Abstract
Anoikis, a detachment-induced apoptosis, is a principal mechanism of inhibition of tumor cell metastasis. Tumor cells can acquire anoikis resistance which is frequently observed in metastatic lung cancer. This phenomenon becomes an important obstacle of efficient cancer therapy. Recently, signaling mediators such as caveolin-1 (Cav-1) and nitric oxide (NO) have garnered attention in metastasis research; however, their role and the underlying mechanisms of metastasis regulation are largely unknown. Using human lung carcinoma H460 cells, we show that NO impairs the apoptotic function of the cells after detachment. The NO donors sodium nitroprusside and diethylenetriamine NONOate inhibit detachment-induced apoptosis, whereas the NO inhibitors aminoguanidine and 2-(4-carboxyphenyl) tetramethylimidazoline-1-oxyl-3-oxide promote this effect. Resistance to anoikis in H460 cells is mediated by Cav-1, which is significantly down-regulated after cell detachment through a non-transcriptional mechanism involving ubiquitin-proteasomal degradation. NO inhibits this down-regulation by interfering with Cav-1 ubiquitination through a process that involves protein S-nitrosylation, which prevents its proteasomal degradation and induction of anoikis by cell detachment. These findings indicate a novel pathway for NO regulation of Cav-1, which could be a key mechanism of anoikis resistance in tumor cells.
Caveolin-1 (Cav-1),3 a 21–24-kDa structural protein component of the plasma membrane microdomains termed caveolae has been shown to function in vesicular trafficking, signal transduction, and cancer progression (1–4). Although up-regulation of this protein normally occurs in a variety of terminally differentiated cells including fibroblasts, adipocytes, smooth muscle cells, endothelial cells, and epithelial cells (5), Cav-1 is greatly reduced in most oncogenically transformed and cancer cells (6–10). Thus, Cav-1 was first explained to function as a tumor suppressor protein (11–13). In contrast, increasing evidence indicates its role as a tumor and metastatic promoter as overexpression or re-expression of Cav-1 was found in many advanced stage and metastatic cancer cells. Up-regulation of Cav-1 was shown to render Rat1A cells more resistant to apoptosis (14). Moreover, antisense-induced down-regulation of Cav-1 caused human prostate cancer cells more sensitive to apoptosis (15, 16). Therefore, the role of Cav-1 in cancer progression remains controversial.
Metastasis is a multistep process composed of cancer cell detachment, migration, extravasation, and adhesion of the detached cells to other target sites. A key mechanism in the regulation of metastasis is anoikis or detachment-induced apoptosis. Previous studies have shown that Cav-1 acts as a negative regulator of anoikis (17, 18), and its elevated expression in lung carcinoma is closely associated with the increased metastasis capacity and poor survival of the patients (19). Likewise, elevated NO and NO synthase levels have been associated in many human metastatic cancers including the lung (20–24), breast (25), colon (26), and cancers of the central nervous system (27). However, the role of NO and its mechanism of metastasis regulation in association with Cav-1 are not well understood. Because resistance to anoikis is a key step in metastasis development and because Cav-1 has been implicated in this process, we investigated the potential regulation of Cav-1 by NO and studied its role in anoikis of human lung carcinoma cells.
NO has been reported to have both pro- and anti-apoptotic effect on cells, depending on a variety of factors, including cell type, cellular redox status, and the flux and dose of local NO (28, 29). In human lung carcinoma cells, we previously reported that NO plays a suppressive role in apoptosis induced by a variety of agents, including Fas death ligand (30), chemotherapeutic agent cisplatin (31), and the metal carcinogen chromium (32). However, the role of NO in cell anoikis and its potential regulation of Cav-1 have not been well investigated. Using molecular and pharmacological approaches, we report here that NO plays an important role in Cav-1 regulation and anoikis function of human lung cancer H460 cells. We also demonstrate for the first time that Cav-1 is down-regulated during cell anoikis through the ubiquitin-proteasomal degradation pathway and that NO regulates this process by inducing S-nitrosylation of the protein which inhibits its ubiquitination and proteasomal degradation. Thus, our study reveals the existence of a novel mechanism of anoikis regulation, which might be exploited in metastasis and cancer therapy.
MATERIALS AND METHODS
Cells and Reagents
Human lung epithelial NCI-H460 cells were obtained from the American Type Culture Collection (Manassas, VA). The cells were cultured in RPMI 1640 medium supplemented with 5% fetal bovine serum, 2 mm l-glutamine, and 100 units/ml penicillin and streptomycin. Cell cultures were maintained in a humidified atmosphere of 5% CO2 at 37 °C. Cells were passaged at preconfluent densities using a solution containing 0.05% trypsin and 0.5 mm EDTA. The NO donors sodium nitroprusside (SNP) and diethylene triamine (DETA) NONOate and the NO inhibitors 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (PTIO) and aminoguanidine (AG) were obtained from Sigma. These NO modulators are water-soluble and were prepared in a sterile culture medium before use. Diaminofluorescein diacetate (DAF-DA) and Hoechst 33342 were obtained from Molecular Probes, Inc. (Eugene, OR). Monoclonal antibody against Cav-1 and protein A-agarose were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against Akt and phospho-Akt Ser-473 and Thr-308 were from Cell Signaling Technology, Inc. (Beverly, MA). Antibodies for ubiquitin, S-nitrosocysteine, β-actin, and peroxidase-conjugated secondary antibodies were from Sigma. The transfecting agent Lipofectamine 2000 was from Invitrogen. All other chemicals and reagents including annexin V-fluorescein isothiocyanate (FITC), propidium iodide (PI), dithiothreitol (DTT), lactacystin, and LY294002 were from Sigma.
NO Detection
Cellular NO production was determined by flow cytometry using DAF-DA as a fluorescent probe and by Griess assay, which measures the stable nitrite byproduct of NO in the culture medium. After detachment, cells (1 × 106/ml) were collected and incubated with 10 μm DAF-DA for 30 min at 37 °C. The cells were then washed, resuspended in phosphate-buffered saline, and analyzed for fluorescence intensity using FACSCaliber (BD Biosciences). Signals were obtained using a 488-nm excitation beam and a 538-nm band-pass filter. In some experiments cells were visualized for fluorescence intensity using a fluorescence microscope (Carl Zeiss Axiovert, Göttingen, Germany). For the Griess assay, cell supernatants were collected, and aliquots (100 μl) were mixed with 100 μl of Griess reagent (1% sulfanilamide, 0.1% naphthyl ethylenediamine dihydrochloride, 2% phosphoric acid) in a 96-well plate. After incubation for 10 min at 25 °C, the absorbance at 550 nm was measured on a microplate reader.
