Abstract
Microtubule plus-end tracking proteins (+TIPs) control microtubule dynamics in fundamental processes such as cell cycle, intracellular transport, and cell motility, but how +TIPs are regulated during mitosis remains largely unclear. Here we show that the endogenous end-binding protein family EB3 is stable during mitosis, facilitates cell cycle progression at prometaphase, and then is down-regulated during the transition to G1 phase. The ubiquitin-protein isopeptide ligase SIAH-1 facilitates EB3 polyubiquitination and subsequent proteasome-mediated degradation, whereas SIAH-1 knockdown increases EB3 stability and steady-state levels. Two mitotic kinases, Aurora-A and Aurora-B, phosphorylate endogenous EB3 at Ser-176, and the phosphorylation triggers disruption of the EB3-SIAH-1 complex, resulting in EB3 stabilization during mitosis. Our results provide new insight into a regulatory mechanism of +TIPs in cell cycle transition.
Microtubule dynamics are essential in many cellular processes, including cell motility, intracellular transport, accurate mitosis, and cytokinesis in all eukaryotes. The regulatory factors for microtubule dynamics can be classified into two main types as follows: microtubule-destabilizing proteins, such as stathmin/Op18 (1) and the Kinesin-13 family (also known as MCAK/KIF2 family) (2), and microtubule-stabilizing proteins, the classic superfamily of microtubule-associated proteins (3). Additionally, the plus-end tracking proteins (+TIPs)3 have recently been identified; this family specifically accumulates at the ends of growing microtubules and regulates the microtubule plus-end targeting to the cell cortex or mitotic kinetochores (4, 5).
The EB1 family is a member of the +TIPs family and consists of three homologs in mammals: EB1, EB2/RP1 (henceforth, EB2), and EB3 (6). As EB1 was originally identified as a protein that interacts with the well characterized tumor suppressor adenomatous polyposis coli (APC) protein (7), the function of EB1 has been investigated extensively. EB1 interacts with other +TIPs, including APC, p150glued, CLIPs, and CLASP1/2, and the interaction network controls microtubule orientation and microtubule-cortex interaction during cell migration (5, 8, 9). EB1 functions not only in the regulation of interphase microtubule dynamics but also in mitotic spindle regulation. For accurate chromosomal segregation, sister chromatids become aligned to the metaphase plate during metaphase, and the alignment requires spindle-kinetochore attachment. Two models have been proposed; in the first, termed the “search-and-capture” model, EB1 localized at the growing microtubule plus-ends searches for binding partners located on kinetochores (10, 11). In the second model proposed recently, EB1 makes kinetochore fibers and centrosomal microtubules connect, and it is essential for the formation of a functional bipolar spindle (12). Thus, EB1 is thought to be a master controller of microtubule plus-ends; however, little is known about other EB1 family members. Given that EB3 is localized on the microtubule network and binds to APC and CLIPs identically to EB1, it is possible that EB3 acts as an EB1 analog in cells (13–15).
Cell division is precisely regulated by several post-translational modifications of proteins, mainly reversible phosphorylation and ubiquitination, which is followed by degradation. Accurate mitotic phase progression requires the appropriate phosphorylation of various proteins by mitotic kinases (16, 17). One of the key mitotic kinases is the Aurora family that has been highly conserved from yeast to humans. There are three homologs (Aurora-A, -B, and -C) in human and mouse (18). Although their homology at the protein level is more than 84%, their functions and subcellular localizations are distinct. Aurora-A is located in the centrosomes and spindle and is required for mitotic entry, centrosome maturation/separation, and spindle assembly (19). Aurora-B is a chromosomal passenger protein that localizes on the inner centromere of the chromosomes until metaphase to regulate the spindle-kinetochore attachment, and from anaphase, it translocates to the central spindle and then accumulates in the midbody for cytokinesis (20, 21). The numerous substrates of the Aurora family include regulatory factors for microtubule dynamics, such as the microtubule-destabilizing proteins MCAK and stathmin, which help to establish the bipolar attachment and spindle assembly, respectively (22–24). It is possible that the Aurora family regulates the EB1 family by phosphorylation.
In this study, we performed yeast two-hybrid screening and obtained the EB1 yeast homolog Bim1 as a protein that interacts with Ipl1, a yeast counterpart of Aurora. Here we demonstrate the novel regulatory mechanisms of EB3 by two cell cycle-dependent post-translational modifications, phosphorylation and ubiquitin-proteasome-mediated degradation.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screening
Screening was performed essentially as described (25). Plasmid pGBDU-IPL1 (URA3, 2 μ), which expresses Ipl1 fused with Gal4 DNA binding domain, was co-transformed with a Gal4 activation domain library (LEU2, 2 μ), which is a collection library containing full-length genes of cytoskeletal components fused with Gal4 activation domain, into Saccharomyces cerevisiae strain PJ69-4A. The transformants were grown on medium lacking leucine, uracil, and histidine. We obtained the BIM1 clones multiple times as a candidate in this screening. The specificity of the two-hybrid interaction was further confirmed by standard two-hybrid protocol, including adenine autotroph and LacZ expression.
Materials
Monoclonal anti-Glu-Glu and anti-Myc tag antibodies were gifts from Dr. Larry A. Feig (Tufts University, Boston). Monoclonal anti-Aurora-A (M11-17) and anti-Aurora-B (H7-4) antibodies have been described (26, 27). Monoclonal anti-pan Aurora (K3-7) antibody recognizes all of the human Aurora family, as described (28). The following commercial antibodies were used: monoclonal anti-EB1 and anti-EB3 (BD Biosciences); anti-α-tubulin (DM1A), anti-β-actin (AC-15), and anti-FLAG (M2) (Sigma); anti-cyclin B1 (GNS1) (Santa Cruz Biotechnology); anti-penta-His (Qiagen); polyclonal anti-p21Cip1/WAF1 (Zymed Laboratories Inc.); anti-SIAH-1 (TransGenic Inc.); horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG (Cell Signaling); and Alexa Fluor 488- or 594-labeled goat anti-mouse or anti-rabbit IgG (Molecular Probes). The peptides were synthesized and purified using high pressure liquid chromatography by Dr. Michael Berne (Tufts University Core Facility, Boston). VX-680 was purchased from Kava Technology.
Expression and Purification of Recombinant Proteins
Human EB1, EB2, and EB3 cDNAs were obtained by reverse transcription-PCR. Ala mutants of EB3 were constructed by PCR mutation. To create a mammalian expression vector, each cDNA was inserted into myc3/pcDNA3, pME4T-2 (29), or into pEGFP-C1 (Clontech). For retrovirus production, cDNA was inserted into altered versions of pMXs, pMXs-GFP or pMXs-mC, that contained GFP or monomeric Cherry (mCherry) at 5′ in the cloning site, respectively. Human SIAH-1, SIAH-1(ΔN), and ubiquitin cDNAs were gifts from Drs. Hiroshi Yamashita (Hiroshima University, Japan) and Dirk Bohmann (EMBL, Germany). The plasmids of human Aurora-A, Aurora-B, and INBOX of INCENP (783–918 amino acids) have been described (27, 29). Bacterial expressions of GST and hexa-His fusion proteins have been described (29).
Cell Culture, Transfection, and Retroviral Infection
All cells were grown in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Cell Culture Technologies). HeLa S3 cells were transfected with each plasmid and siRNA using Lipofectamine 2000 and Oligofectamine (Invitrogen), respectively, according to the manufacturer's instructions. COS-7 cells were transfected by DEAE-dextran, as described previously (29).
Retroviral vector plasmids were used for transfection of Plat-E cells, a ecotropic retrovirus packaging cell line (30). Two days after the transfection, culture supernatants were collected, filtered, and used for infection. The day after the infection, the medium was replaced. One week after the infection, only GFP (or mCherry)-positive cells expressing the lower detectable amounts of fusion protein were isolated by the FACSVantage using CELLQuestTM software (BD Biosciences). Mixed clonal populations of stably expressing cells were used in all the experiments to avoid clonal artifacts. For human HeLa cells, the murine ecotropic retrovirus receptor was first introduced. This procedure made human HeLa cells susceptible to the subsequent infection with ecotropic viral vectors. The infection efficiencies were >80% as judged by using GFP-expressing retroviral vectors.
