Abstract
Parathyroid hormone (PTH) and PTH-related protein (PTHrP) are two related peptides that control calcium/phosphate homeostasis and bone development, respectively, through activation of the PTH/PTHrP receptor (PTH1R), a class B G protein-coupled receptor. Both peptides hold clinical interest for their capacities to stimulate bone formation. PTH and PTHrP display different selectivity for two distinct PTH1R conformations, but how their binding to the receptor differs is unclear. The high resolution crystal structure of PTHrP bound to the extracellular domain (ECD) of PTH1R reveals that PTHrP binds as an amphipathic α-helix to the same hydrophobic groove in the ECD as occupied by PTH, but in contrast to a straight, continuous PTH helix, the PTHrP helix is gently curved and C-terminally “unwound.” The receptor accommodates the altered binding modes by shifting the side chain conformations of two residues within the binding groove: Leu-41 and Ile-115, the former acting as a rotamer toggle switch to accommodate PTH/PTHrP sequence divergence, and the latter adapting to the PTHrP curvature. Binding studies performed with PTH/PTHrP hybrid ligands having reciprocal exchanges of residues involved in different contacts confirmed functional consequences for the altered interactions and enabled the design of altered PTH and PTHrP peptides that adopt the ECD-binding mode of the opposite peptide. Hybrid peptides that bound the ECD poorly were selective for the G protein-coupled PTH1R conformation. These results establish a molecular model for better understanding of how two biologically distinct ligands can act through a single receptor and provide a template for designing better PTH/PTHrP therapeutics.
The parathyroid hormone receptor (PTH1R)3 is a class B G protein-coupled receptor (GPCR) that transduces signals from two related signaling molecules that have distinct functions in biology: parathyroid hormone (PTH) and parathyroid hormone-related protein (PTHrP) (Ref. 1; reviewed in Ref. 2). PTH is an 84-amino acid polypeptide endocrine hormone that is produced by the parathyroid glands and secreted into the circulation in response to low calcium levels (reviewed in Refs. 3–5), to act on bone and kidney cells and thus restore blood calcium to normal levels. In bone, PTH directly stimulates osteoblasts, resulting in bone formation (reviewed in Ref. 6), which in turn activate osteoclasts to induce bone resorption. In the kidney, PTH stimulates the reabsorption of filtered calcium, inhibits the reabsorption of phosphate, and stimulates the synthesis of 1,25-dihydroxyvitamin D3. The paradoxical anabolic/catabolic actions of PTH on bone can be modulated by exogenous PTH, and provide the molecular basis for the clinical use of PTH as an anabolic therapy for osteoporosis (7). Anabolic PTH therapy requires intermittent administration to minimize bone-resorptive effects, which predominate with sustained administration of PTH. PTHrP is a 141-amino acid polypeptide that was originally isolated as the factor responsible for humoral hypercalcemia of malignancy (8–11) and was subsequently shown to be a critical developmental paracrine factor that controls endochondral bone formation (Refs. 12, 13; reviewed in Ref. 14). PTHrP can also mediate bone-anabolic effects when administered to osteoporosis patients (15) and has been suggested to be more anabolic than PTH due to a differential effect on the coupled bone formation and resorptive responses (16).
PTH and PTHrP are encoded by separate genes, each of which is found in vertebrate species ranging from fish to man. How PTH and PTHrP evolved to mediate distinct biological activities: calcium/phosphate homeostasis and tissue development, respectively, via actions upon a single receptor, remains unclear. Amino acid sequence homology is most apparent in the first 34-residue segments of the proteins, and N-terminal 34-residue peptide fragments of PTH and PTHrP are sufficient for high affinity binding to the PTH1R and are generally found to be equally potent for stimulating cAMP formation in PTH1R-expressing cells (1). The interaction of the (1–34)-length ligand with the PTH1R has been postulated to follow a “two-domain” model: residues within the approximate (1–14) segment interact with the 7-transmembrane (7-TM) helical domain embedded in the membrane, and residues within the approximate (15–34) segment interact with the N-terminal extracellular domain (ECD) of the receptor (17, 18). The 1–14 domains of PTH and PTHrP share eight amino acid sequence identities, reflecting a critical role in activating the receptor (18), while the 15–34 domains share only three amino acid identities, despite a critical role in imparting high affinity binding to the receptor.
Recent studies suggest that PTH and PTHrP differ in their relative capacities to bind to two pharmacologically distinguishable high-affinity PTH1R conformations (19–22). One conformation, termed R0, is stable in the presence of GTPγS, but presumably in the absence of G protein coupling, correlates with prolonged signaling responses in vitro and in vivo, and is bound preferentially by PTH-(1–34). The other conformation, termed RG, is sensitive to GTPγS addition, promoted by the overexpression of a high affinity variant of Gαs, and bound preferentially by PTHrP-(1–36). A mechanistic basis for the differing capacities of PTH and PTHrP ligands to bind to these altered PTH1R conformations is not clear at present, although, both the (1–14) and (15–34) portions of PTH contribute importantly to the capacity to bind stably to the proposed R0 conformation (19, 21, 22).
We previously developed a method that allowed us to determine the high resolution crystal structure of recombinant PTH1R ECD in complex with the 15–34 synthetic fragment of PTH (23). The PTH1R ECD adopts a tertiary fold that is conserved among class B GPCR ECDs (24–26), and the PTH(15–34)NH2 domain binds as a continuous and straight amphipathic α-helix to a hydrophobic groove in the ECD. Here we present the high resolution crystal structure of the PTHrP 12–34 fragment in complex with the PTH1R ECD, which reveals a distinct docking conformation toward the C terminus of the PTHrP peptide. Based on the structural differences, we designed hybrid PTH/PTHrP peptides exchanged for residues involved in altered ECD contacts; functional analyses of these peptides confirmed that the altered modes of binding indeed translate into functional consequences in terms of receptor affinity. These results provide critical insights into how PTH and PTHrP can act through a single receptor, and a structural model for designing better PTH/PTHrP analogs for treating osteoporosis.
