Abstract
XplA is a cytochrome P450 of unique structural organization, consisting of a heme- domain that is C-terminally fused to its native flavodoxin redox partner. XplA, along with flavodoxin reductase XplB, has been shown to catalyze the breakdown of the nitramine explosive and pollutant hexahydro-1,3,5-trinitro-1,3,5-triazine (royal demolition explosive) by reductive denitration. The structure of the heme domain of XplA (XplA-heme) has been solved in two crystal forms: as a dimer in space group P21 to a resolution of 1.9 Å and as a monomer in space group P21212 to a resolution of 1.5 Å, with the ligand imidazole bound at the heme iron. Although it shares the overall fold of cytochromes P450 of known structure, XplA-heme is unusual in that the kinked I-helix that traverses the distal face of the heme is broken by Met-394 and Ala-395 in place of the well conserved Asp/Glu plus Thr/Ser, important in oxidative P450s for the scission of the dioxygen bond prior to substrate oxygenation. The heme environment of XplA-heme is hydrophobic, featuring a cluster of three methionines above the heme, including Met-394. Imidazole was observed bound to the heme iron and is in close proximity to the side chain of Gln-438, which is situated over the distal face of the heme. Imidazole is also hydrogen-bonded to a water molecule that sits in place of the threonine side-chain hydroxyl exemplified by Thr-252 in Cyt-P450cam. Both Gln-438 → Ala and Ala-395 → Thr mutants of XplA-heme displayed markedly reduced activity compared with the wild type for royal demolition explosive degradation when combined with surrogate electron donors.
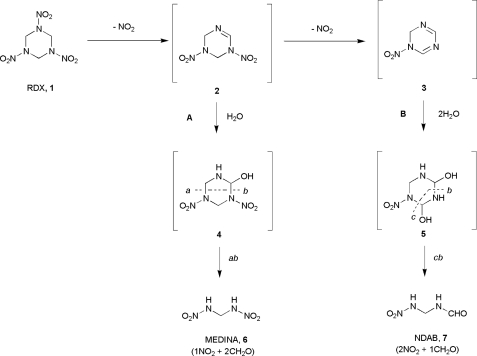
Royal demolition explosive (RDX)2 or cyclotrimethylenetrinitramine 1 (see Fig. 1) is a widely used explosive compound with both military and civil applications. The extensive global usage of RDX has resulted in concerns over environmental contamination, because it is both recalcitrant to degradation, leading to contamination in soil and ground water, and a potent convulsant and possible carcinogen. The bioremediation of RDX has thus been the focus of increasing research in recent years, with a number of bacterial strains reported to catalyze its degradation (1–3). Among these, Bruce and co-workers, using a selective enrichment technique, isolated Rhodococcus rhodochrous strain 11Y from an RDX-contaminated site, which was able to grow on RDX as the sole nitrogen source (4). The products of biotransformation of RDX by this bacterium were shown to be nitrite and formaldehyde. A gene cluster in strain 11Y essential for RDX degradation was identified and shown to contain a novel cytochrome P450, termed XplA, and a redox partner, XplB, which were shown together to be capable of catalyzing the biotransformation of RDX in vitro (5). Near identical xplA and xplB genes have now been identified in strains within the Actinomycetales isolated from geographically distinct sites (6). Interestingly, these genes have been found to be plasmid encoded providing compelling evidence for recent lateral gene transfer. In the interests of developing an efficient technology for the targeted phytoremediation of RDX, the XplA-XplB system was expressed in transgenic strains of Arabidopsis thaliana and shown to successfully remediate RDX from contaminated soil (5, 7).
FIGURE 1.
Biocatalyzed routes proposed for the degradation of the nitramine exposive RDX 1 by cytochrome P450 XplA from Rhodococcus rhodochrous 11Y. Pathway A corresponds to the route proposed under anaerobic conditions to give the cleavage product metabolite methylene dinitramine 6; pathway B is that postulated to occur under aerobic conditions to give cleavage product 4-nitro-2,4-diazabutanal 7.
RDX has an unusual chemical structure, featuring three nitramine (N-NO2) bonds on a saturated six-membered ring, a functionality that has few precedents among natural products (8). The products isolated from XplA-XplB-catalyzed degradation of RDX in vitro were shown to be different under aerobic and anaerobic conditions after initial denitration to imine intermediate 2 (Fig. 1) (5). Under anaerobic conditions A, 2 would undergo hydration to give intermediate 4. Ring cleavage would yield the isolable metabolite methylene dinitramine 6. The aerobic process B was thought to proceed via successive denitration of two nitramine groups, to give the di-imine 3, which would then be twice hydrated to give the unstable intermediate 5, which would undergo spontaneous ring cleavage to give the isolable product 4-nitro-2,4-diazabutanal 7, formaldehyde, and two equivalents of nitrite.
Although the reductive denitration of, for example, glycerol trinitrate (9) and other nitroheterocylic drugs (10), has previously been described for mammalian cytochromes P450, the best known reactions catalyzed by this family of enzymes include a wide range of oxidative chemical reactions, including hydroxylation, heteroatom dealkylation, and C–C bond cleavage reactions most commonly using molecular oxygen and involving cleavage of the O–O bond after dioxygen is bound by the heme-iron (11). A sequence alignment of some cyt-P450 for which the structures have been determined (Fig. 2) shows that the well conserved residues Glu/Asp-Thr/Ser that are thought to be a requirement for oxidative chemistry (12, 13) and to aid cleavage of the scissile O–O bond are not conserved in XplA, being replaced by methionine-394 and alanine-395. In the interests of illuminating the molecular mechanism and substrate specificity in XplA, we were thus interested in obtaining a structure of the heme domain of the enzyme. In this report, the structure of XplA-heme domain in two crystal forms is presented: the first with two molecules in the asymmetric unit, derived from the attempted crystallization of full-length XplA and the other derived from the subcloned, isolated XplA-heme domain, with one molecule in the asymmetric unit and the heme iron ligated to the substrate analogue imidazole. The results reveal a possible access channel for ligand transport and, in combination with mutational studies, demonstrate a highly unusual active site environment for substrate binding, suggesting substrate binding or catalytic roles for Gln-438 and Ala-395.
FIGURE 2.