Cell Cycle Synchronization
Cells were synchronized in S phase by a double-thymidine block method, as described previously (31). To arrest cells in prometaphase, nocodazole (30 ng/ml, final concentration) was added, and mitotic cells were collected by the shake-off procedure 12 h later. The collected cells were then released from the nocodazole arrest into fresh medium. To arrest cells in ana-telophase, dihydrocytochalasin B (5 μg/ml, final concentration) was added 2 h after release from prometaphase arrest, and cells were then released from the dihydrocytochalasin B arrest into a fresh medium.
siRNAs
Stealth siRNAs were purchased from Invitrogen. Each nucleotide sequence corresponding to siRNAs was the following: Aurora-A-(726–750), Aurora-B-(88–112), EB1-(161–185), EB3-(585–609), and SIAH-1 (siRNA2, residues 736–760).
Antibody Generation
The phospho-EB3(Ser-176) rabbit polyclonal antibody for Western blots was raised against the following keyhole limpet hemocyanin-coupled EB3 peptide H2N-TSGRLpSNVAPPC-COOH. Phospho-specific rabbit antibodies were affinity-purified using the phosphorylated and the nonphosphorylated peptide. The phospho-EB3(Ser-176) mouse monoclonal antibody 9.7.5 for immunofluorescence was raised against the following keyhole limpet hemocyanin-coupled EB3 peptide H2N-TSGRLpSNVAPPC-COOH. Clonal populations of fusion cells were screened by enzyme-linked immunosorbent assays for antibody production against phosphorylated peptide and nonphosphorylated peptide conjugated to maleimide-activated bovine serum albumin (Pierce) through a free C-terminal cysteine residue. Productive cells were cloned to monoclonal lines by serial dilution screening. Highly concentrated monoclonal antibodies were isolated from murine ascites after an intraperitoneal injection of hybridoma cells. For making the anti-EB2 mouse monoclonal antibody 5.5, we immunized mice with recombinant GST-tagged EB2, and serum titers were monitored by immunoblotting using lysates of COS cells transfected with myc-EB2/pcDNA3.
Immunofluorescence Microscopy
Cells were simultaneously fixed and permeabilized for 10 min with 3.7% formaldehyde and 0.2% Triton X-100 in PHEM buffer (60 mm PIPES, 25 mm HEPES, 10 mm EGTA, and 2 mm MgCl2, pH 6.9), then washed three times with phosphate-buffered saline (PBS), and then blocked for 30 min with 5% skim milk in PBS. After the blocking, cells were incubated with primary antibodies for 1 h at room temperature. After four washes with PBS, secondary antibody incubations were performed for 1 h at room temperature. After four washes with PBS, the coverslips were mounted (mounting medium; Vector Laboratories) with 4′,6-diamidino-2-phenylindole (DAPI). Cells were observed under a confocal microscope FLUOVIEW FV1000 (Olympus).
Immunoprecipitation and in Vivo Ubiquitination Assays
Preparation of cell lysates and procedures of immunoprecipitation were described previously (29). For detection of ubiquitination in cells, cells were pretreated with MG132 (20 μm, final concentration; Peptide Institute) for 6 h. For GST pulldown, cell lysates were incubated with glutathione-Sepharose beads (Amersham Biosciences).
In Vitro Phosphorylation Assays
In vitro phosphorylation assays were performed using purified GST-H3(5-15), GST-EB1, -EB2, wild type, or mutant EB3s as substrates and immunopurified Glu-Glu-Aurora-B as kinase, as described previously (29).
In Vitro Binding Assays and Biosensor Analysis
In vitro binding assays were performed using purified His-EB1, -EB2, or -EB3 and purified GST-SIAH-1 or -SIAH-1(ΔN), as described (31).
Surface plasmon resonance was measured using a BIAcore 3000 instrument (Biosensor, Uppsala, Sweden) equipped with CM5 sensor chips. All experiments proceeded at 25 °C. The carboxylated dextran matrix of the sensor chip was activated by injecting of 80 μl of 0.2 m N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide and 0.05 m N-hydroxysuccimide according to the manufacturer's instructions. Bacterially expressed recombinant His-SIAH-1(ΔN) was diluted in 10 mm sodium acetate, pH 4.0, and coupled to a CM5 sensor chip, and the remaining binding sites were then blocked with 1 m ethanolamine, pH 8.5, resulting in the immobilization of ∼2000 resonance units. A parallel channel of the sensor chip was derivatized under identical conditions in the absence of His-SIAH-1(ΔN) as a control.
All interactions between SIAH-1(ΔN) and peptides were analyzed in 10 mm HEPES, pH 7.4, 0.15 m NaCl, 1 mm CaCl2, and 0.005% surfactant 20 (v/v), which was also used as the running buffer. Peptide was injected at various concentrations (0.08–2.8 mm) to determine rate constants. Sensor chips were regenerated after each analytical cycle by injecting 20 ml of 50 mm sodium hydroxide. This procedure did not affect the surface binding capacity of the sensor chips.
In Vitro Ubiquitination Assays
GST fusion proteins trapped on glutathione-Sepharose beads were mixed with rabbit ubiquitin-activating enzyme (1 μg), UbcH5c (1.25 μg), biotinylated ubiquitin (15 μg), and 500 ng of His-SIAH-1 or His-SIAH-1(ΔN). The mixture was incubated at 25 °C for 30 min in the presence of 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 2 mm dithiothreitol, and 2 mm ATP in a final volume of 50 μl. The resin was washed three times with 10 mm Tris-HCl, pH 7.4, 3 mm MgCl2, 200 mm NaCl, 0.2 mm phenylmethylsulfonyl fluoride, and 0.1% Nonidet P-40. The ubiquitinated proteins on the resin were subjected to SDS-PAGE and detected by Western blots using peroxidase-conjugated avidin (ExtrAvidin; Sigma).
RESULTS
Auroras Interact with EB1 Family Member Proteins
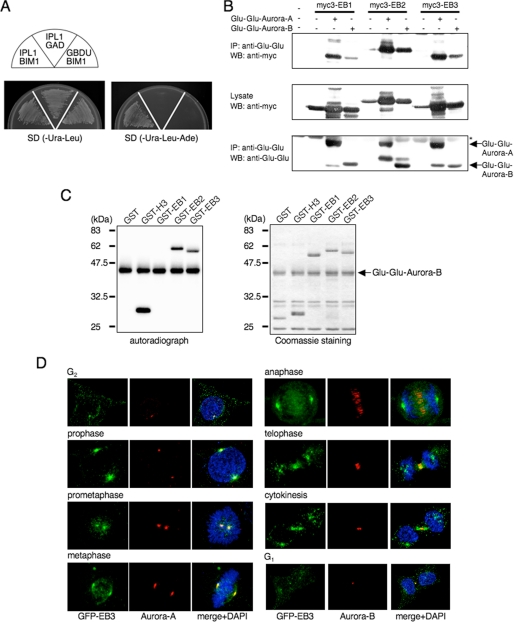
We initially identified Bim1, the sole budding yeast member of the human EB1-related family, in a yeast two-hybrid screen with Ipl1 as the bait (Fig. 1A). Considering that EB1, like Aurora-A and -B, is essential for the formation of a functional and focused spindle and regulates the spindle-kinetochore attachment (4, 5), we investigated the conserved interaction between human EB1 family members and Auroras in detail. COS-7 cells were co-transfected with mammalian expression plasmids encoding Myc-tagged EB1 family members either alone or together with Glu-Glu-tagged human Aurora-A or -B. Anti-Glu-Glu monoclonal antibody was used to immunoprecipitate proteins from the transfected cell lysates, and the immunoprecipitated proteins were examined by Western blots. As shown in Fig. 1B, all three EB1 family members were co-immunoprecipitated with both Glu-Glu-Aurora-A and -B, demonstrating that human EB1 family proteins were specifically associated with Auroras in human cells. In addition, during this experiments we noticed that the Myc end-binding proteins (especially EB2 and EB3) seem to be present at higher levels when Auroras are co-expressed (discussed below).
FIGURE 1.
EB2 and EB3, but not EB1, are substrates of Aurora. A, yeast Aurora homolog, Ipl1, binds Bim1, the sole budding yeast member of the human EB1-related family, in a yeast two-hybrid system. Plasmids transformed into PJ69-4A are indicated in each sector. B, binding of EB1 family members and Aurora family members in COS-7 cells that were transfected with the indicated plasmids. The cell lysates were immunoprecipitated (IP) by using anti-Glu-Glu antibody, and the precipitates were analyzed by Western blots (WB) that were probed sequentially with anti-Myc and anti-Glu-Glu antibodies. Asterisk denotes IgG heavy chain. One Western blot shown is representative of three independent experiments. C, in vitro phosphorylation of the EB1 family proteins. GST-fused EB1, EB2, EB3 or H3-(5–15) as a control was incubated with immunoprecipitated Glu-Glu-Aurora-B in the presence of [γ-32P]ATP. The phosphorylation reactions were detected by autoradiography (left panel). The amounts of GST fusion protein used in the assay were compared by Coomassie Blue staining (right panel). A representative result of three independent experiments is shown. D, representative confocal images of EB3 localization during each stage of mitosis by immunofluorescence staining of HeLa cells that stably expressed GFP-EB3. Green, GFP-EB3; red, Aurora-A (left panels) or Aurora-B (right panels); blue, DAPI.
