Abstract
Berberine, an alkaloid derivative from Berberis vulgaris L., has been used extensively in traditional Chinese medicine to treat diarrhea and diabetes, but the underlying mechanisms for treating diabetes are not fully understood. Recent studies suggested that berberine has many beneficial biological effects, including anti-inflammation. Because type 1 diabetes is caused by T cell-mediated destruction of β cells and severe islet inflammation, we hypothesized that berberine could ameliorate type 1 diabetes through its immune regulation properties. Here we reported that 2 weeks of oral administration of berberine prevented the progression of type 1 diabetes in half of the NOD mice and decreased Th17 and Th1 cytokine secretion. Berberine suppressed Th17 and Th1 differentiation by reducing the expression of lineage markers. We found that berberine inhibited Th17 differentiation by activating ERK1/2 and inhibited Th1 differentiation by inhibiting p38 MAPK and JNK activation. Berberine down-regulated the activity of STAT1 and STAT4 through the suppression of p38 MAPK and JNK activation, and it controlled the stability of STAT4 through the ubiquitin-proteasome pathway. Our findings indicate that berberine targets MAPK to suppress Th17 and Th1 differentiation in type 1 diabetic NOD mice. This study revealed a novel role of ERK in Th17 differentiation through down-regulation of STAT3 phosphorylation and RORγt expression.
Type 1 diabetes, one of the most common autoimmune diseases, is caused by T cell-mediated destruction of insulin-secreting β cells and accounts for ∼5–10% of diagnosed cases of diabetes (1). Th1 and Th17 are two types of inflammatory T cells that play important roles in the development of many autoimmune diseases by producing IFNγ3 (2) and IL-17 (3, 4), respectively. Although Th1 is well known as an important diabetogenic factor in the development of type 1 diabetes (5–7), the role of Th17 in autoimmune diabetes remains debatable. Komiyama et al. (8) reported that IL-17 deficiency did not affect hyperglycemia in NOD mice, whereas other studies have emphasized the role of IL-17 in diabetes development (9–14). For example, IL-17 mRNA was increased during the development of diabetes in NOD mice (11), and Th17 lymphocytes were activated in type 1 diabetic mice (15). Interestingly, the use of glutamic acid decarboxylase-derived peptide 206–220-specific approaches to treat type 1 diabetes in NOD mice revealed that adjuvant-free antigens induced IFNγ and controlled blood glucose via concomitant suppression of IL-17 secretion (9). Of interest, a recent study showed that Th17 cells promote pancreatic inflammation in NOD mice (14). More interestingly, Th17 could be converted to a Th1-like phenotype in vivo and induce diabetes to NOD/SCID recipients (16). Accordingly, the suppression of IL-17 production could be beneficial, if not essential, for the treatment of type 1 diabetes. Therefore, drugs that target Th1 and Th17 differentiation could offer promising candidates for treating autoimmune diabetes.
Many studies are currently exploring the intracellular signaling pathways involved in Th17 differentiation. It has been shown that the orphan nuclear receptor RORγt is essential for Th17 differentiation (17). Subsequent work revealed that RORα is another lineage-specific transcription factor involved in Th17 differentiation (18). Recently, STAT3 was found to act as a novel regulator of cytokine-driven Th17 generation (19). Interestingly, another nuclear receptor, the aryl hydrocarbon receptor, was found to modulate Th17 differentiation (20, 21). In our study, we found that ERK acted as a negative regulator of Th17 differentiation in human and murine cells.
Berberine is used extensively in traditional Chinese medicine to treat diarrhea and diabetes, but the underlying mechanisms for treating diabetes are not fully understood. Recent studies have shown that berberine has many beneficial biological effects, including immunomodulation (22–27), anti-diabetic metabolic effects (28, 29), and chemotherapeutic activity (30, 31). Because type 1 diabetes is associated with islet inflammation, we hypothesized that berberine could ameliorate type 1 diabetes through its immune regulatory properties, which may explain the beneficial use of berberine in the management of type 1 diabetes. In this study, we found that berberine treatment ameliorated type 1 diabetes and decreased the expression of inflammatory cytokines in NOD mice. Berberine also suppressed Th17 and Th1 differentiation via the activation of ERK1/2 and inactivation of p38 MAPK and JNK, respectively. Inhibition of ERK1/2 activity by a selective inhibitor or by retroviral expression of dominant-negative forms of ERK1 or ERK2 promoted Th17 differentiation. Berberine regulated Th1 differentiation by decreasing the activity of STAT1 and STAT4 by suppressing p38 MAPK/JNK and degrading STAT4 through the ubiquitin-proteasome pathway. These findings may help to evaluate the use of natural plant products in drug discovery and to better understand the role of MAPK in T cell differentiation.
MATERIALS AND METHODS
Animals
11-week-old female NOD mice were purchased from the SLAX animal facility at the Shanghai Institutes for Biological Sciences and were treated with phosphate-buffered saline (PBS) or berberine (Sigma, 200 mg/kg body weight), by gavage, daily for 2 weeks (n = 8/group); and the mice were monitored for 33 weeks for the progression of diabetes. This dosage was chosen based on previous studies (28, 29). Nonfasting blood glucose was measured in the tail vein every week between 10 a.m. and 11 a.m. with a glucose meter (Freestyle; TheraSense, Alameda, CA) throughout the study, and mice were considered diabetic when the blood glucose level exceeded 250 mg/dl. The nonfasting blood glucose level of all mice was below 250 mg/dl before starting the study. In the adoptive transfer experiment, in vitro differentiated T cells were transferred into 7-week-old NOD.scid mice by intravenous injection. Nonfasting blood glucose was measured weekly, and it was considered diabetic when the blood glucose was higher than 250 mg/dl. All animal studies were conducted in accordance with the guidelines for animal studies issued by the Institute for Nutritional Sciences, Chinese Academy of Sciences.
Cell Purification
To isolate mouse CD4+ T cells, splenocytes were prepared and depleted of B cells, macrophages, CD8+ T cells, NK cells, dendritic cells, erythrocytes, and granulocytes using a mouse CD4+ negative selection kit (Dynal Biotech). To isolate human CD4+ T cells, peripheral blood mononuclear cells were first isolated from peripheral blood obtained from healthy donors by Ficoll-Paque Plus (Amersham Biosciences), and then CD4+ T cells were purified with a MACS negative selection kit (Miltenyi Biotec). To isolate CD4+CD25− T cells, the purified CD4+ T cell populations were incubated with phycoerythrin-labeled anti-CD25 antibody and anti-phycoerythrin magnetic beads and isolated using a MACS separation column (Miltenyi Biotec). Cell purity was assessed by fluorescence-activated cell sorter and was consistently higher than 90% with an average purity of 96%. A representative result is shown in supplemental Fig. 1.