Amino acid sequence alignment for portions of selected CYP heme domains for which the structures have been solved. P450_pikC (from Streptomyces venezuelae; Uniprot accession no. O87605); P450_cin (from Citrobacter braakii; Q8VQ86); P450_eryF (Saccharopolyspora erythrea; Q00441); P450_moxa (Nonomuraea recticatena; Q2L6S8); P450_staP (Streptomyces sp. TP-A0274; Q83WG3); P450_BioI (B. subtilis; P53554); P450_epok (Sorangium cellulosum; Q9KIZ4); P450_terp (Pseudomonas sp.; P33006); P450_xpla (Rhodococcus sp. 11Y; Q8GPH7); P450_cam (Pseudomonas putida; P00183); and P450_BM3 (Bacillus megaterium; P14779). Each catalyzes oxidative chemistry and most apart from P450_cin contain the (E/D)(S/T) dyad (shown in red) demonstrated to be important in the scission of the dioxygen bond as a prerequisite of substrate mono-oxygenation. XplA possesses Met-394 and Ala-395 (shown in green).
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of XplA-heme
The XplA-heme coding sequence, previously cloned in the pET-16b plasmid as part of full-length XplA (5), was used as template for the sub-cloning of the XplA-heme domain. The PCR amplification of the insert was performed in a 50-μl reaction containing Phusion DNA polymerase (New England BioLabs) with a PCR cycle of 94 °C for 30 s and 35 cycles of 94 °C for 10 s, 65 °C for 30 s, and 72 °C for 1 min. The following oligonucleotides were used for the PCR: forward (5′-CCAGGGACCAGCAATGCGAGGAGGACATGAGATGACCGCTG-3′) and reverse (5′-GAGGAGAAGGCGCGTCAGGACAGGACGATCGGCAGTTTTCG-3′). The PCR product was subcloned into the pET-YSBLIC 3C vector using procedures described previously (14). Full-length XplA was expressed and purified as described previously (5). XplA-heme was expressed in Escherichia coli Rosetta 2 cells (Novagen/Merck). Single colonies were used to inoculate 1-liter cultures of Luria-Bertani broth containing the antibiotics kanamycin and chloramphenicol at concentrations of 100 μg μl−1, respectively. When the optical density (measured at 600 nm) reached 0.8, XplA-heme expression was induced with the addition of 1 mm isopropyl-β-d-thiogalactopyranoside, 1 mm 5-aminolevulinic acid, and 0.5 mm FeCl3. After overnight incubation at 20 °C, cells were harvested by centrifugation, resuspended in 50 mm phosphate buffer, pH 8, containing 300 mm sodium chloride (“buffer”), and disrupted using ultrasonication. The cell debris was removed by centrifugation, and the crude supernatant containing XplA-heme was filtered, before loading onto a 5-ml nickel-agarose affinity column (Amersham Biosciences). The N-terminally His-tagged protein was eluted with an increasing gradient (30–500 mm) of imidazole in buffer, and fractions were analyzed by SDS-PAGE. Fractions containing XplA-heme were pooled, concentrated, and then applied to a SuperdexTM 200 column (Amersham Biosciences) gel-filtration column (equilibrated in 50 mm phosphate buffer, pH 8, containing 300 mm sodium chloride) to yield protein for crystallization and solution assays. Before crystallization, the hexahistidine tag was cleaved from the XplA heme domain using protease 3C (Novagen) using a procedure described previously (15).
Crystallization
Both the full-length XplA and XplA-heme were concentrated to 26 mg ml−1 prior to beginning crystallization trials. Initial screening was carried out using the Mosquito® Crystal robot (TTP LabTech) and commercial crystallization screens from Hampton Research. These trials used 150 nl of protein plus 150 nl of reservoir solution in the crystallization drop. Hits were optimized and scaled up using 24-well Linbro dishes with drops consisting of 1 μl of protein plus 1 μl of reservoir solution, using either the sitting drop technique (full-length XplA) or the hanging drop, vapor-diffusion method (XplA-heme). For full-length XplA, the best crystals were obtained using 0.2 m Bis-Tris propane, pH 6.5, 22% (w/v) polyethylene glycol (Mr 3350) and 0.02 m potassium phosphate. The optimum conditions for XplA-heme crystallization were 0.2 m Bis-Tris propane, pH 6.5, 0.2 m magnesium formate and 20% (w/v) polyethylene glycol (Mr 3350).
Data Collection, Structure Solution, and Refinement
Crystals of both full-length XplA and XplA-heme were transferred into a cryoprotectant solution of mother liquor with 20% ethylene glycol before flash-cooling in liquid nitrogen and stored for data collection. X-ray diffraction data for the crystals derived from full-length XplA were collected on beamline BM-14 at European Synchrotron Radiation Facility (Grenoble, France) at a wavelength of 0.95350 Å. XplA-heme crystals were collected on beamline ID-23-EH2 at the European Synchrotron Radiation Facility at a wavelength of 0.87260 Å (Table 1). In each case, data were integrated and scaled using the HKL2000 suite of programs (16). The structure was solved initially using data from the heme domain derived from full-length XplA (Table 1) using MOLREP (17) using one monomer of the coordinates from pdb entry 2z3t (cytochrome P450 StaP) as a search model, after editing that pdb to remove the heme and converting to a model cut-back to common C-β side chain atoms using CHAINSAW in the ccp4 suite of programs (18). After ten rounds of refinement using the default parameters in REFMAC (19) the difference map clearly showed the presence of a heme ligand and well resolved density for the secondary elements on the proximal face of the heme. The rest of the structure was then built using successive rounds of BUCCANEER (20) and ARP/wARP (21). Model building was then carried out using Coot (22) and refined using REFMAC. Details of data collection and refinement are given in Table 1. The structure of the heme domain of XplA derived from the full-length protein was refined to an Rcryst value of 0.167 and an Rfree value of 0.214. The structure of XplA-heme with imidazole as the ligand was refined to an Rcryst value of 0.147 and an Rfree value of 0.176. The final structures were validated using PROCHECK (23). The coordinates for XplA-heme in P21 and P21212 space groups have been deposited in the Protein Data bank under the accession numbers 2wiv and 2wiy, respectively.
TABLE 1.
