plasmid. Cells were lysed 36 h after transfection, and luciferase activities were measured on a microplate dual-injector luminometer (Veritas) using the Dual-luciferase Assay kit (Promega). All transfections were performed in triplicate, and data are expressed as the mean ± S.D.
Acceptor Photobleaching FRET
Cos-7 cells were transfected with the FRET constructs in 3.5-mm glass-bottom dishes (MatTek Corp.). After 48 h, acceptor photobleaching FRET was performed at the Northwestern University Cell Imaging Facility on a Zeiss LSM510 META system (Carl Zeiss Inc.) equipped with a 63 × 1.4 Plan-Apochromat oil-immersion objective. An argon laser was used to excite CFP and YFP at 458-nm wavelength, and emission signals were recorded in META detector λ detection mode. YFP signals of cells were bleached using a 514-nm laser line at 100% power for 100 iterations, and time series images were captured for both before and after bleach. FRET efficiency was calculated using the equation EFRET = ([CFPafter − CFPbefore] × YFPbefore)/([YFPbefore − YFPafter] × CFPafter) to normalize for the amount of YFP bleached, where CFPbefore and YFPbefore are the mean pre-bleach fluorescence intensities of CFP and YFP, respectively, and CFPafter and YFPafter are the mean post-bleach fluorescence intensities of CFP and YFP. Multiple cells from at least three different transfections were measured for each construct.
RESULTS
The Flexible C-terminal Region of β-Catenin Is Important for Stability
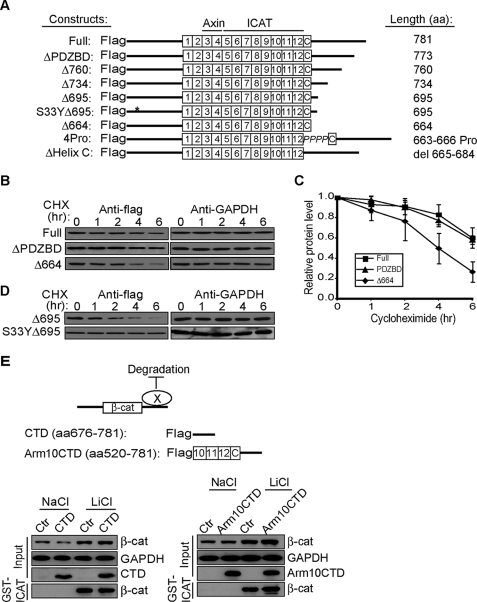
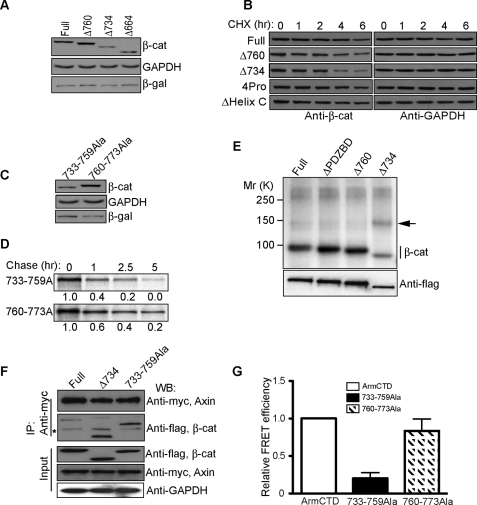
A previous study found that β-catenin lacking its C terminus failed to accumulate in flies (27). To determine whether this reduced accumulation was because of enhanced protein turnover, we generated a panel of truncation mutants (Fig. 1A) and analyzed their half-lives using the cycloheximide chase method. β-Catenin lacking the entire C-terminal region distal to the 12th arm-repeat (amino acids 664–781, Δ664) is degraded more quickly than the full-length protein (Fig. 1, B and C). Similarly, a truncation mutant that retains Helix C (a structured region that “caps” the end of 12th arm repeat) and lacks only the unstructured region of the β-catenin C terminus (29) displays a turnover rate comparable with the Δ664 mutant (Fig. 1D, Δ695 t½ = ∼2 h). Importantly, the instability of Δ695 can be fully rescued by preventing phosphorylation at serine 33 by GSK3β, which is known to comprise a recognition site for the SCFβ-TrCP ubiquitin ligase (3) (Fig. 1D; S33Y Δ695). Of note, the turnover of Δ695 is similar to the Δ664 mutant, and S33Y Δ695 is similar to full-length β-catenin. These data suggest that truncating the C terminus does not generate a grossly misfolded protein that is unstable but, rather, indicates that the flexible C-terminal region of β-catenin is intrinsically required for its stabilization.
FIGURE 1.
The flexible CTD is required for β-catenin stability. A, schematic representation of FLAG-tagged β-catenin mutants. B, turnover rates of β-catenin mutants. HEK293T cells were transfected with 1.0 μg of plasmids 48 h before chasing in 20 μg/ml cycloheximide (CHX) for the times indicated. Total cell lysates were immunoblotted for FLAG tag and endogenous GAPDH. C, quantification of triplicate experiments as shown in panel B. β-Catenin levels were normalized to GAPDH from the same blot. The ratio at time 0 was set to 1.0. D, stabilization of FLAG-β-catenin Δ695 by mutating serine 33 to tyrosine. E, overexpression of β-catenin CTD does not affect stability of endogenous β-catenin. Upper panel, schematic representation of model, where CTD binds protein X to antagonize degradation or promote stability. Unstructured CTD (amino acid (aa) 676–781) and CTD that includes arm repeats 10–12 and Helix C (arm10CTD) are shown. Lower panels, HEK293T cells were transiently transfected with CTD constructs and 48 h later treated with 30 mm NaCl or LiCl overnight. Changes in the cytosolic signaling pool of β-catenin were followed by affinity precipitation with GST-ICAT (31, 38). GAPDH levels show equivalent protein loading.