Plasmid and Transfection
Caveolin-1 plasmid (pEX_Cav-1-YFP) was obtained from the American Type Culture Collection (Manassas, VA). Stable transfectant of Cav-1 was generated by culturing H460 cells in a 6-well plate until they reached 60% confluence. One microgram of cytomegalovirus-neo vector and 15 μl of Lipofectamine reagent with 2 μg of Cav-1 or control pcDNA3 plasmid were used to transfect the cells in the absence of serum. After 12 h the medium was replaced with culture medium containing 5% fetal bovine serum. Approximately 36 h after the beginning of the transfection, the cells were digested with 0.03% trypsin, and the cell suspensions were plated onto 75-ml culture flasks and cultured for 24–28 days with G418 selection (600 μg/ml). The pooled stable transfectant was identified by Western blot analysis of Cav-1 and was cultured in G418-free RPMI 1640 medium for at least two passages before each experiment.
Anoikis Assays
Adherent H460 cells in culture were trypsinized into a single cell suspension and then seeded in 12-well tissue culture plates coated with 200 μl (6 mg/ml in 95% ethanol) of poly-2-hydroxyethylmethacrylate (poly-HEMA; Sigma) at the density of 1 × 105 cells/ml. Suspended cells were incubated at 37 °C for various times up to 24 h. Cells were then harvested, washed, and stained with annexin V-FITC and analyzed for fluorescence intensity by flow cytometry and fluorescence microscopy. For Hoechst 33342 apoptosis assay, cells were incubated with 10 μm the Hoechst dye for 30 min at 37 °C, and the apoptotic cells having intensely condensed chromatin and/or fragmented nuclei were visualized under a fluorescence microscope. For cell survival assay, cells were similarly treated, harvested, washed, and incubated with 20 μm sodium 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) for 4 h at 37 °C. Optical density was then determined by V-max photometer (Molecular Devices, Inc., Menlo Park, CA) at a wavelength of 450 nm.
Western Blot Analysis
Cell extracts were performed by incubating the cells in lysis buffer containing 20 mm Tris-HCl, pH 7.5, 1% Triton X-100, 150 mm sodium chloride, 10% glycerol, 1 mm sodium orthovanadate, 50 mm sodium fluoride, 100 mm phenylmethylsulfonyl fluoride, and a protease inhibitor mixture (Roche Applied Science) for 30 min on ice. Cell lysates were collected and assayed for protein content using the Bradford method (Bio-Rad). Equal amount of proteins per sample (40 μg) were resolved on a 10% SDS-polyacrylamide gel electrophoresis and transferred onto 0.45-μm nitrocellulose membranes (Pierce). The transferred membranes were blocked for 1 h in 5% nonfat dry milk in Tris-buffered saline/Tween 20 (25 mm Tris-HCl, pH 7.4, 125 mm NaCl, and 0.05% Tween 20) and incubated with the appropriate primary antibodies at 4 °C overnight. Membranes were washed 3 times with Tris-buffered saline, Tween 20 for 10 min and incubated with peroxidase-labeled secondary antibodies for 1 h at room temperature. The immune complexes were detected by chemiluminescence (Supersignal West Pico; Pierce) and quantified by imaging densitometry using analyst/PC densitometry software (Bio-Rad).
Immunoprecipitation
Cells were washed after treatments and lysed in lysis buffer for 30 min on ice. Cell lysates were collected and determined for protein content. Equal amounts of proteins per sample (60 μg) were immunoprecipitated with anti-Cav-1 antibody for 6 h at 4 °C. The immune complexes were washed with 30 volumes of lysis buffer, resuspended in 2× Laemmli sample buffer, and boiled at 95 °C for 5 min. The immune complexes were separated by 10% SDS-PAGE and analyzed by Western blotting as described above.
Measurements of Cav-1 S-Nitrosylation
Cells were lysed and immunoprecipitated with anti-Cav-1 antibody as described above. The immunoprecipitated protein was analyzed for S-nitrosylation by Western blot using anti-S-nitrosocysteine antibody and by fluorometric measurements as previously described (32). Briefly, immunoprecipitates were incubated with 200 μm HgCl2 and 200 μm diamino naphthalene in 500 μl of phosphate-buffered saline for 0.5 h at room temperature followed by the addition of 1 m NaOH. The fluorescent triazole product generated from the reaction between diamino naphthalene and NO released from S-nitrosylated Cav-1 was quantified by fluorometry at the excitation and emission wavelengths of 375 and 450 nm, respectively.
Reverse Transcription-PCR
Total RNA was extracted with Trizol (Invitrogen), and reverse transcription-PCR was performed with Access RT-PCR System (Promega, Madison, WI) according to the manufacturer's instructions. Sequences of the PCR primers were: Cav-1 forward, 5′-CGTAGACTCGGAGGGACATC-3′, and reverse, 5′-TTTCGTCACAGTGAAGGTGG-3′; for glyceraldehyde-3-phosphate dehydrogenase, forward, 5′-GCTGAGAACGGGAAGCTTGT-3′, and reverse, 5′-GCCAGGGGTGCTAAGCAG-3′. Reaction products were analyzed after 30 amplification cycles, each of which involved consecutive 1-min steps at 94, 55, and 72 °C. The PCR products were electrophoresed in a 1.5% agarose gel, stained with ethidium bromide, and photographed.
The results obtained by conventional RT-PCR were verified by quantitative real-time PCR. One microgram of Trizol-extracted RNA was reverse-transcribed in a 100-μl reaction mixture containing 500 μm dNTP, 125 units of MultiScribe Reverse Transcriptase (Applied Biosystems, Foster City, CA), 40 units of RNase inhibitor, 2.5 μm oligo(dT), 1× TaqMan reverse transcriptase buffer, and 5 mm MgCl2 at 48 °C for 40 min. The primers used in this study were designed using Primer Express software (Applied Biosystems): Cav-1 (#AI878826) forward 5′-CGAGAAGCAAGTGTACGACGC-3′, and reverse 5′-ACCACGTCATCGTTGAGGTG-3′; glyceraldehyde-3-phosphate dehydrogenase forward, 5′-GAAGGTGAAGGTCGGAGTC-3′, and reverse 5′-GAAGATGGTGATGGGATTTC-3′. Amplification was performed at the following cycling conditions: 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. A SYBR Green PCR Master Mix (Applied Biosystems) was used with 1 ng of cDNA and with 100–400 nm primers. A negative control without any cDNA template was run with every assay. All PCR reactions were performed by using ABI PRISM 7900 Sequence Detection System (Applied Biosystems). Relative mRNA levels were determined by using the comparative CT (threshold cycle) method (33), where the caveolin-1 target is normalized to the control and compared with a reference sample (assigned a relative value of 1) by the equation: 2−ΔΔCT.
Immunofluorescence
Cells (0.5 × 106/well) were seeded in 6-well poly-HEMA-coated plates and treated with NO modulators as described under “Results.” After treatment, the cells were fixed in 3.7% formaldehyde for 10 min at room temperature and then permeabilized and blocked in a solution containing 0.5% saponin, 1% bovine serum, and 1.5% goat serum for 30 min. After primary antibody incubation with Cav-1 monoclonal antibody (BD Biosciences) at 1:100 dilution for 1 h, cells were washed and incubated with Alexa Fluor 488-conjugated secondary antibody (Invitrogen) for 30 min. Cell nucleus was stained with ToPro-3 (Invitrogen), and the actin cytoskeleton was stained with Alexa Fluor 546-conjugated phalloidin (Invitrogen). Cells were cytospun onto a glass slide and mounted using the anti-fade reagent Fluoromont-G (Southern Biotech, Birmingham, AL). Images were acquired by confocal laser scanning microscopy (Zeiss LSM 510).