Data collection and refinement statistics for XplA-heme bound to imidazole
Values in parentheses refer to the highest resolution shell. Rsym = ΣhΣl|Ihl − 〈Ih〉|/ΣhΣl〈Ih〉, where Il is the lth observation of reflection h and 〈Ih〉 is the weighted average intensity for all observations l of reflection h. 〈I/σ(I)〉 indicates the average of the intensity divided by its standard deviation.
| Xpla-heme derived from full-length XplA, P21 | XplA-heme-imidazole complex, P21212 | |
|---|---|---|
| Beamline | BM-14 (ESRF, Grenoble) | ID-23-EH2 (ESRF, Grenoble) |
| Wavelength (Å) | 0.95350 Å | 0.87260 Å |
| Resolution (Å) | 63.89-1.90 (1.95-1.90) | 124.03-1.49 (1.53-1.49) |
| Space Group | P21 | P21212 |
| Unit cell (Å) | a = 54.1, b = 123.0, c = 64.1 | a = 124.3, b = 64.7, c = 54.0 |
| Molecules in asymmetric units | 2 | 1 |
| Unique reflections | 57,099 (1926) | 68,213 (4663) |
| Completeness (%) | 91.5 (41.9) | 99.6 (94.3) |
| Rsym (%) | 0.057 (0.354) | 0.061 (0.191) |
| Multiplicity | 1.8 (1.2) | 7.4 (7.1) |
| 〈I/σ(I)〉 | 10 (6) | 13 (10) |
| Protein atoms | 6,153 | 3,093 |
| Solvent waters | 485 | 709 |
| Rcryst/Rfree | 0.167/0.214 | 0.147/0.176 |
| r.m.s.d. 1–2 bonds (Å) | 0.025 | 0.012 |
| r.m.s.d. 1–3 angles (°) | 1.917 | 1.411 |
| Chiral centre (Å3) | 0.134 | 0.086 |
| Planarity | 0.011 | 0.015 |
| Average main-chain B (Å2) | 24 | 8 |
| Average side-chain B (Å2) | 27 | 12 |
| Average solvent B (Å2) | 32 | 29 |
| Average ligand B (imidazole) | 19 |
Mutagenesis of XplA Heme Domain, Spectral Analysis, and Activity Assays
Five mutants of XplA-heme were generated using the Stratagene QuikChangeTM kit using the wild-type sequence, previously cloned in the pET-YSBLIC 3C plasmid, as template and PCR primers as detailed in Table 2. Each mutant proved to be soluble and was expressed in acceptable yields comparable to that of the isolated XplA-heme domain. CO difference spectra were recorded in each case for the mutants and showed the expected spectral shift to 450 nm upon reduction with dithionite followed by bubbling with carbon monoxide (data not shown). Spectral analyses were performed on a Varian-Cary 50 spectrophotometer scanning between 300 and 600 nm. The data were processed with the Enzyme Kinetics software from SigmaPlot. XplA-heme activity was assayed in 0.5-ml reactions containing 50 mm potassium phosphate, pH 7, 500 μm NADPH, 0.02 unit of spinach ferredoxin reductase (Sigma), 20 μg of spinach ferredoxin (Sigma), different concentrations of heme (wild type, 2.5 nm; M322L, 2.5 nm; M394L, 10 nm; M318L, 20 nm; Q438A, 100 nm; and A395T, 500 nm) and different concentrations of RDX. All reactions were performed aerobically at room temperature (20 °C), and assays were stopped by the addition of 10% trichloroacetic acid. RDX degradation was measured by high-performance liquid chromatography as previously described (5).
TABLE 2.
PCR primers used for construction of XplA-heme mutants
| Mutant | Forward primer |
|---|---|
| A395T | TGGGGCACATGACCATCGGGTACCTGATCGCCTCG |
| Q438A | TGGACCCGCCAGCACTGTCTTTCCTGCGGTTTCCTACC |
| M322L | ATGTTCGAGGCGTTGCTGATGCAAAGCGCGGAGC |
| M318L | TGATGGAGGCGTTGTTCGAGGCGATGCTGATGCAAAGC |
| M394L | CCGTGGGGCACTTGGCCATCGGGTACCTGATCG |
| Mutant | Reverse primer |
|---|---|
| A395T | TACCCGATGGTCATGTGCCCCACGGCATAGAACACC |
| Q438A | CAGGAAAGACAGTGCTGGCGGGTCCATCCGGACCATCTCG |
| M322L | TTGCATCAGCAACGCCTCGAACATCGCCTCCATCACG |
| M318L | ATCGCCTCGAACAACGCCTCCATCACGGCGTCGGCATC |
| M394L | ACCCGATGGCCAAGTGCCCCACGGCATAGAACAC |
RESULTS
Cloning, Expression, and Crystallization of the Heme Domain of XplA
The gene encoding the full-length cytochrome-P450 XplA from R. rhodochrous 11Y, incorporating both flavodoxin and heme-containing domains, had previously been cloned and expressed in E. coli (5). Initial crystallization trials with full-length XplA were successful, and datasets were collected on these crystals, which diffracted routinely to a resolution of ∼2 Å. The structure was solved as described under “Experimental Procedures,” but it was obvious that the solution did not contain the flavodoxin domain of XplA, this having been apparently cleaved during the crystallization process, as confirmed by mass spectrometric analysis of the protein crystals (data not shown). To obtain a more reliable and reproducible system for XplA-heme crystallization, it was decided to sub-clone the heme domain of XplA, choosing as the target a gene sequence encoding amino acids from Arg-154 to Ser-552. Sub-cloning into the pET-YSBLIC vector (14) was employed to introduce an N-terminal hexahistidine tag to facilitate enzyme purification. After expression and purification, it was found that the His-tagged protein gave crystals that diffracted only to a resolution of ∼6 Å. The histidine tag was cleaved for subsequent experiments therefore, leaving an N-terminal residue of three amino acids (in bold) and a protein with the N-terminal sequence GPA-154RGGHEMTA161. This protein was found routinely to give crystals that diffracted beyond 1.5 Å. In the structural solutions obtained from these crystals, clear electron density, including side chains, was observed for residues from Thr-160 to the C terminus Ser-552.