Cell Culture
For Th1 and Th17 differentiation, naive CD4+ T cells from 8- to 10-week-old NOD mice were cultured in complete medium (RPMI 1640 medium and 10% fetal bovine serum) in the presence of α-CD3/28 antibodies (2 μg/ml) to stimulate T cell receptors (TCRs). Th1 mixture (IL-12 (10 ng/ml), α-IL-4 antibody (10 μg/ml)) or Th17 mixture (IL-6 (10 ng/ml), transforming growth factor-β (2 ng/ ml), IL-1β (10 ng/ml), IL-23 (10 ng/ml), α-IL-4 antibody (10 μg/ml), and α-IFNγ antibody (10 μg/ml)) were added to drive Th1 and Th17 polarization, respectively. For human Th17 differentiation, purified CD4+ T cells were cultured under a Th17 condition (IL-6 (20 ng/ml), IL-23 (20 ng/ml), IL-1β (10 ng/ml), α-IL-4 antibody (10 μg/ml) and α-IFNγ antibody (10 μg/ml)). After culture for 4 days, cells were restimulated with 50 ng/ml phorbol 12-myristate 13-acetate (Sigma) and 500 ng/ml ionomycin (Sigma) in the presence of GolgiPlug (Pharmingen) for 5 h before intracellular staining.
To study cytokine production in berberine-treated or untreated mice, NOD mice were sacrificed, and single-cell suspensions of splenocytes or lymphocytes were prepared. To examine cytokine secretion, the splenocytes were activated with α-CD3 antibody for 48 h, and the supernatant was collected and subjected to ELISA. The following chemical inhibitors and activators were added to the cell culture together with the cytokine mixture, at the indicated concentrations: SB-202190 (0.5 μg/ml), PD98059 (10 or 20 μm), SP600125 (1 μm), BADGE (1 μm), rosiglitazone (1 μm), and wortmannin (1 μm).
Histology
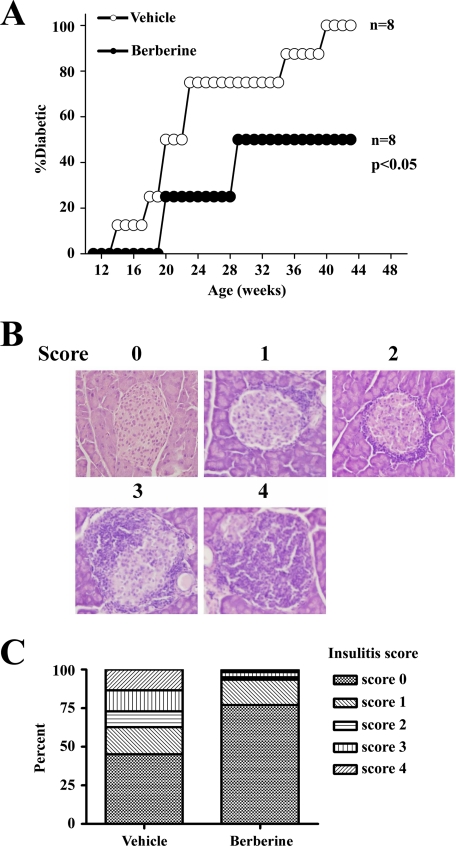
Mice were sacrificed and pancreata were dissected, immediately frozen in liquid nitrogen, and then stored at −80 °C. Pancreata were sectioned (20 μm thick) in a cryostat (Leica), and the sections were transferred to gelatin-coated slides. Slides were stained with hematoxylin and eosin and examined under light microscopy. One hundred islets from vehicle- or berberine-treated mice with equivalent glucose levels were examined. Insulitis was scored using the method illustrated in Fig. 1.
FIGURE 1.
BBR improved type 1 diabetes in NOD mice. 11-week-old female NOD mice were orally administered with vehicle or berberine daily for 14 days. A, nonfasting blood glucose was measured every week for 33 weeks, and the NOD mice were considered diabetic when the blood glucose was higher than 250 mg/dl. B, 14 days after starting berberine administration, berberine-treated or untreated NOD mice were sacrificed, and pancreata were dissected for histology examination. Representative hematoxylin and eosin staining of pancreatic slides with insulitis scores ranging from 0 to 4 are shown. C, insulitis scores of islets from berberine-treated or untreated NOD mice with equivalent glucoses level (100 islets each group).
Western Blotting and Immunoprecipitation
Cells were lysed, and protein was extracted and resolved by 10% SDS-PAGE. Immunoblot analysis was performed by transferring the proteins onto polyvinylidene difluoride membranes (Millipore) using a Mini TransBlot system (Bio-Rad). The membranes were blocked with 5% milk in Tris-buffered saline supplemented with Tween 20 (TBST) and incubated overnight at 4 °C with antibodies against p38 MAPK, JNK, ERK1/2, STAT1, STAT4, actin, phosphorylated p38 MAPK, phosphorylated JNK, phosphorylated ERK1/2, phosphorylated STAT1, or phosphorylated STAT4 (Cell Signaling) followed by incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Signals were detected by enhanced chemiluminescence (Pierce) according to the manufacturer's instructions. To detect STAT4 ubiquitination, protein extracts were incubated overnight with anti-STAT4 antibody (Cell Signaling) and precipitated with protein A/G plus-agarose (Santa Cruz Biotechnology). The immunoprecipitation was resolved by 10% SDS-PAGE and immunoblotted with an anti-ubiquitin antibody (Santa Cruz Biotechnology).
Real Time PCR
RNA (0.5–1 μg) was reverse-transcribed into cDNA using an oligo(dT) primer (Takara), and 1 μl of the synthesized cDNA product was then added to the PCR mixture. Quantitative real time RT-PCR was performed using an ABI Prism 7500 sequence detection system (Applied Biosystems). The sequences of the primers were as follows: IFNγ forward primer 5′-TTGGCTTTGCAGCTCTTCCT-3′ and IFNγ reverse primer 5′-TGACTGTGCCGTGGCAGTA-3′; T-bet forward primer 5′-GCCATTACAGGATGTTTGTGGAT-3′ and T-bet reverse primer 5′-CATGCTGCCTTCTGCCTTTC-3′; IL-17 forward primer 5′-CTCCAGAAGGCCCTCAGACTAC-3′ and IL-17 reverse primer 5′-AGCTTTCCCTCCGCATTGACACAG-3′; IL-17F forward primer 5′-GAGGATAACACTGTGAGAGTTGAC-3′ and IL-17F reverse primer 5′-GAGTTCATGGTGCTGTCTTCC-3′; RORγt forward primer 5′-CCGCTGAGAGGGCTTCAC-3′ and RORγt reverse primer 5′-TGCAGGAGTAGGCCACATTACA-3′; and β-actin forward primer 5′-TGACAGGATGCAGAAGGAGA-3′ and β-actin reverse primer 5′-GTACTTGCGCTCAGGAGGAG-3′. Gene expression was normalized to the level of β-actin in each example.