EXPERIMENTAL PROCEDURES
Expression Plasmid Construction
A DNA fragment encoding human PTH1R residues 23–191 with a C-terminal six histidine residue tag and a stop codon was PCR-amplified from pcDNA3.1/PTH1R (23). The fragment was digested with BamHI and NotI restriction endonucleases and ligated into a pETDuet1 vector (Novagen) encoding maltose-binding protein (MBP) followed by a thrombin cleavage site (Th) and a biotinylation tag sequence (GGLNDIFEAQKIEWHEDT; biotinylation site in bold) in multiple cloning site 1, and the BirA biotin ligase in multiple cloning site 2. The resulting vector co-expresses the MBP-Th-Biotin tag-PTH1R ECD-H6 fusion protein and BirA. The construct was verified by automated DNA sequencing of the coding regions.
Protein Expression and Purification
The MBP-PTH1R ECD-H6 fusion protein used for crystallization, which contains residues 29–187 of human PTH1R fused to the C terminus of bacterial MBP, was previously described (23). The MBP-Th-Biotin tag-PTH1R ECD-H6 fusion protein was expressed in Escherichia coli Origami B (DE3) cells (Novagen). The cells were grown in LB medium at 37 °C to mid-log phase, then the temperature was reduced to 16 °C, biotin was added to 5 μm final concentration, and protein expression was induced with 0.4 mm isopropyl-1-thio-β-d-galactopyranoside for a total induction time of ∼19 h. The cells were harvested by centrifugation, and the fusion protein was purified as previously described for the MBP-PTH1R ECD-H6 fusion protein (23). Native gel electrophoresis of the purified sample suggested that the protein was ∼50% biotinylated as evidenced by two distinct bands of slightly different mobility (data not shown). The protein concentration was determined by the method of Bradford (27).
Peptide Synthesis
Peptides for ECD binding studies and crystallization were custom-synthesized and HPLC-purified by SynBioSci (Livermore, CA). PTH/PTHrP-(1–34)NH2 hybrid peptides were synthesized and HPLC-purified by the Massachusetts General Hospital Biopolymer Core facility.
ECD Peptide Binding Assays
The interaction between biotinylated PTH1R ECD and PTH(1–34)NH2 and PTHrP(1–34)NH2 was analyzed by bio-layer interferometry technology using the Octet Red system (ForteBio). MBP was removed from the MBP-Th-Biotin tag-PTH1R ECD fusion protein by overnight digestion at 4 °C with human α-thrombin (Hematologic Technologies Inc.) at a 1:750 ratio (weight/weight) of protease to fusion protein in Krebs-Ringers-HEPES (KRH) buffer (25 mm HEPES, pH 7.4, 104 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm KH2PO4, 1.2 mm MgSO4) supplemented with 2 mg/ml BSA. Complete cleavage was verified by SDS-PAGE (data not shown). High binding streptavidin sensors were incubated with the digestion reaction diluted to 2 μg/ml protein, followed by a 10 μg/ml biocytin block, and three washes in KRH buffer with 2 mg/ml BSA to wash away non-biotinylated components of the digestion mixture and establish a baseline. The peptides were diluted in KRH buffer with 2 mg/ml BSA, and their association and dissociation were monitored for 20 min each at 25 °C. To account for nonspecific binding, traces obtained with peptide and sensor tips lacking PTH1R ECD were subtracted from the traces obtained with the peptides and immobilized PTH1R ECD. In addition, a baseline drift was subtracted using a trace from a sensor tip that had immobilized PTH1R ECD, but no peptide. Octet Red analysis software was used to analyze the data.
The binding of peptides to the MBP-PTH1R ECD-H6 fusion protein was assessed with an AlphaScreen luminescent proximity assay (PerkinElmer Life Sciences). The reaction mixtures were incubated at room temperature and contained 5 μg/ml each of streptavidin-coated donor beads and nickel chelate-coated acceptor beads, and 23 nm each of N-terminally biotinylated PTH(7–34)NH2 and MBP-PTH1R ECD-H6 in a buffer of 50 mm MOPS pH 7.4, 150 mm NaCl, and 7 mg/ml BSA. The biotinylated PTH and MBP-PTH1R ECD-H6 were separately pre-coupled for 1 h to the streptavidin and nickel-chelate beads, respectively. The pre-coupling reactions were mixed, unlabeled competitor peptides were added as indicated, and the reactions were incubated 4–5 h to reach equilibrium. Photon counts were recorded in 384-well optiplates using an Envision 2104 plate reader (PerkinElmer). Prism 5.0 software (GraphPad Software, San Diego) was used to fit the data to a fixed slope dose-response inhibition equation for determination of IC50 values.
Crystallization and Structure Solution
MBP-PTH1R ECD-H6 in 10 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm maltose, and 1 mm EDTA was mixed with a synthetic PTHrP fragment (residues 12–34) at a 1:1.1 molar ratio (protein/peptide), incubated on ice for 30 min, and spin concentrated to 20 mg/ml for crystallization. Bipyramidal crystals were grown by the sitting drop vapor diffusion method at 20 °C with a reservoir solution of 7.5% PEG 2000, 13% PEG 400. The crystals were transferred into a cryoprotectant solution of 10% PEG 2000, 22% PEG 400, 50 mm NaCl, 1 mm EDTA by dialysis overnight and then were flash-frozen by plunging into liquid nitrogen. A data set was collected from a single crystal at LS-CAT beamline 21-ID-F of the Advanced Photon Source (Argonne, IL) at a wavelength of 0.9785 Å and temperature of 100 K. The HKL2000 package (28) was used to process the data, and the Scalepack intensities were converted to structure factor amplitudes with the CCP4 suite (29). There was no evidence of significant radiation damage in the dataset. The structure was solved by molecular replacement with Phaser (30) using separate search models for MBP and the PTH1R ECD from the PDB coordinate file 3C4M (23). There is one MBP-PTH1R ECD:PTHrP(12–34) complex in the asymmetric unit. Iterative cycles of rebuilding in O (31) and restrained refinement with Refmac5 (32) completed the structure. TLS refinement (33) was included using two TLS groups corresponding to the MBP-maltose complex and the ECD-PTHrP complex. Structure validation with Procheck (34) indicated that 93.6% of the residues were in the most favored region of the Ramachandran plot and no residues were in the disallowed region. The data collection and refinement statistics are listed in Table 1. Analyses of solvent accessible surface area and shape complementarity were performed within the CCP4 suite. Structure figures were prepared with PyMol (35).
TABLE 1.