Overall Fold of XplA-heme
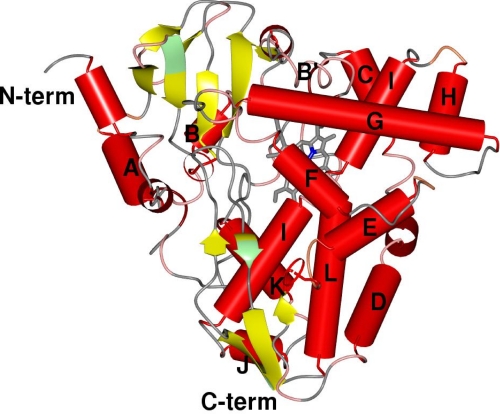
XplA-heme shares amino acid sequence identity of 26%, at most, with other members of the CYP family for which the structures have been determined. Its closest sequence homolog in the SwissProt data base (24), not including the almost identical XplA homologs identified in other species of Rhodococcus isolated from environments worldwide (25), is the CYP from Erwinia carotovora var. atrospetica (Uniprot ID Q6D569; sequence ID 41%). The next best matches have only 30% sequence ID and include CYPs from Streptomyces ambofaciens ATCC 23877 (A0ACI5) and Streptomyces clavuligerus ATCC 27004 (B5GTE2). Data collected on the heme domain grown from full-length XplA were superseded by data collected later on the subcloned XplA-heme domain, so the structural descriptions below are all based on the superior data. The heme domain of cytochrome P450 XplA possesses the canonical cytochrome P450 fold, being a prism structure of dimensions 50 × 50 × 60 Å and a height of 35 Å from the base to the vertex of the G helix. The descriptions below employ secondary structural designations described in previous studies (11, 26) as a guide, with α-helices lettered A–L and β-strands numbered consecutively, but forming part of four β-sheet structures, numbered as for the model of P450cam in a previous study (27). The major structural elements as described for P450cam (CYP101) (27) and typifying the fold, are conserved, including the two-domain structure with the larger domain being predominantly composed of α-helices, including the long I helix which traverses the distal face of the heme (Fig. 3). A similarity search using the DALI server (www2.ebi.ac.uk/dali/) revealed that the closest structural homologues to XplA are the cytP450 BioI (CYP107H1) from Bacillus subtilis (Z-score 45.0; r.m.s.d. 2.2; sequence ID 26%) (28) and P450StaP (CYP245A1) from Streptomyces sp. TP-A0274 (Z-score, 44.7; r.m.s.d., 2.6; and sequence ID, 26%) (29), the latter having been identified by the program BALBES (30) as the most suitable search model for molecular replacement in the original solution (see “Experimental Procedures”). The structure of CYPs StaP and XplA-heme diverge most notably in the sequence between Arg-67 (StaP)/Val-220 (XplA) and Val-93 (StaP)/Val-232 (XplA), which in XplA constitute a condensed helical loop between the B and C helices. In CYP StaP, the helical loop is expanded into two larger loops that protrude from the enzyme surface. One of these, from Arg-67 to Val-81, was proposed to be part of a flexible substrate entry site for the substrate chromopyrrolic acid, a molecule of which was bound at this region (29). The marked divergence of structure may account for the output derived from the molecular replacement solution (see “Experimental Procedures”), in which the protein chains constituting the proximal heme portion of the protein, which overlap very well, were described in the solution, yet the distal secondary elements were not, and hence required tracing using BUCCANEER (20). Other notable features of XplA-heme, which diverged from other structurally characterized P450s, are in the d-helix between Asp-262 and Arg-266 leading to a distortion in the middle of that helix and also, significantly in the I helix, which is kinked in the center in the region above the heme between Phe-388 and Met-394 (Fig. 3).
FIGURE 3.
Topology of XplA-heme (P21212; 2wiy) illustrating the canonical CYP fold from reference 11. In addition to the well conserved secondary elements, XplA-heme possesses an additional short helix near the N terminus, a kinked D-helix, and a significant disruption to the I-helix that traverses the distal face of the heme. The break in helix I is occupied by four residues, including Met-394 and Ala-395.
Active Site and Heme Environment
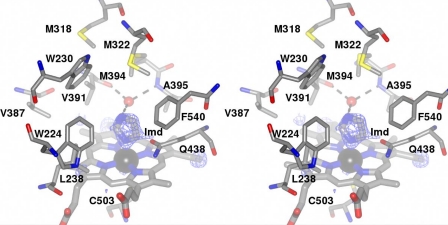
The heme cofactor of XplA-heme is located within the active site, the iron being coordinated in the 5-position by the highly conserved Cys-503 through the side-chain thiolate. The heme propionates are secured by interactions with the side chains of Arg-443 and His-245 and His-501, each of which is conserved in CYP-StaP. The heme itself appears to exist in two distinct conformations as previously observed in the atomic resolution structure of CYP121 from M. tuberculosis (31), evidenced by persistent electron density in the difference map adjacent to the methyl groups of the pyrrole rings bearing the vinyl groups, when refined in either of two possible orientations. On the distal face of the heme iron, occupying the sixth coordination position, clear density was observed in the difference map for the ligand imidazole, which had presumably persisted from the nickel affinity column buffers used during purification of XplA-heme (Fig. 4).
FIGURE 4.
Stereoview of the environment surrounding the heme in XplA-heme. The ceiling of the heme binding site is notably hydrophobic and features an unusual cluster of methionine residues, Met-318, Met-322, and Met-394, the latter held over the heme with the tip of the side chain at a distance of 8.2 Å from the iron. Ligand electron density observed at the distal face of the heme revealed the presence of imidazole. The density corresponds to the omit map (Fo − Fc map contoured at a level of 3.2ς) obtained after structure solution using the native enzyme as a search model in MOLREP (17). H-bonding interactions between the imidazole ligand and the water molecule (shown as a red ball) that occupies the cavity between the peptidic N–H of Ala-395 and the peptidic carbonyl group of Val-391 are shown as dashed black lines. The side chain of Gln-438, hanging over the heme and at a distance of ∼3.5 Å from the imidazole ligand, can also be seen.
