Abstract
Lysosomal neuraminidase-1 (NEU1) forms a multienzyme complex with β-galactosidase and protective protein/cathepsin A (PPCA). Because of its association with PPCA, which acts as a molecular chaperone, NEU1 is transported to the lysosomal compartment, catalytically activated, and stabilized. However, the mode(s) of association between these two proteins both en route to the lysosome and in the multienzyme complex has remained elusive. Here, we have analyzed the hydrodynamic properties of PPCA, NEU1, and a complex of the two proteins and identified multiple binding sites on both proteins. One of these sites on NEU1 that is involved in binding to PPCA can also bind to other NEU1 molecules, albeit with lower affinity. Therefore, in the absence of PPCA, as in the lysosomal storage disease galactosialidosis, NEU1 self-associates into chain-like oligomers. Binding of PPCA can reverse self-association of NEU1 by causing the disassembly of NEU1-oligomers and the formation of a PPCA-NEU1 heterodimeric complex. The identification of binding sites between the two proteins allowed us to create innovative structural models of the NEU1 oligomer and the PPCA-NEU1 heterodimeric complex. The proposed mechanism of interaction between NEU1 and its accessory protein PPCA provides a rationale for the secondary deficiency of NEU1 in galactosialidosis.
Mammalian neuraminidases have been classified as lysosomal (NEU1),4 cytosolic (NEU2), plasma membrane (NEU3), and mitochondria/lysosomal (NEU4) based on their subcellular distributions, pH optimum, kinetic properties, responses to ions and detergents, and substrate specificities (1–3). Of the four sialidases, only NEU1 is ubiquitously expressed at different levels in various tissues and cell types (4–7). The importance of these proteins in normal cellular physiology is illustrated by the numerous metabolic processes that they control, including cell proliferation and differentiation, cell adhesion, membrane fusion and fluidity, immunocyte function, and receptor modification (8–21).
NEU1 initiates the intralysosomal hydrolysis of sialo-oligosaccharides, -glycolipids, and -glycoproteins by removing their terminal sialic acid residues. In human and murine tissues, NEU1 forms a complex with at least two other proteins, β-galactosidase and the protective protein/cathepsin A (PPCA) (22). By virtue of their association with PPCA, NEU1 and β-galactosidase acquire their active and stable conformation in lysosomes. However, PPCA appears to function as a crucial chaperone/transport protein for NEU1. Because NEU1 is poorly mannose 6-phosphorylated, it depends on PPCA for correct compartmentalization and catalytic activation in lysosomes (23–25). Only a small amount of PPCA and β-galactosidase activities is found in the NEU1-PPCA-β-galactosidase complex, which instead contains all of the NEU1 catalytic activity (24–27). By understanding how and when NEU1 and PPCA interact, how they regulate each other in different cell types, and what determinants control their association, we may gain important insight into their significance in physiologic and pathologic conditions.
The absence of NEU1 is associated with two neurodegenerative diseases that involve glycoprotein metabolism; sialidosis, which is caused by structural lesions in the lysosomal NEU1 locus (28), and galactosialidosis (GS), a combined deficiency of NEU1 and β-galactosidase which is caused by the absence of PPCA (22). Patients with sialidosis and those with GS have similar clinical and biochemical features, and both diseases are characterized by multiple phenotypes that are classified according to the age of onset and severity of the symptoms.
Previously, we generated two animal models of primary or secondary NEU1 deficiency, Neu1−/− mice and Ppca−/− mice. Both mouse models have a profound loss of Neu1 activity in multiple tissues and develop clinical, biochemical, and pathologic manifestations resembling those seen in patients with severe sialidosis and GS (29–31). Neu1−/− mice are phenotypically similar but not identical to Ppca−/− mice and, like children with the disease, exhibit a time-dependent splenomegaly associated with extramedullary hematopoiesis (30, 31). We found that the cause of these phenotypic abnormalities is the gradual loss of retention of hematopoietic progenitors within the bone niche due to exacerbated lysosomal exocytosis of bone marrow cells. The latter process is negatively regulated by NEU1 activity (31).
The mode of interaction between PPCA and NEU1 and the mechanism of catalytic activation are not well understood. Here we present biochemical, analytical, and structural analyses of NEU1, PPCA, and the PPCA-NEU1 complex by using purified baculovirus (BV)-expressed wild-type and mutagenized recombinant enzymes and synthetic peptides.
EXPERIMENTAL PROCEDURES
Cell Culture, Antibodies, and Recombinant Proteins
Fibroblasts from a patient with early infantile galactosialidosis were deposited in the European Cell Bank (Rotterdam, The Netherlands). Fibroblasts were maintained in culture in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics (100 units/ml penicillin, 100 μg/ml streptomycin) at 37 °C (5% CO2). Macrophages were isolated from the bone marrow of Ppca-null mice as previously described (32). Antibodies against NEU1 and PPCA were produced and affinity-purified as described previously (32). PPCA and NEU1 were expressed in insect cells and purified to near homogeneity as previously described (32, 33).
Site-directed Mutagenesis and Retroviral Constructs
Wild-type Ppca and Neu1 cDNA constructs have been previously described (4, 34). Ppca cDNA was in vitro mutagenized to introduce the following single amino acid substitutions: R20A, A251E, L354D, K355Q, W382A, and P451A. To generate the cDNA mutants, we performed two rounds of PCR using Ppca oligonucleotide primers with specific nucleotide mutations encoding each of the PPCA amino acid substitutions. The nucleotide exchanges were confirmed by automated sequence analysis. Wild-type and mutant Ppca and Neu1 cDNAs were subcloned into the 5′ internal ribosomal entry sequence (IRES) of the MSCV-IRES-GFP retroviral vector, and retroviruses were generated and expressed in human fibroblasts as previously described (35).
Enzyme Activity Analysis and Immunoblotting
Retrovirally transduced fibroblasts were harvested and lysed in water containing Complete EDTA-free protease inhibitors (Roche Applied Science). NEU1 catalytic activity was measured using the synthetic substrate 4-methylumbelliferyl-α-d-N-acetylneuraminic acid as described earlier (36), and total protein concentration was measured using the BCA assay and bovine serum albumin as a standard (Pierce). Cell lysate samples were resolved on 12.5% polyacrylamide gels under denaturing conditions, transferred onto polyvinylidene difluoride membranes, and incubated with anti-NEU1 and/or anti-PPCA affinity-purified antibodies and peroxidase-conjugated goat-anti-rabbit secondary antibody (Jackson ImmunoResearch, West Grove, PA). NEU1 and PPCA were visualized using Western Lightning Chemiluminescence Reagents (PerkinElmer Life Sciences).
Immunocytochemistry
Fluorescence-sorted retrovirally transduced GFP+ fibroblasts were seeded directly onto Superfrost/Plus microscope glass slides (Fisher) at about 60% confluence. One day later the slides were processed according to the method of van Dongen et al. (37) by using affinity-purified anti-NEU1 antibody and cyanine 3-conjugated goat-anti-rabbit IgG secondary antibody (Jackson ImmunoResearch). The stained slides were examined using a fluorescence microscope as described earlier (4).
Electron Microscopy of NEU1 Protein
Purified BV-NEU1 (1 mg/ml) in 50 mm sodium acetate (pH 5.0, 100 mm NaCl) was adsorbed to carbon-coated grids, negatively stained with 1% aqueous uranyl acetate, and imaged with a Phillips 301 electron microscope operated at 60 kV.