Statistical Analysis
Data were represented as the means ± S.D. from three or more independent experiments. Statistical analysis was performed by Student's t test at a significance level of p < 0.05.
RESULTS
Nitric Oxide Inhibits Detachment-induced Apoptosis of H460 Cells
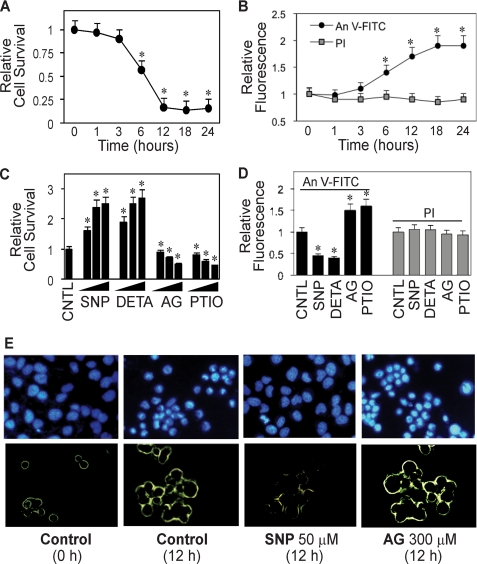
NO has been shown to play an important role in the regulation of cancer cell metastasis; however, the underlying mechanism of this regulation is unclear. To test whether NO might regulate this process by inhibiting detachment-induced apoptosis or anoikis, which is a crucial step in the metastasis of cancer cells, we first investigated anoikis of human lung cancer H460 cells in response to various specific NO donors and inhibitors. Anoikis was induced by detaching the cells and incubating them in attachment-resistant poly-HEMA-coated plates for various times and analyzed for cell viability by XTT assay. Fig. 1A shows that detachment of the cells caused a time-dependent decrease in cell viability with ∼55 and 15% of the cells remaining viable after 6 and 12 h, respectively. Analysis of cell apoptosis by flow cytometry using FITC-labeled annexin V antibody shows a significant increase in annexin V-associated cellular fluorescence as early as 6 h after the detachment and reached a maximum at about 18 h (Fig. 1B). In contrast, analysis of cell necrosis using PI as a probe shows no significant increase in the PI signal over a 24-h period. These results suggest that apoptosis is the primary mode of cell death after detachment of H460 cells. Morphologic analysis of apoptotic cell death by fluorescence microscopy using Hoechst 33342 and annexin V-FITC further confirms the results (Fig. 1E).
FIGURE 1.
Detachment-induced apoptosis and its inhibition by NO. A, effect of cell detachment on cell survival determined by XTT assay. Lung epithelial H460 cells were detached as described under “Materials and Methods” and suspended in HEMA-coated plates for various times (0–24 h). B, effect of cell detachment on apoptosis and necrosis determined by flow cytometry using annexin V-FITC (An V-FITC) and PI assays. C, effect of NO modulators on detachment-induced cell death. Detached cells were treated with various concentrations of NO donor, SNP (10, 50, 100 μm), or DETA NONOate (10, 50, 100 μm) or with NO inhibitor, AG (100, 200, 300 μm), or PTIO (10, 50, 100 μm) for 12 h. Cell survival was then determined by XTT assay. CNTL, control. D, effects of NO modulators on detachment-induced apoptosis and necrosis. Detached cells were treated with SNP (50 μm), DETA NONOate (50 μm), AG (300 μm), or PTIO (50 μm) for 12 h, and cell apoptosis and necrosis were determined as described above. E, upper panel, effect of NO modulators on detachment-induced apoptosis determined by Hoechst 33342 nuclear fluorescence staining. Lower panel, effect of NO modulators on detachment-induced apoptosis determined by annexin V-FITC fluorescence microscopy. Data are the mean ± S.D. (n = 3). *, p < 0.05 versus non-treated control.
To investigate the role of NO in detachment-induced apoptosis, detached H460 cells were treated with various concentrations of NO donors and inhibitors, and their effect on cell survival was determined by XTT assay. Fig. 1C shows that treatment of the cells with NO donor, SNP, or DETA NONOate caused a dose-dependent decrease in cell death, whereas treatment of the cells with NO inhibitor, AG, or PTIO had an opposite effect. Analysis of cell apoptosis by annexin V-FITC and Hoechst 33342 assays similarly shows the inhibitory and promoting effect of the NO donors and inhibitors, respectively, on detachment-induced cell death (Fig. 1, D and E). The NO donors and inhibitors, when used at the indicated concentrations, had no significant effect on cell necrosis as determined by PI assay (Fig. 1D).
Effect of NO Modulators on Cellular NO Level
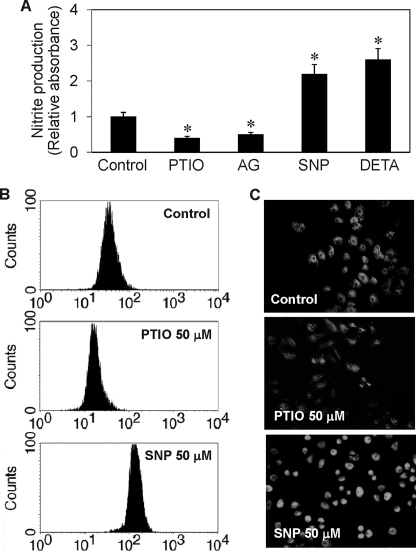
To provide a relationship between cell death and NO modulation induced by the test agents, we analyzed cellular NO levels in response to various NO modulator treatments by colorimetric Griess assay and by flow cytometric and microscopic assays using DAF-DA as a fluorescent probe for NO. Fig. 2A shows the result of the Griess assay which measures the stable nitrite breakdown product of NO. Both NO inhibitors AG and PTIO significantly inhibited cellular nitrite production, whereas the NO donors SNP and DETA NONOate increased the production as compared with non-treated control. These results were confirmed by flow cytometric and microscopic assays of NO (Fig. 2, B and C), which show the induction and inhibition of cellular NO levels by the NO donor SNP and NO inhibitor PTIO, respectively.
FIGURE 2.
Effect of NO modulators on cellular NO and nitrite levels. H460 cells were detached and either left untreated or treated with SNP (50 μm), DETA NONOate (50 μm), AG (300 μm), or PTIO (50 μm) for 2 h. A, nitrite production determined by the Griess assay. B and C, NO production determined by flow cytometry and fluorescence microscopy using DAF-DA as a probe. Data are the mean ± S.D. (n = 3). *, p < 0.05 versus non-treated control.