The ceiling of the heme-binding pocket is hydrophobic, consisting of Val-387, Val-391, Ala-395, Pro-437 (interrupted by Gln-438), Leu-238, Trp-224, Trp-230, and Phe-540 and three clustered methionine residues, Met-318, Met-322 on the F-helix, and Met-394, which is on the central kink in the I-helix (Fig. 4). Significantly, Met-394, the tip of which is 8.7 Å from the heme iron, is one of a pair of amino acid residues, the other of which is Ala-395, which directly replaces the pair, usually an acidic residue followed by threonine (or serine), illustrated by Glu-257/Thr-258 in CYP StaP (29), which is well conserved among CYPs known to catalyze oxygenation reactions, in which they are thought to play a critical role in oxygen activation yet absent, for example, in the nitric-oxide reductase P450nor (32). Met-394 is also isolated on the Ramachandran plot for XplA-heme with values for Φ and Ψ angles of −84.9° and −74.3°, respectively, on the verge of the allowed region. The otherwise well conserved threonine is replaced by alanine, and as with P450eryF (33), the site occupied by the side chain of threonine in the majority of cyt-P450s is occupied by a water molecule, which is H-bonded to the carbonyl oxygen of Val-391, the peptidic NH of Ala-395 itself, and also the N1 atom of the imidazole ligand bound to the heme iron (Fig. 4). The only nonhydrophobic side chain within 12 Å of the heme iron on the distal face of the heme is Gln-438, on the loop following helix K and for which the side-chain amide oxygen is 3.5 Å from the imidazole ligand and 6.0 Å from the heme iron (Fig. 4). The -NH2 of the amide is H-bonded to the carbonyl oxygen of Ser-440. Its proximity to both the heme and the imidazole ligand bound to the heme iron is suggestive of a role in either substrate binding or catalytic activity of XplA as seen below.
A Putative Channel for Ligand Transport
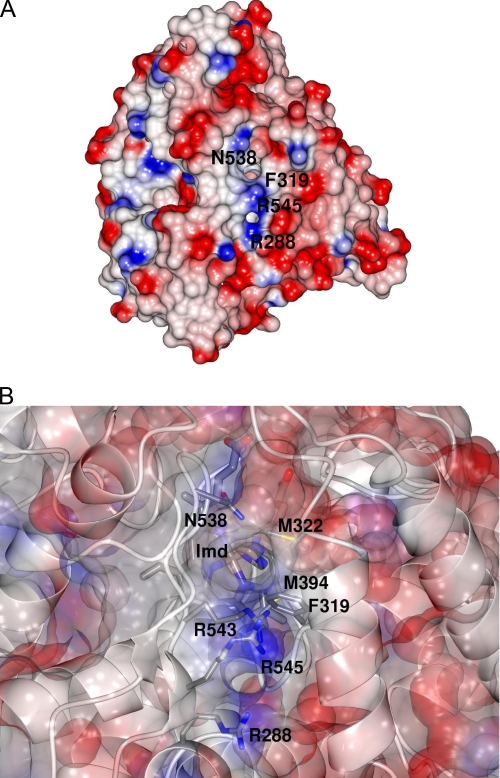
An examination of the electrostatic surface of the XplA molecule revealed a face that presents two channels to the heme, through which the imidazole ligand can be observed (Fig. 5, A and B). The entrance to the first and largest of these channels is bounded by the side chains of Phe-319 on the F helix, Leu-323 on the F-G helix-loop-helix motif, Ala-541 and Gln-438 each on the loop connecting β-strands 7 and 8 in sheet β-4, and with Met-322, also on the F-G loop just beneath (one of the cluster of methionines for which the tip of the side chain forms part of the ceiling over the heme). In this channel, there is a clear chain of seven water molecules from the water H-bonded to the side chain of Gln-438 to the surface of the enzyme. The second channel is bounded by the side chains of Arg-288 at the N-terminal end of the E-helix and Arg-545 on β-strand 8 in β-sheet 3 toward the C terminus of XplA-heme, the two residues forming a positively charged patch on the surface of the protein, which may possibly be involved in capturing the strong electron withdrawing nitro groups on the RDX molecule (Fig. 5, A and B). For crystals of XplA-heme that were soaked with 500 μm RDX for 3 h, electron density above the heme was considerably larger and more diffuse than that observed for imidazole, although the structure of RDX could not be reliably refined in this density, and longer soaks were not successful in improving it. However, these structures did reveal significant mobility in the side chains of several amino acids, leading a path from the heme to the surface of the enzyme through this second channel. These residues were: Arg-288 at the N-terminal end of the E-helix, Met-315, Phe-319, Arg-543, and Arg-545. It was more difficult to trace a complete chain of well resolved hydrogen-bond linked water molecules through this channel to the surface.
FIGURE 5.
A, electrostatic surface of XplA-heme, showing pores to the protein interior bounded by Asn-538 and Phe-319 in the first instance, and Arg-545 and Arg-288 in the second, the latter two in a positively charged patch, which may have a role in recognition and capture of RDX. B, close-up of proposed entrance channel, with partially transparent electrostatic surface in XplA-heme. Residues of relevant side chains are labeled. The putative entrance channel consists of Arg-543, Arg-288, Arg-545, Gln-538, and Phe-319, leading directly to the methionine cluster including Met-394 and Met-322 that is suspended over the heme. The imidazole ligand (Imd) can also be seen through the pore from the enzyme surface.
Mutation Studies
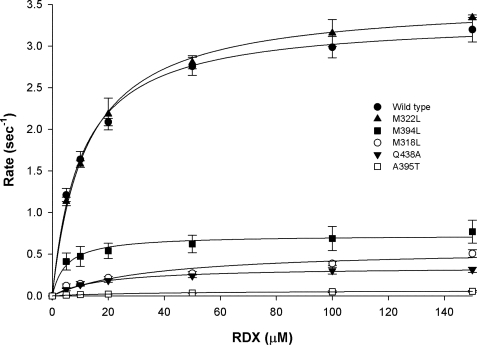
Structural studies of XplA-heme had revealed that, among other residues, Met-322, Met-318, and Met-394, on the kink of the I-helix, and Ala-395, structurally homologous to a threonine residue important for the catalysis of oxygenations in other CYPs, and Gln-438, which projects toward the center of the heme at a short distance from the imidazole ligand, might all have roles in either substrate binding or catalysis by XplA-heme. Five mutants, Met-322 → Leu, Met-318 → Leu, Met-394 → Leu, Ala-395 → Thr, and Gln-438 → Ala were constructed in an attempt to disrupt the activity of XplA. Each variant was assayed for catalytic competence by following RDX consumption by high-performance liquid chromatography against the wild-type enzyme as control, each using surrogate electron donors. The results are shown in Table 3, and a graph showing RDX consumption versus concentration of RDX for each mutant is shown in Fig. 6. The decision to mutate Met-394, Met-322, and Met-394 to leucine was based on previous studies that carried out the systematic substitution of methionines to leucine in calmodulin (34), in an effort to illustrate the contribution of the flexibility of the methionine side chain to relaxed substrate recognition. In terms of catalytic efficiency, there was little effect on the activity of XplA when Met-322 was mutated to leucine, but greater effect for Met-394 and, most notably, for Met-318, for which a 20-fold decrease in kcat/Km was observed (Table 3). Mutation of Gln-438 to alanine resulted in a 17-fold decrease in kcat/Km, and mutation of Ala-395 to threonine gave the most marked decrease in kcat/Km of approximately 200-fold, which was due to both a 4-fold rise in Km and a near 50-fold decrease in kcat. The activity of mutants was also assessed by comparing the spectrophotometrically determined KD (Table 3). The mutation of Met-322 to leucine resulted in a slightly lower KD; the Gln-438 → Ala mutant displayed a 6-fold increase and the relationship between concentration of RDX and the spectroscopic shift for Ala-395 → Thr was linear within the range of RDX concentrations used, suggesting a higher value for the KD that could not be determined.