In Vitro Activation of NEU1
Purified insect cell-expressed recombinant NEU1 and PPCA (mature and precursor forms) were separately dialyzed against phosphate-buffered saline (pH 7) or 50 mm sodium acetate (pH 5.0, 100 mm NaCl). NEU1 and PPCA (pH 5 or 7) were mixed at ∼1:1 (8 μg each) to a 1:7 ratio (8 and 56 μg, respectively) in either 50 mm sodium acetate (pH 5, 100 mm NaCl) or 50 mm MES (pH 6.5, 100 mm NaCl) in a total volume of 100 μl per sample. The protein samples were incubated for 24 h at 4 °C, room temperature, or 37 °C, after which the neuraminidase activity was measured as described above. The denatured and reduced proteins were resolved on a 12% SDS-polyacrylamide gel and stained with 0.1% Coomassie Brilliant Blue (w/v) in 40% methanol, 10% acetic acid (v/v).
Synthetic Peptide Synthesis
A panel of 72 sequential 15-mer NEU1 peptides (4) and 88 sequential 15-mer PPCA peptides (34), each overlapping with the following peptide by 10 residues, were synthesized using an Advanced Chemtech 396 synthesizer. The N-terminal signal peptides of NEU1 and PPCA were not included in the peptide synthesis. The peptide amides were synthesized on 4-hydroxymethylphenoxyacetyl-4′-methylbenzyhydrylamine-amide resin (Applied Biosystems, Inc., Foster City, CA), and the Fmoc (N-(9-fluorenyl)methoxycarbonyl) amino acids were coupled using 1-hydroxybenzotriazole/2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate chemistries. N-terminal acetylation was performed using acetic anhydride and 1-hydroxybenzotriazole. Peptides were cleaved from the resin in 91% trifluoroacetic acid containing 2% phenol, 2% ethanedithiol, and 5% thioanisole. The peptides were precipitated and washed twice with diethyl ether. Peptide concentrations for CD and surface plasmon resonance (SPR) were determined using quantitative amino acid analysis.
Surface Plasmon Resonance
Binding studies were performed using a Biacore 3000 surface plasmon resonance instrument (GE Healthcare). Purified insect cell-expressed PPCA and NEU1 proteins were covalently attached to a CM-5 chip (Biacore) using standard amine coupling methods in 10 mm sodium acetate at pH 5.5. The remaining reactive sites were blocked with ethanolamine. A reference cell was prepared similarly in the absence of protein.
Binding of NEU1, PPCA, or NEU1 synthetic 15-mer peptides was measured by flowing protein or peptides in 10 mm HEPES, 150 mm NaCl, 3 mm EDTA, and 0.005% surfactant P20 (HBS-EP buffer, Biacore) at 20 μl/min in sequence through the reference and protein-containing flow cells at 25 °C. The pH of the HBS-EP buffer was adjusted with 1 m HCl for each pH tested. Synthetic peptides were at a concentration of 0.1 mg/ml, and sample and system buffers were supplemented with 500 mm NaCl, 5% DMSO, and 10 mg/ml dextran sulfate 10 to improve peptide solubility and reduce background binding. The system and sample-diluting buffers were set at the pH being tested. A blank containing only HBS-EP buffer at the appropriate pH was also run. Regeneration of the chip surface to remove bound protein or peptides consisted of two 500-μl injections of HBS-EP buffer at pH 7.4. The flow cells were maintained at 25 °C. Data reported are the maximum differences in SPR signal between the flow cells containing coupled protein and the reference cells. Any contribution of the HBS-EP buffer to the signal was removed by subtracting the blank (HBS-EP buffer) injection from the reference-subtracted signal. The binding level was determined as the maximum signal in the binding phase. Because of the extremes of pH used in these studies, some loss of binding capacity was observed between injections, amounting to ∼1% loss of binding per injection. For measurements of binding levels for pH titration and peptide binding studies, the top value at the end of the binding phase was used as the binding value. Data analysis was done using software and kinetic models supplied with the Biacore 3000 instrument.
Analytical Ultracentrifugation
Purified insect cell-expressed NEU1 and PPCA were dialyzed against the ultracentrifugation buffers, 50 mm sodium acetate, pH 5, or phosphate-buffered saline, 100 mm NaCl, pH 7.0, and subjected to sedimentation velocity in a ProteomeLab XL-I analytical ultracentrifuge with an eight-hole Beckman An-50 rotor and cells containing sapphire windows and charcoal-filled Epon double-sector center pieces (Beckman Coulter, Fullerton, CA). The buffer dialysate was used as optical reference. The loading volume of ∼400 μl was identical for the reference and sample chambers of the double-sector centerpiece, and the final loading protein concentrations were 1.8 to 0.15 mg/ml. The density and viscosity of the ultracentrifugation buffers at 4 °C were calculated from their respective compositions, and the partial specific volume at 4 °C, and molecular weights of the proteins were calculated based on the amino acid composition using the software SEDNTERP (38). After equilibration at 4 °C at rest, the rotor was accelerated to 50,000 rpm and fringe-displacement data at time intervals of 1.0 min were collected with the Rayleigh interference system for 10 h and analyzed with SEDFIT software using the model for continuous sedimentation coefficient distribution c(s) with deconvolution of diffusion effects (39, 40) The sedimentation coefficient distribution c(s) was calculated with maximum entropy regularization at a confidence level of p = 0.7 and at a resolution of sedimentation coefficients of n = 100–120. The positions of the meniscus and bottom as well as time-invariant and radial noises were fitted. A two-dimensional size-shape distribution, c(s, f/f0) (with the first dimension the s distribution and the second the f/f0 distribution) was also calculated with an equidistant f/f0 grid of 0.1 steps that varied from 1.0 to 2.8, a linear s-grid that varied from 0.5 to 15 S, and Tikhonov-Phillips regularization (41). The distributions were transformed to c(s,Mr) and c(s,f/f0) distributions, where Mr is the molecular weight, f/f0 is the frictional ratio, and s is the sedimentation coefficient, and plotted as contour plots. This analysis was with regularization. The dotted lines of the c(s,Mr) distribution plots indicate lines of constant f/f0. The distributions were not normalized (41).
Structural Modeling of the NEU1 Oligomer and the Heterodimeric Complex of PPCA and NEU
A structural model of human NEU1 was generated using the comparative method (42). Initial modeling was performed using the fold-recognition program LOOPP (43, 44), which used an alignment protocol based on the scoring scheme of the structural template of bacterial sialidase from Micromonospora viridifaciens (PDB code 1EUR) (45). Subsequently, an atomic model was generated using MODELLER, Version 9v4 (46, 47).
Data obtained from the surface plasmon resonance peptide scanning from both NEU1 and PPCA sides were used to build the structural models of the NEU1 oligomer and the PPCA-NEU1 complex. Two templates were used to model the structure of the PPCA-NEU1 heterodimeric complex with 1:1 molar ratio; 1) computer-generated model of human NEU1 and 2) crystallographic structure of the precursor PPCA monomer, PDB code 1IVY (48). After visualization of the contacts and preliminary analysis of the surface interactions with the computer graphics program COOT (49), several three-dimensional models were suggested based on the optimal packing of the individual monomers. Attempts were undertaken to discriminate between various models using the protein-protein docking servers ClusPro (50), RosettaDock (51, 52), and Gramm-X (53, 54), with the knowledge-score algorithms based on surface complementarity, energy optimization, and fast Fourier transformation methodologies, respectively.
RESULTS
PPCA-induced Activation of NEU1
NEU1 gains full catalytic activity in the lysosome through its binding to PPCA and assembly within the multienzyme complex, which most likely triggers conformational changes essential for catalysis. The timing of the proteolytic processing of the 54-kDa PPCA zymogen into the mature two-chain, 32/20-kDa enzyme remains unknown, as does whether this process is a prerequisite for activation of the 46-kDa NEU1. No mutations have been identified that prevent the proteolytic processing and activation of PPCA. Instead, all patients with GS present with deficiencies in cathepsin A and NEU1.