Data collection and refinement statistics
| MBP-PTH1R ECD:PTHrP(12–34)a | |
|---|---|
| Data collection | |
| Space group | P41212 |
| Cell dimensions | |
| a, b, c (Å) | 84.04, 84.04, 164.46 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.00-1.94 (2.01-1.94)b |
| Rmerge | 0.091 (0.443) |
| I/σI | 29.66 (2.25) |
| Completeness (%) | 91.2 (55.0)c |
| Redundancy | 17.0 (8.5) |
| Refinement | |
| Resolution (Å) | 50.00-1.94 |
| No. reflections | 38628 |
| Rwork/Rfree | 19.3/23.3 |
| No. atoms | |
| Protein | 3701 |
| Ligand/ion | 214 |
| Water | 237 |
| Mean B-factors | |
| MBP | 42.7 |
| PTH1R ECD | 59.9 |
| PTHrP | 67.2 |
| Maltose | 31.9 |
| Water | 47.1 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.264 |
a Single crystal was used.
b Values in parentheses are for the highest-resolution shell.
c The low completeness in the outer shell is due to anisotropy in the diffraction images. The data are 88.9% complete in the 2.18-2.09 Å shell.
Receptor Binding and Signaling Assays
Peptide binding to the G protein-uncoupled receptor (R0) and G protein-coupled receptor (RG) conformations in COS-7 cell membranes was assessed as previously described (22). Peptide stimulation of cAMP accumulation in COS-1 cells transiently expressing human PTH1R was performed in 96-well tissue culture plates as previously described (36). cAMP was quantitated with a LANCE cAMP kit (PerkinElmer) according to the manufacturer's instructions.
RESULTS
PTH and PTHrP Binding to the PTH1R ECD
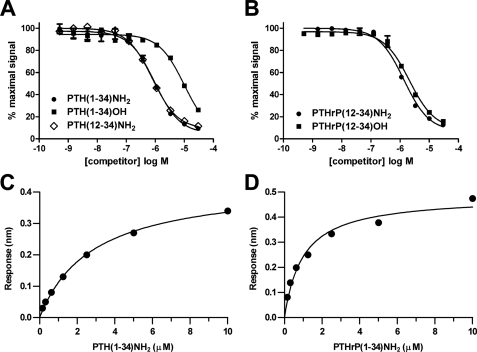
We first determined the binding of PTH and PTHrP ligands to the purified MBP-PTH1R ECD-H6 fusion protein using an AlphaScreen assay. In this assay, the N-terminally biotinylated PTH(7–34)NH2 fragment is bound to streptavidin-coated donor beads, and the fusion protein is bound via its His6 tag to nickel chelate-coated acceptor beads. Association of the PTH-(7–34) ligand with the fusion protein brings the beads into proximity and thus generates a luminescent signal. We previously showed that PTH(15–34)NH2 inhibits this association with an IC50 value of ∼1 μm, which was similar to the Kd value determined for the binding of PTH(15–34)NH2 to the fusion protein by isothermal titration calorimetry (23). PTH(1–34)NH2 and PTH(12–34)NH2 each inhibited the AlphaScreen interaction with an IC50 value of ∼1 μm (Fig. 1A), in agreement with our previous observations. However, PTH(1–34)OH bound the ECD with an affinity ∼10-fold lower than that of the C-terminally amidated peptide (Fig. 1A). This lower binding affinity can be reconciled by the crystal structure of the PTH1R ECD-PTH(15–34)NH2 complex, which shows that two hydrogen bonds are formed between the ECD and the ligand C-terminal amide (23). In contrast, PTHrP(12–34)NH2 and PTHrP(12–34)OH inhibited the association with similar IC50 values of ∼2 μm (Fig. 1B); thus, a C-terminal amide group is not essential for high affinity binding of PTHrP to the ECD.
FIGURE 1.
PTH and PTHrP binding to the PTH1R ECD. A and B, ability of the indicated PTH (A) or PTHrP (B) peptides to compete the association of biotin-PTH(7–34)NH2 (23 nm) and MBP-PTH1R ECD (23 nm) was assessed with an AlphaScreen assay. The data represent the average of duplicate samples. C and D, steady-state analysis of real-time binding data obtained with the Octet Red system. Biotinylated PTH1R ECD was immobilized on streptavidin sensor tips, and PTH(1–34)NH2 (C) and PTHrP(1–34)NH2 (D) binding was assessed. The plots in panels C and D show the response versus peptide concentration curves derived from the raw binding data (supplemental Fig. S1).
We also examined the interaction of the peptides with the isolated ECD using the Octet Red system (ForteBio), which uses biolayer interferometry technology to monitor binding events in real time. The isolated PTH1R ECD (residues 23–191) free of MBP and biotinylated at a single residue at the N terminus was immobilized on the surface of streptavidin sensor tips, and the binding of PTH(1–34)NH2 and PTHrP(1–34)NH2 free in solution was assessed over time. Steady-state analysis of the real time binding curves (supplemental Fig. S1) indicated that PTH and PTHrP bound the ECD with Kd values of 2.8 μm and 0.99 μm, respectively (Fig. 1, C and D). The Kd value for PTH binding to the ECD is similar to that obtained by others using Biacore technology (37). The binding data taken together indicate that PTH and PTHrP bind the PTH1R ECD with similar affinities, but suggest differences in the underlying biochemical mechanisms, because C-terminal amidation affects the ability of PTH, but not PTHrP, to bind the ECD.