TABLE 3.
Kinetic constants determined for wild-type XplA-heme and mutants determined by measurement consumption of RDX by HPLC (kcat and Km) and by spectrophotometry (KD) as detailed under “Experimental Procedures”
| Enzyme | KD | Km | kcat | kcat/Km |
|---|---|---|---|---|
| μm | μm | s−1 | μm−1·s−1 ×103 | |
| Wild type | 11.44 ± 0.39 | 10.23 ± 0.90 | 3.33 ± 0.07 | 325.0 |
| A395T | NDa | 39.85 ± 6.86 | 0.07 ± 0.01 | 1.7 |
| Q438A | 67.28 ± 4.83 | 17.80 ± 1.80 | 0.35 ± 0.01 | 19.6 |
| M322L | 7.97 ± 0.94 | 12.04 ± 0.70 | 3.55 ± 0.05 | 295.0 |
| M318L | 34.32 ± 4.30 | 35.75 ± 13.69 | 0.57 ± 0.08 | 15.9 |
| M394L | 20.46 ± 0.47 | 4.94 ± 1.14 | 0.73 ± 0.03 | 148.0 |
a ND, not determined.
FIGURE 6.
Graph showing rate of RDX consumption as measured by kcat and determined by high-performance liquid chromatography versus concentration of RDX. Methionine-leucine mutants in position 322 displays catalytic behavior comparable to the wild type in this respect, but rates of RDX consumption were much reduced for both Gln-438 → Ala and Ala-395 → Thr mutants.
DISCUSSION
The structure of cyt-P450 XplA-heme provides the first structural insights into a cytochrome P450 heme domain, which catalyzes a reductive denitration reaction. Although it displays the canonical CYP fold overall, XplA-heme contains a number of structural anomalies within the active site that mark it as distinctive from other P450 heme domains for which structures have already been determined. To discuss the potential significance of these structural characteristics with respect to mechanism, the issue of the proposed mechanism of denitration of RDX by XplA must first be considered. It has been suggested that cytochrome-P450 is capable of catalyzing denitration of RDX through two sequential single electron transfer steps (35). It was already demonstrated that one equivalent of NADPH was required to effect anaerobic denitration of one molecule of RDX by the XplB-XplA system (5), but the exact mechanism of electron transfer in the case of XplA remains to be elucidated. Single electron transfer mechanisms have also been established for cyt-P450 acting on a number of substrates (36) and implicated in the abiotic photolytic denitration of, for example, N-nitro-piperidine (37). Given a possible oxygen-independent electron transfer hypothesis as a basis for denitration of RDX, it would not be necessary to consider the structural determinants of a mechanism, which would include, for example, base-catalyzed elimination of HNO2, which is known to occur for nitramines under basic conditions (38); nor is it necessary to consider the positioning of the RDX ligand for C–H hydroxylation, because the denitration is known to be catalyzed in an anaerobic environment. The presence of molecular oxygen bound to the heme iron in cytochromes P450 has been shown to exert profound, if subtle effects on, for example, water structure within the active site that may contribute to the effective delivery of protons to the terminal oxygen atom of the dioxygen bound to heme, as in P450cam (13). Such structural perturbations on oxygen binding may, in turn, have profound effects on the mechanism of RDX breakdown, as is perhaps suggested by the distinctive end products of reaction in either anaerobic or aerobic pathways (Fig. 1). Unfortunately, neither dioxygen nor RDX could be satisfactorily refined into the electron density obtained for either RDX-soaked or native datasets, RDX being insoluble above concentrations of 500 μm in the crystallization drop, but the presence of the imidazole ligand, a nitrogen-containing heterocycle itself, may give some insight into the important interactions in the active site between ligand, active site side chains, and water molecules.
As with P450EryF, XplA-heme lacks the well conserved threonine residue, typified by Thr-252 in P450cam, which in the latter enzyme is thought to accept a hydrogen bond from the hydroperoxy (Fe(III)-OOH) intermediate in the catalytic cycle (13). However, the residue in this position is not thought to always be directly involved in the stabilization of that intermediate: in P450cin, the equivalent asparagine is thought to modulate the regioselectivity of hydroxylation of the substrate cineole, for example (39). Given that XplA catalyzes an oxygen-independent biotransformation, the significance of the lack of a threonine at this position is open to experiment and interpretation. There was a notable decrease in kcat toward RDX when Ala-395 was mutated to threonine, as well as an increase in both Km, and, presumably, KD. In the imidazole complex, the water molecule in the cavity formed between Ala-395 and Val-391 forms an H-bond to the N1 atom of the imidazole (Fig. 4) and this, in conjunction with the kinetic data on the Ala-395 → Thr mutant, may suggest a role therefore in substrate binding by that cavity. Equivalent suppression of activity in the hydroxylation of 6-DEB was observed for Ala-Thr mutants of P450-EryF in this position (40), and this was attributed to steric factors, because P450-EryF-Ala-Thr mutants were still able to hydroxylate the smaller substrate testosterone (41). A role for the Ala-395 to Val-391 water cavity in substrate binding, as opposed to heme-oxygen intermediate stabilization, would also seem reasonable given that XplA does not require oxygen as such, at least in the anaerobic pathway to RDX that culminates in the metabolite methylene dinitramine 6 (Fig. 1).