To better understand the mechanism of NEU1 activation, we examined the ability of purified precursor PPCA and mature PPCA to activate NEU1 in solution. At pH 6.5 and 5.0, we added increasing quantities of PPCA to a constant amount of NEU1, in ratios of 1:0.7 to 1:7, to assess the effect on NEU1 activity. To analyze whether time or temperature was important for catalytic activation, we incubated the samples for 1 or 24 h at room temperature or 37 °C and then assayed NEU1 activity. Increasing amounts of bovine serum albumin added to NEU1 as a control had no effect on the enzyme activity (not shown). At pH 6.5, NEU1 activity increased with the PPCA concentration independent of time, temperature, and whether the precursor or mature PPCA form was present, except that incubation with mature PPCA for 24 h at 37 °C resulted in overall lower NEU1 activity (Fig. 1, A and B).
FIGURE 1.
In vitro catalytic activation of NEU1 by PPCA. A–D, increasing quantities of precursor or mature PPCA (5.6–56 μg) were added to NEU1 (8 μg) at pH 6.5 (A and B) or pH 5 (C and D) in a total volume of 100 μl. The protein mixtures were incubated for 1 h at room temperature (blue plot), 24 h at room temperature (red plot), or 24 h at 37 °C (black plot). NEU1 activity was then measured. NEU1 activity is expressed as nmol substrate/h/mg of NEU1. E, Coomassie Blue-stained SDS gel with precursor PPCA (5.6 μg) mixed with an equal amount of NEU1 (lanes 1 and 2) or mature PPCA (5.6 μg) mixed with an equal amount of NEU1 (lanes 3–5). Protein mixtures were incubated for 0, 1, or 24 h at 37 °C as indicated, before loading on the SDS gel.
At pH 5, a similar temperature-independent increase in NEU1 activity was measured after 1 h of incubation with either form of PPCA (Fig. 1, C and D), but the addition of mature PPCA followed by 24 h of incubation at 37 °C led to a steady decline in NEU1 activity (Fig. 1D). SDS-PAGE analysis showed the increasing appearance of a degradation product (∼36 kDa) in NEU1 samples that had been mixed with an equal amount of mature PPCA and incubated at 37 °C for 1 or 24 h (Fig. 1E). These results may explain the relatively short half-life of endogenous NEU1 in mammalian cells maintained in culture (23) and suggest that mature PPCA may trigger the inactivation and degradation of this protein.
Expression of Mutant PPCA Variants in GS Fibroblasts
The mode of interaction between PPCA and NEU1 and the sites of their binding are unknown. To identify amino acid residues on the PPCA precursor that bind NEU1, we took advantage of the tertiary structure of PPCA (55). We assumed that the solvent-accessible surface of the PPCA structure, as calculated by GRASP (48, 56), would be tailored for its NEU1-protective task. We created single amino acid mutations of surface residues that are conserved in human and mouse PPCA but not in other nonmammalian members of the serine carboxypeptidase family that have no protective function toward other proteins (34, 48). The amino acid substitutions were as follows: R20A, A251E, L354D, K355Q, W382A, and P451A. The individual mutations were modeled onto the structure of precursor PPCA to ascertain that the overall structure of the mutant proteins was preserved (data not shown).
PPCA
Null fibroblasts were transduced with bicistronic MSCV vectors containing GFP, wild-type NEU1 cDNA, and either wild-type or mutant PPCA cDNA. These coexpression experiments were designed to test whether any of the PPCA variants interacted with wild-type NEU1 and whether the interaction promoted transport of NEU1 to the lysosome. PPCA mutants that are correctly compartmentalized and processed in lysosomes into a mature, active enzyme should be structurally uncompromised but may still be defective in binding to NEU1.
Immunoblots showed that all PPCA mutant proteins were processed to the mature subunits and, hence, were efficiently transported to the endosomal/lysosomal compartment, although they were expressed at different levels (Fig. 2A). As expected, NEU1 was not detected in GS fibroblasts transduced with NEU1 alone because of the lack of endogenous PPCA (Fig. 2B). In most cases, however, the level of NEU1 in GS cells transduced with a mutant PPCA exceeded that of cells transduced with wild-type PPCA (Fig. 2B). These results were in agreement with the enzyme activities measured in transduced cells. The cathepsin A activity of four mutants equaled or exceeded that of wild-type PPCA, whereas two mutants (R20A and W382A) showed lower cathepsin A activity. In all cases, NEU1 was at the most only partially activated (Fig. 2C). The inability of these PPCA variants to fully activate NEU1 may have resulted from their reduced affinity toward the protein. However, each PPCA variant coimmunoprecipitated with NEU1 (data not shown and Ref. 23), confirming that none of the amino acid substitutions completely abrogated binding between the two proteins. Overall, these results suggest that our initial structural predictions were correct and that some of the selected residues, alone or in combination, participate in the interaction between NEU1 and PPCA.
FIGURE 2.
Expression of MSCV-PPCA mutants in GS fibroblasts. GS were single- or double-transduced with MSCV-based bicistronic (GFP) retroviral vectors containing wild-type Neu1 cDNA (NEU1) and either wild-type PPCA (PPCA) or mutant PPCA (R20A, A251E, L354D, K355Q, W382A, P451A) cDNAs. The cells were fluorescence-sorted for transduced GFP-expressing cells. Fluorescence-sorted cells were analyzed on immunoblots with anti-PPCA (A) and anti-NEU1 (B) antibodies. C, the activities of cathepsin A (CA; black bars) and NEU1 (gray bars) were measured in homogenates of the transduced GS fibroblasts, nontransduced normal fibroblasts (Control), and nontransduced GS fibroblasts. Cathepsin A activity is expressed as nmol of substrate/min/mg total protein; NEU1 activity is expressed as nmol substrate/h/mg of total protein.
Uptake of BV-expressed Wild-type or Mutant PPCA by Mouse Ppca-deficient Macrophages
To investigate the capacity of three PPCA variants (R20A, L354D, P451A) to correct NEU1 activity via the endocytic route, we overexpressed the mutant proteins in insect cells, purified them to near homogeneity, and used them in uptake experiments in Ppca-deficient macrophages. These variants were selected because they showed different protective activity toward NEU1 in GS fibroblasts: R20A (low cathepsin A activity, no NEU1 activity), L354D (normal cathepsin A activity, intermediate NEU1 activity), and P451A (normal cathepsin A activity, no NEU1 activity). Wild-type and Ppca−/− primary macrophages were used as recipients because they internalize insect cell-produced enzymes (32). Using this approach, we bypassed the requirement for NEU1 and PPCA to interact before their compartmentalization in endosomes/lysosomes and were able to exclusively assess their endosomal/lysosomal behavior.
The activity/stability of wild-type NEU1 endocytosed by Ppca+/+ or Ppca−/− macrophages considerably exceeded the endogenous levels in both recipient cell types, and internalized NEU1 was equally stable for at least 24 h (Fig. 3A). The coadministration of wild-type PPCA and wild-type NEU1 had an adjunctive effect on the NEU1 activity and increased its half-life to more than 24 h (Fig. 3A).
FIGURE 3.
Half-life of endocytosed NEU1 in Ppca−/− and Ppca+/+ macrophages. Macrophages were maintained in culture in 6-well plates in the presence of either purified NEU1 alone (25 μg) or a mixture of BV-NEU1 and BV-PPCA (25 μg each) for 16 h. The medium was then replaced with enzyme-free medium. Cells were harvested at 0, 1, 3, 5, 7, and 24 h and assayed for NEU1 activity (A). NEU1 uptake was measured in Ppca+/+ macrophages (red plot) or Ppca−/−macrophages (blue plot), and PPCA-NEU1 uptake was measured in Ppca−/− macrophages (black plot). NEU1 activity is expressed as nmol of substrate/h/mg of total protein. B–E, harvested cell lysates were also analyzed on immunoblots with anti-PPCA and anti-NEU1 antibodies.
