Abstract
In the heart, autophagy is required for normal cardiac function and also has been implicated in cardiovascular disease. FoxO transcription factors promote autophagy in skeletal muscle and have additional roles in regulation of cell size, proliferation, and metabolism. Here we investigate the role of FoxO transcription factors in regulating autophagy and cell size in cardiomyocytes. In cultured rat neonatal cardiomyocytes, glucose deprivation leads to decreased cell size and induction of autophagy pathway genes LC3, Gabarapl1, and Atg12. Likewise, overexpression of either FoxO1 or FoxO3 reduces cardiomyocyte cell size and induces expression of autophagy pathway genes. Moreover, inhibition of FoxO activity by dominant negative FoxO1 (Δ256) blocks cardiomyocyte cell size reduction upon starvation, suggesting the necessity of FoxO function in cardiomyocyte cell size regulation. Under starvation conditions, endogenous FoxO1 and FoxO3 are localized to the nucleus and bind to promoter sequences of Gabarapl1 and Atg12. In vivo studies show that cellular stress, such as starvation or ischemia/reperfusion in mice, results in induction of autophagy in the heart with concomitant dephosphorylation of FoxO, consistent with increased activity of nuclear FoxO transcription factors. Together these results provide evidence for an important role for FoxO1 and FoxO3 in regulating autophagy and cell size in cardiomyocytes.
The regulation of cell size is finely controlled by the availability of nutrients and growth factors that ultimately control the balance between protein synthesis and degradation. Cardiac myocyte hypertrophy occurs during developmental growth of the heart (1) and is induced in response to exercise or pathological conditions (1–3). Conversely, the heart can undergo reduction in size in response to starvation, as observed in patients with anorexia nervosa (4, 5), or with decreased work load, as occurs with implantation of left ventricular assist device (6, 7). Therefore, multiple stimuli can affect heart size, and maintenance of proper cardiomyocyte size is critical for normal cardiac function. The molecular and physiological regulation of the cardiac hypertrophic response has been studied extensively (3). Much less is known of the pathways that result in the reduction in cardiomyocyte cell size under conditions of starvation or decreased mechanical load.
The autophagic process has been implicated in cellular homeostasis and cell size regulation in multiple organ systems during normal development and in stress conditions, such as starvation (8–10). Autophagy pathway genes, including LC3, Gabarapl1 (homolog to yeast Atg8), Atg12, Atg5, and Atg7, are involved in multiple stages of the formation of the autophagosome (9, 11). Autophagy also is a cellular homeostatic process for normal protein turnover and is necessary to eliminate damaged proteins and organelles, which might be toxic or harmful to the cell (11, 12). During nutrient deprivation, autophagy is induced in several organ systems as a means of generating intracellular nutrients and energy (9, 11, 13). Autophagy also is required in the neonatal period of nutrient deprivation after birth and before feeding, as indicated by neonatal lethality of Atg5-null mice (14).
Autophagy in cardiac myocytes occurs during normal development and homeostasis and also is associated with cardiac disease (11, 15). During the neonatal starvation period, autophagosomes are apparent in the heart muscle of GFP-LC3 transgenic mice (13), and autophagosome formation in the heart is absent in Atg5-null mice (14). In the adult heart, autophagy is required for homeostasis, because cardiac specific loss of Atg5 leads to cardiac hypertrophy, left ventricular dilation, contractile dysfunction, and ultimately heart failure (16). During nutrient deprivation, autophagy is induced in the adult mouse heart in vivo and in cultured neonatal rat cardiomyocytes (13, 17). Increased cardiac autophagy also occurs with dilated cardiomyopathy, heart failure, ischemia/reperfusion, aortic stenosis, and in hibernating myocardium (11, 18–21). It has been suggested that the duration and severity of autophagy may determine whether autophagy is protective or detrimental in response to ischemia/reperfusion in the heart (15). However, factors that are responsible for inducing and regulating autophagy in cardiomyocytes have not been reported previously.
FoxO transcription factors have been implicated in regulating diverse cellular functions, including differentiation, proliferation, metabolism, and survival (22). There are multiple mechanisms that post-translationally regulate transcriptional activity of FoxO transcription factors (23, 24). The FoxO subfamily of transcription factors consists of FoxO1, FoxO3, and FoxO4, which are subject to inhibition by growth factors, including insulin and insulin-like growth factor-1 (25). During growth factor limitation, the inhibition of FoxO activity by phosphatidylinositol 3-kinase/AKT is relieved, and the dephosphorylated FoxO transcription factors are activated and become localized to the nucleus (23, 26, 27). Sirt1, a mammalian ortholog of NAD+-dependent protein deacetylase, Sir2, promotes the activity of FoxO transcription factors by deacetylation under conditions of oxidative stress (24). FoxO target genes are involved in atrophy in skeletal and cardiac myocytes (26, 28), autophagy in skeletal myocytes (29, 30), differentiation in skeletal and smooth muscle (31), or inhibition of cell cycle progression in neonatal cardiomyocytes (27). FoxO1 null mice die at E10.5 from vascular defects, and FoxO3 null mice develop cardiac hypertrophy as adults, suggesting a possible role for FoxO3 in regulating cardiomyocyte cell size (32, 33). Additional evidence for FoxO regulation of cardiomyocyte cell size is that increased expression of FoxO1 or FoxO3 inhibits growth factor-induced cardiac myocyte hypertrophy in cultured cardiomyocytes (26, 33). In skeletal muscle, increased FoxO function leads to decreased myocyte cell size through induction of atrophy and autophagy pathway genes, including LC3, Gabarapl1, and Atg12 (28–30). The function of FoxO1 and FoxO3 in regulation of autophagy pathway genes in cardiomyocytes has not been demonstrated previously.
Here we examine the roles of FoxO1 and FoxO3 transcription factors in regulating autophagy in rat neonatal cardiomyocytes as well as in mice subjected to starvation or cardiac ischemia/reperfusion injury. Glucose deprivation of cultured cardiomyocytes leads to decreased cell size, induces autophagy, and promotes FoxO nuclear localization and binding to autophagy pathway gene regulatory sequences. Increased expression of FoxO1 or FoxO3 results in decreased cardiomyocyte cell size and activation of autophagic vacuole formation and autophagy pathway gene expression. In addition, using dominant negative FoxO1 (Δ256) adenovirus, we show that FoxO activity is necessary to regulate cell size in starved rat neonatal cardiomyocytes. In vivo, starvation or ischemic injury results in FoxO dephosphorylation and induction of autophagy in the heart. Together, these studies provide evidence that FoxO1 and FoxO3 regulate cardiomyocyte cell size and promote autophagy pathway gene expression in cardiomyocytes.
EXPERIMENTAL PROCEDURES
Neonatal Rat Cardiomyocyte Isolation and Culture
Primary neonatal rat cardiomyocytes were isolated from hearts of 1–2-day-old Sprague-Dawley rat pups as described previously (34). After separation from fibroblasts, enriched cardiomyocytes were plated on 2-well chamber slides for immunofluorescence or on 10-cm diameter dishes for Western blot and chromatin immunoprecipitation (ChIP)2 experiments. Cells were grown in DMEM (catalog number 10-017-CV, Cellgro) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. For starvation experiments, cells were grown in DMEM containing FBS and penicillin/streptomycin for 2 days. On the 3rd day, “starved” cells were replenished with glucose-free (catalog number 11966, Invitrogen) FBS-free medium, in parallel with “fed” cells replenished with medium containing 10% FBS and 1% penicillin/streptomycin, as reported previously (18). Additionally, cells were treated with DMEM glucose-supplemented media (4.5 g/liter; catalog number 11965, Invitrogen) and DMEM glucose-depleted media (catalog number 11966, Invitrogen) with no serum for fed and starved experiments with similar results. After 24 h, all cells were harvested either for RNA and protein isolation or fixed for immunofluorescence. In some experiments, the inhibitor of autophagosome formation, 3-methyladenine (3MA, 10 μm), was added to determine the requirement for autophagy in starvation-induced cardiomyocyte cell size reduction. Adenoviral infections were performed as described previously (27) with wild-type (WT), constitutively active (CA) FoxO1 or dominant negative FoxO1 (Δ256) obtained from D. Accili (35) and wild-type (WT) or CA-FoxO3 obtained from K. Walsh (26). Infection with cytomegalovirus β-galactosidase virus was used as a control for all experiments. Cells were infected with the respective viruses in serum-free medium for 6 h, which was then replaced with fresh serum-containing culture medium. After 24 h, cultures were washed with phosphate-buffered saline and harvested for immunostaining or RNA isolation.
Quantitation of Cellular Autophagic Flux
Analysis of autophagic flux was determined as described previously (36). Briefly, cardiomyocytes were transduced with Ad-GFP-LC3-expressing virus (18) (a kind gift from Jeff Robbins, Cincinnati Children's Hospital Medical Center) for 24 h, and medium was changed with either glucose-containing or glucose-depleted medium for another 24 h. In some experiments, cardiomyocytes were infected with Ad GFP-LC3 virus along with β-galactosidase, WT-FoxO1, CA-FoxO1, WT-FoxO3, or CA-FoxO3 virus for 48 h to determine the FoxO-mediated induction of autophagy. Autophagic flux was determined by incubating the cells for the last 4 h either in the absence (steady-state autophagosomes) or in the presence (cumulative autophagosomes) of a mixture of lysosomal inhibitors (bafilomycin A1 (10 nm), E64D (5 μg/ml), and pepstatin A methyl ester (5 μg/ml)) to inhibit autophagosome-lysosome fusion (36). Cardiomyocytes were fixed in 4% paraformaldehyde and observed with fluorescence microscopy. To quantify the autophagic response in a population of cells, the percentage of cells expressing more than five autophagic vacuoles was counted by observing the puncta at ×60 magnification. A minimum of 150 cells was scored for each condition in at least three independent experiments. For quantification of the autophagic response in a single cell, at least 15–20 representative GFP-LC3 puncta-containing cells from each experiment were randomly selected and observed at ×60 magnification to count the total number of LC3 puncta per cell (36).