Gln-438 is unusual in being a hydrophilic residue in an otherwise very hydrophobic heme environment in XplA-heme. The side-chain amide carbonyl of Gln-438 is only 6 Å from the heme iron and in the imidazole complex, is within H-bonding distance of the C5 atom of the imidazole ring, which could also be refined as the N1 atom if the ligand was merely rotated 180° on its vertical axis. The imidazole potentially serves therefore to position a nitrogen atom within bonding distance of suitable H-bonding partners as might be observed for the symmetrical molecule RDX prior to denitration. The side chain of Gln would be unlikely to serve as a catalytic base for the elimination of nitrite from RDX, although general base activity has recently been attributed to an asparagine side chain in a glycosyl hydrolase (42). However, in that enzyme, mutation to alanine abolished activity, and in the Gln-438 → Ala mutant of XplA catalytic efficiency was merely reduced 17-fold. It is unusual to find a glutamine residue in this position in a structurally characterized P450, although a glutamine is present in the active site of the heme-reductase cytochrome c nitrite reductase in which it is thought to support an H-bonding network that assists the stabilization of the distal heme ligand (43).
The cluster of methionine residues, Met-318, Met-322, and Met-394, at the ceiling of the heme binding site in XplA-heme is noteworthy in that one, Met-394, replaces the acidic residue found in oxygenating P450s. Again, in an enzyme that does not require the protonation of dioxygen through a proton relay, the absence of the acidic residue should not be a handicap, at least to anaerobic biotransformation of RDX. It may be that the one or more methionines confer malleability within the putative access channel and also the active site that assists in the transfer and binding of the bulky RDX substrate. The mutation of Met-394 or Met-322 to the less flexible leucine had no marked effect on RDX turnover or binding by XplA-heme compared with the wild-type. Mutation of Met-318 however resulted in a decreased affinity for RDX and a 20-fold decrease in kcat/Km overall. Mutations to Met-220 in the access channel of 14α-sterol demethylase (Cyp51A) of Aspergillus fumigatus have been previously shown to disrupt the binding of itraconazole to that enzyme, conferring drug resistance to the host mutant organisms (44), and, conversely, mutation of an isoleucine residue to methionine in the access channel of the amine oxidase from Aspergillus niger has been shown to increase the rate of turnover, perhaps as a result of improved access to the active site (15). In addition to flexibility, methionine clusters have also been observed to exert electronic effects through their capacity for oxidation to methionine sulfoxide, for example in the potassium channel Slo1 (45), where oxidation of methionines within the channel was thought to enhance the activity of that channel when under oxidative stress, but there is no structural evidence for the oxidation of methionines 318, 322, or 394 in any structures of XplA-heme, that might suggest a role in oxygen sensitivity in this enzyme.
The pores observed leading from the surface of XplA-heme to the heme, in conjunction with the multiple rotamers of relevant amino acid side chains when soaked with RDX, are suggestive of a ligand transport channel in XplA-heme. In a study of the dynamics of substrate access channels in cyt-P450, Winn and co-workers (46) identified the arginine side-chain Arg-47 in cyt-P450 BM3, which was thought to guide negatively charged fatty acid substrates to the active site. In XplA-heme three arginines form part of the putative channel. Of these, Arg-545 is at the surface and is relatively mobile; Arg-288 is also very near the surface and interacts closely with Met-315, which is itself clearly in two conformations in each of the 1.5-Å structures of XplA-heme, these two arginine residues might be ideal for guiding the electronegative RDX to the heme pocket. Arg-543 is buried, but H-bonded within the channel only to water molecules within a large cavity, rather than fixed to backbone carbonyl groups as was noted for Arg-185 in P450-EryF (46). However, the pronounced mobility of Arg-543 strongly suggests a role in substrate transport, perhaps in a gating mechanism, and adds to the hypothesis proposed by Winn, that channel opening mechanisms in CYPs are adjusted to the physicochemical properties of the substrate (46, 47). In their comprehensive review of substrate and solvent access channels in CYPs (47), Winn and co-workers highlight that it is most common in bacterial P450 for the ligand transport channels to be made up of the F-G loop and the B-C loop/B′ helix, yet in XplA-heme, the putative channel would, unusually, be lined by the side chains of the F-G loop, the β-4 loop and the I-helix. The solvent channel as described for other CYPs between E, F, and I helices (47) is also observed in XplA-heme, with a clear chain of well resolved water molecules leading to the surface of the enzyme from the heme propionate bound to Arg-443.
The structure of XplA-heme gives new insight into a cytochrome-P450 that catalyzes the reductive biotransformation of the recalcitrant and potentially carcinogenic xenobiotic compound RDX and will enable detailed mutagenesis studies of the enzyme that will shed light on the molecular determinants of mechanism and substrate specificity. Several XplA-like enzymes are thought to have evolved only recently in response to RDX contamination (25) and have transferred throughout bacterial species all over the world. A detailed understanding of the mechanism and substrate binding in XplA may help explain how these enzymes have been recruited for the detoxification of this unusual xenobiotic.
This work was supported in part by the Strategic and Environmental Research and Development Program of the U.S. Dept. of Defense, by the Biotechnology and Biological Sciences Research Council, and by a studentship from the Centre of Excellence for Biocatalysis, Biotransformation, and Biomanufacture (to F. S.).
The atomic coordinates and structure factors (codes 2wiv and 2wiy) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- RDX
- royal demolition explosive
- XplA
- cytochrome P450 XplA
- XplA-heme
- subcloned heme-domain of XplA
- P450EryF
- cytochrome P450 EryF from S. erythrea
- P450cam
- cytochrome P450cam from P. putida
- r.m.s.d.
- root mean square deviation
- Bis- Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CYP
- cytochrome P450.