Immunoblot analysis of macrophage lysates confirmed the enzyme activity results (Fig. 3, B–E). NEU1 was detected as a doublet when PPCA was either absent (Fig. 3B) or present at endogenous levels (Fig. 3C) but appeared as a single polypeptide when PPCA levels were increased by uptake of exogenous PPCA (Fig. 3E). Thus, PPCA levels are rate-limiting in terms of NEU1 activity and lysosomal stability when excess NEU1 is present (Fig. 3B).
Similar uptake experiments were performed using BV-expressed PPCA variants combined with an equal amount of purified NEU1. Uptake of PPCA-L354D or -P451A restored cathepsin A activity to levels that were similar to those obtained with the wild-type PPCA precursor (Fig. 4A). On immunoblots, the endocytosed mutants appeared to be processed as efficiently as the wild-type protein (Fig. 4B). The expression level and catalytic activity of PPCA-R20A indicated that this variant was unstable in the lysosome, although it still partially activated NEU1 (Fig. 4A).
FIGURE 4.
Uptake of insect cell-expressed wild-type and mutant PPCA by Ppca−/− macrophages. Macrophages were maintained in culture in 6-well plates in the presence of purified wild-type (WT) or mutant BV-PPCA and wild-type BV-NEU1 (25 μg each) for 2 days. Cells were then harvested and assayed for cathepsin A (CA; black bars) and NEU1 (gray bars) activity (A). Cathepsin A activity is expressed as nmol of substrate/min/mg of total protein; NEU1 activity is expressed as nmol of substrate/h/mg of total protein. B, the cell lysates were analyzed on immunoblots for the presence of endocytosed enzymes with anti-PPCA and anti-NEU1 antibodies. Untreated cells are shown in lane 1.
Binding of NEU1 to Wild-type and Mutant PPCAs by Surface Plasmon Resonance
We next tested whether the mature form of BV-expressed wild-type PPCA bound NEU1 by using SPR. We coupled purified NEU1 to a sensor chip and measured the binding of mature PPCA (pH range 7.5-4.5). Binding was minimal at neutral pH but increased steadily as the pH decreased (Fig. 5A). The binding of precursor PPCA (not shown) and that of mature PPCA to NEU1 was similar, albeit the relative binding of the mature form was lower at all tested pH values.
FIGURE 5.
Identification of binding sites between NEU1 and PPCA by SPR-peptide scanning. A, purified NEU1 was covalently coupled to a SPR sensor chip. Binding of PPCA was measured by flowing purified wild-type (black plot) or mutant precursor PPCAs (red, green, and blue plots) at pH ranging from 4.5 to 7.5 through the reference and the NEU1-containing flow cells (A). B, SPR binding of a series of 88 synthetic PPCA peptides (15-mer with 10-residue overlap) was measured by flowing the peptides in sequence through the reference and protein-containing flow cells. C and D, SPR binding of a series of 72 synthetic NEU1 peptides to precursor PPCA (C), mature PPCA (D), and NEU1 (E). Data reported are the differences in SPR signal between the flow cells containing protein and the reference cells. The binding level was determined as the maximum signal in the binding phase. Peptides that gave binding values that were higher than the nonspecific background values are indicated.
We repeated these assays using three PPCA mutants (PPCA-R20A, -L354D, -P451A). All three showed maximum binding to NEU1 at pH 4.5, but their binding affinities were 4–8-fold less than that of wild-type PPCA (Fig. 5A). This finding indicates that either a large surface area or multiple contact sites may be required for interaction of the two proteins. Although these PPCA amino acid substitutions appear to be sufficient to substantially impair the binding of PPCA to NEU1 in a cell-free system, in mammalian cell expression assays the effects of the same mutations was less pronounced. Therefore, under physiological conditions other enzymes that are part of the multienzyme complex, most notably β-galactosidase, may influence the stability of the NEU1-PPCA bond (57).
Identification of PPCA- and NEU1-binding Sites
To complement the SPR results, we performed a SPR-peptide screen against full-length NEU1 protein. This method helps to identify motifs and amino acid residues that are essential for protein-protein interaction (58, 59). We designed 88 synthetic 15-mer peptides covering the entire sequence of PPCA and overlapping each other by 10 amino acids. We then analyzed the ability of each to bind NEU1 coupled to a sensor chip. The screen revealed N-terminal and C-terminal regions of PPCA that contained multiple peptides specific for NEU1 binding. The N-terminal peptide 11 (residues 51–65; VLWLNGGPGCSSLDG) gave a high binding value of 118, whereas the C-terminal peptides 65 (residues 321–335; ALNIPEQLPQWDMCN) and 74 (residues 371–385; VDMACNFMGDEWFVD) had binding values of 110 and 190, respectively (Fig. 5B). Three of the variants that were coexpressed with NEU1 in mammalian cells (PPCA-L354D, -K355Q, -W382A) lay within the C-terminal region of PPCA that shows binding to NEU1. Neither PPCA region involved in binding to NEU1 coincided with the internal 2-kDa “excision peptide” (residues 280–298; LPLKRMWHQALLRSGDKVR) that is proteolytically removed during maturation of the protein (48, 60). Peptide 65 is only 22 amino acids from the excision peptide; therefore, the cleavage of the excision peptide may change the conformation of the adjacent regions in the mature form of PPCA and in turn influence its ability to bind NEU1.
Following a similar strategy, we scanned the NEU1 primary structure for PPCA-binding sites by screening the ability of 72 synthetic 15-mer peptides of NEU1 to bind to a sensor chip containing either mature or precursor PPCA. NEU1 peptides with relative binding values greater than 40 were considered to bind specifically. Overall, the relative binding of the peptides to precursor PPCA was greater than that to the mature enzyme (Fig. 5, C and D). Eight peptides bound to precursor PPCA with a relative binding of 45 to 250, which is severalfold higher than the basal level of peptide binding (Fig. 5C). Peptides 13 (residues 61–75; AKFIALRRSMDQGST) and 48 (residues 23–250; NQNNYHCHCRIVLRS) also bound to mature PPCA (Fig. 5D). Peptide 48 bound slightly better to precursor than mature PPCA, whereas peptide 13 showed the reverse affinity; thus, the NEU1 peptides that interact with the precursor but not with mature PPCA may have recognized a region embedded within the excision peptide.
To identify potential self-binding sites on NEU1, we tested the SPR binding of the 72 synthetic NEU1 peptides to the full-length NEU1 protein. In this assay a relative binding value greater than 80 signified specific binding. Four NEU1 peptides had binding values greater than 90 (Fig. 5D), suggesting that NEU1 competes with PPCA for the same binding site(s) on other NEU1 molecules. This finding supports our earlier report in which we showed the presence of NEU1 in large crystal-like aggregates within the endoplasmic reticulum; however, this occurred only when the enzyme was overexpressed in the absence of PPCA (4). Coexpression of PPCA with NEU1 prevented the formation of these crystal-like bodies and resulted in the lysosomal localization of both proteins (4). Thus, by binding to NEU1 in the endoplasmic reticulum, PPCA prevents the clustering of NEU1 and promotes its transport to the lysosome.