Flow Cytometry
For surface staining of CD4, T cells were resuspended in PBS containing 1% bovine serum albumin and were incubated on ice for 30 min with fluorochrome-conjugated antibodies (eBioscience) for these surface markers. For intracellular staining of IL-17 and IFNγ, cells were fixed, permeabilized, and stained with fluorochrome-conjugated antibodies. The stained cells were then analyzed using a FACSCalibur instrument.
Retroviral Transduction
Wild-type (WT) and the dominant-negative (DN) form of ERK1 and ERK2 (a gift from Dr. Melanie Cobb) were cloned into the retroviral vector pMXs-IRES-GFP (Cell Biolabs). These constructs were transfected into a Platinum-E cell line (Cell Biolabs) to generate retrovirus supernatant. Naive CD4+ T cells (0.5 × 106) were activated under Th17 culture conditions for 2 days before incubation with Platinum-E supernatant containing retroviruses supplemented with 4 μg/ml Polybrene (Sigma) and centrifuged for 1 h at 2000 rpm at 30 °C. Two days after infection, the cells were tested for IL-17 production using intracellular staining.
ELISA
ELISA kits for the detection of murine IL-17, IL-6, IFNγ, and TNFα were purchased from R & D Systems and were used in accordance with the manufacturer's instructions. Microtiter plates were coated overnight at room temperature with mouse monoclonal antibodies (capture antibody). The wells were then blocked for 1 h with PBS containing 1% bovine serum albumin and washed four times with 0.05% Tween 20 in PBS. Duplicate standards and samples were added to adjacent wells and incubated for 2 h at room temperature. Wells were washed, and detection antibodies were added. Two hours later, wells were washed and incubated with a horseradish peroxidase-conjugated antibody in 1% bovine serum albumin/PBS. The plates were washed four times and incubated for 30 min with the substrate 0.0125% tetramethylbenzidine, 0.008% H2O2 in citrate buffer (pH 5.0), and color development was stopped using 2 n H2SO4. Optical densities were measured using a microplate reader, and the concentration of cytokines was calculated using a double standard curve (R & D Systems) on each plate.
Statistical Analysis
The log-rank test was used to calculate differences between diabetes rate curves of NOD mice. Two-tailed Student's t test was used to calculate differences in other assays. p values less than 0.05 were considered statistically significant.
RESULTS
Berberine Delayed the Onset of Type 1 Diabetes in NOD Mice
We first examined the effect of berberine treatment in NOD mice. All (n = 8) of the vehicle-treated mice became diabetic, but only 50% (4:8) of the berberine-treated NOD mice became diabetic (p < 0.05) (Fig. 1A). Insulitis was significantly improved in the berberine-treated mice compared with the vehicle-treated controls. The criteria for insulitis scoring is illustrated in Fig. 1B. One hundred islets from vehicle- or berberine-treated mice with equivalent glucose levels were examined. Overall, 77.0% of the islets from the berberine-treated group did not show lymphocyte infiltration, and only 45.2% of the islets in the vehicle-treated group were free from insulitis (Fig. 1C). As a result, the insulitis scores were markedly lower in the berberine-treated group. To investigate whether berberine suppressed type 1 diabetes development through inhibition of Th1/Th17 cells, in vitro polarized Th1 or Th17 cells with or without berberine treatment were adoptively transferred to NOD.scid mice, respectively. Adoptive transfer of berberine-treated Th1 or Th17 cells both decreased diabetic rate as compared with the berberine-untreated controls, although the suppression of diabetes with berberine-treated Th1 cells was more significant (supplemental Fig. 2). The results suggest that the treatment efficacy of berberine in type 1 diabetes is mediated through the inhibition of both Th1 and Th17 cells differentiation.
Berberine Significantly Decreases the Production of Inflammatory Cytokines in NOD Mice
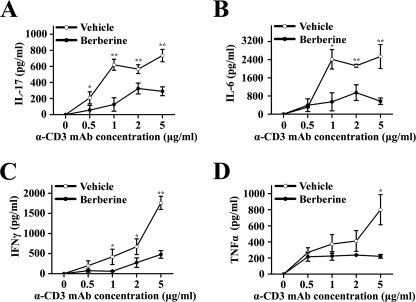
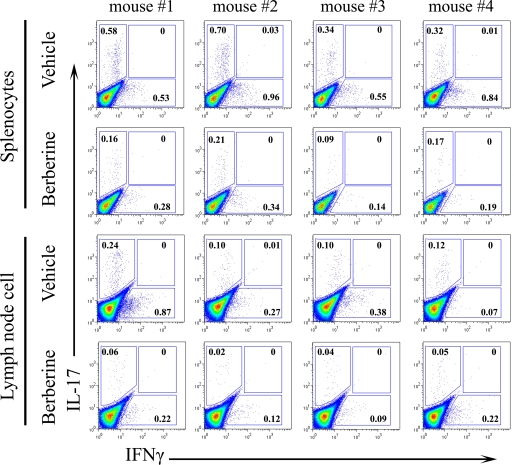
Previous studies have indicated that the administration of berberine up-regulates the expression of IL-10 (25) and down-regulates inflammatory cytokines, including IFNγ, TNFα, and IL-1β, but had limited effect on IL-17 production (23, 24, 27, 32) in various animal models. To examine whether berberine affected the cytokine profile in this murine model of type 1 diabetes, splenocytes were isolated from vehicle- or berberine-treated NOD mice and were stimulated with α-CD3 antibody at the indicated concentration for 48 h. The cytokine profile of the supernatant showed a dramatic decrease in inflammatory cytokines, including IL-17, IL-6, IFNγ, and TNFα (Fig. 2, A–D). To further investigate the effect of berberine administration on IL-17 and IFNγ secretion in vivo, we also performed intracellular staining of these two inflammatory cytokines using CD4+ T cells from both the spleen and lymph nodes (Fig. 3). The level of IL-17 was significantly reduced after berberine treatment from 0.49 ± 0.19% to 0.16 ± 0.05% (p < 0.05, n = 4) in splenic CD4+ T cells and from 0.14 ± 0.07% to 0.04 ± 0.02% in lymph node CD4+ T cells (p < 0.05, n = 4). Similarly, the level of IFNγ was reduced after berberine treatment from 0.72 ± 0.21% to 0.24 ± 0.09% (p < 0.01, n = 4) in splenic CD4+ T cells and from 0.40 ± 0.34% to 0.16 ± 0.07% in lymph node CD4+ T cells (p > 0.05, n = 4). No significant changes were found in berberine-treated NOD regarding NK, NKT, or Treg populations (supplemental Fig. 3).