REFERENCES
- 1.Boopathy R., Gurgas M., Ullian J., Manning J. F. (1998) Curr. Microbiol. 37, 127–131 [DOI] [PubMed] [Google Scholar]
- 2.Binks P. R., Nicklin S., Bruce N. C. (1995) Appl. Environ. Microbiol. 61, 1318–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J.-S., Spain J., Thiboutot S., Ampleman G., Greer C., Hawari J. (2006) FEMS Microbiol. Ecol. 39, 349–357 [DOI] [PubMed] [Google Scholar]
- 4.Seth-Smith H. M., Rosser S. J., Basran A., Travis E. R., Dabbs E. R., Nicklin S., Bruce N. C. (2002) Appl. Environ. Microbiol. 68, 4764–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson R. G., Rylott E. L., Fournier D., Hawari J., Bruce N. C. (2007) Proc. Nat. Acad. Sci. 104, 16822–16827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andeer P. F., Stahl D. A., Bruce N. C., Strand S. E. (2009) Appl. Environ. Microbiol. 75, 3258–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rylott E. L., Jackson R. G., Edwards J., Womack G. L., Seth-Smith H. M., Rathbone D. A., Strand S. E., Bruce N. C. (2006) Nat. Biotechnol. 24, 216–219 [DOI] [PubMed] [Google Scholar]
- 8.Alston T. A., Seitz S. P., Porter D. J., Bright H. J. (1980) Biochem. Biophys. Res. Commun. 97, 294–300 [DOI] [PubMed] [Google Scholar]
- 9.Servent D., Delaforge M., Ducrocq C., Mansuy D., Lenfant M. (1989) Biochem. Biophys. Res. Commun. 163, 1210–1216 [DOI] [PubMed] [Google Scholar]
- 10.Buzukov A. A., Il'asova V. B., Tabak M., Meirelles N. C., Degterev I. A. (1996) Chem. Biol. Interact. 100, 113–124 [DOI] [PubMed] [Google Scholar]
- 11.Munro A. W., Girvan H. M., McLean K. J. (2007) Nat. Prod. Rep. 24, 585–609 [DOI] [PubMed] [Google Scholar]
- 12.Vidakovic M., Sligar S. G., Li H., Poulos T. L. (1998) Biochemistry 37, 9211–9219 [DOI] [PubMed] [Google Scholar]
- 13.Nagano S., Poulos T. L. (2005) J. Biol. Chem. 280, 31659–31663 [DOI] [PubMed] [Google Scholar]
- 14.Bonsor D., Butz S. F., Solomons J., Grant S., Fairlamb I. J., Fogg M. J., Grogan G. (2006) Org. Biomol. Chem. 4, 1252–1260 [DOI] [PubMed] [Google Scholar]
- 15.Atkin K. E., Reiss R., Koehler V., Bailey K. R., Hart S., Turkenburg J. P., Turner N. J., Brzozowski A. M., Grogan G. (2008) J. Mol. Biol. 384, 1218–1231 [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z., Minor A. W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 17.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 18.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 19.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 20.Cowtan K. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 21.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 22.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 23.Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 24.Bairoch A., Apweiler R. (1996) Nucleic Acids Res. 24, 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seth-Smith H. M., Edwards J., Rosser S. J., Rathbone D. A., Bruce N. C. (2008) Appl. Environ. Microbiol. 74, 4550–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson J. A., Graham S. E. (1998) Structure 6, 1079–1085 [DOI] [PubMed] [Google Scholar]
- 27.Poulos T. L., Finzel B. C., Gunsalus I. C., Wagner G. C., Kraut J. (1985) J. Biol. Chem. 260, 16122–16130 [PubMed] [Google Scholar]
- 28.Cryle M. J., Schlichting I. (2008) Proc. Nat. Acad. Sci. 105, 15696–15701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makino M., Sugimoto H., Shiro Y., Asamizu S., Onaka H., Nagano S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11591–11596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long F., Vagin A. A., Young P., Murshudov G. N. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leys D., Mowat C. G., McLean K. J., Richmond A., Chapman S. K., Walkinshaw M. D., Munro A. W. (2003) J. Biol. Chem. 278, 5141–5147 [DOI] [PubMed] [Google Scholar]
- 32.Obayashi E., Shimizu H., Park S. Y., Shoun H., Shiro Y. (2000) J. Inorg. Biochem. 82, 103–111 [DOI] [PubMed] [Google Scholar]
- 33.Cupp-Vickery J. R., Garcia C., Hofacre A., McGee-Estrada K. (2001) J. Mol. Biol. 311, 101–110 [DOI] [PubMed] [Google Scholar]
- 34.Zhang M., Li M., Wang J. H., Vogel H. J. (1994) J. Biol. Chem. 269, 15546–15552 [PubMed] [Google Scholar]
- 35.Bhushan B., Trott S., Spain J. C., Halasz A., Paquet L., Hawari J. (2003) Appl. Environ. Microbiol. 69, 1347–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guengerich F. P. (2001) Chem. Res. Toxicol. 14, 611–650 [DOI] [PubMed] [Google Scholar]
- 37.Lockhart R. W., Snyder R. W., Chow Y. L. (1976) J. Chem. Soc. Chem. Commun. 52 [Google Scholar]
- 38.Hoffsommer J. C., Kubose D. A., Glover D. J. (1977) J. Phys. Chem. 81, 380–385 [Google Scholar]
- 39.Meharenna Y. T., Slessor K. E., Cavaignac S. M., Poulos T. L., De Voss J. J. (2008) J. Biol. Chem. 283, 10804–10812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagano S., Cupp-Vickery J. R., Poulos T. L. (2005) J. Biol. Chem. 280, 22102–22107 [DOI] [PubMed] [Google Scholar]
- 41.Xiang H., Tschirret-Guth R. A., Ortiz de Montellano P. R. (2000) J. Biol. Chem. 275, 35999–36006 [DOI] [PubMed] [Google Scholar]
- 42.Abbott D. W., Macauley M. S., Vocadlo D. J., Boraston A. B. (2009) J. Biol. Chem. 284, 11676–11689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarke T. A., Kemp G. L., Van Wonderen J. H., Tovell N., Doyle R. M., Cheesman M. R., Butt J. N., Cole J. A., Richardson D. J., Hemmings A. M. (2008) Biochemistry 47, 3789–3799 [DOI] [PubMed] [Google Scholar]
- 44.Mellado E., Garcia-Effron G., Alcazar-Fuoli L., Cuenca-Estrella M., Rodriguez-Tudela J. L. (2004) Antimicrob. Agents. Chemother. 48, 2747–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santarelli L. C., Wassef R., Heinemann S. H., Hoshi T. (2006) J. Physiol. 571, 329–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winn P. J., Lüdemann S. K., Gauges R., Lounnas V., Wade R. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cojocaru V., Winn P. J., Wade R. C. (2007) Biochim. Biophys. Acta 1770, 390–401 [DOI] [PubMed] [Google Scholar]