Structural Basis for PTHrP Binding to the PTH1R ECD
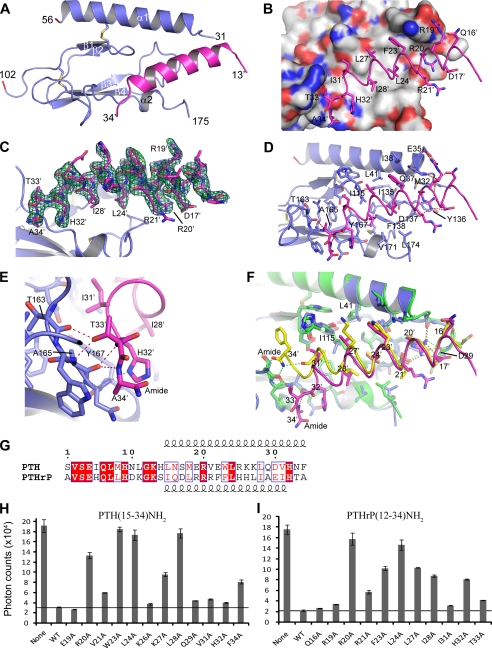
To assess receptor binding mechanisms of PTHrP, we determined the crystal structure of the MBP-PTH1R ECD-H6 fusion protein in complex with PTHrP(12–34)NH2. The structure was solved by molecular replacement and refined at 1.94 Å resolution to an R factor of 19.3% (free R factor 23.3%) (Table 1). Electron density was observed for all PTH1R residues except segments 29–30, 57–101, 176–187, and the C-terminal His6 tag, which were excluded from the final model. The PTH1R ECD forms the conserved class B GPCR ECD fold observed previously (23), and the PTHrP ligand forms an amphipathic α-helix that occupies the hydrophobic groove formed in the ECD at the interfaces of the N-terminal α-helix, the two β-sheets, and the short, C-terminal α-helix (Fig. 2, A and B). Clear electron density was observed for PTHrP residues 13–34 (Fig. 2C). The intermolecular interface is formed largely by hydrophobic interactions involving PTHrP residues Phe-23′, Leu-24′, Leu-27′, and Ile-28′, which contact the hydrophobic groove of the ECD formed by residues Leu-41, Ile-115, Ile-135, Phe-138, and Tyr-167 (Fig. 2, B and D). (Peptide residues are denoted with a prime to distinguish them from receptor residues.) In addition, an extensive network of hydrogen bonds anchors the peptide N and C termini (Fig. 2, D and E). The conserved R20′ forms an intermolecular salt bridge with Asp-137, an intermolecular hydrogen bond to the backbone carbonyl of Met-32, and intramolecular hydrogen bonds with the side chain of Asp-17′ (Fig. 2D). The PTHrP helix “unwinds” at the C terminus after residue Ile-31′ permitting the formation of several additional hydrogen bonds, including those between the δ nitrogen of His-32′ and the backbone amide nitrogen of Tyr-167, between the side chain hydroxyl group of Thr-33′ and the backbone amide nitrogen of Ala-165 and backbone carbonyl of Thr-163, and between the backbone carbonyl of Ala-165 and the backbone amide nitrogens of Thr-33′ and Ala-34′ (Fig. 2E). Consistent with our binding data in Fig. 1B, the C-terminal amide group of PTHrP does not form any interactions with the ECD. Arg-19′ forms a salt bridge with Glu-35, but this interaction may be caused by a crystal packing interaction that limits the mobility of Arg-19′ (not shown). Mutational and cross-linking data suggest that in the intact ligand-receptor complex, Arg-19′ contacts the 7-TM domain of the receptor (38).
FIGURE 2.
Structure of the PTH1R ECD in complex with PTHrP-(12–34) and comparison to the PTH-bound PTH1R ECD. A, overall view of the PTHrP-ECD complex with the ECD in slate blue and PTHrP in magenta. Residues 57–101 were not observed in the electron density maps. MBP is not shown for clarity. B, molecular surface of the ECD showing the hydrophobic groove contacted by Phe-23′, Leu-24′, Leu-27′, and Ile-28′ of PTHrP. The surface is colored according to atom type: gray for carbon, blue for nitrogen, and red for oxygen. C, omit electron density maps for PTHrP. The Fo-Fc omit map is shown as a green mesh contoured at 3 ς and the 2Fo-Fc omit map is shown as a blue mesh contoured at 1 ς. D, detailed view of the PTHrP-ECD interface. PTHrP is shown as a magenta coil, and selected side chains are shown as sticks. The dashed red lines indicate hydrogen bonds, and the red sphere is a water molecule. E, close-up view of the hydrogen bond network involving residues 32–34 of PTHrP. F, structural alignment of the PTHrP-ECD complex and the PTH-ECD complex (PDB code 3C4M). The PTHrP-ECD complex is colored as above, PTH is yellow, and the PTH-bound ECD is green. Selected side chains are shown as sticks. Hydrogen bonds are depicted as red dashes for the PTHrP-ECD complex and green dashes for the PTH-ECD complex. G, amino acid sequence alignment of human PTH and PTHrP residues 1–34 with the length of the α-helices designated above and below the sequence. H, AlphaScreen single point competition assay assessing the ability of PTH(15–34)NH2 alanine-scan peptides (20 μm) to compete the interaction of biotin-PTH (23 nm) with MBP-PTH1R-ECD (23 nm). The results are the average of duplicate samples. I, AlphaScreen assay as in panel H except with PTHrP(12–34)NH2 alanine-scan peptides.
PTH and PTHrP Exhibit Different ECD Binding Modes
Structural alignment of the PTHrP-ECD complex with our previously published PTH-ECD complex highlights the similarities and differences in the binding mechanisms of the two peptides (Fig. 2F). In both cases, the hydrophobic face of the amphipathic peptide binds the hydrophobic groove in the ECD, and the interaction is anchored by the invariant residues Arg-20′ and Leu-24′ (Fig. 2G). The most notable differences occur at the C termini of the peptides. PTH forms a continuous α-helix from Leu-15′ to Phe-34′ where the C-terminal amide group forms important hydrogen bonds to the ECD. In contrast, the PTHrP helix extends from Ile-15′ to Ile-31′, after which it “unwinds”, and the C-terminal amide group is not involved in receptor binding. The PTHrP helix curves gently such that its helical axis diverges from that of PTH after residue Leu-24′, apparently due to the differences in residues at positions 23′, 27′, 28′, and 31′ of the peptides (Fig. 2, F and G). The interface of the PTHrP-ECD complex buries 1,743 Å2 of solvent-accessible surface area, compared with 1,883 Å2 for the PTH-ECD complex. The surface complementarity between peptide and ECD, as measured by the Sc value (39) is slightly lower for the PTHrP-ECD complex (Sc = 0.726) than for the PTH-ECD complex (Sc = 0.782). Thus, the PTH peptide buries slightly more surface area and fits the ECD with slightly “tighter” complementarity than does PTHrP. Nonetheless, the peptides exhibit similar affinities for the ECD, possibly because of the additional hydrogen bonds formed by residues 32–34 of PTHrP (Fig. 2E).