Quantitative Real Time RT-PCR
Total RNA was isolated from fed and starved rat neonatal cardiomyocytes or adult mouse hearts for quantitative real time RT-PCR analysis (27). 1 μg of total RNA was used to generate cDNA with a Superscript II first strand synthesis kit (Invitrogen) according to the manufacturer's instructions. 1 μl of synthesized cDNA was used for analysis by quantitative RT-PCR (MJ Research Opticon 2) as described previously (37). Oligonucleotide primer sequences used for quantification of mouse LC3, Gabarapl1, and Atg12 were described previously (29). Corresponding oligonucleotide primers were designed for rat (r) as follows: rLC3 forward 5′-CGTCCTGGACAAGACCAAGT-3′ and rLC3 reverse 5′-AGTGCTGTCCCGAACGTCTC-3′; rGabarapl1 forward 5′-CATCGTGGAGAAGGCTCCTA-3′ and rGabarapl1 reverse 5′-ATACAGCTGTCCCATGGTAG-3′; and rAtg12 forward 5′-GGCCTCGGAGCAGTTGTTTA-3′ and rAtg12 reverse 5′-CAGCATCAAAACTTCTCTGA-3′. Amplification reactions were performed with 34 cycles of (94 °C for 30 s; 55 °C for 45 s; and 72 °C for 30 s), and normalized to ribosomal protein L7 for mouse (38) or GAPDH for rat (39). Normalized threshold cycle values of samples amplified in triplicate were used to calculate relative gene expression levels. Statistical significance of observed differences was determined by Student's t test and ANOVA (p < 0.05).
Immunofluorescence and Confocal Microscopy
Immunofluorescent staining of rat neonatal cardiomyocytes was performed as described previously (27). Antibodies used for immunofluorescence include the following: FoxO1 (Upstate) (1:100); FoxO3 (Upstate) (1:100); Gabarapl1 (Proteintech Group, Inc.) (1:100); and sarcomeric α-actinin (Sigma) (1:500). Following primary antibody incubation, cells were washed in phosphate-buffered saline and incubated with corresponding Alexa Fluor-488 goat anti-rabbit IgG or goat anti-mouse IgG secondary antibodies (1:200, Molecular Probes). As a negative control, all slides were incubated with only secondary antibody but with no primary to detect the background fluorescence (data not shown). Incubation with To-Pro3 (1:1000, Molecular Probes) for 1 h at room temperature was used to visualize nuclei. Immunofluorescence was detected using a Zeiss LSM 510 confocal microscope. Images were obtained using Zeiss LSM version 3.2 SP2 software. For cell size determination, Image J software was used to calculate cardiomyocyte surface area of at least 200 cells (α-actinin-positive cells) per experimental group. Statistical significance of observed differences in cardiomyocyte cell size was determined by Student's t test and ANOVA (p < 0.05).
Protein Isolation and Western Blotting
Protein lysates were isolated from the ventricles of fed and starved, sham and ischemic/reperfused mouse hearts and rat neonatal cardiomyocytes using ice-cold CelLytic MT tissue lysis buffer or CelLytic-M cell lysis buffer (Sigma), containing protease inhibitor mixture (Pierce) and phosphatase inhibitor (Pierce) according to the manufacturers' instructions. Western blots were performed as described previously (27), except that membranes were incubated with an alkaline phosphatase-conjugated secondary antibody (Thermo Scientific) (1:4000). Immunoblots were developed using chemifluorescent detection with the Vistra ECF reagent (Amersham Biosciences) and scanned using a Storm 860 (Amersham Biosciences). Signal intensities were quantified with ImageQuant 5.0 software (GE Healthcare). Antibodies used for immunoblot analysis included the following: FoxO1 (Santa Cruz Biotechnology) (1:200); phospho-FoxO1 (Ser-256) (Cell Signaling) (1:700); FoxO3 (Santa Cruz Biotechnology) (1:200); phospho-FoxO3 (Ser-318/321) (Cell Signaling) (1:700); LC3 (Nano Tools) (0.5 μg/ml); AKT (Cell Signaling) (1:2000); phospho-AKT (Ser-473) (1:1000); Sirt1 (Millipore) (1:1000). GAPDH (Santa Cruz Biotechnology) (1:10,000) antibody was used as a loading control. Statistical significance was determined by Student's t test (p < 0.05).
ChIP Assay
DNA-protein complexes in fed and starved cultured rat neonatal cardiomyocytes were cross-linked for 10 min by directly adding formaldehyde (Sigma) to the culture medium at a final concentration of 1%. The fixed cells were lysed with lysis buffer (EZ ChIP, Upstate) and sonicated two times for 7 s with output 5 (Virsonic 60; Virtis) with a 2-min refractory period. For immunoprecipitation, cell lysates were incubated with an antibody against FoxO1 (5 μg; Santa Cruz Biotechnology) or FoxO3 (5 μg; Santa Cruz Biotechnology) and incubated overnight at 4 °C. Immunoprecipitation with normal rabbit IgG was used as a negative control. Immunoprecipitation was performed according to the manufacturer's instructions (EZChIP, Upstate) with the exception that protein A-agarose beads were used. The immunoprecipitated and input DNA was subjected to PCR using the following primers: rGabarapl1 5′-TCAAAATGCAAGCACAGGAC-3′ and 5′-TTCCCTCCCCTTCAAGTCTCT-3′ and rAtg12 5′-TCTTCACCATCAAGCCAAAA-3′ and 5′-GTTAATGAGCTCTCATTG-3′ to amplify rat promoter regions corresponding to mouse regulatory sequences that contain previously reported FoxO1- and FoxO3- binding sites (29). Signal intensities were quantified with ImageQuant 5.0 software (GE Healthcare).
Mouse Starvation
To assess the effects of short term nutrient deficiency on induction of cardiomyocyte autophagy, adult FVBN mice (3 months old) were fasted for 48 h (40). Throughout this time, animals had no access to food, but did have free access to water. Control mice had access to food and water ad libitum. After 48 h, the mice were sacrificed by CO2 asphyxiation; the hearts were explanted, and RNA or protein lysates were isolated as described previously (27). All aspects of animal care and use in this study were approved by the Institutional Animal Care and Use Committee (IACUC) of the Cincinnati Children's Medical Hospital Center.
Ischemia Reperfusion in Mice in Vivo
Cardiac ischemia-reperfusion injury was performed in 8–10-week-old mice as described previously (41, 42). Briefly, ischemia was achieved using an 8-0 prolene suture around the left anterior descending coronary artery (LAD) and tied with a slipknot. The thoracotomy was closed, and the mice were revived for a 40-min ischemic injury period, after which the knot was released and the heart was allowed to reperfuse for 30 min. After reperfusion, mice were sacrificed by CO2 asphyxiation, and hearts were harvested for protein isolation.
Statistical Analysis
Statistical analyses between the experimental groups were performed using a Student's t test or one-way ANOVA when comparing multiple groups. Values of p < 0.05 were considered significant.
RESULTS
Nutrient Deprivation Reduces Cell Size and Induces Autophagy in Cultured Rat Neonatal Cardiomyocytes
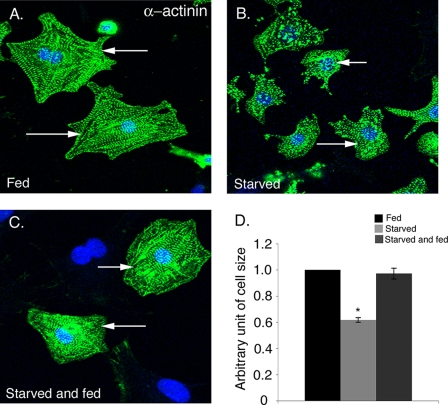
To investigate the effect of nutrient deprivation on cardiomyocyte cell size in vitro, cultured neonatal rat cardiomyocytes were incubated either in medium containing serum and glucose (fed) or in medium deficient in both serum and glucose (starved). After 24 h of nutrient deprivation, fed and starved cardiomyocytes were immunostained with sarcomeric α-actinin, and cell size was determined using ImageJ software. The cardiomyocytes subjected to nutrient deprivation are obviously smaller, and quantification of cell size indicates a 40% reduction compared with fed cardiomyocytes (Fig. 1, A, B, and D). In addition, the α-actinin-positive starved cardiomyocytes had fewer sarcomeres (Fig. 1B) compared with fed cardiomyocytes (Fig. 1A). However, increased cell death was not apparent in starved rat neonatal cardiomyocytes compared with fed cells (data not shown). Cardiomyocytes were also incubated with media containing glucose with no serum (fed) and compared with cells incubated with glucose-depleted media (starved) with comparable results (data not shown). To determine whether the effect of glucose deprivation on cardiomyocytes size is reversible, the starved cardiomyocytes were subsequently incubated in glucose-containing media for 24 h. Starved cells, which were replenished with glucose-containing media, exhibit increased cell size relative to starved (compare Fig. 1, B–D) with cell sizes comparable with fed cardiomyocytes (Fig. 1A). Therefore, the reduction in cell size induced by starvation is completely reversible upon glucose supplementation. Altogether, our results indicate that 24 h of starvation causes a reversible reduction in cardiomyocyte cell size but does not increase cell death.
FIGURE 1.