EB2 and EB3, but Not EB1, Are Phosphorylated by the Aurora Family
To determine whether the EB1 family is a novel substrate of the Aurora family, we performed in vitro phosphorylation by using the recombinant GST fusion proteins containing each of the EB1 family members as substrates and Glu-Glu-immunoprecipitated Auroras from COS-7 cell lysates. Aurora-A and -B efficiently phosphorylated EB2 and EB3, as well as histone H3, a well known substrate of Aurora-B; however, EB1 was not phosphorylated (Fig. 1C, supplemental Fig. 6, and data not shown).
Next, we examined the subcellular localization of EB2 and EB3 during mitosis by using HeLa cells that stably expressed GFP fusion proteins. GFP-EB2 was dispersed in the cytoplasm and did not show any characteristic or dynamic localization pattern in mitotic cells (data not shown); in contrast, GFP-EB3 was predominantly localized to the centrosomes and spindle microtubules throughout mitosis, as well as endogenous human and Drosophila EB1 (10, 32) (Fig. 1D and supplemental Fig. 1). When soluble proteins were removed from cells by simultaneous permeabilization and fixation, residual GFP-EB3 was mainly found on the centrosomes in prometaphase cells, which was co-localized with endogenous Aurora-A, and on the central spindle and the midbody from anaphase to cytokinesis, which was similar to endogenous Aurora-B (Fig. 1D). GFP-EB3 was more broadly located on microtubules than Auroras. Together, these results imply that human EB3 could be a physiologically relevant substrate of the Aurora family during mitosis.
Aurora-A and Aurora-B Phosphorylate EB3 at Ser-176 in a Spatial and Cell Cycle-specific Manner, Respectively
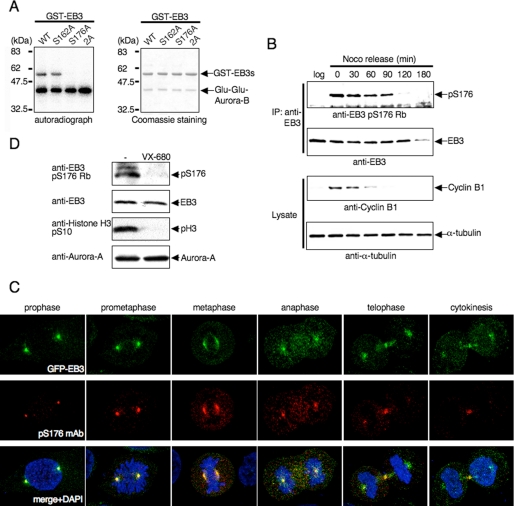
We focused on characterizing the relationship between human EB3 and the Aurora family in more detail. First, we searched for the Aurora phosphorylation site(s) in human EB3. The human Auroras are Arg-directed kinases that preferentially phosphorylate their substrates at the Arg-Xaa-(Ser/Thr) motif (29, 33, 34). Human EB3 has two candidate phosphorylation sites (RTS162 and RLS176). In vitro kinase assays showed that phosphorylation of GST-EB3 by Auroras was abrogated by a Ser-to-Ala substitution at residue 176 (S176A) and the S162A/S176A double mutant, not by S162A mutant (Fig. 2A and data not shown). Thus, Ser-176 appear to be a potential phosphorylation site.
FIGURE 2.
Phosphorylation of EB3 Ser-176 by Aurora-A and Aurora-B during mitosis. A, in vitro phosphorylation of EB3 mutants. The experiment was performed as described in Fig. 1C using GST-fused EB3 mutants and immunoprecipitated Glu-Glu-Aurora-B. The autoradiographs shown are representative of three independent experiments. WT, wild type. B, phosphorylation of endogenous EB3 in mitotic cells. HeLa cells were released from a nocodazole block for the indicated times. Noco and log indicate nocodazole and logarithmic phase, respectively. Immunoprecipitation (IP) was performed with anti-EB3 monoclonal antibody, and the precipitates were analyzed by Western blots probed sequentially with anti-phospho-EB3(Ser-176) rabbit polyclonal antibody (Rb). A representative result of three independent experiments is shown. C, representative confocal images of localization of phospho-EB3(Ser-176) during each stage of mitosis. Immunofluorescence staining of HeLa cells that stably expressed GFP-EB3 was performed using anti-phospho-EB3(Ser-176) mouse monoclonal antibody (mAb). Red, phospho-EB3(Ser-176); green, GFP-EB3; blue, DAPI. D, effects of Aurora inhibitor on EB3 phosphorylation. HeLa cells were arrested at prometaphase with nocodazole and subsequently treated with VX-680 for 4 h, and then the cell lysates were analyzed by Western blots (antibodies indicated at the side of the figure). A representative result of three independent experiments is shown.
To confirm whether Ser-176 of EB3 was phosphorylated in cells, we generated Ser-176-specific phospho-EB3 rabbit polyclonal and mouse monoclonal antibodies (supplemental Fig. 2A). HeLa cells were synchronized at prometaphase by nocodazole treatment. The phosphorylation status of endogenous EB3 was analyzed by Western blots as cells progressed through mitosis into the subsequent G1 phase after their release from the nocodazole block. Cyclin B1 levels decreased within 60 min, indicating that the cells have exited metaphase. The phosphorylation of EB3 peaked in prometaphase and was observed until ∼90 min when the majority of the cells was in transit to cytokinesis (Fig. 2B). The protein level of EB3 was constant throughout mitosis but dramatically declined in early G1 phase (180 min) (discussed in detail below). Thus, human EB3 is indeed phosphorylated on Ser-176 during mitosis.
Antibodies that specifically recognize human EB3 without cross-reactivity to other EB1 family members for immunofluorescence are not currently available. Thus, to examine the subcellular distribution of phospho-EB3(Ser-176) by immunofluorescence, we utilized HeLa cells stably expressing GFP-EB3. In mitotic prophase, phosphorylated EB3 was predominantly localized to the centrosomes, whereas GFP-EB3 was localized to the centrosomes and the mitotic spindle (Fig. 2C). The centrosomal signal of phospho-EB3 dramatically decreased from telophase and eventually disappeared, despite the presence of GFP-EB3 (Fig. 2C). The phospho-EB3 signal also appeared at the midbody during telophase and cytokinesis (Fig. 2C). The specificity of the phospho-EB3 antibodies for immunofluorescence was confirmed by siRNA knockdown of EB3 (supplemental Fig. 2B). Thus, we demonstrated that EB3 phosphorylation indeed occurs at the centrosomes and the midbody.
To examine the contribution of Auroras to EB3 phosphorylation, nocodazole-arrested cells were treated with 600 nm VX-680, an Aurora inhibitor. Phosphorylation of EB3 at Ser-176 as well as histone H3 at Ser-10 was completely inhibited by VX-680 (Fig. 2D). Next, we depleted these proteins by RNA interference in HeLa cells that stably expressed GFP-EB3. As expected, depletion of Aurora-A abolished the centrosomal signal detected by the phospho-EB3(Ser-176) antibody in metaphase cells (supplemental Fig. 2C, left panels), demonstrating that EB3 at Ser-176 on the centrosomes is phosphorylated by Aurora-A during the early stage of mitosis. Given that the Aurora-A kinase activity rapidly decreases from the onset of anaphase (29), it is reasonable that the centrosomal signal of phospho-EB3 was attenuated from anaphase (Fig. 2C). With regard to Aurora-B-depleted cells, the midbody signal (both EB3 and phospho-EB3) was not detected during telophase (supplemental Fig. 2C, right panels) because Aurora-B is essential for the proper midzone/midbody formation and cytokinesis (35). Knockdown of Aurora-A or -B did not change the subcellular localization of GFP-EB3 (supplemental Fig. 2C) except midbody, indicating that the disappearance of the phospho-EB3 signal was because of the loss of Aurora-A or -B expression in each cell. These observations demonstrate that Aurora-A and -B phosphorylate the Ser-176 residue of EB3 on the centrosomes during the early stage and on the midbody during the late stage of mitosis, respectively.