The receptor accommodates the different binding modes of the two peptides with subtle, yet clearly discernable changes in local conformation. The overall structures of the two ECDs are very similar, exhibiting a R.m.s. deviation of their C-α positions of 0.357 Å. A prominent difference in the two structures is seen for the side chain conformations of Leu-41 and Ile-115 (Fig. 2F). In both complexes there is apparent coupling between Leu-41 and Ile-115 of the ECD and positions 23′ and 27′ of the peptides, respectively. In the PTHrP-bound structure L41 adopts the favored χ1 = gauche(+), χ2 = trans rotamer (40) enabling contact between the δ1 methyl group of Leu-41 and the phenyl ring of Phe-23′. In contrast, in the PTH-bound structure Leu-41 adopts the less favored χ1 = trans, χ2 = gauche(−) rotamer, permitting the receptor to accommodate the larger side chain of Trp-23′ and retain contact between the δ2 methyl group of Leu-41 and the indole ring of Trp-23′. This Leu-41 rotamer toggle switch mechanism was predicted in an elegant study by Donnelly and co-workers (41) based on mutagenesis and modeling approaches. I115 adopts the same rotamer conformation in the PTH- and PTHrP-bound structures, but the side chain shifts considerably inward toward the core of the ECD in the PTHrP-bound state, thus permitting the curvature of the PTHrP helix at the position of residue 27′. Thus, the peptide binding site of the ECD exhibits plasticity to accomodate the different binding modes of PTH and PTHrP by changing the conformation of residues Leu-41 and Ile-115.
To validate the different binding modes observed in the crystal structures, we examined the contribution of specific PTH and PTHrP residues to ECD binding by assessing the ability of alanine-scan mutants of PTH(15–34)NH2 and PTHrP(12–34)NH2 to compete the interaction of biotinylated PTH with MBP-PTH1R ECD in the AlphaScreen assay. As shown previously (23), PTH residues Arg-20′, Trp-23′, Leu-24′, and Leu-28′ were most critical for ECD binding, with additional contributions provided by Val-21′, Lys-27′, and Phe-34′ (Fig. 2H). PTHrP residues Arg-20′, Arg-21′, Phe-23′, Leu-24′, Leu-27′, Ile-28′, His-32′, and Thr-33′ were all important for ECD binding, with Arg-20′ and Leu-24′ providing the most critical contacts (Fig. 2I). Notably, Phe-23′ and Ile-28′ of PTHrP are not as critical for ECD binding as Trp-23′ and Leu-28′ of PTH. Conversely, Leu-27′ and His-32′ of PTHrP are more important for ECD binding than Lys-27′ and His-32′ of PTH. These in vitro binding results are consistent with the binding modes observed in the crystal structures.
Binding of Hybrid PTH/PTHrP Peptides to the PTH1R ECD
We next sought to determine which specific residues were responsible for the differing ECD binding modes of PTH and PTHrP. The crystal structures suggest that the residues at positions 23′, 27′, 28′, and 31′ of the peptides largely determine their ECD binding modes. We reasoned that altering the residues at these positions in one peptide to the corresponding residues present in the other peptide might permit conversion between the PTH and PTHrP binding modes. This hypothesis was tested by assessing the binding of hybrid peptides containing swaps of the PTH/PTHrP residues at these four positions to the purified MBP-PTH1R ECD fusion protein in the AlphaScreen assay. The hybrid peptides were made in the PTH(15–34)NH2 and PTHrP(12–34)NH2 scaffolds and contained either a single swap at one of the four positions, a double swap at two of the four positions, or a quadruple swap at all four positions. In principal, the quadruple mutants should adopt the binding properties of the other “donor” peptide, in terms of ECD binding affinity, and sensitivity to C-terminal amidation, according to the binding mode used by the donor.
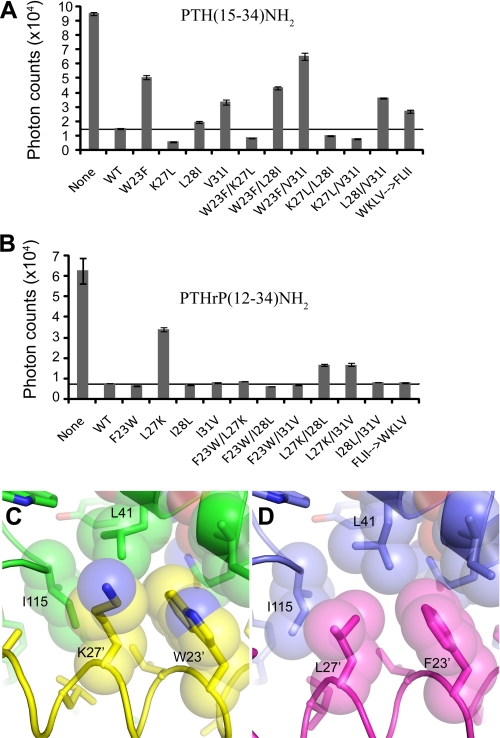
The PTH(15–34)NH2 peptide was relatively intolerant of the corresponding PTHrP residues at positions 23′, 31′, and to a lesser extent 28′, as indicated by the diminished ECD-binding of the W23′F, L28′I, and V31′I singly mutant peptides (Fig. 3A). In contrast, the K27′L mutant retained normal binding affinity, and this single alteration was able to rescue the defects imposed by the W23′F, L28′I and V31′I mutations, as shown by the improved binding of the W23′F/K27′L, K27′L/L28′I, and K27′L/V31′I doubly mutant peptides. The inclusion of all four swap mutations together in the W23′F/K27′L/L28′I/V31′I peptide resulted in somewhat diminished ECD-binding, as compared with either parent peptide, suggesting that the differences in the PTH and PTHrP binding modes are determined by more than just these four positions, at least within the context of the PTH scaffold. The combined effect of the swaps at all four positions in PTH likely direct PTH toward the PTHrP binding mode, but the side chains of Asn-33′ and Phe-34′ of PTH, replaced by Thr and Ala in PTHrP, respectively, would introduce steric incompatibility (Fig. 2E). Indeed, PTH-(15–34) containing the six substitutions W23′F/K27′L/L28′I/V31′I/N33′T/F34′A exhibited wild-type ECD-binding affinity irrespective of C-terminal amidation or carboxylation (supplemental Fig. S2, A and B), suggesting that these six alterations cause the PTH peptide to adopt the PTHrP ECD binding mode.
FIGURE 3.
Binding of hybrid PTH/PTHrP peptides to the MBP-PTH1R ECD protein. A, AlphaScreen single point competition assay assessing the ability of hybrid peptides in the PTH(15–34)NH2 scaffold (20 μm) to compete the interaction of biotin-PTH (23 nm) with MBP-PTH1R-ECD (23 nm). The results are the average of duplicate samples. B, AlphaScreen assay as in panel A except with hybrid peptides in the PTHrP(12–34)NH2 scaffold. C and D, view of the contacts formed by Trp-23′ and Lys-27′ in the PTH-ECD structure (C) and by Phe-23′ and Leu-27′ in the PTHrP-ECD structure (D).