To understand how the C terminus promotes β-catenin stabilization, we first asked whether the C terminus could bind a factor required for β-catenin stability. Although the extreme C terminus of β-catenin contains a highly conserved PDZ binding domain motif, removing this domain had no obvious effects on β-catenin turnover compared with the full-length protein (Fig. 1, B and C). This suggests that the PDZ binding domain motif and its various potential binding partners (32–35) are dispensable for β-catenin stabilization. To rule out the possibility that other regions of the C terminus interact with a factor required for stabilization (Fig. 1E, schematic), we overexpressed the β-catenin CTD or arm repeats 10–12 and the CTD (arm10CTD) in HEK293T cells and looked for a reduction in the levels of endogenous cytosolic β-catenin. Cytosolic β-catenin can be selectively affinity-precipitated with the β-catenin binding partner, ICAT (GST-ICAT). ICAT is an 81-amino acid polypeptide that binds to the cadherin-free, cytosolic pool of β-catenin and restricts its access to TCFs and transcriptional activators (36, 37). We have previously established that the GST-ICAT precipitable pool of β-catenin corresponds to the Wnt-activated signaling pool typically assessed by standard detergent-free fractionation methods (31, 38). Thus, this simple pulldown method can reflect changes in abundance of the cytosolic signaling pool of β-catenin. As seen in Fig. 1E, neither CTD construct reduced the accumulation of cytosolic β-catenin, as would be predicted if the C terminus sequestered a factor required for β-catenin stabilization. These CTD constructs did not affect base-line cytosolic levels of β-catenin (NaCl, control treated) or “activated” β-catenin using the Wnt pathway agonist and GSK3 inhibitor, lithium chloride (LiCl). Furthermore, overexpression of the C-terminal domain appears insufficient to drive interactions with binding partners that can be discerned by immuno- and affinity-precipitation analyses (supplemental Fig. S1 and Ref. 28).
The Flexible C Terminus of β- Catenin Restricts Axin Binding to the Arm Repeat Domain
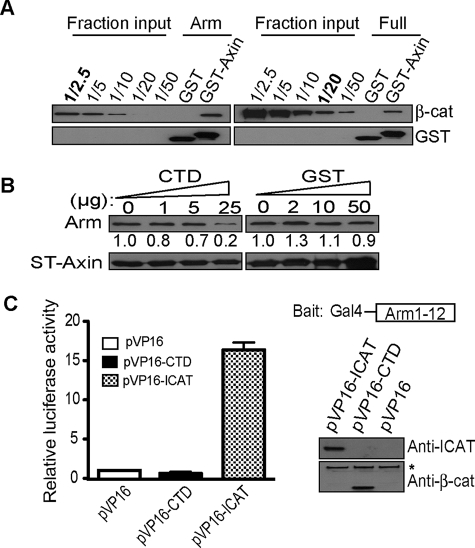
Because the C terminus of β-catenin has been found to bind the central armadillo repeat region of β-catenin in some assays (27, 39) and full-length β-catenin exhibits a lower affinity for axin than β-catenin lacking the CTD (19), we asked whether the C terminus of β-catenin could antagonize the destruction of β-catenin by directly interacting with the arm repeat region of β-catenin and competing arm-repeat binding to axin. In agreement with previous studies (19, 39), the arm repeat region lacking the flanking N- and C-terminal regions (His-arm; amino acids 140–663) can be affinity-precipitated by the CBD of axin (GST-axin-CBD; amino acids 436–498) ∼8 times more efficiently than the full-length β-catenin (His-β-catenin; Fig. 2A). To determine whether this difference in binding is mediated through the flexible C-terminal region of β-catenin, we incubated the arm-repeat region and the GST-axin-CBD in the presence of increasing amounts of the β-catenin CTD (residues 695–781). At high molar excesses, the β-catenin CTD polypeptide can interfere with axin binding to the arm repeat region, whereas an equivalent molar excess of GST protein shows little effect (Fig. 2B). These data raise the possibility that the CTD of β-catenin promotes β-catenin stabilization by directly interfering with the axin/β-catenin binding interaction.
FIGURE 2.
The flexible CTD restricts β-catenin from binding to axin in vitro. A, β-catenin armadillo region alone (His-arm) exhibits better binding to axin compared with full-length β-catenin. 2.0 μg of GST-axin-CBD; amino acids 436–498) pre-coupled glutathione beads were incubated with 0.7 μg of His-arm or 1.0 μg of full-length β-catenin (12 pmol each) purified from baculovirus. Affinity precipitates were separated on SDS-PAGE and loaded in parallel with fractional inputs and immunoblotted for β-catenin to evaluate binding. Note that ∼4.8 pmol of His-arm is pulled down by GST-axin-CBD in comparison with 0.6 pmol of full-length β-catenin. B, CTD, but not GST, competes β-catenin binding to GST-axin-CBD. 2.0 μg of GST-axin-CBD coupled-Sepharose beads were incubated with 1.0 μg of His-β59 in the presence of increasing amounts of β-catenin CTD peptide (amino acids 695–781) or molar equivalents of GST. CTD competes arm-repeat binding to axin at 150-fold molar excess. Affinity precipitates were separated on SDS-PAGE and immunoblotted for the histidine tag. Band intensities calculated with Image J program are shown under corresponding gel bands. The number obtained without competitor peptide was set to 1.0. C, β-catenin CTD does not interact with arm-repeat region of β-catenin in trans. Mammalian two-hybrid assay of pM-arm (amino acid 140–663) and pVP16-CTD. Right, schematic of the Gal4-arm-(1–12) “bait” and immunoblot verifying hybrid proteins. The asterisk (*) denotes endogenous β-catenin.