Hydrodynamic Properties of PPCA and NEU1
Using analytical ultracentrifugation (AUC), we characterized the hydrodynamic properties of PPCA, NEU1, and putative complexes in a mixture of both proteins in solution to investigate their behavior and binding in a more physiological state. These measures included the sedimentation coefficients, molecular masses, and shapes of the proteins in solution. Sedimentation velocity analysis separates particles at high resolution in a size- and shape-dependent manner. The sedimentation coefficient is the rate of sedimentation per unit of centrifugal field for a particle, and the frictional ratio (f/f0) is the ratio of the translational frictional coefficient of the particle to that of a spherical particle of the same mass and volume (61). Although the sedimentation coefficient is related to molecular mass, the frictional ratio is related to molecular shape. Typically, the hydrated frictional ratio ranges from 1.3 (for nearly globular hydrated proteins) to 2.0 (for very elongated or glycosylated proteins) (62).
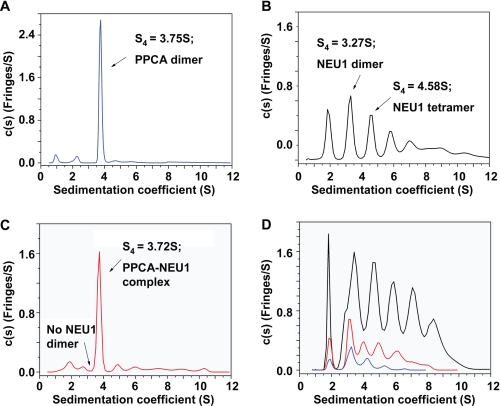
We first assessed the sedimentation properties of precursor PPCA and NEU1 separately and then in a 1:1 mixture at pH 5. The sedimentation profile of PPCA showed a single major peak, indicating that the protein exists as a homodimer in solution (Fig. 6A). In contrast, NEU1 showed a sedimentation profile consisting of as many as seven separate peaks, each representing different-sized species with sedimentation values at 4 °C (s4 values) ranging from 1.5 to 11 S (Fig. 6B). These peaks appeared to be different forms of self-association, i.e. the first peak represents a monomer, and the last, an oligomer containing at least 10 monomers. Furthermore, NEU1 (0.45–1.8 mg/ml) at pH 7 was also distributed among 5 or 6 major overlapping peaks (s4 value range, 1.50–10 S) (Fig. 6D). At lower protein concentration (0.15 mg/ml), only the 4 lower-order species remained. This finding indicates that NEU1 self-associates into oligomers of increasing mass (length) in a reversible, concentration-dependent manner. The best-fit (f/f0)w value decreased slightly with decreasing NEU1 concentration and coincided with lower oligomeric complexity, indicating that the different-sized oligomers dynamically interconverted under thermodynamic control. However, the data did not allow the assignment of accurate molar masses (association states) to each peak.
FIGURE 6.
Sedimentation properties of PPCA, NEU1, and a complex of PPCA and NEU1. c(s) profiles of PPCA (A), NEU1 (B), the PPCA-NEU1 complex (C), and NEU1 at various concentrations (D), i.e. 1.8 (black line), 0.45 (red line), and 0.15 mg/ml (blue line) were obtained. Sedimentation-velocity profiles (fringe displacement) were fitted to a continuous sedimentation coefficient distribution model c(s). Experiments were conducted in 50 mm sodium acetate, pH 5, 100 mm NaCl at 4 °C and at a rotor speed of 50,000 rpm. The best-fit weight-average frictional ratio (f/f0)w was determined from the c(s) analysis, and a single value was assumed. Total loading concentrations for NEU1, PPCA, and the PPCA-NEU1 mixture were 0.58, 0.22, and 0.25 mg/ml, respectively.
Using the same method, we tested the protein binding in the 1:1 mixture of NEU1 and PPCA at pH 5. Remarkably, the mixture of the two proteins resulted in very homogeneous sedimentation profile, compared with that of NEU1 alone. NEU1 oligomers were absent, and only 1 main peak was detected with an s4 value of 3.72 S, corresponding to a molar mass of 104 kDa (Fig. 6C). Strikingly, the 3.27-S NEU1 dimeric species present in the NEU1-alone profile was absent in the PPCA-NEU1 mixture and was presumably consumed in a new PPCA-NEU1 heterodimeric complex, with a predicted molar mass of 102.9 kDa. The values of s4 and mass of the PPCA homodimer were similar to those of the PPCA-NEU1 heterodimer, suggesting that the 104-kDa species in the PPCA-NEU1 profile could be a PPCA homodimer. However, this possibility would not account for the apparent loss of oligomeric NEU1.
To ascertain which proteins are contained in the 104-kDa peak, we integrated the concentration (in fringes) of the material under the specified peaks for PPCA-alone and PPCA-NEU1 mixtures. When compared with the quantified input of the proteins, the 104-kDa species could only be the PPCA-NEU1 heterodimer in a 1:1 ratio. Mass spectrometric analysis of the 104-kDa species showed that PPCA and NEU1 were present in equal molarity (not shown), which is consistent with a PPCA-NEU1 heterodimer. The drastically changed size distribution profile of NEU1 upon the addition of PPCA suggests that NEU1 has higher affinity for PPCA than for other NEU1 molecules. In agreement with the results from the SPR peptide screen, the different association states of NEU1 suggest that this protein contains two different binding sites that can bind to other NEU1 molecules. This is also supported by the analysis of the contour plots of NEU1 (pH 7; 1.80 mg/ml), which identified two NEU1 homodimers of different shapes, one extended (s4 = 3.05 S; f/f0 1.46; molecular mass = 94 kDa) and the other more globular (s4 = 3.65 S; f/f0 = 1.25; molecular mass = 96 kDa).
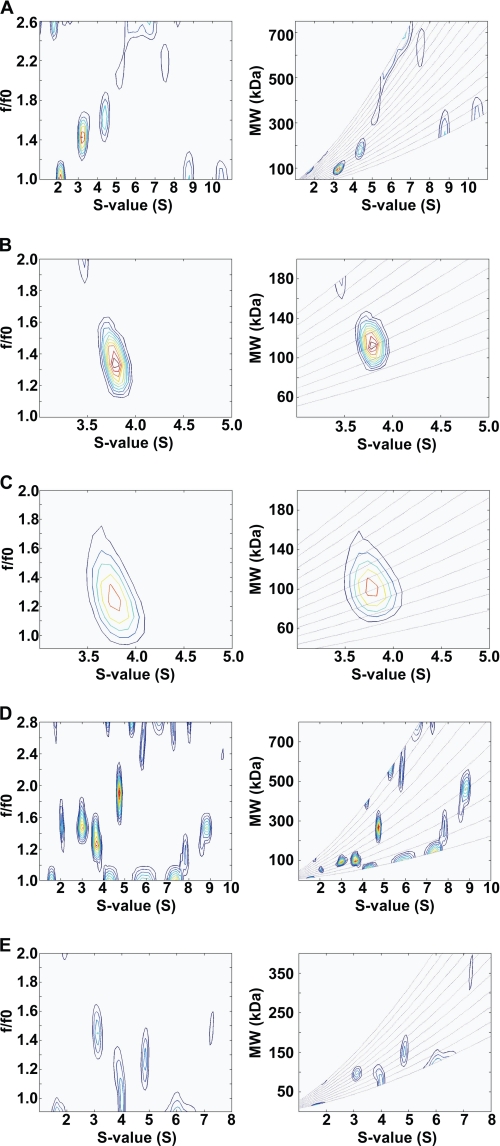
To determine the molar masses of multiple and single species in solution, we calculated a more reliable two-dimensional size-and-shape distribution, c(s,f/f0) (with one dimension being the s distribution and the other being the f/f0 distribution) with the velocity data of NEU1, PPCA, and the mixture of NEU1 and PPCA (Fig. 7, A–E). The best-fit values and estimates of the c(s,f/f0) analyses are shown in Table 1. Contour plots of s versus f/f0 and s versus Mr showed that at pH 5, PPCA (Fig. 7B) and the PPCA-NEU1 complex (Fig. 7C) resolved in single major peaks, with an identical s4 value of 3.77 S. This analysis also revealed that the mass value for the PPCA homodimer was 110 kDa and that of the PPCA-NEU1 heterodimer was 100 kDa; both values are in agreement with their predicted molar masses of 110.8 and 102.9 kDa, respectively (Table 1). Similar results were obtained at pH 7 (not shown). We observed that the molecular shape of the PPCA homodimer was more extended (f/f0 = 1.34) than that of the PPCA-NEU1 heterodimer (f/f0 = 1.24).