Cell Cycle-dependent Regulation of EB3 Protein Level
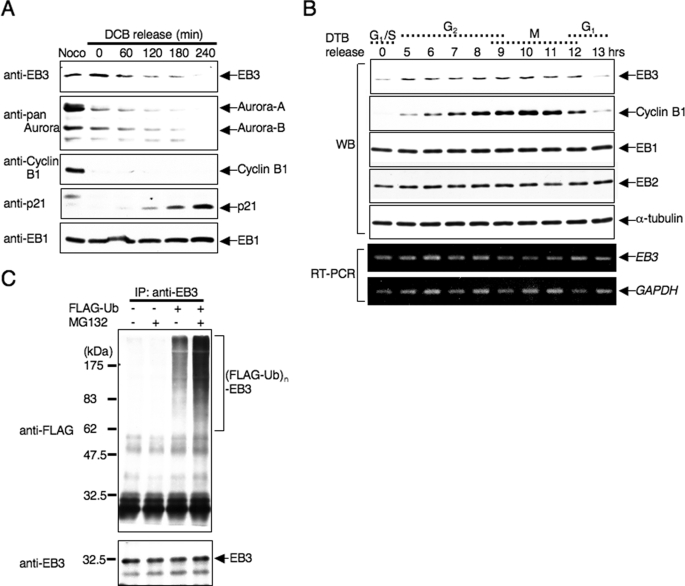
During time course studies of endogenous EB3 phosphorylation, we observed that the total protein level of EB3 decreased 180 min after release from nocodazole arrest, i.e. at a time when most cells have achieved mitotic exit and reached the following G1 phase (Fig. 2B). Thus, we examined the alteration of the EB3 protein level through mitosis into the subsequent G1 phase in more detail. HeLa cells were arrested at ana-telophase by dihydrocytochalasin B (DCB) after release from prometaphase arrest (36); the cells were then harvested at the indicated time points after DCB release, and endogenous EB3 was detected by Western blots of the cell lysates. As shown in Fig. 3A, the time-dependent down-regulation of EB3 was observed again; we noticed that the degradation of Aurora-A and -B preceded the down-regulation of EB3 (Fig. 3A, DCB 0 min compared with Noco). In contrast, p21, which is a marker protein during G1 phase progression (37), actually increased in parallel with the down-regulation of EB3, indicating that EB3 is down-regulated during the transition from mitosis to the G1 phase.
FIGURE 3.
EB3 protein level is regulated by the ubiquitin-proteasome system. A, down-regulation of EB3 during the mitosis-to-G1 transition. Lysates from HeLa cells that were released from a DCB block for the indicated times were subjected to Western blots (antibodies indicated on the side of the figure). Noco indicates nocodazole. B, cell cycle-dependent protein expression of EB3. HeLa cells were synchronized at G1/S phase by double thymidine block (DTB) release, and then cells were collected at the indicated times after release from arrest. The cell lysates were used for Western blots (WB) to determine each protein level, and RNAs extracted from each cell were used for reverse transcription-PCR (RT-PCR). C, polyubiquitination of endogenous EB3 in cells. HeLa cells were transfected with plasmids encoding FLAG-ubiquitin. After 24 h, cells were treated with or without 20 μm MG132 for 6 h. Immunoprecipitation (IP) was performed with anti-EB3 antibody, and the precipitates were analyzed by Western blots probed sequentially with anti-FLAG and anti-EB3 antibodies. The results shown are from one representative experiment out of at least three independent experiments.
Furthermore, the protein levels of EB3 over a wide range of the cell cycle were analyzed by using HeLa cells synchronized by double thymidine block-release boundary, and the cell cycle was concomitantly monitored by fluorescence-activated cell sorter analysis (data not shown). The EB3 protein level increased in late S and G2 phase, peaked in mitosis, and then decreased into the subsequent G1 phase (Fig. 3B). This expression pattern distinguishes EB3 from other EB1 family members, because the level of EB1 and EB2 showed no significant changes during the cell cycle (Fig. 3, A and B). In addition, the mRNA level of EB3 was not significantly altered (Fig. 3B). Thus, these results suggest that only EB3 within the EB1 family is actively degraded in vivo in a cell cycle-dependent manner.
Cell cycle control (including mitotic progression) depends on the degradation of key regulatory proteins by the ubiquitin-proteasome system (38, 39). To examine the possible involvement of the ubiquitin-proteasome system in EB3 degradation, we first determined the half-life of endogenous EB3 protein in cells treated with the protein synthesis inhibitor cycloheximide (CHX). As shown in supplemental Fig. 3, a time-dependent decrease in EB3 protein levels was observed, and its half-life was about 12 h, whereas EB1 levels did not change. Treatment with the proteasome inhibitor MG132 abolished the decrease (supplemental Fig. 3, right panel), demonstrating that EB3 degradation requires the function of the 26 S proteasomal pathway.
The dependence of 26 S proteasomal degradation suggests that EB3 might be polyubiquitinated in cells. Therefore, we performed in vivo ubiquitination assays for EB3. HeLa cells were transfected with an expression plasmid encoding FLAG-tagged ubiquitin. Endogenous EB3 was immunoprecipitated from the transfected cell lysates by using anti-EB3 monoclonal antibody and then examined by Western blots. Endogenous EB3 was polyubiquitinated in cells that overexpressed FLAG-ubiquitin. MG132 treatment elicited a robust increase in EB3 polyubiquitination (Fig. 3C). Taken together, we conclude that the EB3 protein level is regulated by the ubiquitin-proteasome system during the cell cycle.
Identification of SIAH-1 as an E3 for EB3
Given that the anaphase-promoting complex/cyclosome (APC/C) is an E3 and especially controls the completion of mitosis in all eukaryotes (40), the timing of EB3 degradation allows us to determine whether the APC/C functions as an E3 for EB3. Activation of the APC/C requires association with the WD-repeat protein, hCdc20 or hCdh1. We therefore investigated whether or not WD-repeat proteins activate APC/C for EB3 degradation in vivo. Overexpression of hCdc20 or hCdh1 with EB3 in HeLa cells did not affect EB3 protein level (data not shown) (28). Furthermore, using anti-Cdc27 antibody to immunoprecipitate the APC/C from cells transfected with hCdc20 or hCdh1, EB3 was not polyubiquitinated in vitro (data not shown) (26). Thus, we conclude that neither Cdc20 nor Cdh1-activated APC/C enhanced EB3 polyubiquitination.
Recently, a map of the interactome network in Caenorhabditis elegans, which is constructed by a comprehensive analysis of protein-protein interactions in C. elegans, was reported (41). According to the interaction list, 10 proteins are identified as interactive proteins with ebp-2 (sequence name VW02B12L.3), which is a putative ortholog of the human EB1 family in Caenorhabditis elegans. One of these proteins, sia-1 (sequence name Y37E11AR.2), which is an ortholog of Drosophila Sina (seven in absentia) and is apparently a RING finger-type E3. Thus, it is possible that SIAH-1, the human ortholog of Drosophila Sina, might be an E3 for human EB3.
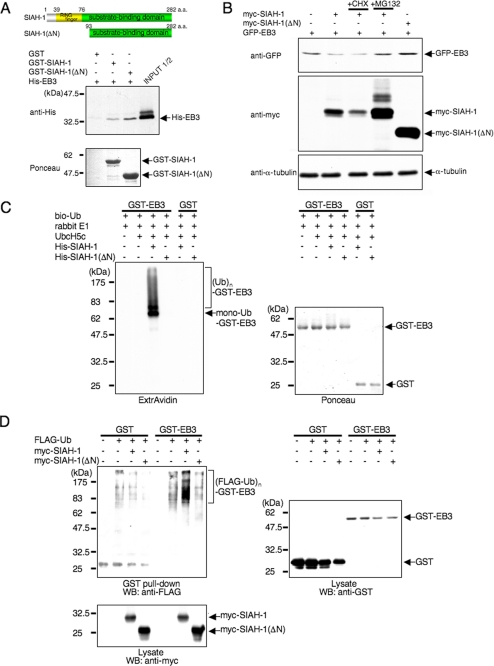
First, to define a functional link between EB3 and SIAH-1, GST pulldown assays were performed with recombinant His6-EB3 and GST-fused wild-type SIAH-1 or the deletion mutant SIAH-1(ΔN) retaining the C-terminal substrate binding domain but lacking the N-terminal RING finger domain (Fig. 4A, upper panel). EB3 was co-precipitated with both full-length SIAH-1 and SIAH-1(ΔN) but not with GST alone (Fig. 4A, lower panel), indicating that EB3 directly binds to SIAH-1 through its substrate binding domain.
FIGURE 4.