PTHrP(12–34)NH2 exhibited normal ECD-binding when altered with the corresponding PTH residue at positions 23′, 28′, and 31′, but diminished binding with the L27′K alteration (Fig. 3B). The defect of the L27′K alteration was fully rescued by the F23′W alteration and partially rescued by the I28′L or I31′V alterations in the F23′W/L27′K, L27′K/I28′L, and L27′K/I31′V doubly mutant peptides. The quadruply mutant PTHrP peptide exhibited normal binding to the ECD, consistent with the possibility that this peptide adopts the PTH binding mode; however, the C-terminally carboxylated version did not exhibit diminished ECD affinity (supplemental Fig. S2C). In contrast, PTHrP-(12–34) containing the five substitutions F23′W/L27′K/I28′L/I31′V/A34′F retained wild-type ECD affinity when C-terminally amidated, but had reduced ECD affinity when C-terminally carboxylated (supplemental Fig. S2D), suggesting that these five alterations are required to cause PTHrP to adopt the PTH ECD-binding mode, presumably because Phe-34′ contributes to the ECD binding affinity of PTH (Fig. 2H).
The most striking feature of the data in Fig. 3, A and B is the functional coupling of positions 23′ and 27′ that is evident for both peptides. In either scaffold, the combination of Phe at position 23′ and Lys at position 27′ (F23′/K27′) resulted in severely diminished ECD binding, while the W23′/K27′, W23′/L27′, and F23′/L27′ combinations were without detriment. The simplest explanation is that the F23′/K27′ combination does not bury as much hydrophobic surface area as the other combinations (Fig. 3, C and D). The W23′F alteration in PTH would result in less hydrophobic contact to the receptor because of the smaller Phe side chain, and this would be compensated by the additional contacts available from a Leu at position 27′ (Fig. 3C). Similarly, the L27′K alteration in PTHrP would result in less hydrophobic contact to the receptor because, unlike Leu, Lys does not branch at the γ carbon (Fig. 3D). The L27′K defect could be compensated by the additional hydrophobic contacts available from a Trp at position 23′.
Binding of the Hybrid PTH/PTHrP Peptides to the Intact Receptor and Effects on R0 versus RG Conformation
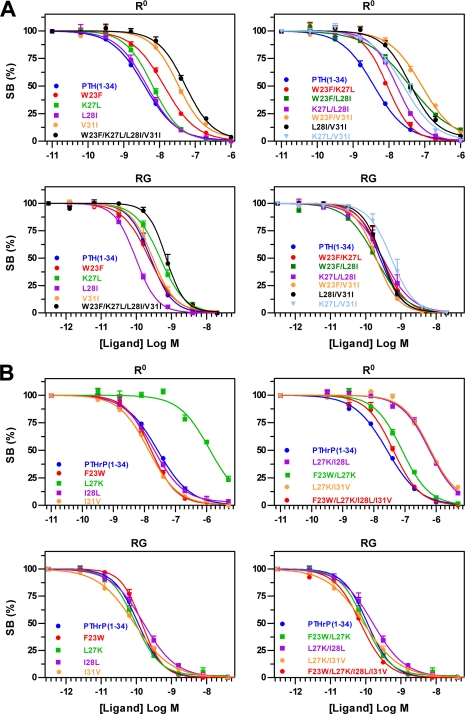
We then assessed the effects of the ligand swap substitutions on the capacity of the ligands to bind to the intact PTH1R expressed in COS-7 cell membranes. We first assessed binding in assays performed using 125I-PTH-(1–34) as a tracer radioligand, for which binding depends critically on interactions to the receptor ECD, and in the presence of GTPγS, which promotes the G protein-uncoupled PTH1R conformation, R0 (19, 22). Each swap mutation that resulted in diminished binding to the isolated ECD also resulted in diminished binding to the intact PTH1R (R0) (Fig. 4). We then assessed binding in competition assays performed utilizing 125I-M-PTH-(1–15) as a tracer radioligand, which binds predominantly to the receptor transmembrane domain region, and membranes prepared from cells that co-expressed the receptor with a high affinity (dominant negative) form of Gαs, so as to promote the G protein-coupled receptor conformation (RG). The same substitutions that strongly diminished binding to R0, had little or no effect on binding to RG (Figs. 3 and 4 and supplemental Table S1). PTH(1–34)NH2 [W23′F/V31′I] and PTHrP(1–34)NH2 [L27′K] were particularly striking in this regard as both peptides exhibited large reductions in binding to the ECD as well as to the intact R0 conformation, but retained near normal affinity for the RG conformation. In addition, the rescue effects of certain swap alterations observed on the isolated ECD, were also apparent on the intact PTH1R (Figs. 3 and 4). These results indicate that the binding interactions mediated by the ECD play a key role in determining the capacity of the ligand to bind to the intact PTH1R in the R0 conformation, but not to the intact PTH1R in the RG conformation. Consistent with a more direct role for the RG conformation, versus R0, in mediating agonist-induced transmembrane signaling, peptides that exhibited reduced affinity for R0, but nevertheless retained high affinity for RG, exhibited high potency for stimulating cAMP accumulation in COS cells transiently expressing PTH1R (Fig. 5).
FIGURE 4.
Binding of hybrid PTH/PTHrP(1–34)NH2 peptides to R0 and RG conformations of PTH1R in COS-7 cell membranes. Competition binding curves for the indicated hybrid peptides in the PTH(1–34)NH2 (A) or PTHrP(1–34)NH2 (B) scaffolds are shown. For the R0 state, the tracer radioligand was [125I]-PTH(1–34), and 10 μm GTPγS was included in the reactions. For the RG state, the tracer radioligand was [125I]-M-PTH(1–15) and the membranes were prepared from cells co-expressing the receptor and a high affinity dominant negative version of Gαs.
FIGURE 5.