FIGURE 2.
Cytokine profiles in berberine-treated NOD mice. 11-week-old female NOD mice were orally administered with vehicle or berberine daily for 14 days. Splenocytes were prepared and cultured with α-CD3 antibodies at the indicated concentration for 48 h. The supernatant was harvested, and the concentration of IL-17 (A), IL-6 (B), IFNγ (C), and TNFα (D) was measured by ELISA (*, p < 0.05; **, p < 0.01, n = 3).
FIGURE 3.
Berberine treatment decreased IL-17 and IFNγ secretion in NOD mice. 11-week-old female NOD mice were orally administered with vehicle or berberine daily for 14 days. Freshly isolated splenocytes and lymph nodes cells were stimulated with 50 ng/ml phorbol 12-myristate 13-acetate and 500 ng/ml ionomycin in the presence of GolgiPlug for 5 h before intracellular staining (n = 4).
Berberine Suppresses Th17 Differentiation in Vitro by Activating ERK
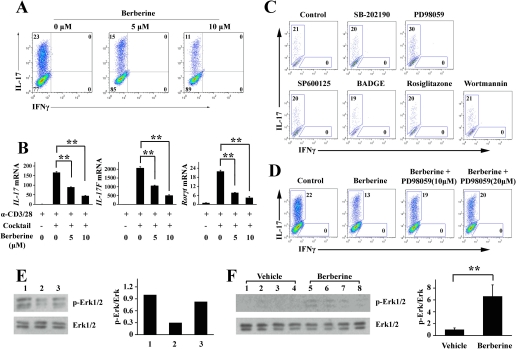
Next we investigated whether berberine affected cytokine-driven Th17 differentiation in an in vitro setting. After 4 days of culture under the Th17 condition, about 20% of the naive CD4+ T cells were polarized into IL-17-secreting Th17 cells, although the addition of berberine dose-dependently inhibited this polarization (Fig. 4A). The result was confirmed by real time PCR. As shown in Fig. 4B, the expression of genes associated with Th17 differentiation such as IL-17A, IL-17F, and Rorγt was suppressed by berberine in a dose-dependent fashion.
FIGURE 4.
Berberine suppressed Th17 differentiation in vitro. A, naive CD4+ T cells from NOD mice were cultured under the Th17 condition in the absence or presence of berberine at the indicated concentration for 4 days. Intracellular staining profiles for IL-17 and IFNγ are shown. B, naive CD4+ T cells were isolated from NOD mice and cultured with α-CD3/28 to stimulate TCR in the absence of presence of the Th17 differentiation mixture (transforming growth factor-β, IL-6, IL-1β, IL-23, α-IL-4, and α-IFNγ). Berberine was added at the indicated concentration. Three days later, RNA was extracted and reverse-transcribed into cDNA and subjected to real time PCR. The gene expression is presented relative to that of β-actin (**, p < 0.01, n = 3). C, naive CD4+ T cells from NOD mice were cultured under the Th17 condition in the presence of the indicated drugs for 4 days. Intracellular staining profiles for IL-17 and IFNγ are shown. D, naive CD4+ T cells from NOD mice were cultured under the Th17 condition in the absence or presence of berberine or PD98059 for 4 days. Intracellular staining profiles for IL-17 and IFNγ are shown. E, naive CD4+ T cells from NOD mice were cultured in the presence of α-CD3/28 alone (lane 1), with the Th17 mixture alone (lane 2), or with the Th17 mixture plus berberine (lane 3) for 24 h. The cell lysates were subjected to Western blotting for ERK and phosphorylated ERK. Quantification of the optical density of the bands is shown in the right panel. F, CD4+ T cells were purified from vehicle- or berberine-treated mice, and cell lysates were subjected to Western blotting for ERK and phosphorylated ERK (four mice per group). Quantification of the optical density of the bands is shown in the right panel (**, p < 0.01, n = 4).
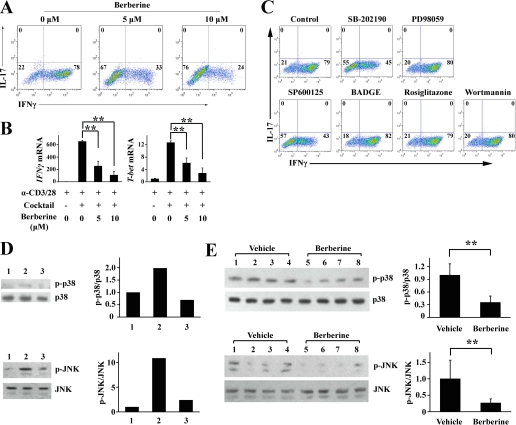
To characterize the mechanisms underlying the suppression of Th17 by berberine, we examined the activity of signaling pathways, which were reported to be involved in the biochemical effects of berberine (22, 28, 29, 33–35), using a series of specific inhibitors and activators, including SB-202190 (an inhibitor of p38 MAPK), PD98059 (an inhibitor of MEK1/2), SP600125 (an inhibitor of JNK/SAPK), BADGE (an inhibitor of PPARγ), rosiglitazone (an activator of PPARγ), and wortmannin (an inhibitor of PI3K/Akt). We found that PD98059 increased the proportion of IL-17+ T cells from 20 to 30% (Fig. 4C), and the inhibition of Th17 differentiation by berberine was restored by adding PD98059 (Fig. 4D). We found that berberine treatment activated ERK1/2 in both in vitro and in vivo settings (Fig. 4, E and F), suggesting that berberine inhibited Th17 differentiation by increasing ERK activity, which implies that ERK may regulate Th17 differentiation.