Starvation of cardiomyocytes results in a reversible reduction in cell size. Rat neonatal cardiomyocytes were incubated in media containing 10% FBS (Fed) (A) or in serum/glucose depleted (Starved) media (B) for 24 h. C, starved cardiomyocytes were then replenished with fed media for 24 h demonstrating reversibility of cell size reduction. D, cardiomyocyte cell size was quantified for fed (black bar), starved (light gray bar), and starved and fed (dark gray bar) cells (n = 200) for three independent experiments. Cardiomyocytes are immunostained with sarcomeric α-actinin (cardiac muscle actinin, green) and To-Pro3 (nucleus, blue). The average cardiomyocyte area in the fed condition is 1128 μm2 and starved condition is 565 μm2 (at least 200 cells from two independent experiments). Representative cells from each group are indicated with white arrows. Significance (*) was determined by one-way ANOVA (p < 0.05; n = 3).
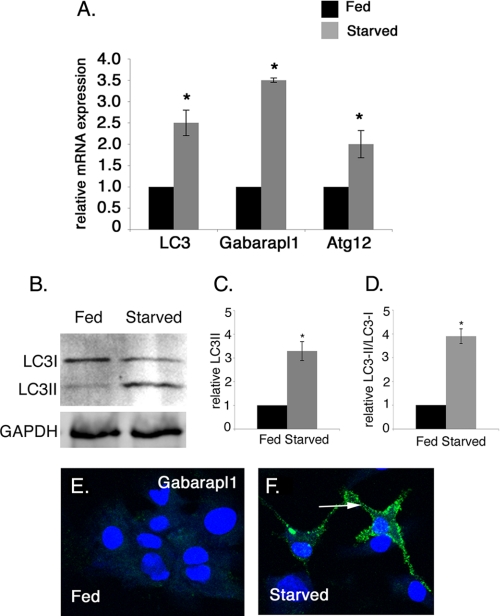
Multiple assays were used to determine whether autophagy is induced in starved cardiomyocytes with reduced cell size (Fig. 2 and Fig. 3). The expression of autophagy pathway genes LC3, Gabarapl1, Atg12 in fed and starved cardiomyocytes was quantified by real time RT-PCR. In these experiments, starved cardiomyocytes have increased gene expression of LC3 (2.5-fold), Gabarapl1 (3.5-fold), and Atg12 (2-fold) compared with fed cardiomyocytes, indicating starvation induces autophagy pathway gene expression in cultured cardiomyocytes (Fig. 2A). To further confirm the activation of autophagy in starved cardiomyocytes, LC3 protein modification was evaluated by Western blot analysis. During autophagy, conjugation of phosphatidylethanolamine to a glycine residue converts LC3I from its soluble form to the autophagic vesicle-associated form LC3II (43). In starved cardiomyocytes, relative LC3II only and the ratio of LC3II/LC3I are elevated (3.3- and 3.9-fold, respectively) compared with fed cardiomyocytes, further demonstrating induction of autophagy (Fig. 2, B–D). Likewise Gabarapl1 protein expression is increased in starved rat neonatal cardiomyocytes as detected by immunostaining, providing additional evidence for the induction of autophagy in starved cardiomyocytes (Fig. 2, E and F). Together these studies show that the reduction in cell size of starved cardiomyocytes is coincident with the up-regulation of multiple markers of the autophagy pathway in these cells.
FIGURE 2.
Starvation induces autophagy-related gene expression in rat neonatal cardiomyocytes. A, expression of autophagy pathway genes LC3 (2.5-fold), Gabarapl1 (3.5-fold), and Atg12 (2.0-fold) mRNA levels is increased as detected by real time RT-PCR of 24-h starved versus fed rat neonatal cardiomyocytes. B, expression of the LC3II protein isoform is increased as detected by Western blot in starved cardiomyocytes compared with fed cardiomyocytes. GAPDH was used as loading control. C and D, densitometric analysis shows 3.4- and 3.9-fold induction in the level of LC3II and the ratio of LC3II/LC3I in starved cardiomyocytes compared with fed cardiomyocytes. E and F, cardiomyocytes were immunostained with Gabarapl1 antibody (green) and To-Pro3 (blue) for nuclear staining. Gabarapl1 protein expression in starved cardiomyocytes is indicated by an arrow. Significance (*) was determined by Student's t test (p < 0.05; n = 3).
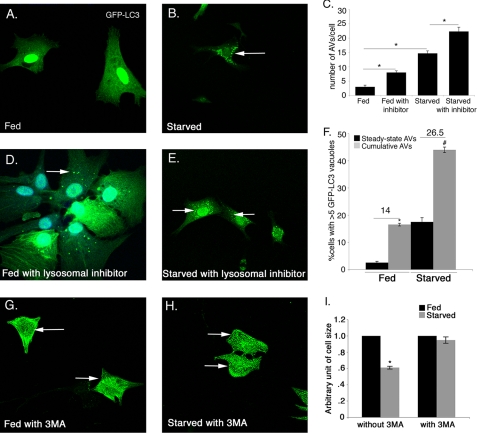
FIGURE 3.
Autophagic flux is increased in starved cardiomyocytes. Cardiomyocytes infected with Ad-GFP-LC3 adenovirus were analyzed for induction of autophagy in fed and starved conditions. Autophagic flux in cardiomyocytes was determined in the absence or presence of lysosomal inhibitors. A, fed cardiomyocytes show diffuse, uniform expression of GFP-LC3 with no detectable LC3 puncta. B, starvation results in increased GFP-LC3 puncta in cardiomyocytes. Increased GFP-LC3 puncta were observed in the presence of lysosomal inhibitors both fed (D) and starved (E) cardiomyocytes compared with the absence of inhibitors (A and B). C, number of AVs/cell was significantly increased in starved cardiomyocytes compared with fed cardiomyocytes and also in presence of inhibitors compared with their respective controls in both conditions. F, significant increase in autophagic flux was observed in starved compared with fed cardiomyocytes. GFP-LC3 puncta are indicated by white arrows in B, D, and E. G–I, cardiomyocytes were incubated in fed and starved media either in the presence or in the absence of the specific autophagy inhibitor 3MA (10 μm) for 24 h. Cardiomyocytes are immunostained with sarcomeric α-actinin (cardiac muscle actinin, green) and cell size was measured. Inhibition of autophagy with 3MA completely rescues the starvation-induced cell size reduction. Significance (*) was determined by one-way analysis of variance (ANOVA) (p < 0.05; n = 3).
To quantify the induction of autophagy in fed and starved conditions, cardiomyocytes were infected with Ad-GFP-LC3. Induction of autophagy was determined by counting the fluorescent GFP-LC3 puncta indicative of autophagic vacuoles (AV). Autophagy was quantified as either the percentage of cells displaying numerous GFP-LC3 puncta (more than five per cell) or total number of GFP-LC3 puncta present per cell. In addition, autophagic flux was determined in Ad-GFP-LC3-infected cardiomyocytes by comparing cultures that were fed or starved either in the absence of a mixture of lysosomal inhibitors to show steady-state autophagy or in the presence of inhibitors to determine the cumulative AVs. The difference between the percentage of cells with cumulative AVs and steady-state AVs is indicative of the autophagic flux (36). Under normal fed conditions, GFP-LC3 is uniformly/diffusely distributed throughout the cell with very few detectable AVs (Fig. 3A). In contrast, upon glucose deprivation, there is a significant increase in both the number of autophagic vacuoles per cell (15 AVs/cell) and also in the percentage of cells with numerous AVs (17.5% in starved compared with 3% in fed) (Fig. 3, A–C and F). Interestingly, in the presence of lysosomal inhibitors, both in the fed and starved conditions, the GFP-LC3 puncta/cell is significantly increased (3 in fed versus 8 in fed with inhibitor and 14 in starved versus 23 in starved with inhibitor) (Fig. 3, A–E). Moreover, the percentage of cells expressing numerous (>5) GFP-LC3 puncta is also increased in the presence of lysosomal inhibitors both in fed and starved cardiomyocytes (3% in fed (steady state) versus 17% in fed with inhibitor (cumulative) and 17.5% in starved (steady state) versus 44% in starved with inhibitor (cumulative)) compared with the absence of inhibitors (Fig. 3F). The difference between the percentage of cells with cumulative and steady-state AVs in fed versus starved conditions is 14 and 26.5%, respectively (Fig. 3F). These data show fed cells have normal low level autophagy, whereas there is a significant increase in the autophagy in starved cardiomyocytes compared with fed cardiomyocytes.
To test whether the autophagy is required for cardiomyocyte cell size reduction upon starvation, autophagy was inhibited in starved cardiomyocytes using the specific inhibitor of early autophagosome formation (3MA), and then the cell size was measured. Interestingly, inhibition of autophagy with 3MA completely rescues the starvation-induced cell size reduction compared with fed cardiomyocytes (Fig. 3, G–I). In contrast, the treatment with more general lysosomal inhibitors (bafilomycin A1, E64D, and pepstatin A methyl ester) that inhibit all lysosomal processing does not rescue the reduction in cell size in starved cardiomyocytes (Fig. 3E). This is likely due to more specific inhibition of autophagosome formation by 3MA rather than the inhibition of more generalized lysosome-associated processing used to measure autophagic flux. This result indicates that inhibition of autophagy early in the pathway is sufficient to prevent starvation-induced cell size reduction. Therefore, autophagy contributes to cell size regulation in starved cardiomyocytes.
Activation of FoxO1 or FoxO3 Reduces Cardiomyocyte Cell Size and Induces Autophagy-related Gene Expression
The function of FoxO transcription factors in regulation of cardiomyocyte cell size and induction of autophagy pathway gene expression was examined in cardiomyocytes infected with recombinant adenovirus expressing FoxO1 or FoxO3. Previous studies demonstrated that increased activity of FoxO3 leads to decreased cardiomyocyte cell size, and loss of FoxO3 leads to cardiac hypertrophy in mice (26, 33). However, the ability of FoxO1 to regulate cardiomyocyte cell size has not been reported previously. In addition, neither FoxO1 nor FoxO3 has previously been associated with induction of autophagy pathway gene expression in cardiomyocytes. FoxO1 gain of function was achieved using adenovirus-expressing wild-type FoxO1 (WT-FoxO1) or constitutively active-FoxO1 (CA-FoxO1) (35). Similarly, viruses expressing wild-type FoxO3 (WT-FoxO3) or constitutively active-FoxO3 (CA-FoxO3) were used for FoxO3 gain of function (44). CA-FoxO1 and CA-FoxO3 cannot be phosphorylated by AKT and therefore maintain nuclear activity in the unphosphorylated form. Increased FoxO nuclear function also has been observed with WT-FoxO1 and WT-FoxO3 adenovirus infection (27). Together these viruses were used to examine the effects of increased FoxO1 activity on cardiomyocyte cell size regulation and to determine whether increased FoxO1 or FoxO3 function is sufficient to induce autophagy pathway gene expression in cardiomyocytes.