SIAH-1 is E3 for EB3. A, schematic representation of full-length SIAH-1 and SIAH-1(ΔN) (upper panels) and in vitro binding of EB3 and SIAH-1 (lower panels). Purified His-EB3 was incubated with GST-fused, full-length SIAH-1 and SIAH-1(ΔN) trapped on glutathione-Sepharose beads. The bound EB3 was then eluted and detected by Western blots with anti-penta-His antibody (middle panel). The bottom panel shows Ponceau staining of the GST fusion protein samples used in the assay. a.a., amino acids. B, SIAH-1 targets EB3 for proteasomal degradation. COS-7 cells were transfected with the indicated plasmids. After 48 h, cells were treated with or without 10 μg/ml CHX or 20 μm MG132 for 6 h. Using these cell lysates, Western blots were performed with the indicated antibodies. C, SIAH-1-mediated ubiquitination (Ub) of EB3 in vitro (left panel). The ubiquitination reaction was performed as described under “Experimental Procedures.” The ubiquitinated proteins in the reaction were detected by Western blots using ExtrAvidin. The right panel shows Ponceau staining of the GST fusion protein samples used in the assay. D, SIAH-1-mediated EB3 ubiquitination in cells. COS-7 cells were transfected with the indicated plasmids. After 48 h, cells were treated with 20 μm MG132 for 6 h. GST pulldown was performed, and the precipitates were analyzed by Western blots (WB) probed with anti-FLAG antibody. The results shown are from one representative experiment out of at least three independent experiments.
To explore the functional significance of SIAH-1 on EB3 degradation, we performed in vivo degradation assays of EB3. As expected, the co-expression of EB3 and full-length SIAH-1, but not SIAH-1(ΔN), led to decreased expression levels of transfected EB3 in COS-7 cells (Fig. 4B). This result indicates that ligase activity is required for SIAH-1-mediated EB3 degradation. SIAH-1-dependent EB3 depletion was inhibited by MG132, indicating that SIAH-1 facilitates proteasomal degradation of EB3. The EB3 level also slightly increased in the presence of SIAH-1(ΔN) compared with the absence of SIAH-1(ΔN); this increase is probably caused by the dominant-negative effect of SIAH-1(ΔN) (Fig. 4B, lane 5).
Next, we tested whether SIAH-1 functions as an E3 toward EB3 in vitro. EB3 was efficiently polyubiquitinated by full-length SIAH-1, but not by SIAH-1(ΔN) lacking the RING finger domain (Fig. 4C). Interestingly, although SIAH-1 could bind to all three EB1 family members, only EB3 was polyubiquitinated by SIAH-1 (supplemental Fig. 4, A and B). Therefore, we investigated whether EB3 is polyubiquitinated by SIAH-1 in cells. Fig. 4D shows that polyubiquitination of GST-EB3 was facilitated when COS-7 cells were co-transfected with full-length SIAH-1, not with SIAH-1(ΔN), which is consistent with the in vitro results. We also purified His-ubiquitin under denaturing conditions from cells transfected with His-ubiquitin and GST-EB3 by pulldown using nickel-nitrilotriacetic acid beads, and the elute was immunoblotted with anti-GST. The polyubiquitin-specific signals were efficiently augmented by full-length SIAH-1, but not by SIAH-1(ΔN), to ensure that the signals are indeed caused by EB3 polyubiquitination and not by autoubiquitinated SIAH-1 (supplemental Fig. 4C). These results indicate that SIAH-1 acts as an E3 for EB3 through its RING finger domain.
SIAH-1 Is Required for EB3 Degradation during the Transition from Mitosis to G1 Phase
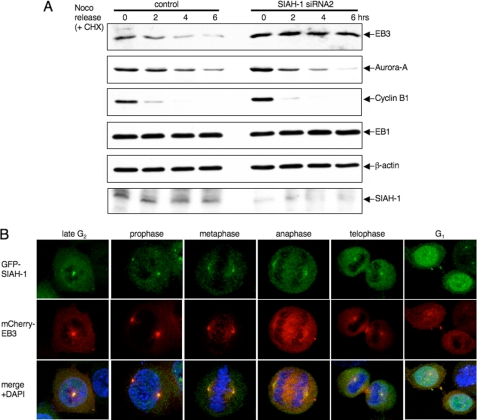
Next, we used siRNA to deplete SIAH-1 expression in cells. siRNAs directed against SIAH-1 resulted in a significant accumulation of EB3, without affecting the EB1 level (supplemental Fig. 5), demonstrating that EB3 is a physiologically relevant target protein of SIAH-1 in cells. Therefore, we investigated the effects of SIAH-1 down-regulation on EB3 degradation during the transition from mitosis to G1 phase. In control siRNA-transfected cells, the EB3 level was down-regulated after release from a nocodazole block, and its degradation rate was also significantly higher than during asynchronous growth (compare with 6 h as in supplemental Fig. 3 and Fig. 5A). In contrast, siRNA to SIAH-1 caused no decline in EB3 expression and a decrease of EB3 degradation rate (Fig. 5A). Both cells showed normal transition to anaphase, as judged by the disappearance of cyclin B1 and Aurora-A (Fig. 5A), thereby ruling out the possibility that the prolonged retention of EB3 in SIAH-1 knockdown cells was because of the blockage of cell cycle progression. These data indicate that SIAH-1 mediates EB3 degradation during the transition from mitosis to G1 phase.
FIGURE 5.
Degradation of EB3 by SIAH-1 at the mitosis to G1 transition. A, effects of SIAH-1 knockdown on EB3 stability during the mitosis-to-G1 transition. HeLa cells were transfected with siRNA for SIAH-1. After 48 h, nocodazole was added for 16 h; subsequently, cells were released from a nocodazole (Noco) block for the indicated times in the presence of 10 μg/ml CHX. The cell lysates were subjected to Western blots with the indicated antibodies. One Western blot shown is representative of three independent experiments. B, representative confocal images of localization of SIAH-1 and EB3 during each stage of mitosis. Immunofluorescence staining is shown of HeLa cells that stably expressed GFP-SIAH-1 and mCherry-EB3. Green, GFP-SIAH-1; red, mCherry-EB3; blue, DAPI.
To clarify the location of EB3 degradation mediated by SIAH-1 in cells, we examined the subcellular localization of SIAH-1 and EB3 during mitosis. We utilized HeLa cells co-expressing GFP-tagged SIAH-1 along with a monomeric Cherry (mCherry) (42) fusion EB3, as antibodies against SIAH-1 for immunofluorescence were not available. As reported previously (43), GFP-SIAH-1 was localized to the nucleus and cytoplasm during interphase and dispersed throughout the mitotic cytoplasm when cells were fixed with cold methanol (data not shown). When we analyzed cells from which the bulk of soluble proteins was removed by extraction, residual SIAH-1 assembled at the centrosome during late G2 phase and maintained the centrosomal localization until telophase, co-localizing with EB3 (Fig. 5B). As described above, EB3 also accumulated in the central spindle from anaphase to cytokinesis, whereas SIAH-1 was not localized to the central spindle localization (Fig. 5B). Interestingly, in early G1 phase, SIAH-1 appeared in the midbody where EB3 was already concentrated (Fig. 5B). Thus, the timing of EB3 degradation (Fig. 2B and Fig. 3A) with these observations suggests that EB3 may be degraded by SIAH-1 in the midbody.
SIAH-1-mediated EB3 Polyubiquitination and Subsequent Degradation Are Regulated by EB3 Phosphorylation at Ser-176
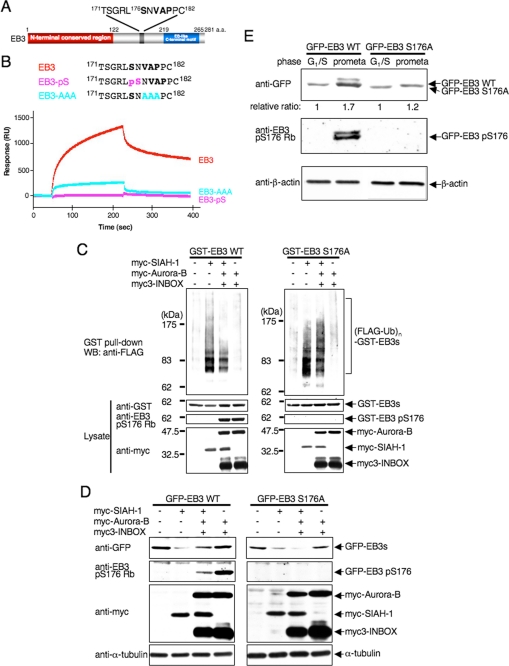
End-binding proteins contain highly conserved N- and C-terminal domains that are separated by a less conserved linker sequence (5). We explored the binding region of EB3 in detail. The consensus motif for SIAH-binding proteins is Pro-Xaa-Ala-Xaa-Val-Xaa-Pro with Val-Xaa-Pro constituting the core residues (44, 45). We noted that EB3 contains a Val-Xaa-Pro motif (residues 178–180) that is adjacent to the phosphorylated Ser-176 (Fig. 6A). Thus, the phosphorylation may influence the binding of EB3 to SIAH-1 through the Val-Xaa-Pro motif. We examined the ability of peptides derived from EB3-(171–182) to bind purified recombinant SIAH-1(ΔN) by surface plasmon resonance assays. The short wild-type peptide was competent enough to bind to SIAH-1 (Kd; 7–8 μm), whereas the corresponding phosphopeptide at Ser-176 completely abolished binding to SIAH-1 (Fig. 6B). The mutation of VAP-(178–180) into AAA markedly attenuated binding to SIAH-1. These results indicate that the core Val-Xaa-Pro motif and the phosphorylation of the evolutionally conserved Ser in the proximity of the motif within EB3 is crucial for binding to SIAH-1 in vitro. It is noteworthy that the replacement of Ser-176 by aspartic acid or glutamic acid did not inhibit in vitro binding of EB3 to SIAH-1 (data not shown), suggesting that the additional negative charge introduced by this substitution cannot mimic phosphorylation of EB3.