In Drosophila the C-terminal region of Armadillo was found to physically interact with the central arm repeat domain in a yeast two-hybrid screen (27). Because direct “trans” binding between the CTD and arm-repeat domain has been difficult to observe using in vitro binding assays (19), we sought to determine whether this interaction could be confirmed using a mammalian two-hybrid assay. By fusing the arm repeat region of β-catenin to the Gal4-DNA binding domain (bait) and the CTD to the widely used transcriptional activator sequence VP16 (VP16-CTD), we asked whether co-transfection of these components could reconstitute a functional transcription factor through direct, trans-association of arm repeat and CTD. VP16 fused to ICAT, a strong binding partner for the arm-repeat region of β-catenin, was used as a robust positive control (36). In contrast to what has been previously reported (27, 39), no direct interaction between the arm-repeat domain and CTD was observed using this assay (Fig. 2C). Given that the β-catenin CTD may only compete the axin/arm repeat interaction at very high molar ratios (Fig. 2B), any trans interaction between the β-catenin CTD and the arm repeat region appears to be extremely inefficient. Whether interactions between the CTD and arm repeat region may be more significant in the context of a native β-catenin molecule, where the CTD is directly attached to the arm-repeat region, has remained unexamined.
β-Catenin CTD Is Spatially Close to the N-terminal Arm Repeat Region by FRET Analysis
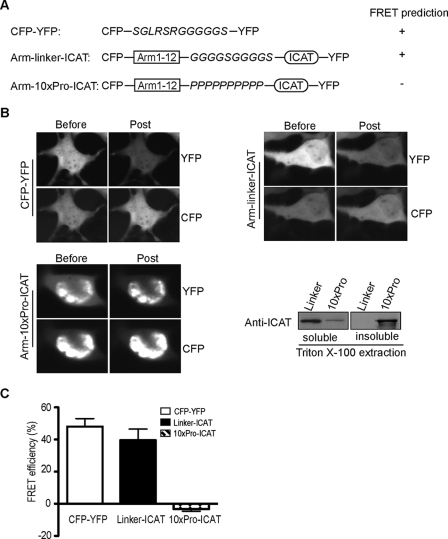
To interrogate spatial relationships between the CTD and arm repeat region of β-catenin within a single molecule, we established a system that would allow a FRET-based technique using live cell imaging. Similar approaches have been used to explore the regulated conformations of proteins, such as kinesin and myosin light chain kinase activation (40, 41). Before testing our model on β-catenin, robust positive and negative control constructs were designed to assess whether predicted spatial relationships could be quantified and confirmed (Fig. 3A). For example, as ICAT is a strong binding partner for β-catenin (37) and is similar in size to the CTD of β-catenin (81 amino acids for ICAT; ∼100 amino acids for CTD), we predicted that a β-catenin arm region/ICAT chimera, separated by a 10-amino acid flexible linker (G4SG4S) and terminally tagged with cyan or yellow fluorescent Protein (CFP or YFP), would allow ICAT to fold back and engage the arm repeat region, enabling the CFP/YFP pair to exhibit robust FRET activity (Fig. 3A; CFP-arm-linker-ICAT-YFP). Indeed, Cos-7 cells transiently transfected with the CFP-arm-linker-ICAT-YFP chimera showed a FRET efficiency of 40%, which is comparable with that observed when CFP and YFP are directly fused through a short 12-amino acid flexible linker (48%; Fig. 3, B and C). Importantly, the FRET signal in our CFP-arm-linker-ICAT-YFP chimera is likely because of an intramolecular interaction that depends on a flexible linker, as replacement of this linker with a rigid helical peptide consisting of 10 prolines (CFP-arm-10×Pro-ICAT-YFP) completely abolishes the FRET activity (Fig. 3, B and C). In contrast to the uniform distribution of CFP-arm-linker-ICAT-YFP, most of the CFP-arm-10Pro-ICAT-YFP protein is severely aggregated and largely Triton X-100 inextractable (Fig. 3B, right), demonstrating that replacement of the flexible linker with a more rigid structure dramatically changes the solubility properties of this chimera. These data demonstrate that the FRET activity associated with these chimeric constructs strongly correlates with their predicted conformations and suggests that we could use this approach to interrogate the relationship between the CTD and arm repeat region within a native β-catenin molecule.
FIGURE 3.
FRET analysis of β-catenin structure in cells. A, schematic of control constructs for single molecule acceptor photobleaching FRET. A construct containing CFP and YFP fluorescent proteins separated by 12 amino acids was designed to optimize acceptor photobleaching FRET conditions. ICAT protein was linked to the β-catenin arm-repeat region by using either a flexible linker (GGGGSGGGGS) or a rigid peptide consisting of 10 proline residues to serve as positive and negative controls, respectively. B, representative FRET images and immunoblot of CFP-YFP, arm-linker-ICAT, and arm-10Pro-ICAT. Cos-7 cells were solubilized in 1% Triton X-100 lysis buffer after imaging, and the relative solubility of arm-linker-ICAT and arm-10Pro-ICAT proteins was assessed by immunoblotting using antibody for ICAT. C, FRET efficiencies for CFP-YFP, Linker-ICAT, and 10xPro-ICAT were calculated from at least 100 cells imaged from multiple transfections and are expressed as the mean ± S.D.