ERK Inhibits Th17 Differentiation by Down-regulation of STAT3 and RORγt Signaling Pathway
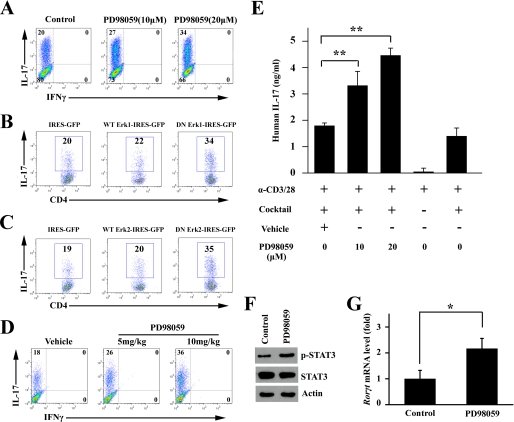
Our findings described above support a negative regulatory role of ERK in Th17 differentiation. To confirm this finding, ERK selective inhibitor and the dominant-negative (DN) form of ERK1/2 were used in vitro, and PD98059 was administered to NOD mice in vivo. As shown in Fig. 5A, inhibition of ERK expression with PD98059 promoted Th17 polarization from 20 to 27% at 10 μm and to 34% at 20 μm. Next, we expressed the wild-type (WT) or the DN form of ERK1/2 in naive CD4+ T cells to investigate their effects on Th17 differentiation. As shown in Fig. 5B, ERK1-DN promoted Th17 differentiation from 20 to 34%, whereas WT ERK1 had little effect. Similarly, ERK2-DN increased IL-17+ T cells from 19% to 35%, whereas WT ERK2 had little effect (Fig. 5C). To determine the role of ERK in Th17 differentiation in vivo, PD98059 at 5 and 10 mg/kg was administered to NOD mice. After 3 consecutive days of administration of PD98059 or vehicle, NOD mice were sacrificed. CD4+ T cells from the PD98059-treated mice were more susceptible to Th17 induction in a dose-dependent fashion (Fig. 5D). The negative regulatory role of ERK in Th17 differentiation was also found in human CD4+ T cells as demonstrated in Fig. 5E, and inhibition of ERK by PD98059 dose-dependently promoted IL-17 secretion. The inhibition of ERK activity by PD98059 increased phosphorylated STAT3 level and the expression of RORγt in CD4+ T cells cultured under the Th17 conditions (Fig. 5, F and G). These results collectively indicate that ERK plays a negative regulatory role in Th17 programming upstream to STAT3 and RORγt.
FIGURE 5.
ERK negatively regulated in vitro Th17 differentiation. A, naive CD4+ T cells from NOD mice were cultured under the Th17 condition in the absence or presence of PD98059 at the indicated concentration for 4 days. Intracellular staining profiles for IL-17 and IFNγ are shown. B and C, retroviral expression of the DN form of ERK1 (B) and ERK2 (C) promoted Th17 differentiation. The WT and DN forms of ERK1 and ERK2 were cloned into the retroviral vector pMXs-IRES-GFP. Plasmids were transfected into Platinum-E to generate a retrovirus supernatant. Naive CD4+ T cells were activated with TCR stimulants and the Th17 mixture for 2 days before infection with the retrovirus-containing Platinum-E supernatant and then cultured for a further 2 days. The gate was set on GFP+ CD4+ T cells to study IL-17 production. D, 11-week-old female NOD mice were intraperitoneally injected with vehicle or PD98059 (5 or 10 mg/kg) for 3 consecutive days (one time each day) and were then sacrificed. Naive CD4+ T cells were isolated and cultured under the Th17 condition for 4 days. Intracellular staining profiles of IL-17 and IFNγ are shown. E, human CD4+ T cells were isolated from healthy donors and cultured under the Th17 condition in the presence of PD98059 at the indicated concentration for 3 days. The supernatant was collected for ELISA tests. The concentration of IL-17 was normalized for cell number (**, p < 0.01, n = 3). F and G, naive CD4+ T cells from NOD mice were cultured under the Th17 condition in the absence or presence of PD98059 for 4 days. Cell lysates were subjected to Western blotting (F) and real time PCR after RNA extraction and reverse transcription to cDNA (G). The gene expression is presented relative to that of β-actin (*, p < 0.05).
Berberine Suppresses Th1 Differentiation in Vitro by Inhibiting p38 MAPK and JNK
Because Th1 is well known as a diabetogenic factor for the development of type 1 diabetes, we determined whether berberine affects Th1 differentiation. As shown in Fig. 6A, although the Th1 differentiation mixture induced the differentiation of about 80% of naive CD4+ T cells into Th1, the addition of berberine decreased the proportion of IFNγ+ Th1 cells to 33% at a low dose (5 μm) and to 24% (10 μm) at a high dose. We also examined the lineage markers of the CD4+ effector T subsets, IFNγ and T-bet. As expected, the mRNA levels of both genes were down-regulated by berberine (Fig. 6B).
FIGURE 6.
Berberine suppress Th1 differentiation in vitro. A, naive CD4+ T cells from NOD mice were cultured under the Th1 condition in the absence or presence of berberine at the indicated concentration for 4 days. Intracellular staining profiles of IL-17 and IFNγ are shown. B, naive CD4+ T cells were isolated from NOD mice and cultured with α-CD3/28 to stimulate TCR in the absence or presence of Th1 differentiation mixture (IL-12 and α-IL-4). Berberine was then added at the indicated concentration. Three days later, RNA was extracted and reverse-transcribed into cDNA and subjected to real time PCR. Gene expression is presented relative to that of β-actin (**, p < 0.01, n = 3). C, naive CD4+ T cells from NOD mice were cultured under the Th1 condition in the presence of the indicated drugs for 4 days. Intracellular staining profiles of IL-17 and IFNγ are shown. D, naive CD4+ T cells from NOD mice were cultured in the presence of α-CD3/28 alone (lane 1), with the Th1 mixture alone (lane 2), or with the Th1 mixture plus berberine (lane 3) for 24 h. Cell lysates was subjected to Western blotting for p38 MAPK, JNK, phosphorylated p38 MAPK and phosphorylated JNK. Quantification of the optical density of the bands is shown in the right panel. E, CD4+ T cells were purified from vehicle- or berberine-treated mice, and cell lysates were subjected to Western blotting for p38 MAPK, JNK, phosphorylated p38 MAPK, and phosphorylated JNK. Quantification of the optical density of the bands is shown in the right panel (**, p < 0.01, n = 4).
To study the mechanisms underlying the suppressive effect of berberine on Th1 differentiation, we next examined the signaling pathways as described above. Interestingly, we found that SB-202190 and SP600125 could inhibit Th1 differentiation, which was consistent with the earlier studies showing that p38 MAPK and JNK played an important role in Th1 differentiation (Fig. 6C). We also found that berberine treatment down-regulated the phosphorylation status of p38 MAPK and JNK (Fig. 6, D–G), suggesting that berberine inhibited Th1 differentiation by decreasing p38 MAPK and JNK activity.