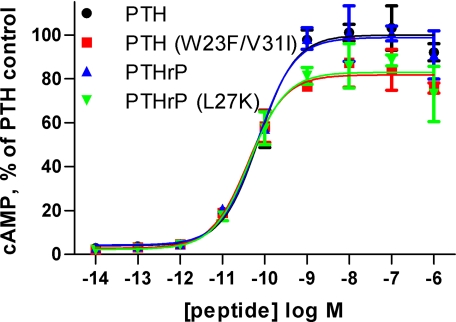
Stimulation of cAMP signaling by RG-selective hybrid PTH/PTHrP(1–34)NH2 peptides. COS-1 cells transiently expressing PTH1R were stimulated with the indicated peptides for 30 min at 37 °C after which the cells were lysed, and cAMP content was assessed. The data are the average of duplicate samples and are normalized to the maximal cAMP level observed in the presence of PTH(1–34)NH2 for which basal cAMP was 0.24 pmol per well and maximal cAMP was 6.1 pmol per well. The EC50 values for PTH, PTH(W23′F/V31′I), PTHrP, and PTHrP(L27′K) were 72, 42, 68, and 44 pm, respectively.
DISCUSSION
PTH and PTHrP elicit distinct biological actions by activating their common receptor, PTH1R. A high resolution structural understanding of how the peptides bind the receptor is important to elucidate the biochemical mechanisms responsible for the regulation of mineral ion homeostasis and bone remodeling by PTH and bone development by PTHrP, and also has practical application for aiding the rational design of peptide analogs for therapeutic purposes. In this report we present a structure of PTHrP in complex with the PTH1R ECD. Combined with our previous structure of the PTH-PTH1R ECD complex (23), we now have more complete views of the structural mechanisms used by the PTH1R ECD to recognize PTH and PTHrP.
The peptides bind to the ECD as amphipathic α-helices that contact the hydrophobic groove in the ECD, consistent with NMR solution structures of the isolated peptides that showed α-helical structure in the 15–34 fragments of the ligands (42, 43). Two of the three invariant residues in the 15–34 fragments, Arg-20′ and Leu-24′, anchor the interaction with the ECD. After residue Leu-24′, the peptides diverge such that their C termini make significantly different contacts to the ECD (Fig. 2F), a result foreshadowed by our ECD-binding data showing that the C-terminal amide group of PTH, but not PTHrP, is important for ECD binding (Fig. 1). The third invariant residue in the 15–34 fragments, His-32′, provides a critical hydrogen bond to the receptor in the PTHrP-bound structure, but is solvent-exposed in the PTH-bound structure. Interestingly, Arg-20′ and Leu-24′ are conserved in TIP39, a peptide that activates the PTH type 2 receptor (44), but His-32′ is not, consistent with our observations that the N-terminal portion of the 15–34 fragments provides the anchor point of the peptide-ECD interactions and the C-terminal portions vary in their interaction with the receptor.
The receptor accommodates the two different peptides with relatively minor changes in the ECD conformation, including a spatial shift in the Ile-115 side chain in response to the PTHrP curvature, and a rotamer toggle switch change in the Leu-41 side chain to maintain van der Waals contact with the divergent aromatic side chain volumes at ligand position 23 (Fig. 3, C and D). PTHrP was generally more tolerant to alteration than PTH with respect to ECD binding. Three out of the four single swap mutants of PTHrP retained normal binding to the ECD, but only one of the PTH single swap mutants did the same. The PTH interaction is probably more sensitive to the conservative substitutions because of its higher surface complementarity to the ECD and larger buried surface area, which leave less room for adjustments to the interface in response to mutation. This suggests that PTHrP may provide a better scaffold in which to introduce alterations for therapeutic analog development.
The remarkable correlation of the ECD affinity of PTH/PTHrP swap peptides with their R0/RG selectivity has important implications for the development of PTH/PTHrP analogs for treating osteoporosis. Recent studies on PTH analogs indicate that the relative capacity of a ligand to bind to the receptor R0 conformation correlates with the duration of calcium and phosphate responses induced in animals, as well as with the net effects on bone turnover (22). Indeed, PTHrP, which is more RG selective, appears to be a more purely anabolic agent than PTH with less side effects of adverse hypercalcemia (16). The availability of the high resolution structures guided our rational design of the swap alterations that ultimately yielded new peptide analogs, particularly PTHrP(1–34) [L27K], that are highly selective for the G protein-coupled receptor, and thus might have more favorable properties in terms of therapeutic applications, as these would avoid the prolonged signaling and hence bone-resorptive effects observed with highly R0-selective PTH analogs (22). Additional in vitro pharmacological studies and in vivo animal studies will be required to test these hypotheses.
Supplementary Material
Acknowledgments
We thank S. Anderson and J. Brunzelle for assistance with data collection at sector 21 (LS-CAT) of the Advanced Photon Source, Kelly Suino-Powell for advice on the Octet Red experiments, and Kaleeckal Harikumar for advice on the cAMP assay.
This work was supported, in whole or in part, by National Institutes of Health Grants DK071662, DK066202, and HL089301 (to H. E. X.), and DK-11794 (to T. J. G.). This work was also supported by the Jay and Betty Van Andel Foundation.
The atomic coordinates and structure factors (code 3H3G) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- PTH1R
- PTH/PTHrP receptor
- PTH
- parathyroid hormone
- PTHrP
- parathyroid hormone-related protein
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- ECD
- extracellular domain
- BSA
- bovine serum albumin
- MBP
- maltose-binding protein
- PEG
- polyethyleneglycol
- TM
- transmembrane
- MOPS
- 4-morpholinepropanesulfonic acid
- R.m.s.
- root mean-squared
- R0
- PTH1R conformation in cell membranes in the presence of GTPγS
- RG
- PTH1R conformation in cell membranes in the presence of a high affinity, dominant negative Gαs protein.