Cav-1 Overexpression Renders H460 Cells Resistant to Detachment-induced Apoptosis
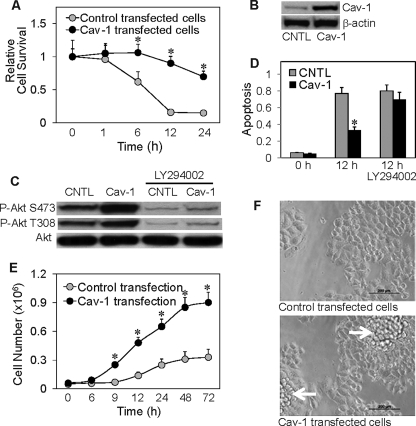
The role of Cav-1 in the regulation of cancer cell anoikis is unclear. We studied this role by stably transfecting H460 cells with Cav-1 or control plasmid and evaluated their effect on detachment-induced cell death. Transfected cells were detached, suspended in poly-HEMA-coated plates, and analyzed for cell survival at various times by XTT assay. Fig. 3A shows that Cav-1-transfected cells exhibited resistance to detachment-induced cell death as compared with control-transfected cells. Western blot analysis of Cav-1 expression in the transfected cells shows an increased expression of Cav-1 protein in the Cav-1-transfected cells compared with control-transfected cells (Fig. 3B). These results indicate the role of Cav-1 as a negative regulator of detachment-induced cell death in lung epithelial H460 cells. Because Cav-1 has been shown to regulate cell death and survival through a PI3K/Akt-dependent mechanism (16, 35, 36), we tested the effect of Cav-1 overexpression on Akt activity and examined its effect on detachment-induced cell death. Our results show that Cav-1 overexpression induced Akt activation as indicated by the increased phosphorylation of Akt (Thr-308 and Ser-473), whereas it had no effect on total Akt level (Fig. 3C). The induction of Akt phosphorylation was inhibited by the PI3K inhibitor LY294002, suggesting that this induction was mediated through the PI3K pathway. Analysis of apoptosis shows that Cav-1 overexpression decreased detachment-induced cell death and that this effect was inhibited by the PI3K inhibitor LY294002 (Fig. 3D). These results suggest that Cav-1 exerts its anti-apoptotic effect during cell detachment through a mechanism that is dependent on PI3K/Akt activation.
FIGURE 3.
Cav-1 overexpression increases cell death resistance and Akt phosphorylation, alters growth pattern, and increases growth rate. A, H460 cells were stably transfected with Cav-1 or control plasmid as described under “Materials and Methods.” Transfected cells were grown in culture medium and analyzed for cell survival at various times after detachment using XTT assay. Attached cells showed no significant apoptosis during the test period of 24 h (data not shown). B, Western blot analysis of Cav-1 expression in control and Cav-1-transfected cells. Cell extracts were prepared and separated on 10% polyacrylamide-SDS gels, transferred, and probed with Cav-1 antibody. β-Actin was used as a loading control. CNTL, control. C, effect of Cav-1 overexpression on Akt phosphorylation. Transfected cells were detached and incubated in HEMA-coated plates for 12 h in the presence or absence of LY294002 (10 μm). Cell lysates were prepared and analyzed for Akt phosphorylation by Western blotting. Blots were probed with antibodies specific to phospho-Akt (Ser-473 and Thr-308) and Akt. D, apoptosis of the treated cells was analyzed by Hoechst 33342 assay and expressed as the ratio of apoptotic nuclei to total nuclei. E, effect of Cav-1 overexpression on cell proliferation. Cav-1 and control-transfected cells were grown in normal tissue culture plates and analyzed for cell proliferation at various times by hemocytometry. F, morphology of control and Cav-1-transfected cells in culture. Data are the mean ± S.D. (n = 3). *, p < 0.05 versus vector-transfected control.
Cav-1 Overexpression Alters Cell Growth and Morphology of H460 Cells
Fig. 3E shows that under a normal growth condition that allows cell attachment, Cav-1-overexpressing cells exhibited an increased growth rate over control-transfected cells. The lag phase before cell growth was significantly reduced in Cav-1-overexpressing cells. As compared with control-transfected cells, which grew as an epithelial monolayer, Cav-1-overexpressing cells formed cell mounds and grew as multilayer epithelial cells (Fig. 3F). This multilayer growth pattern is consistent with the increased growth rate of Cav-1-overexpressing cells. These results suggest that Cav-1 may function as a tumor promoter by enhancing cell growth.
Detachment Induces Cav-1 Down-regulation through a Non-transcriptional Proteasome-dependent Mechanism
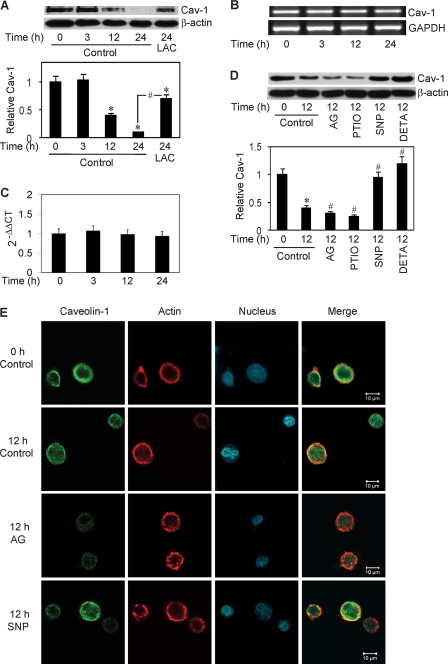
Having demonstrated the role of Cav-1 as a negative regulator of cell anoikis in H460 cells, we next investigated the expression profile of Cav-1 after cell detachment. Detached cells were suspended in attachment-resistant plates for various times and analyzed for Cav-1 protein and mRNA expression by Western blotting and RT-PCR, respectively. Fig. 4A shows that Cav-1 protein levels were significantly reduced in cells after detachment in a time-dependent manner. The reduction was strongly inhibited by lactacystin, a specific proteasome inhibitor, suggesting that detachment-induced Cav-1 down-regulation was mediated through proteasomal degradation. This result was confirmed by the observation that another proteasome inhibitor, MG132, also inhibited the decrease in Cav-1 protein expression (data not shown). Analysis of Cav-1 mRNA levels by RT-PCR shows that Cav-1 transcripts were relatively unchanged after cell detachment (Fig. 4B), whereas the protein levels were substantially reduced. The RT-PCR result was confirmed by quantitative real-time RT-PCR, which shows no significant changes in the Cav-1 mRNA level after cell detachment (Fig. 4C). Thus, cell detachment appears to cause Cav-1 protein reduction through a transcription-independent mechanism. These results along with subsequent data showing the effect of cell detachment on Cav-1 ubiquitination support the role of ubiquitin-proteasomal degradation as an important mechanism of Cav-1 down-regulation induced by cell detachment.
FIGURE 4.