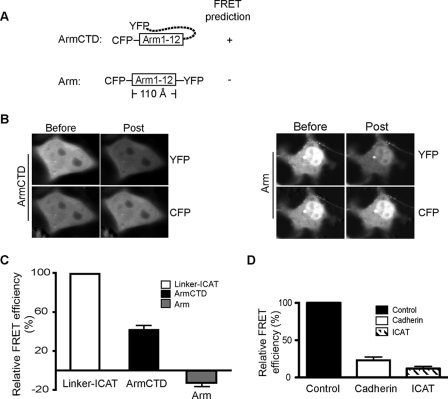
As expected, no FRET activity is detected when the arm repeat region is flanked by CFP and YFP (CFP-arm-YFP; Fig. 4, A–C), as the arm repeat superhelical core structure spans more than 110 Å (15), which is beyond the FRET detection limit. When the CTD was present (CFP-arm-(1–12)-CTD-YFP), FRET activity was restored to ∼40% of our CFP-arm-linker-ICAT-YFP positive control (Fig. 4C). Importantly, co-transfection of CFP-arm-(1–12)-CTD-YFP with the cytoplasmic domain of E-cadherin or ICAT markedly reduces the FRET signal to 10% of control levels (Fig. 4D). These data indicate that the flexible CTD of β-catenin is required for the CFP-YFP pair to adopt a close enough proximity to FRET. Because FRET activity is blocked by ligands known to directly engage the arm repeat region of β-catenin, the YFP-C terminus of β-catenin only approaches the CFP-N-terminal arm repeats in an unliganded β-catenin molecule.
FIGURE 4.
β-Catenin CTD is required for FRET activity. A, schematic of β-catenin FRET constructs designed to detect the relationship between the CTD and arm-repeat region. B, representative images obtained from FRET measurement performed in Cos-7 cells transfected with CFP-arm-CTD-YFP and CFP-arm-YFP constructs. C, FRET efficiencies were calculated from at least 100 cells imaged from multiple transfections and compared with FRET efficiency measured in CFP-arm-linker-ICAT-YFP fusion protein, where efficiency of CFP-arm-linker-ICAT-YFP was set to 100%. D, β-catenin binding partners reduce FRET activity. 0.1 μg of CFP-arm-CTD-YFP construct was co-transfected with 1.0 μg of a plasmid expressing the cadherin cytoplasmic domain as a cytosolic protein or FLAG-ICAT. An empty vector was co-transfected as a control, and the FRET efficiency was set to 1.0.
Amino Acids 733–759 Are Critical for β-Catenin Stability and FRET Activity
To determine whether the CTD sequences required for stability are also those required for FRET activity, we generated additional truncation and point mutants. Deletion of amino acids 734–781 (Δ734), but not 760–781 (Δ760), reduced β-catenin accumulation and stability (Fig. 5, A and B), indicating that amino acids 733–759 are most critical for β-catenin stabilization. Consistent with this interpretation, replacement of the sequence between amino acids 733 and 759 with a stretch of alanines results in a β-catenin that can be degraded faster than a β-catenin where amino acids 760–773 have been changed to alanines (Fig. 5, C and D). Deletion of Helix C (amino acids 665–684) or forcing the orientation of Helix C away from the arm repeat domain by replacing residues between the 12th arm repeat and Helix C with 4 proline residues did not affect β-catenin turnover (Fig. 5B). These data suggest that information encoded within the 733–759 amino acid sequence contributes to β-catenin stabilization.
FIGURE 5.
Amino acids 733–759 are critical for FRET activity and stability of wild-type β-catenin. A, Δ734 and Δ664 accumulate to a lesser extent than full-length or Δ760 in transiently transfected HEK293T cells. 0.2 μg of a plasmid expressing β-galactosidase was cotransfected as an internal control along with 1.0 μg of the β-catenin constructs indicated. B, turnover of full-length and truncated (Δ760, Δ734) β-catenins. Introducing 4 prolines (4Pro; changing amino acids 663–666 to prolines) just after Helix C or deleting Helix C did not affect β-catenin stability. Protein turnover study was carried out as in Fig. 1. β-gal, β-galactosidase; CHX, cycloheximide. C and D, mutation of amino acids 733–759 to alanines reduces β-catenin accumulation (C, Western blot) and stability (D, [35S]methionine/cysteine pulse-chase analysis). Autoradiography bands were quantified using phosphorimage analysis (FujiFilm FLA-5100 Imager), and the corresponding numbers were normalized to time 0. E, Δ734 truncation exhibits increased binding to a ∼140-kDa 32P-labeled protein. F, Δ734 truncation and 733–759Ala mutation show increased binding to axin compared with full-length β-catenin. FLAG-tagged β-catenin proteins and Myc-tagged axin were transiently expressed in HEK293T cells for 48 h, immunoprecipitated (IP) with an antibody against Myc-axin, and blotted (WB) with the FLAG antibody. The asterisk (*) denotes nonspecific band. G, 733–759Ala, but not 760–773Ala mutation, decreases β-catenin FRET efficiency. FRET efficiencies were measured as in Fig. 3. FRET efficiency of CFP-arm-CTD-YFP was set to 1.0, and other constructs were quantified relative to the CFP-arm-CTD-YFP efficiency. Data were expressed as the mean ± S.D.