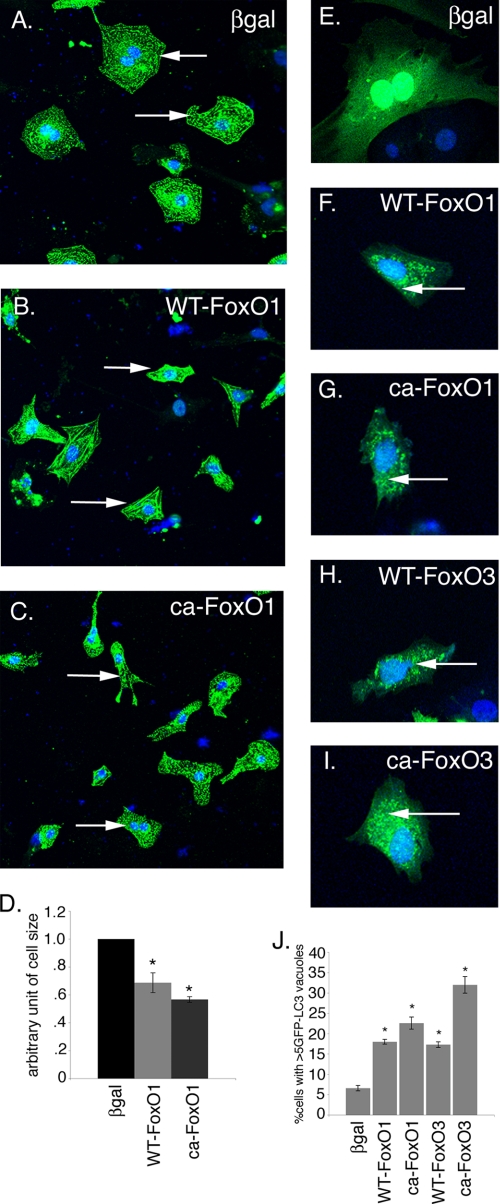
To test whether increased activity of FoxO1 affects cardiomyocyte cell size, neonatal rat cardiomyocytes were infected with adenoviral vectors expressing either WT-FoxO1 or CA-FoxO1. Increased expression of either WT-FoxO1 (Fig. 4B) or CA-FoxO1 (Fig. 4C) leads to a statistically significant decrease in cell size compared with β-galactosidase-infected (Fig. 4A) cardiomyocytes. 30 and 40% reduction in cell size was observed in WT-FoxO1 and CA-FoxO1, respectively, compared with β-galactosidase-infected cardiomyocytes (Fig. 4D). These data demonstrate that FoxO1 is sufficient to reduce cardiomyocyte cell size, as was demonstrated previously for FoxO3. Decreased expression of genes associated with hypertrophy also was observed in WT-FoxO1- and CA-FoxO1-infected cardiomyocytes (data not shown). In contrast to starved cardiomyocytes, striations were observed in WT-FoxO1- and CA-FoxO1-infected cardiomyocytes, and the reduction in cell size was not as severe as was observed in starved cardiomyocytes (Fig. 1). No evidence for increased apoptosis was observed in cardiomyocytes infected with adenovirus expressing either WT-FoxO1 or CA-FoxO1, as was also reported for embryonic cardiomyocyte cultures (27). Together these studies suggest that both FoxO1 and FoxO3 are sufficient for decreased cardiomyocyte cell size.
FIGURE 4.
Increased FoxO1 activation reduces cell size and induces autophagy in cardiomyocytes. Cardiomyocytes were infected with WT-FoxO1, CA-FoxO1, or β-galactosidase adenovirus for 24 h and immunostained with sarcomeric α-actinin (green) and To-Pro3 nuclear stain (blue). A–C, infection of cardiomyocytes with either WT-FoxO1 (B) or CA-FoxO1 adenovirus (C) results in decreased cell size compared with β-galactosidase-infected cells (βgal) (A). Arrows indicate representative cardiomyocytes from each treated group. D, quantitative analysis indicates significant reduction (>30%) in WT-FoxO1 and CA-FoxO1-infected cardiomyocyte cell size compared with β-galactosidase-infected cardiomyocytes. E–I, Ad-GFP-LC3-infected cardiomyocytes were co-infected with β-galactosidase, WT-FoxO1, CA-FoxO1, WT-FoxO3, or CA-FoxO3 adenovirus. Increased in GFP-LC3 puncta are apparent compared with β-galactosidase-infected cardiomyocytes (E–I, arrows). J, quantitative analysis indicates a significant increase in the percentage of cells expressing more than five GFP-LC3 puncta in FoxO-infected cardiomyocytes compared with β-galactosidase-infected cardiomyocytes. Significance (*) was determined by one-way analysis of variance (p < 0.05; n = 3).
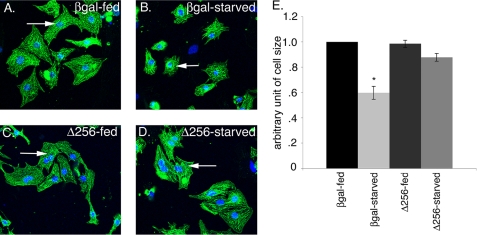
To determine whether FoxO is necessary for cell size reduction in starved cardiomyocytes, cells were infected with adenovirus expressing Δ256-FoxO (dominant negative) to inhibit FoxO activity, followed by cell size determination in fed and starved conditions. The dominant negative FoxO1 (Δ256) protein contains the entire forkhead DNA binding domain but lacks the transactivation domain, thus inhibiting the transcriptional activity of FoxO1, FoxO3, and FoxO4 (45). Cardiomyocytes infected in parallel with β-galactosidase serve as a control. As expected, β-galactosidase-infected starved cardiomyocytes show 40% reduction in cell size compared with β-galactosidase-infected fed cardiomyocytes (Fig. 5, A, B, and E). Interestingly, starved cardiomyocytes infected with dominant negative Δ256-FoxO fail to reduce cell size and have comparable cell size with fed cardiomyocytes (compare Fig. 5, C with D and E). These results suggest that during starvation FoxO activity is required for regulating cardiomyocyte cell size, and inhibition of FoxO activity with dominant negative FoxO virus prevents cardiomyocyte cell size reduction upon starvation. Together these studies indicate that FoxO activity is necessary for reduction in cell size during starvation.
FIGURE 5.
FoxO transcriptional function is necessary to regulate cell size in starved cardiomyocytes. Cardiomyocytes were infected with β-galactosidase or dominant negative FoxO1 (Δ256) adenovirus for 24 h, and then cells were either replenished with fed or starved media for another 24 h. A and B, β-galactosidase (βgal)-infected cardiomyocytes subjected to starvation have a significant reduction in cell size (B) compared with fed (A) cardiomyocytes. C and D, dominant negative FoxO1 (Δ256)-infected cardiomyocytes subjected to starvation (D) fail to reduce cell size compared with fed cardiomyocytes (C). E, cardiomyocyte cell size (n = 200) was quantified for β-galactosidase and Δ256-infected cardiomyocytes in both fed and starved conditions for three independent experiments. Cardiomyocytes are immunostained with sarcomeric α-actinin (cardiac muscle actinin, green) and To-Pro3 (nucleus, blue). Representative cells for each condition were shown with white arrows. Significance (*) was determined by one-way analysis of variance (ANOVA) (p < 0.05; n = 3).
Because increased activity of FoxO1 or FoxO3 leads to decreased cell size, the ability of FoxO1 or FoxO3 to induce autophagy and autophagy pathway gene expression in cardiomyocytes was examined. To determine whether FoxO transcription factors are able to induce autophagy in cardiomyocytes, rat neonatal cardiomyocytes were infected with Ad GFP-LC3 virus along with β-galactosidase, WT-FoxO1, CA-FoxO1, WT-FoxO3, or CA-FoxO3 virus for 48 h. The induction of the autophagic response was determined by counting the percentage of cells expressing more than five GFP-LC3 puncta. A significant increase in the percentage of cells expressing numerous GFP-LC3 puncta was observed for all FoxO gain of function viruses tested (WT-FoxO1, CA-FoxO1, WT-FoxO3, and CA-FoxO3) compared with β-galactosidase (Fig. 4, E–J). The most robust autophagic response was observed in cardiomyocytes infected with CA-FoxO3, but significant increases in autophagy were observed with WT-FoxO1, WT-FoxO3, and CA-FoxO3 viruses compared with β-galactosidase (Fig. 4J). Together these studies demonstrate increased activity of FoxO1 or FoxO3 is sufficient to induce autophagy in cardiomyocytes.