Effect of cell detachment on Cav-1 expression and its regulation by NO. A, H460 cells were detached and seeded in HEMA-coated plates for various times (0–24 h) in the presence or absence of lactacystin (LAC) (10 μm). Cells extracts were prepared and analyzed for Cav-1 protein expression by Western blotting. Blots were reprobed with β-actin antibody to confirm equal loading of samples. The immunoblot signals were quantified by densitometry, and mean data from independent experiments (one of which is shown here) were normalized to the results in control cells at 0 h. B, RT-PCR analysis of Cav-1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression at various times (0–24 h) after cell detachment. C, relative Cav-1 mRNA expression determined by quantitative real-time PCR. The relative mRNA expression was determined by using the comparative CT method as described under “Materials and Methods.” D, detached cells were treated with NO inhibitor, AG (300 μm) or PTIO (50 μm), or with NO donor, SNP (50 μm), or DETA NONOate (50 μm) for 12 h, after which they were analyzed for Cav-1 expression by Western blotting. E, detached cells were either left untreated or treated with SNP (50 μm) or AG (300 μm) for 12 h and analyzed for Cav-1 by immunofluorescence confocal microscopy. Cells were also stained with Alexa Fluor 546-conjugated phalloidin and ToPro-3 to aid visualization of actin cytoskeleton and nucleus. Data are the mean ± S.D. (n = 3). *, p < 0.05 versus attached cell control; #, p < 0.05 versus the indicated control or 12-h detached cell control.
Nitric Oxide Prevents Detachment-induced Cav-1 Down-regulation
We further investigated the potential regulation of Cav-1 by NO. Cells were detached and suspended in HEMA-coated plates in the presence or absence of NO donors and inhibitors. Cav-1 protein expression was then determined by Western blotting. Fig. 4D shows that the NO donors SNP and DETA NONOate strongly inhibited detachment-induced Cav-1 down-regulation at the concentrations shown to induce an increase in cellular NO levels (Fig. 2). In contrast, the NO inhibitors AG and PTIO promoted this down-regulation (Fig. 4D). These results were confirmed by confocal immunofluorescence microscopy which shows that Cav-1 fluorescence intensity was reduced after cell detachment and that the NO donor SNP was able to inhibit this reduction, whereas the NO inhibitor AG further reduced the signal intensity (Fig. 4E). Although these results are non-quantitative, they support the Western blot data and provide additional evidence for the suppressive role of NO in detachment-induced Cav-1 down-regulation. Analysis of the fluorescence pattern in the treated and control cells shows no apparent changes in the cellular localization of Cav-1, which is largely membrane-bound.
Detachment Induces Cav-1 Ubiquitination and Its Inhibition by NO
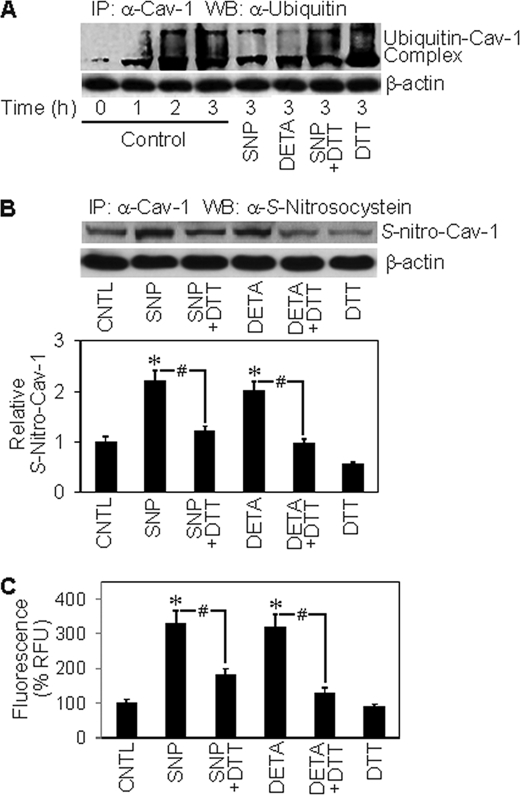
The observation that detachment-induced Cav-1 down-regulation was inhibited by proteasome inhibitors suggests that this down-regulation could be mediated by protein ubiquitination and subsequent degradation by the proteasome. Because ubiquitination of Cav-1 has not been reported, we examined whether cell detachment could induce Cav-1 ubiquitination and whether or not this process is regulated by NO. Cells were detached and suspended in HEMA-coated plates in the presence or absence of NO donors for various times. Cell lysates were prepared and immunoprecipitated with anti-Cav-1 antibody, and the resulting immune complexes were analyzed for ubiquitin by Western blotting. Fig. 5A shows that Cav-1 was rapidly ubiquitinated as early as 1 h after cell detachment and peaked at about 3 h. The NO donors SNP and DETA NONOate strongly inhibited this ubiquitination, suggesting that NO-mediated inhibition of protein ubiquitination could be a key mechanism of Cav-1 stabilization by NO, and our subsequent study supports this notion.
FIGURE 5.
Effects of NO modulators on Cav-1 ubiquitination and S-nitrosylation. A, H460 cells were detached and either left untreated or treated with SNP (50 μm) or DETA NONOate (50 μm) in the presence or absence of DTT (1 mm) in HEMA-coated plates. Cells lysates were prepared and immunoprecipitated (IP) with anti-Cav-1 antibody. The resulting immune complexes were then analyzed for ubiquitin by Western blotting (WB) at various times. Maximum ubiquitination of Cav-1 was observed at ∼3 h after cell detachment. Lysate input was determined by probing β-actin. CNTL, control. B, detached cells were similarly treated with the test agents, and Cav-1 S-nitrosylation was determined by immunoprecipitation using anti-Cav-1 antibody followed by Western blot analysis of the immunoprecipitated protein using anti-S-nitrosocysteine antibody. Densitometry was performed to determine the relative S-nitrosocysteine/β-actin levels. C, Cav-1 S-nitrosylation was determined by fluorometry. Immunoprecipitates from above were incubated with 200 μm HgCl2 and 200 μm diaminonaphthalene in phosphate-buffered saline. NO released from S-nitrosylated Cav-1 was quantified at 375/450 nm. Plots are the mean ± S.D. (n = 3). *, p < 0.05 versus non-treated control; #, p < 0.05 versus the indicated treatment controls.