Evidence suggests that amino acids 733–759 may be specifically required to displace β-catenin from the axin-scaffold complex. For example, transient expression of Δ734 in [32P]orthophosphate-labeled Cos cells shows increased association with a ∼140-kDa phosphoprotein compared with Δ760 and full-length β-catenin proteins (Fig. 5E). This phosphoprotein may be composed of axin, as axin is reported to run in the 120–140-kDa range (42), and β-catenin lacking the last 47 amino acids and, more specifically, 733–759 could be coimmunoprecipitated more efficiently with exogenously expressed myc-axin (Fig. 5F). Endogenous axin/β-catenin mutant complexes were not detected, likely because axin levels are known to be limiting in cells (43), and currently available antibodies inefficiently precipitate the axin complex (not shown). Nevertheless, reduced association between β-catenin and axin may depend on an orientation of the CTD that approaches the N-terminal arm repeats, as a β-catenin where amino acids 733–759 are replaced with alanines (arm-CTD-(733–759)-Ala) shows reduced FRET activity compared with arm-CTD or arm CTD-(760–773)-Ala (Fig. 5G). These data indicate that the sequence and/or charged nature of amino acids 733–759 are critical for full FRET activity and protein stability.
To determine whether the 733–759 sequence is important for Wnt-activated β-catenin stabilization within the native protein, we took advantage of mono-specific antibodies that recognize the CTD of β-catenin. Interestingly, a monoclonal antibody that recognizes β- catenin between amino acids 751 and 781 (monoclonal antibody M.5; supplemental Fig. S2, A and B) fails to recognize the cytoplasmic pool of β-catenin in Wnt3a-expressing L cells despite evidence that an N-terminal monoclonal antibody detects both nuclear and cytoplasmic pools of β-catenin (monoclonal antibody 1.1.1; supplemental Fig. S2C). Altogether, these data indicate that amino acids 733–759 appear to be masked in the cytoplasmic pool of Wnt-activated β-catenin, where this masking may be related to β-catenin FRET activity and stabilization through restricting β-catenin binding to axin.
Differential Signaling of Zebrafish β-Catenins May Be Because of Different Turnover Rates
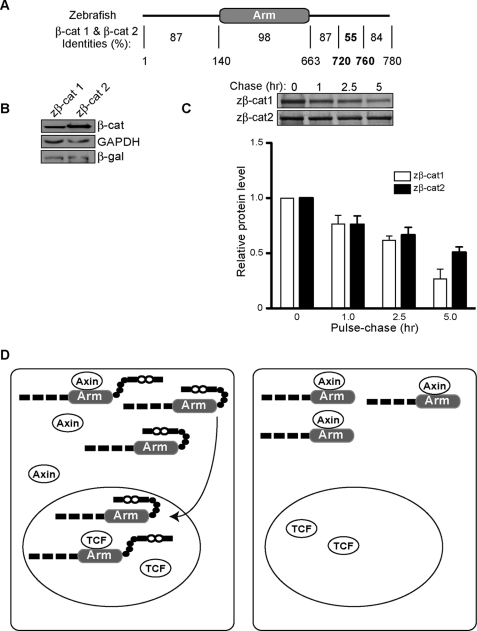
A role for CTD sequences in modulating β-catenin stability raises the possibility that species with multiple β-catenin gene products with divergent C-terminal sequences may have evolved β-catenins with different turnover rates. Ichabod was previously identified as a spontaneous maternal effect mutation in zebrafish, which results in a severe ventralization phenotype because of a failure of the endogenous dorsal zebrafish β-catenin to localize to the nucleus (44). Rather than encode a component that regulates β-catenin nuclear accumulation and signaling, ichabod encodes a second, highly homologous β-catenin gene (zβ-cat-2 (30)). Remarkably, even though zβ-cat-2 is 92% identical to the previously described zβ-cat-1 and both zβ-catenins are present in the wild-type embryo, it is the reduction of zβ-cat-2 in the mutant that causes failure to form the primary axis. This raises the possibility that the small differences in amino acid sequence may contribute to the differential accumulation of zβ-catenins in the cell. Because the greatest differences between zβ-cat-1 and -2 are found between amino acids 106–110 and 722–757 (Fig. 6A), the latter of which corresponds to a region that contributes to β-catenin stabilization (733–759), we sought to determine whether inherent differences in zβ-cat turnover could explain their different biological activities. Although the difference is modest, zβ-cat-1 is consistently turned over more quickly than zβ-cat-2 in 35[S]methionine/cysteine-label pulse-chase experiments (Fig. 6B). These data suggest that zebrafish evolved two β-catenin proteins with different signaling capacities, at least in part through selecting for amino acid changes in the CTD that affect the rate of β-catenin disappearance.
FIGURE 6.
Zebrafish β-catenins display different turnover rates. A, protein alignment of zebrafish β-cat-1 (zβ-cat-1, amino acid 780) and β-cat-2 (zβ-cat-2, amino acid 778) using the Blast program. Amino acid numbering is according to zβ-cat-1. B, zβ-cat-1 accumulates less than zβ-cat-2. β-Galactosidase (-β-gal) expression plasmid (0.2 μg) was cotransfected with zβ-catenins (1.0 μg) as an internal control. C, 35[S]methionine/cysteine pulse-chase result of zβ-cat-1 and zβ-cat-2. Lower panel, quantification of triplicate pulse-chase experiments by phosphorimage analysis (FujiFilm FLA-5100 Imager) (mean ± S.D.). D, proposed model for a role of the CTD in β-catenin stability. Left panel, CTD of β-catenin effectively shields the arm region from the axin degradation complex, promoting β-catenin stabilization. Right panel, loss of CTD abolishes the protective effects of CTD on arm repeats and results in more rapid turnover of β-catenin.