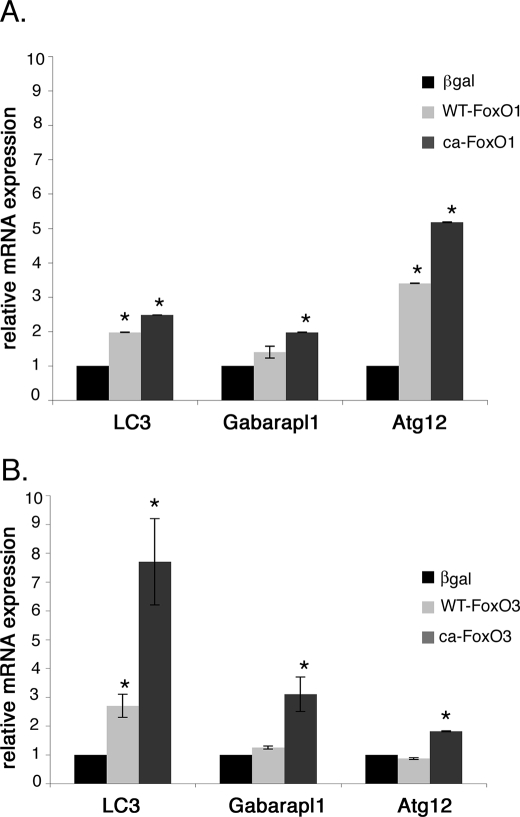
Rat neonatal cardiomyocytes were infected with WT-FoxO1, WT-FoxO3, CA-FoxO1, or CA-FoxO3 viruses, and induction of autophagy pathway gene expression was determined by real time quantitative RT-PCR. Gene expression of autophagy pathway genes, LC3, Gabarapl1, and Atg12, was determined by real time RT-PCR. Overexpression of either CA-FoxO1 or CA-FoxO3 in cardiomyocyte results in significantly increased gene expression of LC3, Gabarapl1, and Atg12 relative to levels in β-galactosidase-infected cells (Fig. 6, A and B). Increased autophagy pathway gene expression also was observed for WT-FoxO1 or WT-FoxO3 infected cells, but wild-type proteins were not as efficient as CA-FoxO1 or CA-FoxO3. It is interesting to note that Atg12 is the most highly up-regulated gene with increased FoxO1 function (3.5-fold with WT-FoxO1 and 5-fold with CA-FoxO1), whereas LC3 was most highly up-regulated gene with increased FoxO3 function (2.7-fold with WT-FoxO3 and 7.7-fold with CA-FoxO3). This could be due to the differential activity of FoxO1 and FoxO3 in controlling early and late stages of autophagosome formation, because Atg12 functions in the formation of the phagophore precursor, and LC3 is involved later in membrane elongation (11). Together these studies provide evidence that increased activity of either FoxO1 or FoxO3 is sufficient for induction of autophagy pathway gene expression associated with decreased cardiomyocyte cell size.
FIGURE 6.
FoxO1 and FoxO3 activate autophagy-related gene expression in cardiomyocytes. Expression of autophagy pathway genes LC3, Gabrapl1, and Atg12 was quantified by real time RT-PCR in cardiomyocytes infected with WT- or CA-FoxO1 (A) and WT- or CA-FoxO3 (B) recombinant adenovirus for 24 h. LC3, Gabarapl1, and Atg12 expression was significantly up-regulated with overexpression of FoxO1 or FoxO3 in cardiomyocytes. Atg12 is the most highly up-regulated gene in FoxO1-infected cardiomyocytes, whereas LC3 is the most highly up-regulated gene in FoxO3-infected cardiomyocytes. The base-line levels of LC3, Gabarapl1, and Atg12 relative to GAPDH expression are 0.96, 2.18, and 2.6, respectively. Significance (*) was determined by one-way ANOVA (p < 0.05; n = 3).
Endogenous FoxO1 and FoxO3 Are Localized to the Nucleus and Bind Autophagy Pathway Gene Regulatory Sequences in Starved Cardiomyocytes
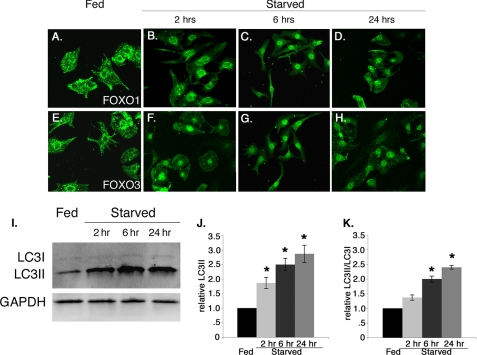
Phosphorylation of FoxO transcription factors by AKT under growth conditions results in nuclear export and subsequent inactivation of their transcriptional activity (25). Under conditions of growth factor deprivation or stress, unphosphorylated FoxO transcription factors remain in the nucleus to activate target gene expression. Therefore, we examined the subcellular localization of endogenous FoxO1 and FoxO3 in fed versus starved cultured rat neonatal cardiomyocytes. In starved cardiomyocytes, nuclear localization of endogenous FoxO1 and FoxO3 was observed within 2 h and was maintained for at least 24 h of starvation (Fig. 7, B–D and F–H) with a concomitant significant increase in autophagic induction as shown by the increased level of LC3II only and the ratio of LC3II/LC3I (Fig. 7, I–K). By contrast in fed cardiomyocytes, endogenous FoxO1 and FoxO3 are both predominantly localized in the cytoplasm or the perinuclear region of the cell at all time points examined (Fig. 7, A and E). Therefore, starvation of cardiomyocytes leads to increased nuclear localization of endogenous FoxO1 and FoxO3 proteins, which is consistent with increased transcriptional function and a role in up-regulation of autophagy pathway genes. In addition, both FoxO nuclear localization and LC3 accumulation are apparent within 2 h of starvation conditions.
FIGURE 7.
Starvation promotes nuclear localization of endogenous FoxO1 and FoxO3 in cardiomyocytes. A–H, fed (DMEM plus glucose, no serum) and starved (DMEM alone) treated rat neonatal cardiomyocytes were immunostained with FoxO1 (green, A–D) and FoxO3 (green, E–H) antibodies. In fed cardiomyocytes FoxO1 (A) and FoxO3 (E) are localized to the cytoplasm and perinuclear region. Starvation causes predominant nuclear localization of FoxO1 (B–D) and FoxO3 (F–H) within 2 h, and nuclear localization is maintained at least for 24 h. I–K, starvation induces autophagy within 2 h, which is maintained for 24 h as measured by the increased level of LC3II and the ratio of LC3II/LC3I compared with fed cardiomyocytes. Significance (*) was determined by one-way ANOVA (p < 0.05; n = 3).
ChIP was performed to determine whether starvation promotes FoxO transcription factor binding to the promoter regions of autophagy pathway genes in cardiomyocytes (Fig. 8). In mouse skeletal muscle, FoxO3 can bind to the regulatory sequences of LC3, Gabarapl1, and Atg12 genes that contain FoxO consensus binding sites (29). However, the binding of FoxO1 to these elements or FoxO interactions with corresponding sequences in rat autophagy pathway genes in cardiac muscle have not been reported previously. FoxO transcription factors share a characteristic DNA binding domain that recognizes the specific consensus sequence (T/C/G)(G/A/T)AAA(C/A)A (29, 46). For these experiments, the region amplified from the promoter region of rat Gabarapl1 is 94% identical to the corresponding mouse promoter region (29) and contains the FoxO consensus DNA-binding site “GGAAACA” (data not shown). Similarly, the amplified region of the rat Atg12 promoter is 42% identical to the corresponding mouse promoter (29) and contains the FoxO consensus DNA-binding site “CAAAACA” (data not shown).
FIGURE 8.
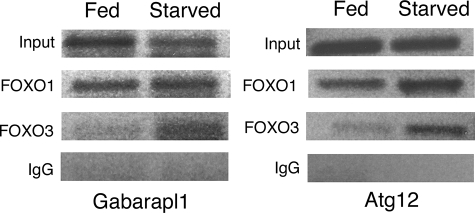
Increased binding of FoxO1 and FoxO3 to autophagy gene regulatory sequences in starved cardiomyocytes. Chromatin immunoprecipitation was performed on fed and starved cardiomyocytes. Cross-linked DNA-protein complexes from fed and starved cardiomyocytes were immunoprecipitated with either FoxO1, FoxO3, or IgG (negative control) antibodies. Precipitated DNA was then PCR-amplified using primers specific for Gabarpl1 and Atg12 gene regulatory sequences containing identified FoxO-binding sites. Increased binding of FoxO3 to Atg12 (2.7-fold) and Gabarapl1 (1.7-fold) promoters was observed under starvation conditions relative to fed controls. Increased FoxO1 binding was observed in both conditions (1.5-fold for Gabarapl1 and 1.65-fold for Atg12), but induction was reduced relative to FoxO3 compared with fed cardiomyocytes. Input represents cross-linked DNA amplified without immunoprecipitation with respective antibodies.
Cross-linked DNA-protein complexes were isolated from fed and starved cultured rat neonatal cardiomyocytes and immunoprecipitated with FoxO1- and FoxO3-specific antibodies. Immunoprecipitated DNA fragments were used as a template to amplify regulatory sequences using primers that span regions of rat Gabarapl1 and Atg12 promoters containing reported FoxO1- and FoxO3-binding sites (46). Increased binding of FoxO3 to Atg12 (2.7-fold) and Gabarapl1 (1.7-fold) promoters was observed under starvation conditions relative to fed controls. FoxO1 binding was apparent in both conditions (1.5-fold for Gabarapl1 and 1.65-fold for Atg12), but induction was reduced relative to FoxO3. The observed FoxO1 and FoxO3 binding to Gabarapl1 and Atg12 regulatory sequences in fed cardiomyocytes is likely due to base-line level of autophagy required for cellular homeostasis as observed in Fig. 3, A and D. Likewise, the increased FoxO1 binding to the Gabrapl1 and Atg12 promoter regions relative to that of FoxO3 in normal fed cardiomyocytes could be related to induction of early stage autophagy genes, as demonstrated in Fig. 6. Together these data provide evidence that both FoxO1 and FoxO3 interact with regulatory sequences of Gabarapl1 and Atg12 genes that are up-regulated in cardiomyocytes during starvation-induced autophagy (Fig. 2A).