NO-mediated S-Nitrosylation of Cav-1 as a Potential Mechanism of Protein Stability Regulation
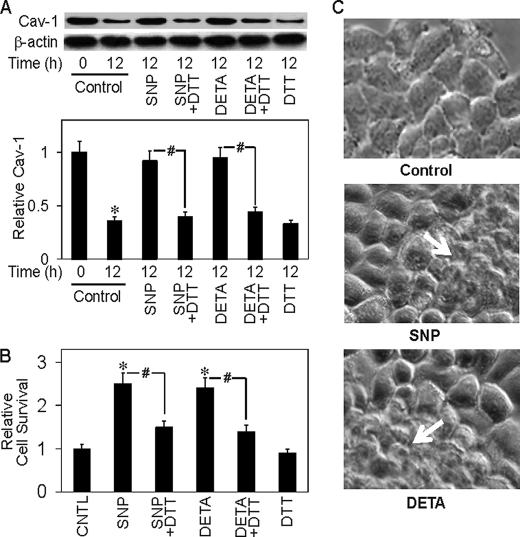
Previous studies have shown that NO induced S-nitrosylation of some apoptosis regulatory proteins, such as c-FLIP and Bcl-2, and prevented their ubiquitination and degradation by the proteasome (30–32). To test whether S-nitrosylation could be involved in the regulation of Cav-1 by NO, we performed immunoprecipitation experiments examining the effect of NO on Cav-1 S-nitrosylation and protein expression. The results show that treatment of the cells with NO donors, SNP, and DETA NONOate induced S-nitrosylation of Cav-1 as determined by Western blot analysis of the immunoprecipitated protein using S-nitrosocysteine antibody (Fig. 5B). Similar results were obtained when the immunoprecipitated protein was analyzed for S-nitrosylation by fluorometric measurements of the released NO product (Fig. 5C). Cav-1 S-nitrosylation by the NO donors was inhibited by DTT, a known inhibitor of S-nitrosylation (37, 38) (Fig. 5, B and C), supporting the specificity of S-nitrosylation detection. To test whether S-nitrosylation of Cav-1 affects its stability, we analyzed the effect of NO donors and DTT on Cav-1 protein expression. Fig. 6A shows that the NO donors SNP and DETA NONOate were able to stabilize the protein after cell detachment and that the S-nitrosylation inhibitor DTT inhibited the stabilizing effect of NO donors. Together, these results indicate that NO regulates Cav-1 expression at least in part by inducing protein S-nitrosylation which interferes with its ubiquitination and subsequent degradation by the proteasome. This new finding provides a mechanistic insight into the regulation of Cav-1 by NO, which could be important in the control of cancer cell anoikis and metastasis.
FIGURE 6.
NO inhibits detachment-induced Cav-1 down-regulation and cell death. A, H460 cells were detached and either left untreated or treated with SNP (50 μm) or DETA NONOate (50 μm) in the presence or absence of DTT (1 mm) in HEMA-coated plates. A, cell lysates were prepared and analyzed for Cav-1 protein expression by Western blotting after 12 h. Densitometry was performed to determine the relative levels of Cav-1 after reprobing the membranes with β-actin antibody. B, cells survival was determined by XTT assay after 12 h. C, morphology of cells treated with SNP (50 μm) or DETA NONOate (50 μm) for 12 h. Data are the mean ± S.D. (n = 3). *, p < 0.05 versus non-treated control; #, p < 0.05 versus the indicated treatment controls.
Nitric Oxide Induces Anoikis Resistance and Multilayer Formation
To provide supporting evidence for the role of NO in the regulation of cell anoikis through protein S-nitrosylation, H460 cells were detached and incubated with NO donors in the presence or absence of DTT. Cell viability was then determined after 12 h using XTT assay. Fig. 6B shows that the NO donors SNP and DETA NONOate significantly increased cell viability after detachment and that DTT inhibited this effect of NO donors. In the earlier study we showed that cells overexpressing Cav-1 exhibited multilayer formation. Because NO up-regulates Cav-1 expression, we examined whether NO could induce a similar multilayer formation in H460 cells. Cells were grown in culture plates in the presence or absence of NO donors, and cell morphology was examined by microscopy after 24 h. Fig. 6B shows that the NO donors SNP and DETA NONOate were able to induce multilayer formation of H460 cells as compared with non-treated control. Because multilayer formation is a key characteristic of malignant tumor cells, this finding suggests that NO may regulate tumorigenesis by promoting malignant transformation through Cav-1 up-regulation.
DISCUSSION
Lung cancer is the leading cause of cancer mortality worldwide, and most death is associated with tumor metastasis. To metastasize, a malignant cell must detach from its primary tumor, invade the nearby circulatory or lymph system, and establish itself in a new site. Once in the bloodstream, most of the cells die by anoikis, which is an important mechanism that prevents metastasis. However, some cancer cells develop resistance to anoikis and consequently survive to establish new metastases. Several anoikis-regulatory proteins including Cav-1 have been investigated in recent years. However, the role of Cav-1 in metastatic cancer progression and the underlying mechanisms are unclear. Both pro- and anti-carcinogenic effect of Cav-1 have been described. Recombinant Cav-1 overexpression was shown to inhibit cell proliferation by inducing cell cycle arrest at G0/G1 phase (1). Genomic analysis of Cav-1−/− null mice and human breast cancer mutations (P132L) supported the role of Cav-1 as a negative regulator of cell transformation and tumorigenesis (39). Likewise, stable expression of Cav-1 in human breast cancer MCF-7 cells attenuated cell proliferation and inhibited anchorage-independent growth (17). In contrast to its suppressive role in cancer cell growth, an elevated expression of Cav-1 has been reported in several human tumors including prostate, colon, and breast (23, 24). Overexpression of Cav-1 also prevented detachment-induced apoptosis and p53 activation in cancer cells (18). Furthermore, in lung cancer cells Cav-1 overexpression promoted metastasis (19). Consistent with the pro-survival role of Cav-1, we found that Cav-1 positively regulated cell growth and inhibited detachment-induced apoptosis of lung cancer H460 cells (Fig. 3). The anti-apoptotic effect of Cav-1 was found to be associated with its ability to activate PI3K/Akt because inhibition of Akt activation by the PI3K inhibitor LY294002 abrogated this effect. Cav-1 has been shown to interact with and inhibit serine/threonine protein phosphatases PP1 and PP2A, leading to sustained Akt activation and inhibition of apoptosis by thapsigargin in prostate cancer cells (16). Cav-1 has also been shown to interact with PI3K and regulate ceramide-induced cell death in fibroblasts (35). Likewise, Cav-1 overexpression in fibroblasts and HeLa cells induces Akt activation; however, such activation promotes cell death induced by arsenite (36). Thus, although there is a general agreement that Cav-1 can activate PI3K/Akt, its consequence on cell death can be different and is dependent on cellular context, cell type, and apoptotic stimuli used.
In this study we observed that Cav-1 is rapidly down-regulated after cell detachment and that overexpression of Cav-1 or stabilization of the protein by NO protected the cells from apoptosis (Figs. 3 and 6). NO induction and Cav-1 overexpression also promoted cell transformation facilitating multilayer formation (Figs. 3 and 6). The expression of Cav-1 is tightly regulated at various levels, including transcriptional and post-transcriptional (for review, see Ref. 40). Although the importance of transcriptional regulation of Cav-1 has been emphasized in numerous studies, post-translational modifications such as ubiquitination and phosphorylation have emerged as important regulators of protein stability and function (for reviews, see Refs. 29 and 41). In the present study we found that Cav-1 was rapidly ubiquitinated and degraded by the proteasome after cell detachment in concomitant with anoikis (Figs. 4 and 5). Cav-1 transcripts were relatively unchanged during this process (Fig. 4), indicating a non-transcriptional regulation of Cav-1 expression after cell detachment. These results support the ubiquitin-proteasomal degradation as a primary mechanism of detachment-induced Cav-1 down-regulation. This finding adds Cav-1 to the growing list of cellular proteins that are subjected to regulation by the ubiquitin-proteasomal degradation pathway.