Berberine Decreases STAT4 Protein Expression through the Ubiquitin-Proteasome Pathway
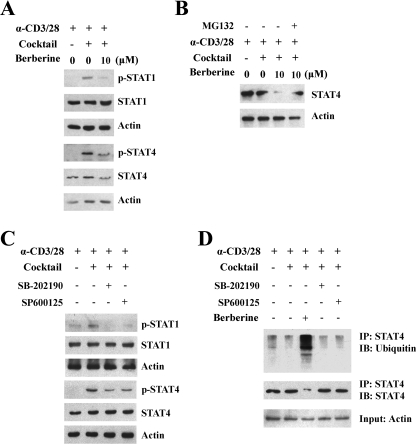
We next examined the phosphorylation status of STAT1 and STAT4, two transcription factors associated with Th1 differentiation. As shown in Fig. 7A, berberine treatment markedly attenuated the phosphorylation of STAT1 and STAT4. Interestingly, the protein level of STAT4 was also decreased after berberine treatment (Fig. 7A). We hypothesized that berberine treatment promoted the degradation of STAT4. Next we found that MG132, a proteasome inhibitor, could reverse the degradation of STAT4 induced by berberine (Fig. 7B). Further experiments revealed that inhibition of p38 MAPK and JNK decreased phosphorylation of STAT1 and STAT4 but did not affect their protein level, suggesting that p38 MAPK and JNK were not involved in the degradation of STAT4 (Fig. 7C). Instead, berberine degraded the STAT4 protein through the ubiquitin-proteasome pathway, which was independent of p38 MAPK and JNK (Fig. 7D).
FIGURE 7.
Berberine decreased the STAT4 protein level via the ubiquitin-proteasome pathway. A, naive CD4+ T cells from NOD mice were cultured at the indicated conditions in the absence or presence of berberine for 24 h, and cell lysates were subjected to Western blotting. B, naive CD4+ T cells from NOD mice were cultured in the absence or presence of the indicated conditions of berberine or MG132 for 24 h, and cell lysates were subjected to Western blotting. C, naive CD4+ T cells from NOD mice were cultured in the absence or presence of the indicated conditions of SB-202190 and SP600125 for 24 h, and cell lysates were subjected to Western blotting. D, naive CD4+ T cells from NOD mice were cultured in the presence of berberine or SB-202190 or SP600125 at the indicated conditions for 24 h, and cell lysates were immunoprecipitated (IP) with a monoclonal antibody against STAT4 followed by Western blotting (IB) against ubiquitin and STAT4. Actin was used as a loading control.
DISCUSSION
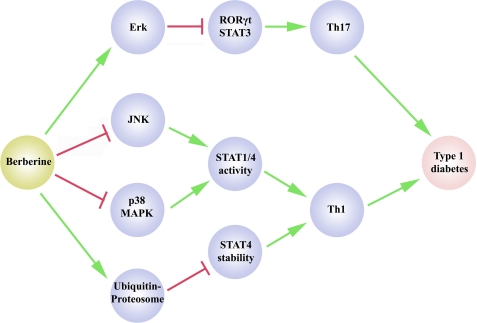
In this study, we showed that berberine differentially regulated MAPK activity to decrease IL-17 and IFNγ production, which was associated with the protection against the development of type 1 diabetes in NOD mice as illustrated in Fig. 8. This conclusion is supported by the following experimental evidence. (i) The improved outcome of diabetes is correlated with decreased IL-17 and IFNγ production. (ii) Berberine treatment activated ERK1/2 activation and inactivated p38 MAPK/JNK in vitro and in vivo. (iii) Inhibition of ERK1/2 using the selective inhibitor PD98059 or by inducing the expression of dominant-negative forms of ERK promoted Th17 differentiation, whereas inhibition of p38 MAPK and JNK decreased the expression of IFNγ during Th1 differentiation.
FIGURE 8.
Schematic illustration of berberine in the control of Th17 and Th1 differentiation in type 1 diabetes. In this model, berberine inhibits Th17 and Th1 differentiation to suppress the development of type 1 diabetes. On the one hand, berberine activates ERK to suppress Th17 differentiation. On the other hand, berberine inhibits p38 MAPK and JNK to down-regulate STAT1 and STAT4 activity to suppress Th1 polarization. Berberine also regulates the Th1 program by degrading STAT4 through the ubiquitin-proteasome pathway.
The MAPK superfamily, which includes p38 MAPK, ERK, and JNK, plays important roles in cell proliferation, differentiation, and cell death. Recent studies have also implicated MAPK in immune responses. p38 MAPK, JNK, and ERK are reported to be involved in CD4+ effector T cell differentiation. The expression of the dominant-negative form of p38 MAPK selectively impaired Th1 responses, suggesting a key regulatory role of the p38 MAPK pathway (36). The Th1 response was attenuated in JNK-deficient mice, which was primarily due to reduced IL-12-stimulated IFNγ production by CD4+ T cells (37). In contrast, JNK-deficient CD4+ T cells preferentially differentiated into Th2 subset effector T cells (38). ERK1 and ERK2 specifically affect T cell development in the thymus (39, 40). Moreover, ERK activation favored Th2 development through stabilizing GATA3 (41). Our study also showed that ERK plays a role in the program of Th17, another subset of CD4+ T cells, which enhanced our understanding of the relationship between MAPK and T cell differentiation (Fig. 8). A previous study showed that PD98059 slightly inhibited IL-17 production by lymph node cells activated by concanavalin A (42), a difference that can be attributed to the cell culture conditions used. Lymph node cells stimulated with concanavalin A secreted various cytokines other than IL-17, and these cytokines could have various effects on IL-17 production. However, under the Th17 culture condition, the T cells mainly secreted cytokines belonging to the Th17 family, including IL-17, IL-17F, and IL-22 (43). Therefore, T cells cultured with the Th17 mixture were a much more homogenous population compared with those simulated with concanavalin A and may be a better system to study the effect of PD98059 on Th17 differentiation.