Starvation Activates FoxO Transcription Factors and Induces Autophagy Gene Expression in the Mouse Heart in Vivo
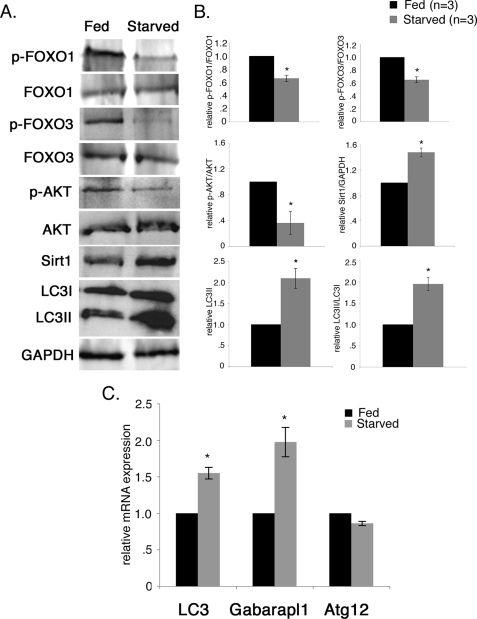
We observed activation of FoxO transcription factors and induction of autophagy pathway gene expression in starved cultured rat neonatal cardiomyocytes (Figs. 2, 7, and 8). Therefore, we examined whether FoxO1 and FoxO3 are activated, and autophagy pathway gene expression is induced in starved mouse hearts in vivo. To assess the effects of short term nutrient deficiency on induction of cardiomyocyte autophagy in vivo, adult FVBN mice (3 months) were fasted for 48 h (40). Throughout this time, animals had no access to food but did have free access to water. There was no change in the heart weight/body weight ratio during short term nutrient deprivation (data not shown). Induction of the autophagic response and autophagy pathway gene expression was examined in starved mouse hearts. The level of LC3II and the ratio of LC3II/LC3I from the starved mouse heart are both increased by 2-fold compared with the fed mouse heart, demonstrating that autophagy is induced in the starved mouse heart (Fig. 9, A and B).
FIGURE 9.
Starvation activates FoxO transcription factors and induces autophagy pathway gene expression in mouse hearts in vivo. To assess the effects of short term nutrient deficiency on induction of cardiomyocyte autophagy in vivo, adult FVBN mice (3 months) were fasted for 48 h. The phosphorylation status of FoxO transcription factors in heart cell extracts from fed and starved mice was examined by Western blot (A) and quantified by densitometric analysis in B. In fed mice, phosphorylated p-FoxO1 and p-FoxO3 are increased relative to starved mice, whereas the total FoxO1 and FoxO3 protein level was unchanged. In the starved mouse heart, p-FoxO1 and p-FoxO3 was reduced by 30% compared with the fed mouse heart. Decreased AKT phosphorylation and increased Sirt1 also were observed in starved mouse heart. The level of LC3II and the ratio of LC3II/LC3I from the starved mouse heart are both increased by 2.0-fold compared with the fed mouse heart, demonstrating that autophagy is induced in the starved mouse heart. Significance (*) was determined by Student's t test (p < 0.05; n = 3). C, expression of autophagy pathway genes in fed and starved mouse hearts was determined by quantitative real time RT-PCR. Significantly increased gene expression of LC3 (1.6-fold) and Gabrapl1 (2-fold) was observed in the starved mouse heart compared with the fed mouse heart. No significant change in Atg12 gene expression was observed between fed and starved mouse hearts. Significance (*) was determined by Student's t test (p < 0.05; n = 4).
The phosphorylation status of FoxO transcription factors was examined as an indicator of FoxO activation in heart cell lysates from fed and starved mice. Under normal fed conditions, phosphorylated forms of both FoxO1 and FoxO3 are increased relative to the levels observed in starved mouse hearts, whereas the total FoxO1 and FoxO3 protein levels were unchanged (Fig. 9A). In the starved mouse heart, phosphorylated FoxO1 and FoxO3 are reduced by 30% compared with the fed mouse heart (Fig. 9B). FoxO activation with growth factor deprivation can occur with reduced AKT activity (26). In support of this mechanism in vivo, we observed 70% reduction in the level of AKT phosphorylation in starved mouse heart compared with fed mouse heart (Fig. 9, A and B). Likewise, nutrient depletion activates Sirt1, an upstream activator of FoxO transcription factors both in vitro and in vivo (47, 48). Sirt1 protein was significantly increased (1.5-fold) in the starved mouse heart compared with fed mouse heart (Fig. 9, A and B). Together, these data show that the decreased phosphorylation of FoxO1 and FoxO3 in the starved mouse heart is consistent with increased FoxO activation in cardiomyocytes resulting from nutrient deprivation in vivo. The combination of decreased AKT activation and increased Sirt1 likely regulates increased FoxO activity with starvation. Expression of FoxO target genes related to autophagy was assessed in starved mouse hearts. LC3 (1.6-fold) and Gabarapl1 (2-fold) gene expression is significantly up-regulated in the starved mouse heart compared with the fed mouse heart (Fig. 9C). However, the level of Atg12 is not significantly changed in the starved mouse heart compared with fed mouse heart (Fig. 9C), which could be due to late stage of autophagic response after 48 h. Together these studies demonstrate that FoxO1 and FoxO3 transcription factors are activated with induction of cardiac autophagy that occurs as a result of starvation at the organismal level.
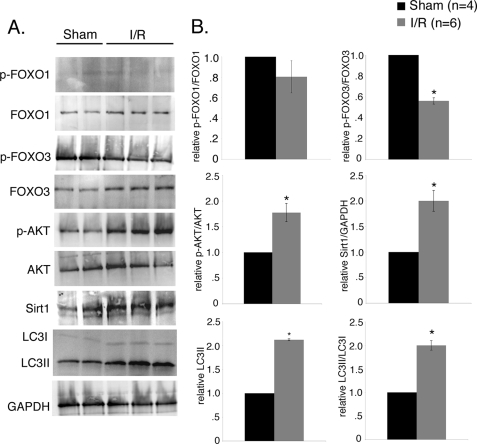
Ischemia/Reperfusion Injury Induces Autophagy with Concomitant Increase in FoxO3 Activity in Mouse Heart
Previous studies have shown that ischemia/reperfusion injury induces autophagy in mouse cardiomyocytes in vitro and in vivo (18, 19, 21, 49). Therefore, we examined the induction of FoxO transcription factors during ischemia/reperfusion in mouse hearts in vivo (Fig. 10). Induction of autophagy was confirmed by increased level of LC3II and the ratio of LC3II/LC3I by 2-fold in mouse hearts subjected to ischemia/reperfusion compared with sham (Fig. 10, A and B). The phosphorylation status of FoxO transcription factors was examined as an indicator of FoxO activation in sham and ischemia/reperfused mouse hearts. In these studies FoxO3 phosphorylation was reduced by 50% in the ischemia/reperfused heart compared with sham, although no significant change was observed in the levels of phosphorylated FoxO1 between ischemia/reperfusion and sham mouse hearts (Fig. 10, A and B). To test the upstream activators of FoxO transcription factors in ischemia/reperfused mouse hearts, we examined the levels of phosphorylated AKT and Sirt1 protein. We observed increased phosphorylation of AKT (2-fold) after ischemia/reperfusion injury compared with sham, which could indicate a cardioprotective effect to inhibit cell death and increase the viability of surviving cardiomyocytes during reperfusion (Fig. 10, A and B) (50, 51). Interestingly, Sirt1 protein level was significantly increased (2-fold) in ischemia/reperfused mouse heart compared with sham (Fig. 10, A and B) indicating oxidative stress. Similarly, Sirt1 deacetylates FoxO3 and activates transcriptional activity during oxidative stress, suggesting a possible link of Sirt1 and increased FoxO3 activity upon ischemia/reperfusion injury (24). Together these data support a mechanism in the ischemia/reperfused heart whereby Sirt1 regulates the activity of FoxO3 to induce autophagy.
FIGURE 10.
Ischemia/reperfusion injury induces autophagy with concomitant increase in FoxO3 activation in mouse hearts. FVBN mice were subjected to 40 min of ischemia plus 30 min of reperfusion (I/R) and compared with sham. A, Western blot analysis of heart homogenates with quantification by densitometric analysis in B. Increased levels of LC3II (2.1-fold) and the ratio of LC3II/LC3I (2-fold) in I/R indicates induction of autophagy in ischemia/reperfusion injured mouse heart. Decreased phosphorylated p-FoxO3 (∼45%) was observed in I/R injured mouse compared with sham control indicating increased activity of FoxO3 in I/R. No significant change in the level of p-FoxO1 was observed between the two groups. The level of Sirt1 was also increased (2-fold) in ischemia/reperfusion injured mouse hearts compared with sham control. B, densitometric and statistical analysis of the Western blot shown in A. Significance (*) was determined by Student's t test (p < 0.05; sham (n = 4) and I/R (n = 6).
DISCUSSION
Here we demonstrate the functions of FoxO1 and FoxO3 transcription factors in regulating autophagy in cultured cardiomyocytes as well as in mice subjected to starvation or cardiac ischemia/reperfusion. FoxO transcription factors are both sufficient and necessary for autophagy in rat neonatal cardiomyocytes. Under conditions of glucose deprivation, FoxO1 and FoxO3 are dephosphorylated and localized to the nucleus, where they activate autophagy pathway gene expression. FoxO1 and FoxO3 directly bind to the promoter regions of Gabarapl1 and Atg12 in cardiomyocytes, and this binding is increased upon starvation. Although both FoxO1 and FoxO3 promote reduced cell size in cardiomyocytes, differential regulation of autophagy pathway gene expression and binding to autophagy gene regulatory elements was observed for FoxO1 and FoxO3. Increased FoxO1 activity preferentially activates Atg12 gene expression, whereas FoxO3 preferentially activates expression of the LC3 gene, which acts later to complete the autophagosome formation. Interestingly, a higher percentage of cardiomyocytes with autophagic vacuoles was observed in CA-FoxO3-infected cardiomyocytes, compared with FoxO1-infected cardiomyocytes, suggesting FoxO3 is a more potent inducer of autophagy in cardiomyocytes than FoxO1. Furthermore, differential regulation of FoxO1 and FoxO3 activity was also observed between starvation and ischemia/reperfusion injury in mouse heart in vivo. Upon starvation, both FoxO1 and FoxO3 are dephosphorylated with induction of autophagy, whereas only FoxO3 is activated during induction of autophagy in ischemia/reperfusion injury. Therefore, FoxO3 has a predominant role in induction of autophagy in the heart, but FoxO1 also can regulate autophagy in cardiomyocytes.