The results of this study also demonstrated that Cav-1 stability and function is regulated by NO. NO has been shown to regulate apoptosis under various physiologic and pathologic conditions (28, 42, 43); however, its roles in anoikis and metastasis are unclear. Recent evidences suggest that depending on its expression level, NO can exert either promoting or inhibitory effects on tumor growth and metastasis. The promoting effects of NO are generally observed at relatively low but sustained levels of NO, whereas the inhibitory effects of NO are seen at high and acute concentrations that induce tumor cell death (44–46). The results of this study are consistent with previous reports showing that low (micromolar) levels of NO inhibited cell death and increased cell migration (47–49). The promoting role of NO in tumorigenesis is also supported by the observations that both inducible and constitutive forms of NO synthase are elevated in several human tumors (50, 51) and that human and murine carcinomas expressing NO synthase are very aggressive when implanted into mice (52). Furthermore, there seems to be a direct correlation between the expression of NO synthase and the tumor grade, suggesting a causative role for NO in promoting metastasis (52).
We showed that NO negatively regulates anoikis of lung epithelial H460 cells as demonstrated by the ability of NO donors to suppress detachment-induced apoptosis and the reversal effect induced by NO inhibitors (Fig. 1). The NO donors also had a stabilizing effect on detachment-induced Cav-1 down-regulation, whereas the NO inhibitors showed an opposite effect (Figs. 4 and 6). These results support NO-mediated stabilization of Cav-1 as a key mechanism of anoikis regulation. The mechanism by which NO stabilizes Cav-1 was shown to involve inhibition of protein ubiquitination, as the NO donors inhibited the ubiquitination of Cav-1 by cell detachment (Fig. 5).
Recent evidence indicate that S-nitrosylation is an important mechanism by which NO modulates the function of cellular proteins (for reviews, see Refs. 29 and 53). S-Nitrosylation can either attenuate or accentuate protein functions (34, 54, 55), and our previous studies have shown that it plays an important role in stability and function of apoptosis regulatory proteins such as c-FLIP and Bcl-2 (30–32). In the present study we found that Cav-1 was rapidly S-nitrosylated by NO after cell detachment, and inhibition of this S-nitrosylation by DTT blocked the effect of NO on Cav-1 ubiquitination (Fig. 5). The inhibition of S-nitrosylation by DTT also led to a decrease in Cav-1 protein expression and cell survival (Fig. 6), supporting the role of S-nitrosylation in Cav-1 stability and function. The mechanism by which S-nitrosylation promotes Cav-1 stability was shown to involve protein ubiquitination inhibition, although the precise mechanism of this inhibition remains to be investigated. It is possible that S-nitrosylation of Cav-1 may alter the protein conformation such that it could not be recognized by the enzyme ubiquitin ligases that serve to tag the protein for subsequent degradation by the proteasome.
In conclusion, we demonstrated that Cav-1 plays an important role as a negative regulator of anoikis in human lung carcinoma H460 cells. We also demonstrated for the first time that Cav-1 is down-regulated during cell anoikis through a non-transcriptional mechanism involving ubiquitin-proteasomal degradation. NO regulates this process at least in part through protein S-nitrosylation, which prevents its ubiquitination and degradation by the proteasome. In demonstrating the effect of NO on Cav-1 stability and function, we documented a novel mechanism of anoikis regulation, which could be important in the control of cancer metastasis. Because NO and Cav-1 have been shown to be overexpressed in many human tumors, NO-mediated regulation of Cav-1 could be a common mechanism of anoikis resistance and metastasis. This novel finding of the regulation of Cav-1 by NO may have important implications in cancer therapy.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-HL076340. This work was also supported by Thailand Research Fund and Commission on Higher Education (MRG G5080134).
- Cav-1
- caveolin-1
- SNP
- sodium nitroprusside
- AG
- aminoguanidine
- PTIO
- 2-(4-carboxyphenyl)-4,4,5,5-tetramethyl-imidazoline-1-oxyl-3-oxide
- DTT
- dithiothreitol
- DAF-DA
- diaminofluorescein diacetate
- DETA
- diethylene triamine
- FITC
- fluorescein isothiocyanate
- PI
- propidium iodide
- XTT
- sodium 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide
- RT
- reverse transcription
- PI3K
- phosphatidylinositol 3-kinase.
REFERENCES
- 1.Galbiati F., Volonté D., Liu J., Capozza F., Frank P. G., Zhu L., Pestell R. G., Lisanti M. P. (2001) Mol. Biol. Cell 12, 2229–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glenney J. R., Jr., Zokas L. (1989) J. Cell Biol. 108, 2401–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothberg K. G., Heuser J. E., Donzell W. C., Ying Y. S., Glenney J. R., Anderson R. G. (1992) Cell 68, 673–682 [DOI] [PubMed] [Google Scholar]
- 4.Scherer P. E., Okamoto T., Chun M., Nishimoto I., Lodish H. F., Lisanti M. P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 131–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razani B., Schlegel A., Liu J., Lisanti M. P. (2001) Biochem. Soc. Trans. 29, 494–499 [DOI] [PubMed] [Google Scholar]
- 6.Koleske A. J., Baltimore D., Lisanti M. P. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1381–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman J. A., Chu C., Lin A., Jo H., Ikezu T., Okamoto T., Kohtz D. S., Lisanti M. P. (1998) FEBS Lett. 428, 205–211 [DOI] [PubMed] [Google Scholar]
- 8.Lee S. W., Reimer C. L., Oh P., Campbell D. B., Schnitzer J. E. (1998) Oncogene 16, 1391–1397 [DOI] [PubMed] [Google Scholar]
- 9.Hurlstone A. F., Reid G., Reeves J. R., Fraser J., Strathdee G., Rahilly M., Parkinson E. K., Black D. M. (1999) Oncogene 18, 1881–1890 [DOI] [PubMed] [Google Scholar]
- 10.Racine C., Bélanger M., Hirabayashi H., Boucher M., Chakir J., Couet J. (1999) Biochem. Biophys. Res. Commun. 255, 580–586 [DOI] [PubMed] [Google Scholar]