FIGURE 6.
Auroras regulate SIAH-1-dependent EB3 degradation through Ser-176 phosphorylation of EB3. A, schematic diagram of a potential SIAH-binding motif (V178AP180) in EB3. Ser-176 is an Aurora phosphorylation site. a.a., amino acids. B, top, schematic diagram of the sequences of wild-type and mutant peptides derived from EB3. Bottom, a BIAcore sensorgram shows that wild-type EB3 peptides, but not mutant EB3 peptides, bind to immobilized SIAH-1. A representative result of four independent experiments is shown. C, effects of Aurora phosphorylation on SIAH-1-mediated ubiquitination of EB3 in cells. COS-7 cells were transfected with each plasmid (as indicated above the figure). The ubiquitinated proteins were detected as described in Fig. 4D. WT, wild type. D, effects of Aurora phosphorylation on SIAH-1-mediated EB3 degradation. COS-7 cells were transfected with each plasmid (as indicated above the figure). After 48 h, cells were harvested, and Western blots (WB) were performed with the indicated antibodies. E, effects of Aurora phosphorylation on the stability of the EB3 proteins in mitosis. HeLa cells that stably expressed each GFP-wild-type EB3 or EB3 S176A were synchronized at each phase. Subsequently, cells were treated with CHX for 6 h, and the cell lysates were subjected to Western blots with the indicated antibodies. The relative protein level of GFP-EB3 at G1/S phase against the prometaphase is shown below each lane. C–E are representative of three independent experiments.
To assess whether Aurora-dependent phosphorylation of EB3 interferes with the polyubiquitination and subsequent degradation of EB3, we investigated the effect using in vivo ubiquitination assays. As expected, overexpression of Aurora-B remarkably compromised the polyubiquitination of GST-tagged EB3 mediated by SIAH-1 (Fig. 6C, left panel). Under the same experimental condition, EB3 was phosphorylated. The nonphosphorylatable EB3 S176A mutant was also SIAH-1-mediated polyubiquitinated, even with the co-expression of Aurora-B (Fig. 6C, right panel); thus, decreased polyubiquitination of wild-type EB3 with overexpression of Aurora-B is not because of altered SIAH-1 E3 activity. The effect within living cells was also observed in cell-free in vitro ubiquitination assays (supplemental Fig. 6). Furthermore, in vivo degradation assays revealed that the SIAH-1-mediated down-regulation of the wild-type EB3 protein level was partially restored when EB3 was co-expressed with Aurora-B (as compared with SIAH-1 only), whereas that of the EB3 S176A mutant protein level was not restored even by the overexpression with Aurora-B (Fig. 6D). These findings suggest that Auroras interfere with the interaction between EB3 and SIAH-1 through EB3 phosphorylation and therefore block SIAH-1-mediated polyubiquitination and the subsequent degradation of EB3. These results also provide an unprecedented mechanism of phospho-dependent substrate selection by SIAH-1.
As mentioned above, EB3 phosphorylation at Ser-176 occurred during mitosis, and the protein level of EB3 was higher during mitosis than in the G1 phase (Fig. 2B and Fig. 3B). Given that Aurora phosphorylation of EB3 Ser-176 resulted in stabilization of EB3 protein by preventing SIAH-1-mediated degradation, it is conceivable that a nonphosphorylatable form of EB3 might be unstable in mitotic cells. To examine this possibility, HeLa cells that stably expressed GFP-wild-type EB3 or EB3 S176A were synchronized at the G1/S phase or prometaphase (with CHX treatment for 6 h), and then the protein levels were detected by Western blots. The level of wild-type EB3 protein, but not the S176A mutant EB3 protein, in prometaphase-arrested cells increased more than that at the G1 phase (Fig. 6E). These results indicate that EB3 phosphorylation at Ser-176 regulates the steady-state level of EB3 during mitosis.
Abundance of EB3 Might Facilitate Cell Cycle Progression at Prometaphase
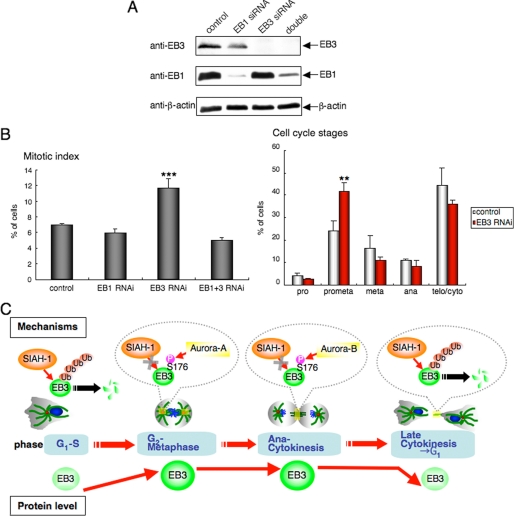
Inhibition of Drosophila EB1 induces an accumulation of metaphase cells by the activation of the spindle checkpoint because of defects in the mitotic spindle structure (10), whereas siRNA-mediated inhibition of human EB1 causes similar defects without arresting cells in mitosis (46). Therefore, we explored the consequences of human EB3 knockdown on mitotic progression. Immunostaining with α-tubulin antibody and DAPI revealed that EB3 knockdown cells exhibited a 2-fold increase in their mitotic index as compared with EB1 alone or together with EB3 knockdown, or control cells (Fig. 7B, left panel), and that EB3 knockdown cells accumulated in prometaphase with a concomitant decrease of cells in later mitotic stages, i.e. metaphase, anaphase, and telophase/cytokinesis (Fig. 7B, right panel).
FIGURE 7.
Effects of EB3 knockdown on mitotic progression. A, knockdown of endogenous EB1 and/or EB3. HeLa cells were treated with siRNAs for EB1 and/or EB3 for 72 h, and then the cell lysates were subjected to Western blots with the indicated antibodies. One Western blot shown is representative of three independent experiments. B, left panel, quantitation of the mitotic index of HeLa cells treated with each siRNA after 72 h. The mitotic index is expressed as the percentage of the total population of cells in mitosis (n = 1000). Right panel, quantitation of the cell cycle stages of HeLa cells treated with control or EB3 siRNA after 72 h. Cell cycle stages were determined by scoring all mitotic cells by immunofluorescence for α-tubulin and chromosome staining (n = 1000). C, schematic representation of our results.
As mentioned before, siRNA-mediated inhibition of human EB1 causes chromosomal mis-segregation and/or cytokinesis defects. When the endogenous EB1 protein level was reduced, multinuclear, heteromorphic, and micronuclear interphase cells were frequently observed (22%, supplemental Fig. 7). In contrast, cells treated with siRNA against EB3 alone exhibited less frequent defects (10.7%, supplemental Fig. 7) than that of cells treated with siRNA against EB1 alone or together with EB3. These results suggest that depletion of endogenous EB3 has little effect on accurate chromosomal segregation and/or cytokinesis as compared with that of EB1.
In summary, SIAH-1-mediated EB3 polyubiquitination and subsequent degradation were regulated by two mitotic kinases, Aurora-A and Aurora-B, through EB3 phosphorylation at Ser-176 during mitosis, and the abundance of EB3 might facilitate cell cycle progression at prometaphase.