Under basal conditions, a low level of autophagy is necessary to degrade and recycle long lived or toxic proteins and damaged organelles to maintain cell viability and cellular homeostasis (52). Our data demonstrate a low level of constitutive autophagy in neonatal cardiomyocytes, and upon starvation, autophagy is significantly induced. Under stress conditions such as nutrient deprivation, autophagy is necessary to mobilize protein degradation and nutrient production (8). Insufficient autophagy results in accumulation of defective proteins and damaged organelles, resulting in cell death (11). Therefore, a critical threshold of autophagy is necessary for the normal physiological function of a cell. In the mouse heart, autophagy is induced under conditions of starvation or ischemia as well with dilated cardiomyopathy or heart failure (18, 19, 21, 40). The heart is particularly sensitive to dysregulation of autophagy (11, 53). Mice with cardiomyocyte-specific knock-out of the autophagy-related gene 5 (Atg5) develop left ventricular enlargement, cardiac hypertrophy, contractile dysfunction, and abnormal calcium transients leading to heart failure (16). Therefore, autophagy in cardiomyocytes is required for normal cellular function in the basal state. Inhibition of autophagy in cultured adult rat ventricular cardiomyocytes results in cell death, further supporting the essential role of autophagy in cardiomyocyte cell survival (17, 18). Overall, autophagy is required for normal cellular function and for response to multiple types of stress in the neonatal and adult heart.
Cardiac autophagy also has been implicated in the pathogenesis of heart failure and nonapoptotic mechanisms of cardiomyocyte cell death (53). Increased cardiac autophagy resulting from transgenic Beclin1 overexpression leads to pathologic remodeling and heart failure (40). Incomplete autophagy and protein degradation in LAMP-2 (lysosome-associated membrane protein-2)-deficient mice lead to cardiomyocyte death and heart failure (54). In human failing hearts with idiopathic dilated cardiomyopathy, cells undergoing autophagy were 40-fold more abundant than apoptotic cells (55). Increased numbers of autophagosomes in cardiomyocytes also occur in patients with a variety of cardiovascular disorders, including dilated cardiomyopathy, aortic stenosis, ischemia, and heart failure, implicating cardiac autophagy in disease pathology (53, 55–57). Autophagy during ischemia/reperfusion injury may serve as a protective mechanism by eliminating damaged organelles to minimize the oxidative stress and cardiac dysfunction (18, 36). Conversely, inhibition of autophagy reduces cardiomyocyte cell death in cell culture and decreases infarct size in vivo, suggesting the activation of autophagy may be detrimental for the heart during ischemia/reperfusion (18, 58). Interestingly, here we have seen increased FoxO3 activity along with increased expression of autophagy markers in the ischemia/reperfused mouse heart. However, it is unclear whether induction of autophagy is a cause of pathophysiology or a compensatory mechanism to prevent the disease progression. It is increasingly apparent that autophagy can have both beneficial and detrimental effects on cardiomyocytes.
In this study, we show that increased FoxO1 or FoxO3 activation leads to decreased cardiomyocyte cell size and induction of autophagy pathway gene expression. Likewise inhibition of autophagy prevents cardiomyocyte cell size reduction under starvation conditions. Loss of FoxO3 in mice results in cardiac hypertrophy, which further supports a role for FoxO in regulating cell size in the heart in vivo (33). The mechanism by which FoxO regulates cardiomyocyte cell size is complex. Here we show that activation of FoxO is necessary and sufficient to induce cardiomyocyte autophagy. Previous studies showed FoxO1 and FoxO3 can promote expression of the atrophy-associated ubiquitin ligase gene atrogin-1, and Atrogin-1 can also activate FoxO1 function in cardiomyocyte cell cycle control (26, 33, 59). Similarly in skeletal muscle, FoxO3 induces both atrophy and autophagy, leading to decreased cell size through direct regulation of atrophy and autophagy-associated genes (29, 30). In skeletal muscle, the regulation of proteosomal (atrogene-mediated) and lysosomal (autophagy related gene-mediated) protein degradation by FoxO transcription factors occurs independently (30). It is likely that FoxO1 and FoxO3 have similar complex roles in cell size regulation and autophagy in cardiomyocytes. Our study suggests that the decreased cardiomyocyte cell size that occurs with activation of FoxO transcription factors is mainly due to induction of autophagy, but it is possible that proteasomal proteolysis (atrophy) also may play a role.
In the presence of growth factors such as insulin or insulin-like growth factor-1, FoxO is inactivated by AKT phosphorylation, but with starvation or growth factor deprivation, dephosphorylated FoxO is translocated to the nucleus where they are transcriptionally active (23). In this study we demonstrate the FoxO transcription factors are activated concomitantly with inactivation of AKT in starved mouse hearts, but AKT is active with ischemia/reperfusion injury consistent with its reported role in cell survival (51). Sirt1, a stress-responsive protein, is significantly up-regulated in mouse hearts subjected to either starvation or ischemia/reperfusion injury compared with respective controls. In human embryonic kidney cells, Sirt1 regulates FoxO3 through deacetylation, and Sirt1 activity modulates the transcriptional activity of FoxO1 and FoxO3 in cardiomyocytes subjected to glucose deprivation (24). Glucose deprivation also results in the activation of AMP-activated protein kinase, which has been demonstrated to directly phosphorylate FoxO3 to induce the transcriptional activity of FoxO3 in mouse embryonic fibroblast cells (18, 60, 61). Together, these studies provide evidence for activation of FoxO1 and FoxO3 by multiple mechanisms in starved cardiomyocytes and support a predominant role for Sirt1 in FoxO3 activation during cardiac ischemia/reperfusion injury.
Multiple roles for FoxO transcription factors in the neonatal and adult heart have been demonstrated (26, 27, 33). In the adult heart, loss of FoxO3 results in cardiac hypertrophy, and increased FoxO1 expression is associated with heart failure (33, 62). In neonatal cardiomyocytes, FoxO1 becomes activated and localized to the nucleus, whereas it is present in the cytoplasm of embryonic cardiomyocytes (27). However, the functions of FoxO1 in the neonatal and adult heart have not yet been fully determined, because mice lacking FoxO1 do not survive beyond embryonic day 10.5 (32). In transgenic mice, increased cardiac FoxO1 function leads to decreased cardiomyocyte cell proliferation through regulation of the cell cycle inhibitor genes p21Cip1 and p27Kip2 (27). The increased activation of FoxO1 during the neonatal period also could be involved in regulation of autophagy with starvation that occurs between birth and lactation. Autophagy is necessary for neonatal life in mammals as indicated by early postnatal lethality of Atg5 null mice (14). Further targeted studies are necessary to determine the functions of FoxO factors in the neonatal transition period and to determine whether FoxO1 and FoxO3 have distinct and overlapping functions in cardiac development, homeostasis, and disease.
The precise regulation of cardiomyocyte cell size has important implications for normal heart function and cardiovascular disease. In addition, the availability of nutrients is absolutely critical for cardiomyocyte contractility and survival. There is accumulating evidence that autophagy has important functions in both of these processes in normal heart function and in cardiac disease. Evidence from this study and previous reports implicates FoxO1 and FoxO3 transcription factors as critical regulators of autophagy and cell size in the developing and adult heart. Pharmacological manipulation of autophagy or the signaling pathways that regulate FoxO activation are attractive therapeutic strategies for cardiovascular disease. However, the complexity of these regulatory mechanisms with both beneficial and detrimental functions in the heart should be taken into account when considering potential therapeutic approaches. In the past few years, significant advances have been made in the understanding of autophagy, as well as FoxO function, in a variety of normal and pathologic biological processes. It is likely that these discoveries will have important applications in future clinical efforts directed toward the treatment of cardiovascular disease.
Acknowledgments
We thank Michelle Sargeant for performance of cardiac ischemia in mice, Christina Alfieri for technical support, and Heather Evans-Anderson for preliminary studies.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 HL069779.
- ChIP
- chromatin immunoprecipitation
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- WT
- wild type
- CA
- constitutively active
- 3-MA
- 3-methyladenine
- RT
- reverse transcription
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ANOVA
- analysis of variance
- AV
- autophagic vacuole.