Th17 is critically involved in the development of many autoimmune diseases in both human and mouse models, including experimental autoimmune encephalomyelitis and collagen-induced arthritis. However, the role of Th17 in the development of type 1 diabetes remains debatable. A study by Komiyama et al. (8) indicated that IL-17 deficiency did not affect hyperglycemia in NOD mice. On the other hand, other studies have emphasized the role of IL-17 in diabetes development in NOD mice. For example, it was shown that IL-17 transcripts increased during the evolution of diabetes in NOD mice (11) and that Th17 lymphocytes are activated in type 1 diabetic mice (15). Interestingly, in multiple low dose streptozotocin-induced diabetic mice, another murine model of type 1 diabetes, streptozotocin-stimulated IL-17 secretion promoted nitric oxide-dependent toxic inflammatory responses toward MIN6, a mouse insulinoma cell line (10). Meanwhile, another study showed that, in the same model, IL-23 is involved in the development of type 1 diabetes by inducing the production of IL-17 (12). Of greater interest, the use of glutamic acid decarboxylase-derived peptide 206–220-specific approaches to treat type 1 diabetes in NOD mice showed that adjuvant-free antigen induced IFNγ and controlled blood glucose via the suppression of IL-17 secretion (9). A recent study using IL-21r knock-out mice suggested that IL-17 may play an important role in the development of type 1 diabetes (13). Meanwhile, studies by Dong and co-workers (14) suggested that Th17 promoted pancreatic inflammation. Our study has shown that berberine strongly inhibited IL-17 production in vivo and in vitro, which could be beneficial, if not responsible, for its therapeutic effect in protecting against the development of type 1 diabetes in NOD mice.
A couple of studies have described the anti-inflammatory properties of alkaloid derivatives from Berberis vulgaris L., including berbamine and berberine. Ren et al. (32) found that berbamine targets STAT4 via a ubiquitin-protein isopeptide ligase resulting in decreased production of IFNγ in CD4+ T cells of experimental autoimmune encephalomyelitis mice, whereas the secretion of IL-17 was not markedly altered. In our study, berberine inhibited the production of both IFNγ and IL-17 in NOD mice probably as a result of structural differences between berberine and berbamine. Another possible reason why berberine could suppress the development type 1 diabetes is its antimicrobial activity (44, 45). A previous study revealed that intestinal microbes are a critical epigenetic factor that modifies the predisposition toward type 1 diabetes (46). Therefore, berberine could also suppress the development of type 1 diabetes by influencing microbial flora in the gut.
Taken together, our study provides new evidence that berberine differentially regulates MAPK activity to control Th17 and Th1 differentiation. This could be favorable in terms of its therapeutic potential in type 1 diabetes. This study also provides new insights into the anti-inflammatory properties of Chinese herbs.
Supplementary Material
Acknowledgments
We thank Dr. Melanie Cobb for providing the plasmids containing wild-type and dominant-negative forms of ERK1 and ERK2. We thank Daisy L. Zhou and Zhijun Xiang for help in measuring human IL-17 cytokine concentrations.
This work was supported in part by grants from the Hundred Talent Project, Chinese Academy of Sciences Grants KSCX2-YW-R-116 and KSCX1-YW-R-44, and Ministry of Science and Technology of China Grants 2009CB919000 and STCSM 07JC14011.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- IFNγ
- interferon-γ
- IL
- interleukin
- PBS
- phosphate-buffered saline
- DN
- dominant-negative
- MAPK
- mitogen-activated protein kinase
- JNK
- c-Jun NH2-terminal kinase
- ERK
- extracellular signal-regulated kinase
- TCR
- T cell receptor
- ELISA
- enzyme-linked immunosorbent assay
- WT
- wild type
- BADGE
- bisphenol A diglycidyl ether.
REFERENCES
- 1.Tisch R., McDevitt H. (1996) Cell 85, 291–297 [DOI] [PubMed] [Google Scholar]
- 2.Mosmann T. R., Coffman R. L. (1989) Annu. Rev. Immunol. 7, 145–173 [DOI] [PubMed] [Google Scholar]
- 3.Harrington L. E., Hatton R. D., Mangan P. R., Turner H., Murphy T. L., Murphy K. M., Weaver C. T. (2005) Nat. Immunol. 6, 1123–1132 [DOI] [PubMed] [Google Scholar]
- 4.Park H., Li Z., Yang X. O., Chang S. H., Nurieva R., Wang Y. H., Wang Y., Hood L., Zhu Z., Tian Q., Dong C. (2005) Nat. Immunol. 6, 1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hultgren B., Huang X., Dybdal N., Stewart T. A. (1996) Diabetes 45, 812–817 [DOI] [PubMed] [Google Scholar]
- 6.Wang B., André I., Gonzalez A., Katz J. D., Aguet M., Benoist C., Mathis D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13844–13849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellison C. A., Fischer J. M., HayGlass K. T., Gartner J. G. (1998) J. Immunol. 161, 631–640 [PubMed] [Google Scholar]
- 8.Komiyama Y., Nakae S., Matsuki T., Nambu A., Ishigame H., Kakuta S., Sudo K., Iwakura Y. (2006) J. Immunol. 177, 566–573 [DOI] [PubMed] [Google Scholar]
- 9.Jain R., Tartar D. M., Gregg R. K., Divekar R. D., Bell J. J., Lee H. H., Yu P., Ellis J. S., Hoeman C. M., Franklin C. L., Zaghouani H. (2008) J. Exp. Med. 205, 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miljkovic D., Cvetkovic I., Momcilovic M., Maksimovic-Ivanic D., Stosic-Grujicic S., Trajkovic V. (2005) Cell. Mol. Life Sci. 62, 2658–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vukkadapu S. S., Belli J. M., Ishii K., Jegga A. G., Hutton J. J., Aronow B. J., Katz J. D. (2005) Physiol. Genomics 21, 201–211 [DOI] [PubMed] [Google Scholar]