REFERENCES
- 1.Jüppner H., Abou-Samra A. B., Freeman M., Kong X. F., Schipani E., Richards J., Kolakowski L. F., Jr., Hock J., Potts J. T., Jr., Kronenberg H. M. (1991) Science 254, 1024–1026 [DOI] [PubMed] [Google Scholar]
- 2.Gardella T. J., Juppner H. (2000) Rev. Endocr. Metab. Disorders 1, 317–329 [DOI] [PubMed] [Google Scholar]
- 3.Murray T. M., Rao L. G., Divieti P., Bringhurst F. R. (2005) Endocr. Rev. 26, 78–113 [DOI] [PubMed] [Google Scholar]
- 4.Potts J. T. (2005) J. Endocrinol. 187, 311–325 [DOI] [PubMed] [Google Scholar]
- 5.Potts J. T., Gardella T. J. (2007) Ann. NY Acad. Sci. 1117, 196–208 [DOI] [PubMed] [Google Scholar]
- 6.Jilka R. L. (2007) Bone 40, 1434–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neer R. M., Arnaud C. D., Zanchetta J. R., Prince R., Gaich G. A., Reginster J. Y., Hodsman A. B., Eriksen E. F., Ish-Shalom S., Genant H. K., Wang O., Mitlak B. H. (2001) N. Engl. J. Med. 344, 1434–1441 [DOI] [PubMed] [Google Scholar]
- 8.Burtis W. J., Wu T., Bunch C., Wysolmerski J. J., Insogna K. L., Weir E. C., Broadus A. E., Stewart A. F. (1987) J. Biol. Chem. 262, 7151–7156 [PubMed] [Google Scholar]
- 9.Moseley J. M., Kubota M., Diefenbach-Jagger H., Wettenhall R. E., Kemp B. E., Suva L. J., Rodda C. P., Ebeling P. R., Hudson P. J., Zajac J. D. (1987) Proc. Natl. Acad. Sci. U. S. A. 84, 5048–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suva L. J., Winslow G. A., Wettenhall R. E., Hammonds R. G., Moseley J. M., Diefenbach-Jagger H., Rodda C. P., Kemp B. E., Rodriguez H., Chen E. Y., et al. (1987) Science 237, 893–896 [DOI] [PubMed] [Google Scholar]
- 11.Klein R. F., Strewler G. J., Leung S. C., Nissenson R. A. (1987) Endocrinology 120, 504–511 [DOI] [PubMed] [Google Scholar]
- 12.Karaplis A. C., Luz A., Glowacki J., Bronson R. T., Tybulewicz V. L., Kronenberg H. M., Mulligan R. C. (1994) Genes Dev. 8, 277–289 [DOI] [PubMed] [Google Scholar]
- 13.Lanske B., Karaplis A. C., Lee K., Luz A., Vortkamp A., Pirro A., Karperien M., Defize L. H., Ho C., Mulligan R. C., Abou-Samra A. B., Jüppner H., Segre G. V., Kronenberg H. M. (1996) Science 273, 663–666 [DOI] [PubMed] [Google Scholar]
- 14.Kronenberg H. M. (2006) Ann. NY Acad. Sci. 1068, 1–13 [DOI] [PubMed] [Google Scholar]
- 15.Horwitz M. J., Tedesco M. B., Gundberg C., Garcia-Ocana A., Stewart A. F. (2003) J. Clin. Endocrinol. Metab. 88, 569–575 [DOI] [PubMed] [Google Scholar]
- 16.Plotkin H., Gundberg C., Mitnick M., Stewart A. F. (1998) J. Clin. Endocrinol. Metab. 83, 2786–2791 [DOI] [PubMed] [Google Scholar]
- 17.Bergwitz C., Gardella T. J., Flannery M. R., Potts J. T., Jr., Kronenberg H. M., Goldring S. R., Jüppner H. (1996) J. Biol. Chem. 271, 26469–26472 [DOI] [PubMed] [Google Scholar]
- 18.Luck M. D., Carter P. H., Gardella T. J. (1999) Mol. Endocrinol. 13, 670–680 [DOI] [PubMed] [Google Scholar]
- 19.Dean T., Vilardaga J. P., Potts J. T., Jr., Gardella T. J. (2008) Mol. Endocrinol. 22, 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean T., Linglart A., Mahon M. J., Bastepe M., Jüppner H., Potts J. T., Jr., Gardella T. J. (2006) Mol. Endocrinol. 20, 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoare S. R., Gardella T. J., Usdin T. B. (2001) J. Biol. Chem. 276, 7741–7753 [DOI] [PubMed] [Google Scholar]
- 22.Okazaki M., Ferrandon S., Vilardaga J. P., Bouxsein M. L., Potts J. T., Jr., Gardella T. J. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 16525–16530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pioszak A. A., Xu H. E. (2008) Proc. Natl. Acad. Sci. U. S. A. 105, 5034–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parthier C., Kleinschmidt M., Neumann P., Rudolph R., Manhart S., Schlenzig D., Fanghänel J., Rahfeld J. U., Demuth H. U., Stubbs M. T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 13942–13947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pioszak A. A., Parker N. R., Suino-Powell K., Xu H. E. (2008) J. Biol. Chem. 283, 32900–32912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Runge S., Thøgersen H., Madsen K., Lau J., Rudolph R. (2008) J. Biol. Chem. 283, 11340–11347 [DOI] [PubMed] [Google Scholar]
- 27.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol Crystallogr. 50, 760–76315299374 [Google Scholar]
- 30.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleywegt G. J., Jones T. A. (1997) Methods Enzymol. 277, 208–230 [DOI] [PubMed] [Google Scholar]
- 32.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 33.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 34.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 35.DeLano W. (2002) DeLano Scientific, Palo Alto, CA [Google Scholar]
- 36.Harikumar K. G., Pinon D. I., Miller L. J. (2007) J. Biol. Chem. 282, 30363–30372 [DOI] [PubMed] [Google Scholar]
- 37.Grauschopf U., Lilie H., Honold K., Wozny M., Reusch D., Esswein A., Schäfer W., Rücknagel K. P., Rudolph R. (2000) Biochemistry 39, 8878–8887 [DOI] [PubMed] [Google Scholar]
- 38.Gensure R. C., Shimizu N., Tsang J., Gardella T. J. (2003) Mol. Endocrinol. 17, 2647–2658 [DOI] [PubMed] [Google Scholar]
- 39.Lawrence M. C., Colman P. M. (1993) J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 40.Penel S., Doig A. J. (2001) J. Mol. Biol. 305, 961–968 [DOI] [PubMed] [Google Scholar]
- 41.Mann R., Wigglesworth M. J., Donnelly D. (2008) Mol. Pharmacol. 74, 605–613 [DOI] [PubMed] [Google Scholar]
- 42.Marx U. C., Adermann K., Bayer P., Forssmann W. G., Rösch P. (2000) Biochem. Biophys. Res. Commun. 267, 213–220 [DOI] [PubMed] [Google Scholar]
- 43.Weidler M., Marx U. C., Seidel G., Schäfer W., Hoffmann E., Esswein A., Rösch P. (1999) FEBS Lett. 444, 239–244 [DOI] [PubMed] [Google Scholar]
- 44.Hoare S. R., Clark J. A., Usdin T. B. (2000) J. Biol. Chem. 275, 27274–27283 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.