DISCUSSION
Here, we report that the E3 SIAH-1 interacts with EB3 and mediates EB3 polyubiquitination and degradation through the ubiquitin-proteasome system. During mitosis two kinases, Aurora-A and Aurora-B, phosphorylate EB3 at Ser-176, and the resulting phosphorylation disrupts the EB3-SIAH-1 complex. Indeed, EB3 is stabilized during mitosis and facilitates cell cycle progression. Our results on the mitotic regulation of EB3 are summarized in Fig. 7C.
Recent publications provide a molecular basis of the recognition/specificity as the hydrophobic interaction between SIAH-1 and its substrates (45, 47). Our structure-based modeling reveals that the peptide derived from EB3 forms hydrophobic interactions with SIAH-1 mainly through Val-178 and Pro-180 and that the peptide binding groove is lined by Phe-165 and Trp-178 (supplemental Fig. 8). Ser-176 of EB3 lies close to Met-180 of SIAH-1; its γ-hydroxyl oxygen is located less than 4.0 Å from the sulfur atom. A phosphate group on the Ser residue would clearly prevent a close approach of the peptide to the apolar groove on SIAH-1. Given that most, if not all, SIAH-binding proteins contain phosphorylatable residues within the consensus motif (supplemental Fig. 9) and that conversely the SIAH-1 protein itself is subjected to regulatory phosphorylation (48–50), it is important to elucidate whether post-translational modifications such as phosphorylation can modulate the interactions between SIAH-1 and its binding partners, and how the modifications regulate the functions of SIAH-binding proteins individually.
Our results demonstrate that two mitotic kinases, Aurora-A and Aurora-B, phosphorylate EB3 at the conserved Ser-176 to disrupt the EB3-SIAH-1 complex and thereby protect EB3 from SIAH-1-dependent degradation during mitosis, and EB3 is then down-regulated during the transition to G1 phase. As the mitotic up-regulation of endogenous SIAH-1 expression (51) and SIAH-1 as an established p53 target gene (52, 53) have been previously reported, mitotic and/or unscheduled degradation of EB3 most likely is prevented by the post-translational modification. Two major E3s, APC/C and SCF (Skp1-Cullin-F-box protein complex), have a role in cell cycle control (54). SIAH-1 may be a third E3 in switch-like cellular events such as cell cycle transitions.
EB1 degradation is ubiquitin-proteasome-dependent and regulated by the COP9 signalsome (55). Our findings demonstrate that in contrast to EB3, EB1 and EB2 protein levels are not regulated during the cell cycle, and these two proteins are not substrates for SIAH-1 (Fig. 3 and supplemental Fig. 4B). As reported previously (56), EB1 is not a substrate of two mitotic kinases, Aurora-A and Aurora-B (Fig. 1C and data not shown). As EB1 family proteins are involved in microtubule dynamics in concert with their binding partners +TIPs, such as APC (57), and SIAH-1 also binds to APC, EB3 might regulate microtubule dynamics and organization, especially during mitosis, coordinating with other functionally redundant EB1 family members.
In our EB3 siRNA experiments, we only observe a 2-fold but significant increase in their mitotic index but no further observable phenotype. EB3 can interact to known EB1 partners, such as APC (13). EB3 and EB1 can form heterodimers in cells. In HeLa cells EB1 appears to be expressed at higher levels than EB3 (data not shown), in line with the recently published data (58). Thus, EB1 may compensate for the lack of EB3. This probably explains why the single depletion of EB3 has little effect on mitosis in these cells, although in the knockdown of EB1 or of both EB1 and EB3, inhibition of EB1 alone could compromise the spindle checkpoint function without arresting cells in mitosis.
The question of why EB3 needs to be protected from its degradation during mitosis is an important one. The simplest explanation is that EB3 may be needed for microtubule dynamics and organization in mitosis, depending on tissue specificity or EB1 expression level. Alternatively, it is possible that the regulated destruction of EB3 is required during mitotic exit. The EB3 class harboring both Aurora phosphorylation sites and a potential SIAH-1-binding motif is found only in mammals. EB3 has initially been described to be mostly expressed in brain and muscles (13). More recently roles have been attributed to EB3 during myotube differentiation with an EB3 protein up-regulation (59) and for the regulation of neuritogenesis (60). In addition SIAH-1 has also been shown to ubiquitinate α-synuclein and synphilin-1 involved in Parkinson disease (61). The physiological relevance of the regulation of EB3 by SIAH-1 and Aurora kinases might exist in noncycling cells. Future studies are necessary to clarify the detailed cell type- and tissue-specific regulation.
Recently, EB1 was reported to be an Aurora-B-interacting activator that prevents Aurora-B from dephosphorylation by protein phosphatase 2A (56). Our results demonstrate that EB1 family proteins interact with not only Aurora-B but also with Aurora-A. Additionally, EB1 family proteins cannot activate Aurora-B to prevent its dephosphorylation by protein phosphatase 2A in our hands (supplemental Fig. 10). Given that microtubules directly stimulate Aurora-B kinase activity in vitro (62, 63), this mechanism needs further investigation.
In summary, we report on two mitotic kinases, Aurora-A and Aurora-B, that regulate the EB3-SIAH-1 interaction through site-specific phosphorylation of EB3 to protect EB3 from SIAH-1-mediated degradation during mitosis.
Supplementary Material
Acknowledgments
We are grateful to K. Ida (Olympus) for technical assistance with the confocal time-lapse microscopy. We thank L. A. Feig for critical reading of the manuscript, valuable suggestions, and for the anti-Glu-Glu and anti-Myc tag monoclonal antibody; T. Kitamura for the Plat-E cells and pMXs retroviral vectors; and R. Y. Tsien for mCherry cDNA. We also thank F. Ishikawa, T. Matsumoto, and K. Kaibuchi for helpful scientific comments.
This work was supported in part by a grant-in-aid for scientific research (to G. S., K. F., and T. U.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10.
- +TIPs
- microtubule plus-end tracking proteins
- APC
- tumor suppressor adenomatous polyposis coli protein
- DCB
- dihydrocytochalasin B
- CHX
- cycloheximide
- APC/C
- anaphase-promoting complex/cyclosome
- mCherry
- monomeric Cherry
- E3
- ubiquitin-protein isopeptide ligase
- PBS
- phosphate-buffered saline
- DAPI
- 4′,6-diamidino-2-phenylindole
- GFP
- green fluorescent protein
- siRNA
- small interfering RNA
- GST
- glutathione S-transferase
- PIPES
- 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Steinmetz M. O. (2007) J. Struct. Biol. 158, 137–147 [DOI] [PubMed] [Google Scholar]
- 2.Howard J., Hyman A. A. (2007) Curr. Opin. Cell Biol. 19, 31–35 [DOI] [PubMed] [Google Scholar]
- 3.Cassimeris L. (1999) Curr. Opin. Cell Biol. 11, 134–141 [DOI] [PubMed] [Google Scholar]
- 4.Morrison E. E. (2007) Cell. Mol. Life Sci. 64, 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akhmanova A., Steinmetz M. O. (2008) Nat. Rev. Mol. Cell Biol. 9, 309–322 [DOI] [PubMed] [Google Scholar]
- 6.Juwana J. P., Henderikx P., Mischo A., Wadle A., Fadle N., Gerlach K., Arends J. W., Hoogenboom H., Pfreundschuh M., Renner C. (1999) Int. J. Cancer 81, 275–284 [DOI] [PubMed] [Google Scholar]
- 7.Su L. K., Burrell M., Hill D. E., Gyuris J., Brent R., Wiltshire R., Trent J., Vogelstein B., Kinzler K. W. (1995) Cancer Res. 55, 2972–2977 [PubMed] [Google Scholar]
- 8.Vaughan K. T. (2005) J. Cell Biol. 171, 197–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lansbergen G., Akhmanova A. (2006) Traffic 7, 499–507 [DOI] [PubMed] [Google Scholar]