DISCUSSION
Although the stabilization of β-catenin is an established key step during Wnt signaling, whether and how β-catenin is released from the phospho-destruction complex and the features of β-catenin that are intrinsic to its stabilization are incompletely understood. In this study we show that the flexible C-terminal region of β-catenin is important for promoting β-catenin stability by restricting its association with axin, a key scaffold component required for the destruction of β-catenin. This model is based on evidence that forms of β-catenin lacking the entire C terminus, and in particular, a stretch of negatively charged amino acids between amino acids 733 and 759 show faster turnover than full-length or minimally truncated β-catenins (Figs. 1 and 5). Mutants that exhibit faster turnover also show enhanced association with axin in transfected cells (Fig. 5). These results are consistent with in vitro biochemical evidence that full-length β-catenin exhibits a lower affinity for axin than β-catenin lacking the C-terminal domain using the isothermal titration calorimetry method (19).
Because the structure of the arm repeat domain appears identical between partial, full-length, and β-catenin/ligand co-crystals (for review, see Refs. 45 and 46), it has remained unclear how the C terminus of β-catenin might restrict axin binding to β-catenin. Some studies indicate that the CTD may directly contact the arm-repeat region of β-catenin, effectively competing this domain from certain β-catenin binding partners. For example, the CTD of β-catenin was isolated in a yeast two-hybrid screen for arm domain binding partners in the Drosophila armadillo protein (27). This interaction was also observed using purified arm domain and CTD components followed by semiquantitative Western and blot overlay analysis (39). With similar methods, two studies found that the CTD of β-catenin could compete its binding to the cadherin (39, 47), but not TCF (47), raising the possibility that in a full-length protein the CTD may fold back along the arm domain in a closed conformation that restricts binding to some partners (27, 47). However, specific binding between the CTD and the arm domain was not supported when Coomassie-stained gels were employed to quantify binding (19) or NMR spectroscopy was used to detect transient weak interactions (29). As these experiments are done in trans, it has remained untested whether a CTD tethered in cis could interact more effectively. In this regard, although β-catenin lacking the CTD did not improve binding affinity to tight ligands such as E-cadherin, Lef1, and phosphorylated APC, the CTD was found to decrease the affinity for weaker ligands such as unphosphorylated APC and axin (19). Indeed, a high molar excess of the CTD polypeptide appears to compete the β-catenin arm domain from binding to axin (Fig. 2B).
Our FRET analysis reveals that the C terminus of β-catenin can become spatially close to the N-terminal arm repeat region of β-catenin. Although this does not prove intramolecular interactions between the CTD and arm repeat domain, evidence that the amino acid chemistry between residues 733–759 is critical for FRET activity strongly suggests that something other than length and flexibility is important for the CTD orientation. Because the CTD is highly negatively charged and the arm repeat groove is lined with positive charges (15), it is possible that the fixed negative charges along the 733–759 segment make low affinity interactions with the ligand-binding groove, effectively stabilizing the CFP-YFP FRET pair. Consistent with this model, β-catenin FRET activity is strongly diminished by the coexpression of β-catenin binding partners (Fig. 4D), indicating that an unliganded groove is absolutely required for an orientation that allows FRET. Although no FRET activity is observed when a full-length β-catenin is terminally tagged with CFP and YFP (not shown), the N-terminal deletion construct that exhibits FRET activity in this study is similar to mutants typically used to drive Wnt signaling (e.g. Ref. 48) as well as a form of β-catenin generated in neurons after glutamate-induced calpain cleavage (49).
Evidence that amino acids 733–759 are critical for both β-catenin FRET activity and stability strongly suggests that the spatial orientation of the CTD is required for the stabilization of β-catenin. These data suggest a model where the CTD of β-catenin extends toward the N-terminal arm repeat region and protects these repeats from the axin degradation complex (Fig. 6C).
It has been previously speculated that low affinity charge interactions between the basic arm repeat domain and acidic terminal regions may effectively shield the arm repeats from nonspecific interactions (29). Consistent with this model, β-catenin lacking its entire CTD was found to coimmunoprecipitate numerous 32P radioactively labeled species (supplemental Fig. S3). Moreover, in contrast to the uniform distribution of CFP-arm-linker-ICAT-YFP, most of the CFP-arm-10Pro-ICAT-YFP protein is severely aggregated and largely Triton X-100 inextractable (Fig. 3B), suggesting that an arm repeat domain that cannot be shielded tends to aggregate. A similar phenomenon is observed when unliganded β-catenin is overexpressed in cells (50). This indicates that the CTD of β-catenin may play a more fundamental role in catenin structure and function. Beyond its established function in recruiting factors required for gene expression (51), the CTD is critical for the post-translational stabilization and solubility of β-catenin through this shielding function.
This model may be also relevant to species that rely on distinct β-catenin gene products during development, where the CTD may determine different rates of β-catenin disappearance after a Wnt signal. In this regard, sequence differences in the terminal region of zebrafish β-catenins result in proteins with distinct in vivo activities (30), which may be explained by their different stabilities (Fig. 6B). In addition, flies generate a splice variant of β-catenin that lacks the CTD and is used exclusively in neuronal cell-cell adhesion (52). We speculate that this neural-specific form of β- catenin may ensure that dynamic changes in adhesion do not result in β-catenin stabilization and signaling.
Whether β-catenin FRET activity and CTD shielding function can be regulated is unclear, although an increasing number and variety of molecular components are known to impact β-catenin signaling at the level of the phospho destruction complex (for review, see Ref. 20). For example, the prolyl-isomerase, Pin1 was previously shown to recognize specific phospho-Ser/Pro residues in β-catenin and activate signaling (53), but whether the mechanism is through simply competing β-catenin from the APC-axin complex or kinking the polypeptide backbone of β-catenin remains unknown. Our CFP-arm CTD-YFP construct may be a useful tool for interrogating conformational changes in β-catenin in a cellular context. Moreover, given recent evidence that cell-cell adhesion limits Wnt signaling by enhancing the N-terminal phosphorylation and degradation of cytosolic β-catenin (38), it will be interesting to learn whether adhesion signaling reduces β-catenin stability by antagonizing this CTD orientation and FRET activity.