- 12.Mensah-Brown E. P., Shahin A., Al-Shamisi M., Wei X., Lukic M. L. (2006) Eur. J. Immunol. 36, 216–223 [DOI] [PubMed] [Google Scholar]
- 13.Spolski R., Kashyap M., Robinson C., Yu Z., Leonard W. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14028–14033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin-Orozco N., Chung Y., Chang S. H., Wang Y. H., Dong C. (2009) Eur. J. Immunol. 39, 216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srinivasan S., Bolick D. T., Lukashev D., Lappas C., Sitkovsky M., Lynch K. R., Hedrick C. C. (2008) Diabetes 57, 484–493 [DOI] [PubMed] [Google Scholar]
- 16.Bending D., Pena H., Veldhoen M., Phillips J. M., UYttenhove C., Stockinger B., Cooke A. (2009) J. Clin. Invest. 119, 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov I. I., McKenzie B. S., Zhou L., Tadokoro C. E., Lepelley A., Lafaille J. J., Cua D. J., Littman D. R. (2006) Cell 126, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 18.Yang X. O., Pappu B. P., Nurieva R., Akimzhanov A., Kang H. S., Chung Y., Ma L., Shah B., Panopoulos A. D., Schluns K. S., Watowich S. S., Tian Q., Jetten A. M., Dong C. (2008) Immunity 28, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X. O., Panopoulos A. D., Nurieva R., Chang S. H., Wang D., Watowich S. S., Dong C. (2007) J. Biol. Chem. 282, 9358–9363 [DOI] [PubMed] [Google Scholar]
- 20.Quintana F. J., Basso A. S., Iglesias A. H., Korn T., Farez M. F., Bettelli E., Caccamo M., Oukka M., Weiner H. L. (2008) Nature 453, 65–71 [DOI] [PubMed] [Google Scholar]
- 21.Veldhoen M., Hirota K., Westendorf A. M., Buer J., Dumoutier L., Renauld J. C., Stockinger B. (2008) Nature 453, 106–109 [DOI] [PubMed] [Google Scholar]
- 22.Kang B. Y., Chung S. W., Cho D., Kim T. S. (2002) Biochem. Pharmacol. 63, 1901–1910 [DOI] [PubMed] [Google Scholar]
- 23.Hsiang C. Y., Wu S. L., Cheng S. E., Ho T. Y. (2005) J. Biomed. Sci. 12, 791–801 [DOI] [PubMed] [Google Scholar]
- 24.Yang J., Wang H. D., Lu D. X., Wang Y. P., Qi R. B., Li J., Li F., Li C. J. (2006) Acta Pharmacol. Sin. 27, 173–178 [DOI] [PubMed] [Google Scholar]
- 25.Li F., Wang H. D., Lu D. X., Wang Y. P., Qi R. B., Fu Y. M., Li C. J. (2006) Acta Pharmacol. Sin. 27, 1199–1205 [DOI] [PubMed] [Google Scholar]
- 26.Choi B. H., Ahn I. S., Kim Y. H., Park J. W., Lee S. Y., Hyun C. K., Do M. S. (2006) Exp. Mol. Med. 38, 599–605 [DOI] [PubMed] [Google Scholar]
- 27.Enk R., Ehehalt R., Graham J. E., Bierhaus A., Remppis A., Greten H. J. (2007) J. Ethnopharmacol. 109, 170–175 [DOI] [PubMed] [Google Scholar]
- 28.Kong W., Wei J., Abidi P., Lin M., Inaba S., Li C., Wang Y., Wang Z., Si S., Pan H., Wang S., Wu J., Wang Y., Li Z., Liu J., Jiang J. D. (2004) Nat. Med. 10, 1344–1351 [DOI] [PubMed] [Google Scholar]
- 29.Lee Y. S., Kim W. S., Kim K. H., Yoon M. J., Cho H. J., Shen Y., Ye J. M., Lee C. H., Oh W. K., Kim C. T., Hohnen-Behrens C., Gosby A., Kraegen E. W., James D. E., Kim J. B. (2006) Diabetes 55, 2256–2264 [DOI] [PubMed] [Google Scholar]
- 30.Nishino H., Kitagawa K., Fujiki H., Iwashima A. (1986) Oncology 43, 131–134 [DOI] [PubMed] [Google Scholar]
- 31.Pandey M. K., Sung B., Kunnumakkara A. B., Sethi G., Chaturvedi M. M., Aggarwal B. B. (2008) Cancer Res. 68, 5370–5379 [DOI] [PubMed] [Google Scholar]
- 32.Ren Y., Lu L., Guo T. B., Qiu J., Yang Y., Liu A., Zhang J. Z. (2008) J. Immunol. 181, 1491–1498 [DOI] [PubMed] [Google Scholar]
- 33.Lin S., Tsai S. C., Lee C. C., Wang B. W., Liou J. Y., Shyu K. G. (2004) Mol. Pharmacol. 66, 612–619 [PubMed] [Google Scholar]
- 34.Lee S., Lim H. J., Park J. H., Lee K. S., Jang Y., Park H. Y. (2007) Biochem. Biophys. Res. Commun. 362, 853–857 [DOI] [PubMed] [Google Scholar]
- 35.Huang C., Zhang Y., Gong Z., Sheng X., Li Z., Zhang W., Qin Y. (2006) Biochem. Biophys. Res. Commun. 348, 571–578 [DOI] [PubMed] [Google Scholar]
- 36.Rincón M., Enslen H., Raingeaud J., Recht M., Zapton T., Su M. S., Penix L. A., Davis R. J., Flavell R. A. (1998) EMBO J. 17, 2817–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang D. D., Conze D., Whitmarsh A. J., Barrett T., Davis R. J., Rincón M., Flavell R. A. (1998) Immunity 9, 575–585 [DOI] [PubMed] [Google Scholar]
- 38.Dong C., Yang D. D., Wysk M., Whitmarsh A. J., Davis R. J., Flavell R. A. (1998) Science 282, 2092–2095 [DOI] [PubMed] [Google Scholar]
- 39.Pagès G., Guérin S., Grall D., Bonino F., Smith A., Anjuere F., Auberger P., Pouysségur J. (1999) Science 286, 1374–1377 [DOI] [PubMed] [Google Scholar]
- 40.Fischer A. M., Katayama C. D., Pagès G., Pouysségur J., Hedrick S. M. (2005) Immunity 23, 431–443 [DOI] [PubMed] [Google Scholar]
- 41.Yamashita M., Shinnakasu R., Asou H., Kimura M., Hasegawa A., Hashimoto K., Hatano N., Ogata M., Nakayama T. (2005) J. Biol. Chem. 280, 29409–29419 [DOI] [PubMed] [Google Scholar]
- 42.Stojanoviæ I., Cvjetiæanin T., Lazaroski S., Stosiæ-Grujiciæ S., Miljkoviæ D. (2009) Immunology 126, 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nurieva R. I., Chung Y., Hwang D., Yang X. O., Kang H. S., Ma L., Wang Y. H., Watowich S. S., Jetten A. M., Tian Q., Dong C. (2008) Immunity 29, 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu H. H., Kim K. J., Cha J. D., Kim H. K., Lee Y. E., Choi N. Y., You Y. O. (2005) J. Med. Food 8, 454–461 [DOI] [PubMed] [Google Scholar]
- 45.Higaki S., Hasegawa Y., Morohashi M., Takayoshi Y. (1995) J. Dermatol. 22, 4–9 [DOI] [PubMed] [Google Scholar]
- 46.Wen L., Ley R. E., Volchkov P. Y., Stranges P. B., Avanesyan L., Stonebraker A. C., Hu C., Wong F. S., Szot G. L., Bluestone J. A., Gordon J. I., Chervonsky A. V. (2008) Nature 455, 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.