- 10.Rogers S. L., Rogers G. C., Sharp D. J., Vale R. D. (2002) J. Cell Biol. 158, 873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tirnauer J. S., Grego S., Salmon E. D., Mitchison T. J. (2002) Mol. Biol. Cell 13, 3614–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goshima G., Nédélec F., Vale R. D. (2005) J. Cell Biol. 171, 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakagawa H., Koyama K., Murata Y., Morito M., Akiyama T., Nakamura Y. (2000) Oncogene 19, 210–216 [DOI] [PubMed] [Google Scholar]
- 14.Bu W., Su L. K. (2001) Oncogene 20, 3185–3192 [DOI] [PubMed] [Google Scholar]
- 15.Komarova Y., Lansbergen G., Galjart N., Grosveld F., Borisy G. G., Akhmanova A. (2005) Mol. Biol. Cell 16, 5334–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nigg E. A. (2001) Nat. Rev. Mol. Cell Biol. 2, 21–32 [DOI] [PubMed] [Google Scholar]
- 17.Ferrari S. (2006) Cell. Mol. Life Sci. 63, 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carmena M., Earnshaw W. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 842–854 [DOI] [PubMed] [Google Scholar]
- 19.Marumoto T., Zhang D., Saya H. (2005) Nat. Rev. Cancer 5, 42–50 [DOI] [PubMed] [Google Scholar]
- 20.Andrews P. D., Knatko E., Moore W. J., Swedlow J. R. (2003) Curr. Opin. Cell Biol. 15, 672–683 [DOI] [PubMed] [Google Scholar]
- 21.Ruchaud S., Carmena M., Earnshaw W. C. (2007) Nat. Rev. Mol. Cell Biol. 8, 798–812 [DOI] [PubMed] [Google Scholar]
- 22.Moore A., Wordeman L. (2004) Trends Cell Biol. 14, 537–546 [DOI] [PubMed] [Google Scholar]
- 23.Kinoshita K., Noetzel T. L., Arnal I., Drechsel D. N., Hyman A. A. (2006) J. Muscle Res. Cell Motil. 27, 107–114 [DOI] [PubMed] [Google Scholar]
- 24.Gadea B. B., Ruderman J. V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4493–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.James P., Halladay J., Craig E. A. (1996) Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda K., Mihara H., Kato Y., Yamaguchi A., Tanaka H., Yasuda H., Furukawa K., Urano T. (2000) Oncogene 19, 2812–2819 [DOI] [PubMed] [Google Scholar]
- 27.Li X., Sakashita G., Matsuzaki H., Sugimoto K., Kimura K., Hanaoka F., Taniguchi H., Furukawa K., Urano T. (2004) J. Biol. Chem. 279, 47201–47211 [DOI] [PubMed] [Google Scholar]
- 28.Taguchi S., Honda K., Sugiura K., Yamaguchi A., Furukawa K., Urano T. (2002) FEBS Lett. 519, 59–65 [DOI] [PubMed] [Google Scholar]
- 29.Ohashi S., Sakashita G., Ban R., Nagasawa M., Matsuzaki H., Murata Y., Taniguchi H., Shima H., Furukawa K., Urano T. (2006) Oncogene 25, 7691–7702 [DOI] [PubMed] [Google Scholar]
- 30.Morita S., Kojima T., Kitamura T. (2000) Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 31.Ban R., Irino Y., Fukami K., Tanaka H. (2004) J. Biol. Chem. 279, 16394–16402 [DOI] [PubMed] [Google Scholar]
- 32.Berrueta L., Kraeft S. K., Tirnauer J. S., Schuyler S. C., Chen L. B., Hill D. E., Pellman D., Bierer B. E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiyama K., Sugiura K., Hara T., Sugimoto K., Shima H., Honda K., Furukawa K., Yamashita S., Urano T. (2002) Oncogene 21, 3103–3111 [DOI] [PubMed] [Google Scholar]
- 34.Ferrari S., Marin O., Pagano M. A., Meggio F., Hess D., El-Shemerly M., Krystyniak A., Pinna L. A. (2005) Biochem. J. 390, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C. L., Peters J. M. (2003) J. Cell Biol. 161, 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martineau S. N., Andreassen P. R., Margolis R. L. (1995) J. Cell Biol. 131, 191–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherr C. J., Roberts J. M. (1999) Genes Dev. 13, 1501–1512 [DOI] [PubMed] [Google Scholar]
- 38.Gutierrez G. J., Ronai Z. (2006) Trends Biochem. Sci. 31, 324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sumara I., Maerki S., Peter M. (2008) Trends Cell Biol. 18, 84–94 [DOI] [PubMed] [Google Scholar]
- 40.Peters J. M. (2006) Nat. Rev. Mol. Cell Biol. 7, 644–656 [DOI] [PubMed] [Google Scholar]
- 41.Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P. O., Han J. D., Chesneau A., Hao T., Goldberg D. S., Li N., Martinez M., Rual J. F., Lamesch P., Xu L., Tewari M., Wong S. L., Zhang L. V., Berriz G. F., Jacotot L., Vaglio P., Reboul J., Hirozane-Kishikawa T., Li Q., Gabel H. W., Elewa A., Baumgartner B., Rose D. J., Yu H., Bosak S., Sequerra R., Fraser A., Mango S. E., Saxton W. M., Strome S., Van Den Heuvel S., Piano F., Vandenhaute J., Sardet C., Gerstein M., Doucette-Stamm L., Gunsalus K. C., Harper J. W., Cusick M. E., Roth F. P., Hill D. E., Vidal M. (2004) Science 303, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 43.Bruzzoni-Giovanelli H., Faille A., Linares-Cruz G., Nemani M., Le Deist F., Germani A., Chassoux D., Millot G., Roperch J. P., Amson R., Telerman A., Calvo F. (1999) Oncogene 18, 7101–7109 [DOI] [PubMed] [Google Scholar]
- 44.House C. M., Frew I. J., Huang H. L., Wiche G., Traficante N., Nice E., Catimel B., Bowtell D. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3101–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.House C. M., Hancock N. C., Möller A., Cromer B. A., Fedorov V., Bowtell D. D., Parker M. W., Polekhina G. (2006) Structure 14, 695–701 [DOI] [PubMed] [Google Scholar]
- 46.Green R. A., Wollman R., Kaplan K. B. (2005) Mol. Biol. Cell 16, 4609–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santelli E., Leone M., Li C., Fukushima T., Preece N. E., Olson A. J., Ely K. R., Reed J. C., Pellecchia M., Liddington R. C., Matsuzawa S. (2005) J. Biol. Chem. 280, 34278–34287 [DOI] [PubMed] [Google Scholar]
- 48.Winter M., Sombroek D., Dauth I., Moehlenbrink J., Scheuermann K., Crone J., Hofmann T. G. (2008) Nat. Cell Biol. 10, 812–824 [DOI] [PubMed] [Google Scholar]
- 49.Xu Z., Sproul A., Wang W., Kukekov N., Greene L. A. (2006) J. Biol. Chem. 281, 303–312 [DOI] [PubMed] [Google Scholar]
- 50.Yun S., Möller A., Chae S. K., Hong W. P., Bae Y. J., Bowtell D. D., Ryu S. H., Suh P. G. (2008) J. Biol. Chem. 283, 1034–1042 [DOI] [PubMed] [Google Scholar]
- 51.Frew I. J., Dickins R. A., Cuddihy A. R., Del Rosario M., Reinhard C., O'Connell M. J., Bowtell D. D. (2002) Mol. Cell. Biol. 22, 8155–8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fiucci G., Beaucourt S., Duflaut D., Lespagnol A., Stumptner-Cuvelette P., Géant A., Buchwalter G., Tuynder M., Susini L., Lassalle J. M., Wasylyk C., Wasylyk B., Oren M., Amson R., Telerman A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3510–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsuzawa S., Takayama S., Froesch B. A., Zapata J. M., Reed J. C. (1998) EMBO J. 17, 2736–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morgan D. O. (2007) in The Cell Cycle: Principles of Control (Lawrence E.) pp. 46–49, New Science Press Ltd, London [Google Scholar]
- 55.Peth A., Boettcher J. P., Dubiel W. (2007) J. Mol. Biol. 368, 550–563 [DOI] [PubMed] [Google Scholar]
- 56.Sun L., Gao J., Dong X., Liu M., Li D., Shi X., Dong J. T., Lu X., Liu C., Zhou J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7153–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akhmanova A., Hoogenraad C. C. (2005) Curr. Opin. Cell Biol. 17, 47–54 [DOI] [PubMed] [Google Scholar]
- 58.Komarova Y., De Groot C. O., Grigoriev I., Gouveia S. M., Munteanu E. L., Schober J. M., Honnappa S., Buey R. M., Hoogenraad C. C., Dogterom M., Borisy G. G., Steinmetz M. O., Akhmanova A. (2009) J. Cell Biol. 184, 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Straube A., Merdes A. (2007) Curr. Biol. 17, 1318–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geraldo S., Khanzada U. K., Parsons M., Chilton J. K., Gordon-Weeks P. R. (2008) Nat. Cell Biol. 10, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 61.Engelender S. (2008) Autophagy 4, 372–374 [DOI] [PubMed] [Google Scholar]
- 62.Fuller B. G., Lampson M. A., Foley E. A., Rosasco-Nitcher S., Le K. V., Tobelmann P., Brautigan D. L., Stukenberg P. T., Kapoor T. M. (2008) Nature 453, 1132–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosasco-Nitcher S. E., Lan W., Khorasanizadeh S., Stukenberg P. T. (2008) Science 319, 469–472 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.