The strong correlation between FRET activity and stability suggests that our CFP-arm CTD-YFP construct may serve as FRET-based indicator of β-catenin stability, which may be useful for screening compounds that can negatively modulate β-catenin protein levels. Given evidence that β-catenin nuclear signaling activity is not exclusively determined by LEF/TCF-family transcription factors (e.g. Refs. 54–56), the ability to screen for chemicals that reduce β-catenin levels may broaden the identification of agents that target these other functions of β-catenin.
Supplementary Material
Acknowledgments
We thank Jean-Luc Teillard (Jussieu University, France) and Sylvie Robine (Curie Institute, France) for N- and C-terminal-specific β-catenin monoclonal antibodies, Emelyn Shroff for contributing supplemental Fig. S2C, Bill Weis (Stanford University) for the GST-axin-CBD plasmid, and Rebecca L. Daugherty for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants T32 GM08061 (to M. T. M.), P30 CA060553 (to the Northwestern Imaging Facility and Robert H. Lurie Comprehensive Cancer Center), R01 HD39272 (to E. S. W. and G. B.), and GM076561 (to C. J. G.). This work was also supported by American Heart Association Pre-doctoral Award 09PRE2261197 (to M. T. M.) and the American Heart Association Grant 0530304Z (to C. J. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- TCF
- T cell factor
- GSK
- glycogen synthase kinase
- APC
- adenomatous polyposis coli
- FRET
- fluorescent resonance energy transmission
- arm
- armadillo repeat domain
- GST
- glutathione S-transferase
- ICAT
- inhibitor of catenin and TCF
- CTD
- C-terminal domain
- CBD
- catenin binding domain
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. (2002) Cell 108, 837–847 [DOI] [PubMed] [Google Scholar]
- 2.Yost C., Torres M., Miller J. R., Huang E., Kimelman D., Moon R. T. (1996) Genes Dev. 10, 1443–1454 [DOI] [PubMed] [Google Scholar]
- 3.Hart M., Concordet J. P., Lassot I., Albert I., del los Santos R., Durand H., Perret C., Rubinfeld B., Margottin F., Benarous R., Polakis P. (1999) Curr. Biol. 9, 207–210 [DOI] [PubMed] [Google Scholar]
- 4.Jho E., Lomvardas S., Costantini F. (1999) Biochem. Biophys. Res. Commun. 266, 28–35 [DOI] [PubMed] [Google Scholar]
- 5.Willert K., Shibamoto S., Nusse R. (1999) Genes Dev. 13, 1768–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo W., Peterson A., Garcia B. A., Coombs G., Kofahl B., Heinrich R., Shabanowitz J., Hunt D. F., Yost H. J., Virshup D. M. (2007) EMBO J. 26, 1511–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dajani R., Fraser E., Roe S. M., Yeo M., Good V. M., Thompson V., Dale T. C., Pearl L. H. (2003) EMBO J. 22, 494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubinfeld B., Albert I., Porfiri E., Fiol C., Munemitsu S., Polakis P. (1996) Science 272, 1023–1026 [DOI] [PubMed] [Google Scholar]
- 9.Rubinfeld B., Tice D. A., Polakis P. (2001) J. Biol. Chem. 276, 39037–39045 [DOI] [PubMed] [Google Scholar]
- 10.Xing Y., Clements W. K., Kimelman D., Xu W. (2003) Genes Dev. 17, 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha N. C., Tonozuka T., Stamos J. L., Choi H. J., Weis W. I. (2004) Mol. Cell 15, 511–521 [DOI] [PubMed] [Google Scholar]
- 12.Xing Y., Clements W. K., Le Trong I., Hinds T. R., Stenkamp R., Kimelman D., Xu W. (2004) Mol. Cell 15, 523–533 [DOI] [PubMed] [Google Scholar]
- 13.Su Y., Fu C., Ishikawa S., Stella A., Kojima M., Shitoh K., Schreiber E. M., Day B. W., Liu B. (2008) Mol. Cell 32, 652–661 [DOI] [PubMed] [Google Scholar]
- 14.Riggleman B., Wieschaus E., Schedl P. (1989) Genes Dev. 3, 96–113 [DOI] [PubMed] [Google Scholar]
- 15.Huber A. H., Nelson W. J., Weis W. I. (1997) Cell 90, 871–882 [DOI] [PubMed] [Google Scholar]
- 16.von Kries J. P., Winbeck G., Asbrand C., Schwarz-Romond T., Sochnikova N., Dell'Oro A., Behrens J., Birchmeier W. (2000) Nat. Struct. Biol. 7, 800–807 [DOI] [PubMed] [Google Scholar]
- 17.Graham T. A., Weaver C., Mao F., Kimelman D., Xu W. (2000) Cell 103, 885–896 [DOI] [PubMed] [Google Scholar]
- 18.Huber A. H., Weis W. I. (2001) Cell 105, 391–402 [DOI] [PubMed] [Google Scholar]
- 19.Choi H. J., Huber A. H., Weis W. I. (2006) J. Biol. Chem. 281, 1027–1038 [DOI] [PubMed] [Google Scholar]