- 11.Galbiati F., Volonte D., Engelman J. A., Watanabe G., Burk R., Pestell R. G., Lisanti M. P. (1998) EMBO J. 17, 6633–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell L., Hollins A. J., Al-Eid A., Newman G. R., von Ruhland C., Gumbleton M. (1999) Biochem. Biophys. Res. Commun. 262, 744–751 [DOI] [PubMed] [Google Scholar]
- 13.Mikol D. D., Hong H. L., Cheng H. L., Feldman E. L. (1999) Glia 27, 39–52 [PubMed] [Google Scholar]
- 14.Timme T. L., Goltsov A., Tahir S., Li L., Wang J., Ren C., Johnston R. N., Thompson T. C. (2000) Oncogene 19, 3256–3265 [DOI] [PubMed] [Google Scholar]
- 15.Li L., Yang G., Ebara S., Satoh T., Nasu Y., Timme T. L., Ren C., Wang J., Tahir S. A., Thompson T. C. (2001) Cancer Res. 61, 4386–4392 [PubMed] [Google Scholar]
- 16.Li L., Ren C. H., Tahir S. A., Ren C., Thompson T. C. (2003) Mol. Cell. Biol. 23, 9389–9404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiucci G., Ravid D., Reich R., Liscovitch M. (2002) Oncogene 21, 2365–2375 [DOI] [PubMed] [Google Scholar]
- 18.Ravid D., Maor S., Werner H., Liscovitch M. (2005) Oncogene 24, 1338–1347 [DOI] [PubMed] [Google Scholar]
- 19.Ho C. C., Huang P. H., Huang H. Y., Chen Y. H., Yang P. C., Hsu S. M. (2002) Am. J. Pathol. 161, 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C. Y., Wang C. H., Chen T. C., Lin H. C., Yu C. T., Kuo H. P. (1998) Br. J. Cancer 78, 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arias-Díaz J., Vara E., Torres-Melero J., García C., Baki W., Ramírez-Armengol J. A., Balibrea J. L. (1994) Cancer 74, 1546–1551 [DOI] [PubMed] [Google Scholar]
- 22.Fujimoto H., Ando Y., Yamashita T., Terazaki H., Tanaka Y., Sasaki J., Matsumoto M., Suga M., Ando M. (1997) Jpn. J. Cancer Res. 88, 1190–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang G., Truong L. D., Timme T. L., Ren C., Wheeler T. M., Park S. H., Nasu Y., Bangma C. H., Kattan M. W., Scardino P. T., Thompson T. C. (1998) Clin. Cancer Res. 4, 1873–1880 [PubMed] [Google Scholar]
- 24.Thompson T. C. (1998) Cancer Metastasis Rev. 17, 439–442 [DOI] [PubMed] [Google Scholar]
- 25.Thomsen L. L., Miles D. W., Happerfield L., Bobrow L. G., Knowles R. G., Moncada S. (1995) Br. J. Cancer 72, 41–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambs S., Merriam W. G., Bennett W. P., Felley-Bosco E., Ogunfusika M. O., Oser S. M., Klein S., Shields P. G., Billiar T. R., Harris C. C. (1998) Cancer Res. 58, 334–341 [PubMed] [Google Scholar]
- 27.Cobbs C. S., Brenman J. E., Aldape K. D., Bredt D. S., Israel M. A. (1995) Cancer Res. 55, 727–730 [PubMed] [Google Scholar]
- 28.Heigold S., Sers C., Bechtel W., Ivanovas B., Schäfer R., Bauer G. (2002) Carcinogenesis 23, 929–941 [DOI] [PubMed] [Google Scholar]
- 29.Iyer A. K., Azad N., Wang L., Rojanasakul Y. (2008) Nitric Oxide 19, 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chanvorachote P., Nimmannit U., Wang L., Stehlik C., Lu B., Azad N., Rojanasakul Y. (2005) J. Biol. Chem. 280, 42044–42050 [DOI] [PubMed] [Google Scholar]
- 31.Chanvorachote P., Nimmannit U., Stehlik C., Wang L., Jiang B. H., Ongpipatanakul B., Rojanasakul Y. (2006) Cancer Res. 66, 6353–6360 [DOI] [PubMed] [Google Scholar]
- 32.Azad N., Vallyathan V., Wang L., Tantishaiyakul V., Stehlik C., Leonard S. S., Rojanasakul Y. (2006) J. Biol. Chem. 281, 34124–34134 [DOI] [PubMed] [Google Scholar]
- 33.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 34.Xu L., Han C., Lim K., Wu T. (2008) J. Biol. Chem. 283, 3077–3087 [DOI] [PubMed] [Google Scholar]
- 35.Zundel W., Swiersz L. M., Giaccia A. (2000) Mol. Cell. Biol. 20, 1507–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shack S., Wang X. T., Kokkonen G. C., Gorospe M., Longo D. L., Holbrook N. J. (2003) Mol. Cell. Biol. 23, 2407–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryu S. D., Yi H. G., Cha Y. N., Kang J. H., Kang J. S., Jeon Y. C., Park H. K., Yu T. M., Lee J. N., Park C. S. (2004) Life Sci. 75, 2559–2572 [DOI] [PubMed] [Google Scholar]
- 38.Moon K. H., Kim B. J., Song B. J. (2005) FEBS Lett. 579, 6115–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams T. M., Lee H., Cheung M. W., Cohen A. W., Razani B., Iyengar P., Scherer P. E., Pestell R. G., Lisanti M. P. (2004) J. Biol. Chem. 279, 24745–24756 [DOI] [PubMed] [Google Scholar]
- 40.Williams T. M., Lisanti M. P. (2005) Am. J. Physiol. Cell Physiol. 288, C494–C506 [DOI] [PubMed] [Google Scholar]
- 41.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 42.Borutaite V., Brown G. C. (2003) Free Radic. Biol. Med. 35, 1457–1468 [DOI] [PubMed] [Google Scholar]
- 43.Souici A. C., Mirkovitch J., Hausel P., Keefer L. K., Felley-Bosco E. (2000) Carcinogenesis 21, 281–287 [DOI] [PubMed] [Google Scholar]
- 44.Mocellin S., Bronte V., Nitti D. (2007) Med. Res. Rev. 27, 317–352 [DOI] [PubMed] [Google Scholar]
- 45.Lala P. K., Chakraborty C. (2001) Lancet Oncol. 2, 149–156 [DOI] [PubMed] [Google Scholar]
- 46.Monteiro H. P., Silva E. F., Stern A. (2004) Nitric Oxide 10, 1–10 [DOI] [PubMed] [Google Scholar]
- 47.Antonova G. N., Snead C. M., Antonov A. S., Dimitropoulou C., Venema R. C., Catravas J. D. (2007) Am. J. Physiol. Heart Circ. Physiol. 292, H893–H903 [DOI] [PubMed] [Google Scholar]
- 48.Choi B. M., Pae H. O., Chung H. T. (2003) Free Radic. Biol. Med. 34, 1136–1145 [DOI] [PubMed] [Google Scholar]
- 49.Dimmeler S., Zeiher A. M. (1999) Cell Death Differ. 6, 964–968 [DOI] [PubMed] [Google Scholar]
- 50.Park S. W., Lee S. G., Song S. H., Heo D. S., Park B. J., Lee D. W., Kim K. H., Sung M. W. (2003) Int. J. Cancer 107, 729–738 [DOI] [PubMed] [Google Scholar]
- 51.Ambs S., Bennett W. P., Merriam W. G., Ogunfusika M. O., Oser S. M., Khan M. A., Jones R. T., Harris C. C. (1998) Br. J. Cancer 78, 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jenkins D. C., Charles I. G., Thomsen L. L., Moss D. W., Holmes L. S., Baylis S. A., Rhodes P., Westmore K., Emson P. C., Moncada S. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4392–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 54.Li J., Billiar T. R., Talanian R. V., Kim Y. M. (1997) Biochem. Biophys. Res. Commun. 240, 419–424 [DOI] [PubMed] [Google Scholar]
- 55.Kim Y. M., Talanian R. V., Billiar T. R. (1997) J. Biol. Chem. 272, 31138–31148 [DOI] [PubMed] [Google Scholar]