FIGURE 7.
Molar masses of PPCA, NEU1, and a complex of PPCA and NEU1. Velocity data for NEU1 at pH 5 (A), PPCA at pH 5 (B), the PPCA-NEU1 mixture at pH 5 (C), NEU1 at pH 7 and 1.8 mg/ml (D), and NEU1 at pH 7 and 0.45 mg/ml (E) were fitted to a two-dimensional size-and-shape distribution model, (c(s,f/f0)). Contour plots, frictional ratio versus sedimentation coefficient (left panels), and molecular weight versus sedimentation coefficient (lines represent constant f/f0 values) (right panels) were generated from the c(s,f/f0) analysis. The differently colored contours represent c(s,f/f0) values from 0.0 fringes/S (white) to maximum-value fringes/S (red), with increasing color temperature indicating higher values.
TABLE 1.
Best-fit values and estimates of the c(s,f/f0) analyses of NEU1, PPCA, and the PPCA-NEU1 heterodimeric complex
| Sample (mg/ml) | pH | s4a (Svedberg) | s20,w0b (Svedberg) | f/f0c (s20,w0) | Mrd | Protein complex |
|---|---|---|---|---|---|---|
| kDa | ||||||
| NEU1 (0.25) | 5 | 3.20 (0.11) | 5.15 | 1.43 (1.43) | 93 (95) | Dimer |
| 4.40 (0.08) | 7.08 | 1.65 (1.65) | 185 (190) | Tetramer | ||
| PPCA (0.22) | 5 | 3.77 (0.16) | 6.07 | 1.34 (1.30) | 110 (111) | Dimer |
| PPCA-NEU1 (0.25) | 5 | 3.77 (0.147) | 6.07 | 1.24 (1.28) | 100 (103) | Heterodimer |
| NEU1 (0.45) | 7 | 3.05 (0.10) | 4.94 | 1.46 (1.49) | 94 (95) | Dimer |
| 3.65 (0.07) | 5.91 | 1.25 (1.25) | 96 (95) | Dimer | ||
| 4.72 (0.07) | 7.77 | 1.90 (2.00) | 270 (285) | Hexamer |
a Sedimentation coefficient at 4 °C (s4) taken from the ordinate maximum of each peak in the best-fit c(s,f/f0) distribution. The relative amount of each species is given in parenthesis.
b Converted s values of the best-fit c(s,f/f0) distribution data to s20,w0 values. Standard conditions: 20 °C, water as the solvent, and zero protein concentration were used.
c Frictional ratio (f/f0) obtained from the calculated c(s,f/f0) distribution, and the ratio was calculated with SENTERP.
d Molar mass values were obtained from the calculated c(s,f/f0) distribution and transformed to the c(s,Mr) distribution (41).
Structural Models of the NEU1 Oligomer and the PPCA-NEU1 Heterodimeric Complex
The SPR data showed that two of the 5 NEU1 peptides that had the highest affinity for binding to PPCA also bound NEU1. Thus, PPCA and NEU1 compete for the same binding site(s). This is also supported by the AUC results. Furthermore, because NEU1 contains two binding sites that can bind to other NEU1 monomers, it preferentially forms oligomers in the absence of PPCA. Theoretically, both ends of the oligomers are accessible to other NEU1 molecules, which potentially allows for unlimited growth. Using the empiric model of the NEU1 monomer, we have generated a new model of NEU1 oligomers that closely matches the analytical data. We first determined that the two binding sites on the surface of NEU1 are not opposite to each other but rather are at an angle of less than 90°. Therefore, the oligomers are predicted to form a spiral-like structure (Fig. 8A). We then translated this hypothetical spiral packing of the NEU1 oligomer into a structural model without clashes or overlaps (Fig. 8B). Electron microscopic analysis of purified NEU1 protein gave validity to this model by showing elongated chains of as many as 10 NEU1 monomers (Fig. 8D). Electron microscopy of PPCA alone showed no chain-like structures, whereas the addition of PPCA to NEU1 led to a near-complete disappearance of the chain-like NEU1 structures (data not shown).
FIGURE 8.
Mechanism of NEU1 oligomerization. A, each NEU1 monomer (red circles) has two binding sites on its surface (1 and 2). Monomers bind to each other and form a spiral chain that varies in length. PPCA homodimers (green spheres) compete for binding to NEU1-binding sites 1 or 2 at the ends of the NEU1 chain. B, this results in the formation of PPCA-NEU1 heterodimers and a shorter NEU1 oligomeric chain. C, a structural model of the NEU1 oligomer. Peptide 1 (cyan) of the first molecule interacts with peptide 2 (dark blue) of the second molecule. A free peptide 1 of the second molecule interacts with a free peptide 2 of the third molecule and so on. The distance between the tips of these peptides and the center of the molecule is 25–26 Å, and they are oriented at roughly 70° in relation to each other. D, electron micrograph of purified BV-NEU1 protein confirms the spiral chain structure of the NEU1 oligomers. Scale bar, 7 nm.
To ascertain whether the synthetic PPCA- and NEU1-interacting NEU1 peptides were solvent-accessible, we located their positions in an empiric structural model of NEU1. This model was based on the structure of M. viridifaciens sialidase, which is predicted to share a high degree of structural similarity with the mammalian enzyme (4, 45). An initial model of the NEU1 monomer was obtained using the fold-recognition server LOOPP (43), with the bacterial sialidase (45) as a template. The model was then converted into an atomic model of NEU1 using the program MODELLER (47). On this model, at least a portion of each peptide is exposed to the surface, indicating that the peptides are potentially in position to bind PPCA or NEU1 (Fig. 9A). Interestingly, the peptides appear to form two attachment bands on the surface; these include peptides 33, 34, 42, 48, and 70 and peptides 13 and 19 (Fig. 9B).
FIGURE 9.
Structural models of PPCA-NEU1 heterodimers. A, structural model of NEU1 based on the crystal structure of M. viridifaciens sialidase. Peptides that showed positive binding to PPCA and/or NEU1 in the SPR-peptide screens are indicated in blue or red. B, a portion of each peptide is exposed to the surface, indicating that they are potentially in position to bind PPCA or NEU1. The peptides form two attachment bands on the surface; peptides 33, 34, 42, 48, and 70 (blue) form one band, and peptides 13 and 19 (red) form the other. C, structural model 1 of the PPCA-NEU1 heterodimer; NEU1 peptides 33, 34, and 42 (blue) interact with the PPCA excision peptide, and NEU1 peptide 48 (blue) interacts with the PPCA peptide 65 (red). D, structural model 2 of the PPCA-NEU1 heterodimer; NEU1 peptides 13 (red) and 70 (blue) interact with the PPCA peptide 74 (red) and the PPCA residue Pro-451.
NEU1 peptides 33, 34, and 42 bind only to precursor PPCA, whereas NEU1 peptides 13 and 48 bind only to mature PPCA. This suggests that peptides 33, 34, and 42 should be placed in close proximity to the excision peptide of PPCA in a hypothetical model of the PPCA-NEU1 heterodimer.