- 20.Daugherty R. L., Gottardi C. J. (2007) Physiology 22, 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cselenyi C. S., Jernigan K. K., Tahinci E., Thorne C. A., Lee L. A., Lee E. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 8032–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piao S., Lee S. H., Kim H., Yum S., Stamos J. L., Xu Y., Lee S. J., Lee J., Oh S., Han J. K., Park B. J., Weis W. I., Ha N. C. (2008) PLoS ONE 3, e4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu G., Huang H., Garcia Abreu J., He X. (2009) PLoS ONE 4, e4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peifer M., Sweeton D., Casey M., Wieschaus E. (1994) Development 120, 369–380 [DOI] [PubMed] [Google Scholar]
- 25.Guger K. A., Gumbiner B. M. (2000) Dev. Biol. 223, 441–448 [DOI] [PubMed] [Google Scholar]
- 26.Staal F. J., Noort Mv M., Strous G. J., Clevers H. C. (2002) EMBO Rep. 3, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox R. T., Pai L. M., Kirkpatrick C., Stein J., Peifer M. (1999) Genetics 153, 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sierra J., Yoshida T., Joazeiro C. A., Jones K. A. (2006) Genes Dev. 20, 586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing Y., Takemaru K., Liu J., Berndt J. D., Zheng J. J., Moon R. T., Xu W. (2008) Structure 16, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellipanni G., Varga M., Maegawa S., Imai Y., Kelly C., Myers A. P., Chu F., Talbot W. S., Weinberg E. S. (2006) Development 133, 1299–1309 [DOI] [PubMed] [Google Scholar]
- 31.Gottardi C. J., Gumbiner B. M. (2004) Am. J. Physiol. Cell Physiol. 286, C747–CC756 [DOI] [PubMed] [Google Scholar]
- 32.Kanamori M., Sandy P., Marzinotto S., Benetti R., Kai C., Hayashizaki Y., Schneider C., Suzuki H. (2003) J. Biol. Chem. 278, 38758–38764 [DOI] [PubMed] [Google Scholar]
- 33.Nishimura W., Yao I., Iida J., Tanaka N., Hata Y. (2002) J. Neurosci. 22, 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perego C., Vanoni C., Massari S., Longhi R., Pietrini G. (2000) EMBO J. 19, 3978–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobrosotskaya I. Y., James G. L. (2000) Biochem. Biophys. Res. Commun. 270, 903–909 [DOI] [PubMed] [Google Scholar]
- 36.Tago K., Nakamura T., Nishita M., Hyodo J., Nagai S., Murata Y., Adachi S., Ohwada S., Morishita Y., Shibuya H., Akiyama T. (2000) Genes Dev. 14, 1741–1749 [PMC free article] [PubMed] [Google Scholar]
- 37.Daniels D. L., Weis W. I. (2002) Mol. Cell 10, 573–584 [DOI] [PubMed] [Google Scholar]
- 38.Maher M. T., Flozak A. S., Stocker A. M., Chenn A., Gottardi C. J. (2009) J. Cell Biol. 186, 219–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castaño J., Raurell I., Piedra J. A., Miravet S., Duñach M., García de Herreros A. (2002) J. Biol. Chem. 277, 31541–31550 [DOI] [PubMed] [Google Scholar]
- 40.Cai D., Hoppe A. D., Swanson J. A., Verhey K. J. (2007) J. Cell Biol. 176, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chew T. L., Wolf W. A., Gallagher P. J., Matsumura F., Chisholm R. L. (2002) J. Cell Biol. 156, 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang L., Lin C., Liu Z. R. (2006) Cell 127, 139–155 [DOI] [PubMed] [Google Scholar]
- 43.Lee E., Salic A., Kirschner M. W. (2001) J. Cell Biol. 154, 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelly C., Chin A. J., Leatherman J. L., Kozlowski D. J., Weinberg E. S. (2000) Development 127, 3899–3911 [DOI] [PubMed] [Google Scholar]
- 45.Gottardi C. J., Gumbiner B. M. (2001) Curr. Biol. 11, R792–794 [DOI] [PubMed] [Google Scholar]
- 46.Gottardi C. J., Peifer M. (2008) Structure 16, 336–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottardi C. J., Gumbiner B. M. (2004) J. Cell Biol. 167, 339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chenn A., Walsh C. A. (2002) Science 297, 365–369 [DOI] [PubMed] [Google Scholar]
- 49.Abe K., Takeichi M. (2007) Neuron 53, 387–397 [DOI] [PubMed] [Google Scholar]
- 50.Giannini A. L., Vivanco M. M., Kypta R. M. (2000) Exp. Cell Res. 255, 207–220 [DOI] [PubMed] [Google Scholar]
- 51.Willert K., Jones K. A. (2006) Genes Dev. 20, 1394–1404 [DOI] [PubMed] [Google Scholar]
- 52.Loureiro J., Peifer M. (1998) Curr. Biol. 8, 622–632 [DOI] [PubMed] [Google Scholar]
- 53.Ryo A., Nakamura M., Wulf G., Liou Y. C., Lu K. P. (2001) Nat. Cell Biol. 3, 793–801 [DOI] [PubMed] [Google Scholar]
- 54.Essers M. A., de Vries-Smits L. M., Barker N., Polderman P. E., Burgering B. M., Korswagen H. C. (2005) Science 308, 1181–1184 [DOI] [PubMed] [Google Scholar]
- 55.Shah S., Islam M. N., Dakshanamurthy S., Rizvi I., Rao M., Herrell R., Zinser G., Valrance M., Aranda A., Moras D., Norman A., Welsh J., Byers S. W. (2006) Mol. Cell 21, 799–809 [DOI] [PubMed] [Google Scholar]
- 56.Kidd A. R., 3rd, Miskowski J. A., Siegfried K. R., Sawa H., Kimble J. (2005) Cell 121, 761–772 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.