REFERENCES
- 1.Li F., Wang X., Capasso J. M., Gerdes A. M. (1996) J. Mol. Cell. Cardiol. 28, 1737–1746 [DOI] [PubMed] [Google Scholar]
- 2.Frey N., Katus H. A., Olson E. N., Hill J. A. (2004) Circulation 109, 1580–1589 [DOI] [PubMed] [Google Scholar]
- 3.Heineke J., Molkentin J. D. (2006) Nat. Rev. Mol. Cell Biol. 7, 589–600 [DOI] [PubMed] [Google Scholar]
- 4.de Simone G., Scalfi L., Galderisi M., Celentano A., Di Biase G., Tammaro P., Garofalo M., Mureddu G. F., de Divitiis O., Contaldo F. (1994) Br. Heart J. 71, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senzaki H., Kurihara M., Masutani S., Sasaki N., Kyo S., Yokote Y. (2006) Circulation 113, e759–e761 [DOI] [PubMed] [Google Scholar]
- 6.Papadimitriou J. M., Hopkins B. E., Taylor R. R. (1974) Circ. Res. 35, 127–135 [DOI] [PubMed] [Google Scholar]
- 7.Zafeiridis A., Jeevanandam V., Houser S. R., Margulies K. B. (1998) Circulation 98, 656–662 [DOI] [PubMed] [Google Scholar]
- 8.Meléndez A., Neufeld T. P. (2008) Development 135, 2347–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vellai T., Bicsák B., Tóth M. L., Takács-Vellai K., Kovács A. L. (2008) Autophagy 4, 507–509 [DOI] [PubMed] [Google Scholar]
- 11.Gustafsson A. B., Gottlieb R. A. (2008) J. Mol. Cell. Cardiol. 44, 654–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shintani T., Klionsky D. J. (2004) Science 306, 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 15.Nishida K., Kyoi S., Yamaguchi O., Sadoshima J., Otsu K. (2009) Cell Death Differ. 16, 31–38 [DOI] [PubMed] [Google Scholar]
- 16.Nakai A., Yamaguchi O., Takeda T., Higuchi Y., Hikoso S., Taniike M., Omiya S., Mizote I., Matsumura Y., Asahi M., Nishida K., Hori M., Mizushima N., Otsu K. (2007) Nat. Med. 13, 619–624 [DOI] [PubMed] [Google Scholar]
- 17.Maruyama R., Goto K., Takemura G., Ono K., Nagao K., Horie T., Tsujimoto A., Kanamori H., Miyata S., Ushikoshi H., Nagashima K., Minatoguchi S., Fujiwara T., Fujiwara H. (2008) Am. J. Physiol. Heart. Circ. Physiol. 295, H1599–H1607 [DOI] [PubMed] [Google Scholar]
- 18.Matsui Y., Takagi H., Qu X., Abdellatif M., Sakoda H., Asano T., Levine B., Sadoshima J. (2007) Circ. Res. 100, 914–922 [DOI] [PubMed] [Google Scholar]
- 19.Gustafsson A. B., Gottlieb R. A. (2009) Circ. Res. 104, 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsui Y., Kyoi S., Takagi H., Hsu C. P., Hariharan N., Ago T., Vatner S. F., Sadoshima J. (2008) Autophagy 4, 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gustafsson A. B., Gottlieb R. A. (2008) Autophagy 4, 416–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greer E. L., Brunet A. (2005) Oncogene 24, 7410–7425 [DOI] [PubMed] [Google Scholar]
- 23.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 24.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 25.Huang H., Tindall D. J. (2007) J. Cell Sci. 120, 2479–2487 [DOI] [PubMed] [Google Scholar]
- 26.Skurk C., Izumiya Y., Maatz H., Razeghi P., Shiojima I., Sandri M., Sato K., Zeng L., Schiekofer S., Pimentel D., Lecker S., Taegtmeyer H., Goldberg A. L., Walsh K. (2005) J. Biol. Chem. 280, 20814–20823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans-Anderson H. J., Alfieri C. M., Yutzey K. E. (2008) Circ. Res. 102, 686–694 [DOI] [PubMed] [Google Scholar]
- 28.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffiano S., Lecker S. H., Goldberg A. L. (2004) Cell 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J., Brault J. J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S. H., Goldberg A. L. (2007) Cell Metab. 6, 472–483 [DOI] [PubMed] [Google Scholar]
- 30.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S. J., Di Lisi R., Sandri C., Zhao J., Goldberg A. L., Schiaffino S., Sandri M. (2007) Cell Metab. 6, 458–471 [DOI] [PubMed] [Google Scholar]
- 31.Hribal M. L., Nakae J., Kitamura T., Shutter J. R., Accili D. (2003) J. Cell Biol. 162, 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosaka T., Biggs W. H., 3rd, Tieu D., Boyer A. D., Varki N. M., Cavenee W. K., Arden K. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni Y. G., Berenji K., Wang N., Oh M., Sachan N., Dey A., Cheng J., Lu G., Morris D. J., Castrillon D. H., Gerard R. D., Rothermel B. A., Hill J. A. (2006) Circulation 114, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Windt L. J., Lim H. W., Haq S., Force T., Molkentin J. D. (2000) J. Biol. Chem. 275, 13571–13579 [DOI] [PubMed] [Google Scholar]
- 35.Nakae J., Kitamura T., Silver D. L., Accili D. (2001) J. Clin. Invest. 108, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamacher-Brady A., Brady N. R., Gottlieb R. A. (2006) J. Biol. Chem. 281, 29776–29787 [DOI] [PubMed] [Google Scholar]
- 37.Shelton E. L., Yutzey K. E. (2007) Dev. Biol. 302, 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hemmerich P., von Mikecz A., Neumann F., Sözeri O., Wolff-Vorbeck G., Zoebelein R., Krawinkel U. (1993) Nucleic Acids Res. 21, 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsujita Y., Muraski J., Shiraishi I., Kato T., Kajstura J., Anversa P., Sussman M. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11946–11951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu H., Tannous P., Johnstone J. L., Kong Y., Shelton J. M., Richardson J. A., Le V., Levine B., Rothermel B. A., Hill J. A. (2007) J. Clin. Invest. 117, 1782–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaiser R. A., Bueno O. F., Lips D. J., Doevendans P. A., Jones F., Kimball T. F., Molkentin J. D. (2004) J. Biol. Chem. 279, 15524–15530 [DOI] [PubMed] [Google Scholar]
- 42.Kaiser R. A., Liang Q., Bueno O., Huang Y., Lackey T., Klevitsky R., Hewett T. E., Molkentin J. D. (2005) J. Biol. Chem. 280, 32602–32608 [DOI] [PubMed] [Google Scholar]
- 43.Ohsumi Y. (2001) Nat. Rev. Mol. Cell Biol. 2, 211–216 [DOI] [PubMed] [Google Scholar]
- 44.Skurk C., Maatz H., Kim H. S., Yang J., Abid M. R., Aird W. C., Walsh K. (2004) J. Biol. Chem. 279, 1513–1525 [DOI] [PubMed] [Google Scholar]
- 45.Nakae J., Kitamura T., Kitamura Y., Biggs W. H., 3rd, Arden K. C., Accili D. (2003) Dev. Cell 4, 119–129 [DOI] [PubMed] [Google Scholar]
- 46.Barthel A., Schmoll D., Unterman T. G. (2005) Trends Endocrinol. Metab. 16, 183–189 [DOI] [PubMed] [Google Scholar]
- 47.Nemoto S., Fergusson M. M., Finkel T. (2004) Science 306, 2105–2108 [DOI] [PubMed] [Google Scholar]
- 48.Kanfi Y., Peshti V., Gozlan Y. M., Rathaus M., Gil R., Cohen H. Y. (2008) FEBS Lett. 582, 2417–2423 [DOI] [PubMed] [Google Scholar]
- 49.Hamacher-Brady A., Brady N. R., Logue S. E., Sayen M. R., Jinno M., Kirshenbaum L. A., Gottlieb R. A., Gustafsson A. B. (2007) Cell Death Differ. 14, 146–157 [DOI] [PubMed] [Google Scholar]
- 50.Fujio Y., Nguyen T., Wencker D., Kitsis R. N., Walsh K. (2000) Circulation 101, 660–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsui T., Tao J., del Monte F., Lee K. H., Li L., Picard M., Force T. L., Franke T. F., Hajjar R. J., Rosenzweig A. (2001) Circulation 104, 330–335 [DOI] [PubMed] [Google Scholar]
- 52.Levine B., Yuan J. (2005) J. Clin. Invest. 115, 2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishida K., Yamaguchi O., Otsu K. (2008) Circ. Res. 103, 343–351 [DOI] [PubMed] [Google Scholar]
- 54.Tanaka Y., Guhde G., Suter A., Eskelinen E. L., Hartmann D., Lüllmann-Rauch R., Janssen P. M., Blanz J., von Figura K., Saftig P. (2000) Nature 406, 902–906 [DOI] [PubMed] [Google Scholar]
- 55.Kostin S., Pool L., Elsässer A., Hein S., Drexler H. C., Arnon E., Hayakawa Y., Zimmermann R., Bauer E., Klövekorn W. P., Schaper J. (2003) Circ. Res. 92, 715–724 [DOI] [PubMed] [Google Scholar]
- 56.Elsässer A., Vogt A. M., Nef H., Kostin S., Möllmann H., Skwara W., Bode C., Hamm C., Schaper J. (2004) J. Am. Coll. Cardiol. 43, 2191–2199 [DOI] [PubMed] [Google Scholar]
- 57.Kunapuli S., Rosanio S., Schwarz E. R. (2006) J. Card. Fail. 12, 381–391 [DOI] [PubMed] [Google Scholar]
- 58.Valentim L., Laurence K. M., Townsend P. A., Carroll C. J., Soond S., Scarabelli T. M., Knight R. A., Latchman D. S., Stephanou A. (2006) J. Mol. Cell. Cardiol. 40, 846–852 [DOI] [PubMed] [Google Scholar]
- 59.Li H. H., Kedar V., Zhang C., McDonough H., Arya R., Wang D. Z., Patterson C. (2004) J. Clin. Invest. 114, 1058–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greer E. L., Oskoui P. R., Banko M. R., Maniar J. M., Gygi M. P., Gygi S. P., Brunet A. (2007) J. Biol. Chem. 282, 30107–30119 [DOI] [PubMed] [Google Scholar]
- 62.Hannenhalli S., Putt M. E., Gilmore J. M., Wang J., Parmacek M. S., Epstein J. A., Morrisey E. E., Margulies K. B., Cappola T. P. (2006) Circulation 114, 1269–1276 [DOI] [PubMed] [Google Scholar]