Initial three-dimensional visualization of the contacts between the two proteins, removing clashes and optimization of packing with the computer program COOT (49), resulted in several potential models of the PPCA-NEU1 heterodimer. Further selections were made using the information on the interacting NEU1 peptides and the PPCA variants identified above. The position of the binding peptides at the surface of the NEU1 model suggests that there are potentially two contact sites between PPCA and NEU1. Therefore, two models were finally selected. The first model shows peptides 33, 34, and 42 interacting with the PPCA excision peptide and peptide 48 interacting with the PPCA peptide 65 (Fig. 9C). The second model is based on contacts between the NEU1 peptides 13 and 70 with the PPCA peptide 74 and the Pro-451 residue on PPCA, which was the only residue that consistently interfered with PPCA-NEU1 binding in the mutation studies (Fig. 9D). The 2 models were within the top 10 of the most probable structures after running the Rosetta docking server (52). The AUC analysis of the PPCA-NEU1 mixture suggests the existence of only one PPCA-NEU1 heterodimeric complex. However, because it is uncertain whether AUC is sensitive enough to discriminate between the two heterodimeric models, either or both heterodimeric forms may coexist physiologically.
DISCUSSION
Using purified proteins in a cell-free system, we showed that precursor PPCA and mature, active PPCA are both capable of activating NEU1 in a concentration-dependent manner. In the absence of PPCA, NEU1 bound to β-galactosidase; however, in this case the two enzymes did not form a high Mr complex (57). NEU1 also bound to a deletion mutant of PPCA that corresponded to the 20-kDa C-terminal part of PPCA; yet again, no high Mr complex was formed. In contrast, another PPCA deletion mutant corresponding to the 32-kDa N-terminal portion of the protein failed to bind NEU1 (57). Thus, the C-terminal portion of PPCA, which includes the Pro-451 residue that is important for the interaction with NEU1, appeared to contain an important NEU1 binding domain, and both the 20- and 32-kDa PPCA subunits are essential for the formation of a high Mr complex.
Our SPR peptide-scanning and AUC experiments revealed a new and complex mechanism in which PPCA and NEU1 compete for the same binding sites on other NEU1 molecules. Differences between the precursor and mature forms of PPCA toward binding NEU1 suggest that stable binding already occurs en route to the lysosome but may conformationally change in the acidic environment of this organelle. There is evidence that NEU1 is present in the multienzyme complex in an oligomeric conformation while remaining associated with PPCA and that oligomerization is required for acquisition of NEU1 catalytic activity (57). Oligomerization or multimerization is not a common feature among the nonviral sialidases, but it is not unique either. A trans-sialidase from the protozoan parasite Trypanosoma cruzi is present in host organisms in two distinct catalytically active forms; that is, a monomeric form with low molecular mass and a multimeric form with a molecular mass greater than 400 kDa (63). Only the multimeric enzyme contains a large C-terminal domain that is essential for oligomerization. Interestingly, although NEU1 is homologous to this trans-sialidase, it lacks homology to the C-terminal oligomerization domain. Therefore, we conclude that there is no similarity between the mechanisms of oligomerization of these two enzymes.
Most lysosomal hydrolases are synthesized as zymogens or precursors with low catalytic activity that are proteolytically processed in the lysosome into a mature, fully active enzyme. In contrast, NEU1 does not get processed, which suggests that another mechanism may prevent its premature catalytic activation (4). The identification of binding sites on NEU1 that have affinity for both PPCA and other NEU1 molecules would support a model in which heterodimerization with PPCA is essential not only for the routing of the enzyme to the lysosome but also could prevent premature self-association and catalytic activation in an early biosynthetic compartment. However, the 1:1 heterodimeric conformation between PPCA and NEU1 may not be maintained in lysosomes, because the multienzyme complex appears to contain unequal amounts of the two enzymes; it contains all NEU1 activity but very little CA activity (64). Our structural models of the NEU1 oligomer and the PPCA-NEU1 heterodimeric complex suggest that in the high Mr multienzyme complex PPCA could bind to both ends of NEU1 oligomers. This mode of association could prevent the uncontrolled growth of NEU1 oligomers or the premature disassembly into inactive low molecular mass forms of the enzyme.
In closing, our study gives new insight into the unique interactions between the serine carboxypeptidase/chaperone PPCA and the lysosomal sialidase NEU1. We also provide plausible structural-interaction models of these two enzymes. Although our computer-generated structural models do not contradict the analytical and biochemical data, the precise orientation of the NEU1 monomers remains uncertain. A thorough crystallographic study will be necessary to confirm or reject these putative models.
Acknowledgments
We are grateful to Dr. K. G. Murti and Linda Mann for electron microscopy of NEU1 and to Dr. Angela McArthur for scientific editing of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM60905 and DK52025. This work was also supported by the Assisi Foundation of Memphis and the American Lebanese Syrian Associated Charities.
- NEU1
- lysosomal neuraminidase-1
- AUC
- analytical ultracentrifugation
- BV
- baculovirus
- GFP
- green fluorescent protein
- PPCA
- protective protein/cathepsin A
- SPR
- surface plasmon resonance
- GS
- galactosialidosis
- MES
- 4-morpholineethanesulfonic acid
- MSCV
- murine stem cell virus.
REFERENCES
- 1.Saito M., Yu R. K. (1995) in Biology of the Sialic Acids (Rosenberg A. ed) Plenum Press, New York [Google Scholar]
- 2.Comelli E. M., Amado M., Lustig S. R., Paulson J. C. (2003) Gene 321, 155–161 [DOI] [PubMed] [Google Scholar]
- 3.Monti E., Bassi M. T., Bresciani R., Civini S., Croci G. L., Papini N., Riboni M., Zanchetti G., Ballabio A., Preti A., Tettamanti G., Venerando B., Borsani G. (2004) Genomics 83, 445–453 [DOI] [PubMed] [Google Scholar]
- 4.Bonten E., van der Spoel A., Fornerod M., Grosveld G., d'Azzo A. (1996) Genes Dev. 10, 3156–3169 [DOI] [PubMed] [Google Scholar]
- 5.Rottier R. J., Bonten E., d'Azzo A. (1998) Hum. Mol. Genet. 7, 313–321 [DOI] [PubMed] [Google Scholar]
- 6.Seyrantepe V., Landry K., Trudel S., Hassan J. A., Morales C. R., Pshezhetsky A. V. (2004) J. Biol. Chem. 279, 37021–37029 [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi K., Hata K., Koseki K., Shiozaki K., Akita H., Wada T., Moriya S., Miyagi T. (2005) Biochem. J. 390, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ando K., Kikugawa K., Beppu M. (1996) J. Biochem. 119, 639–647 [DOI] [PubMed] [Google Scholar]
- 9.Domingues R. M., Cavalcanti S. M., Andrade A. F., Ferreira M. C. (1992) Zentralbl. Bakteriol. 277, 340–344 [DOI] [PubMed] [Google Scholar]
- 10.Fisher K. J., Aronson N. N., Jr. (1991) J. Biol. Chem. 266, 12105–12113 [PubMed] [Google Scholar]
- 11.Haddad E. B., Gies J. P. (1992) Eur. J. Pharmacol. 211, 273–276 [DOI] [PubMed] [Google Scholar]
- 12.Hechler V., Mersel M., Dreyfus H., Maitre M. (1990) Mol. Cell. Biochem. 93, 87–94 [DOI] [PubMed] [Google Scholar]
- 13.Igarashi M., Bando Y. (1990) J. Immunol. Methods 135, 91–93 [DOI] [PubMed] [Google Scholar]
- 14.Karasuno T., Kanayama Y., Nishiura T., Nakao H., Yonezawa T., Tarui S. (1992) Eur. J. Immunol. 22, 2003–2008 [DOI] [PubMed] [Google Scholar]
- 15.Nakamura M., Kirito K., Yamanoi J., Wainai T., Nojiri H., Saito M. (1991) Cancer Res. 51, 1940–1945 [PubMed] [Google Scholar]
- 16.Nilsson J., Ksiazek T., Thyberg J. (1982) Exp. Cell Res. 142, 333–339 [DOI] [PubMed] [Google Scholar]
- 17.Ohyashiki T., Taka M., Mohri T. (1987) Biochim. Biophys. Acta 905, 57–64 [DOI] [PubMed] [Google Scholar]
- 18.Puri A., Grimaldi S., Blumenthal R. (1992) Biochemistry 31, 10108–10113 [DOI] [PubMed] [Google Scholar]
- 19.Sato K., Miyagi T. (1996) Biochem. Biophys. Res. Commun. 221, 826–830 [DOI] [PubMed] [Google Scholar]
- 20.Stenberg P. E., Levin J., Baker G., Mok Y., Corash L. (1991) J. Cell. Physiol. 147, 7–16 [DOI] [PubMed] [Google Scholar]
- 21.Wu G., Ledeen R. W. (1991) J. Neurochem. 56, 95–104 [DOI] [PubMed] [Google Scholar]
- 22.d'Azzo A., Andria G., Strisciuglio P., Galjaard H. (2001) in (Scriver C., Beaudet A., Sly W., Valle D. eds) The Metabolic and Molecular Bases of Inherited Disease, 8th Ed., pp. 3811–3826, McGraw-Hill Publishing Co., New York [Google Scholar]
- 23.van der Spoel A., Bonten E., d'Azzo A. (1998) EMBO J. 17, 1588–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Spoel A., Bonten E., d'Azzo A. (2000) J. Biol. Chem. 275, 10035–10040 [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto Y., Nishimura K. (1987) Int. J. Biochem. 19, 435–442 [DOI] [PubMed] [Google Scholar]
- 26.Potier M., Michaud L., Tranchemontagne J., Thauvette L. (1990) Biochem. J. 267, 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheibe R., Hein K., Wenzel K. W. (1990) Biomed. Biochim. Acta 49, 547–556 [PubMed] [Google Scholar]
- 28.Thomas G. H. (2001) in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A. L., Sly W. S., Valle D. eds) 8th Ed., pp. 3507–3534, McGraw Hill, Inc., New York [Google Scholar]
- 29.Zhou X. Y., Morreau H., Rottier R., Davis D., Bonten E., Gillemans N., Wenger D., Grosveld F. G., Doherty P., Suzuki K., Grosveld G. C., d'Azzo A. (1995) Genes Dev. 9, 2623–2634 [DOI] [PubMed] [Google Scholar]
- 30.de Geest N., Bonten E., Mann L., de Sousa-Hitzler J., Hahn C., d'Azzo A. (2002) Hum. Mol. Genet 11, 1455–1464 [DOI] [PubMed] [Google Scholar]
- 31.Yogalingam G., Bonten E. J., van de Vlekkert D., Hu H., Moshiach S., Connell S. A., d'Azzo A. (2008) Dev. Cell. 15, 74–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonten E. J., Wang D., Toy J. N., Mann L., Mignardot A., Yogalingam G., d'Azzo A. (2004) FASEB J. 9, 971–973 [DOI] [PubMed] [Google Scholar]
- 33.Wang D., Bonten E. J., Yogalingam G., Mann L., d'Azzo A. (2005) Mol. Genet. Metab. 85, 181–189 [DOI] [PubMed] [Google Scholar]
- 34.Galjart N. J., Gillemans N., Harris A., van der Horst G. T., Verheijen F. W., Galjaard H., d'Azzo A. (1988) Cell 54, 755–764 [DOI] [PubMed] [Google Scholar]
- 35.Wang D., Zaitsev S., Taylor G., d'Azzo A., Bonten E. J. (2009) Biochim. Biophys. Acta 1790, 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galjaard H. (1979) Ann. Clin. Biochem. 16, 343–353 [DOI] [PubMed] [Google Scholar]
- 37.van Dongen J. M., Willemsen R., Ginns E. I., Sips H. J., Tager J. M., Barranger J. A., Reuser A. J. (1985) Eur. J. Cell Biol. 39, 179–189 [PubMed] [Google Scholar]
- 38.Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S., Rowe A., Horton J. eds) Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 39.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., Schubert D. (2002) Biophys. J. 82, 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown P. H., Schuck P. (2006) Biophys. J. 90, 4651–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 43.Teodorescu O., Galor T., Pillardy J., Elber R. (2004) Proteins 54, 41–48 [DOI] [PubMed] [Google Scholar]
- 44.Meller J., Elber R. (2001) Proteins 45, 241–261 [DOI] [PubMed] [Google Scholar]
- 45.Gaskell A., Crennell S., Taylor G. (1995) Structure 3, 1197–1205 [DOI] [PubMed] [Google Scholar]
- 46.Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 47.Eswar N., Eramian D., Webb B., Shen M. Y., Sali A. (2008) Methods Mol. Biol. 426, 145–159 [DOI] [PubMed] [Google Scholar]
- 48.Rudenko G., Bonten E., d'Azzo A., Hol W. G. J. (1995) Structure 3, 1249–1259 [DOI] [PubMed] [Google Scholar]
- 49.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 50.Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) Bioinformatics 20, 45–50 [DOI] [PubMed] [Google Scholar]
- 51.Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. (2003) J. Mol. Biol. 331, 281–299 [DOI] [PubMed] [Google Scholar]
- 52.Chaudhury S., Sircar A., Sivasubramanian A., Berrondo M., Gray J. J. (2007) Proteins 69, 793–800 [DOI] [PubMed] [Google Scholar]
- 53.Tovchigrechko A., Vakser I. A. (2005) Proteins 60, 296–301 [DOI] [PubMed] [Google Scholar]
- 54.Tovchigrechko A., Vakser I. A. (2006) Nucleic Acids Res. 34, W310–W314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rudenko G., Bonten E., Hol W. G., d'Azzo A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 621–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicholls A., Sharp K. A., Honig B. (1991) Proteins 11, 281–296 [DOI] [PubMed] [Google Scholar]
- 57.Bonten E. J., d'Azzo A. (2000) J. Biol. Chem. 275, 37657–37663 [DOI] [PubMed] [Google Scholar]
- 58.Laune D., Molina F., Ferrieres G., Mani J. C., Cohen P., Simon D., Bernardi T., Piechaczyk M., Pau B., Granier C. (1997) J. Biol. Chem. 272, 30937–30944 [DOI] [PubMed] [Google Scholar]
- 59.Johansson A., Sandström P., Ullén A., Behravan G., Erlandsson A., Levi M., Sundström B., Stigbrand T. (1999) Cancer Res. 59, 48–51 [PubMed] [Google Scholar]
- 60.Bonten E. J., Galjart N. J., Willemsen R., Usmany M., Vlak J. M., d'Azzo A. (1995) J. Biol. Chem. 270, 26441–26445 [DOI] [PubMed] [Google Scholar]
- 61.Harding S. E., Chowdhry B. Z. (2001) Protein-Ligand Interactions: Hydrodynamics and Calorimetry, Oxford University Press, Oxford [Google Scholar]
- 62.Brown P. H., Balbo A., Schuck P. (2008) Curr. Protoc. Immunol., pp. 18.15.1–18.15.39, John Wiley & Sons, Inc., Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 63.Schenkman S., Chaves L. B., Pontes de Carvalho L. C., Eichinger D. (1994) J. Biol. Chem. 269, 7970–7975 [PubMed] [Google Scholar]
- 64.Vinogradova M. V., Michaud L., Mezentsev A. V., Lukong K. E., El-Alfy M., Morales C. R., Potier M., Pshezhetsky A. V. (1998) Biochem. J. 330, 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]