Abstract
We analyzed genetic linkage and association of measures of language, speech and reading phenotypes to candidate regions in a single set of families ascertained for SLI. Sib-pair and family-based analyses were carried out for candidate gene loci for Reading Disability (RD) on chromosomes 1p36, 3p12-q13, 6p22, and 15q21, and the speech-language candidate region on 7q31 in a sample of 322 participants ascertained for Specific Language Impairment (SLI). Replication or suggestive replication of linkage was obtained in all of these regions, but the evidence suggests that the genetic influences may not be identical for the three domains. In particular, linkage analysis replicated the influence of genes on chromosome 6p for all three domains, but association analysis indicated that only one of the candidate genes for reading disability, KIAA0319, had a strong effect on language phenotypes. The findings are consistent with a multiple gene model of the comorbidity between language impairments and reading disability and have implications for neurocognitive developmental models and maturational processes.
Electronic supplementary material
The online version of this article (doi:10.1007/s11689-009-9031-x) contains supplementary material, which is available to authorized users.
Keywords: Gene linkage; Language, reading, speech phenotypes; Language impairments; Specific language impairment; Gene associations
Introduction
Although there has been substantial progress recently in the genetics of language impairment and there is strong support for localization to candidate regions on chromosomes 16 and 19 [1–3], the search for candidate genes remains inconclusive [4] with the exception of a recently identified candidate, CNTNAP2 [5]. In contrast, candidate genes are identified for the closely related clinical conditions of Speech Sound Disorder (SSD) and Reading Disability/Dyslexia (RD), including ROBO1, DCDC2, KIAA0319, and DYX1C1 [6]. A significant limitation of the available studies is that the evidence for overlapping genetic etiology is emerging from different samples, ascertained by SSD or RD, with investigations of single dimension phenotypes per sample. One exception is a recent study [7] which investigated multiple phenotypes in a sample ascertained for language impairment using a multivariate variance-components approach to define phenotypes. This study focused on two quantitative trait loci (QTLs) on chromosomes 16q (SLI1) and 19q (SLI2). The authors reported different effects for the two QTLs, such that SLI1 had equally strong effects on a non-word repetition phenotype as on reading and spelling phenotypes, while SLI2 influenced non-word repetition and language phenotypes but not literacy phenotypes. The outcomes draw attention to the need for investigations of possible overlapping gene effects in the domain of language and literacy. In the study reported here we pursue possible overlap of language impairment, SSD, and RD with the sites linked to SSD and RD in a sample of children ascertained as language impaired using concurrent measurements of language, speech and reading abilities for probands, siblings, and other family members.
Specific Language Impairment (SLI) is a condition characterized by late emerging and protracted language acquisition relative to age expectations, without intellectual disability, autism diagnosis, hearing loss, or other obvious contributing conditions. The prevalence is estimated as 7% of 6-year-old children [8]. The impairments involve both receptive and expressive language and include late talking and deficits in grammar, vocabulary and discourse [9]. There is a significant effect on a child’s ability to communicate. Although some of the problems appear to resolve with age, other difficulties persist. Recent evidence has shown that children who are late talkers are at higher risk for continued language problems, particularly in syntax [10]. If the condition is not resolved by school age, children are not likely to “outgrow” it. Instead, language impairments are likely to remain into adolescence and adulthood [11–13].
Twin-based heritabilities of between 0.50 and 0.97 have been reported for measures of SLI [14, 15], particularly in populations which sought therapy [16]. Family aggregation studies document increased risk for SLI among siblings and parents of affected children. Twenty-two percent of nuclear family members of SLI probands are reported with a positive history compared to 7% of control families [17], with a similar range of affectedness across studies [18, 19].
Recent linkage studies from the SLI Consortium of Great Britain [1, 2] report genome-wide linkage screens of quantitative measures of language that implicate chromosomes 16q (SLI1) and 19q (SLI2). A follow-up study [3]confirmed linkage to chromosomes 16 and 19 in a subset of the SLI Consortium full sample.
Based on the finding that a complex speech-language disorder is due to mutation in the FOXP2 gene on chromosome 7q [20], this gene became a candidate for SLI. Microsatellite markers in the FOXP2 gene and surrounding region yielded association with one of the markers about 5 Mb proximal to the gene as well as one marker in the CFTR gene, distal to FOXP2 [21]. Further study of the SLI Consortium identified a down-stream regulatory effect of the FOXP2 gene on chromosome seven on the CNTNAP2 neurexin gene that in turn is known to regulate cortical development [5] and has been linked to late appearance of first words in a sample of children with autism [22].
Interpretation of these advances requires consideration of the behavioral phenotypes of the linkage studies. Language is multidimensional and various measures are utilized in the investigations to date. With the exception of the investigation of Monaco and colleagues [7], the phenotypes have been examined unidimensionally. Omnibus language assessments are full-scale tests that include items across multiple dimensions, adjusted for age expectations. Such broad measures are often used to define probands as well as the phenotype in linkage studies. For example, the SLI Consortium studies used the Clinical Evaluation of Language Fundamentals (CELF) test [23]. Two other more narrowly defined phenotypes are of interest; one is an index of morphosyntax in the domain of tense-marking (TNS) and the second is performance on non-word repetition tasks (NWR). Tense-marking and non-word repetition performance have been identified as strong candidates for clinical markers [24]. Significant heritability in twins is reported for NWR and TNS [25] with a tense marking task originally developed in the lab of Rice as an experimental precursor to a standardized test [26] The SLI Consortium used another experimental TNS task [27]. Non-word repetition tasks are measures of phonological short-term memory that have been suggested as “core deficits” in SLI [28] or as a “key contributory trait of SLI” [29] . The SLI Consortium has consistently used an experimental task [30]. More recently, Bishop [31] cautions that the evidence for non-word repetition deficit as a cause of syntactic deficits (such as the TNS marker) is quite limited; she proposes instead that if both abilities are weak then language impairment is more likely to be evident. Recent studies [3–5]treat non-word repetition as an endophenotype that functions as a marker of SLI when language impairments are not present.
Most studies of genotype/phenotype correspondence in linkage and association analyses reported to date focus on SLI1 and SLI2, with mixed outcomes for phenotypes. The SLI consortium [1] found linkage for SLI1 for NWR and linkage for SLI2 for a CELF measure, outcomes replicated with a second cohort of families [2] . Falcaro et al [3]used the Manchester sample portion of the SLI Consortium, highlighting linkage to SLI1 for nonword rep (N = 33 families) and to SLI2 for TNS (N = 32 families). Results were less strong for the CELF measure (N = 24 for SLI1 and N = 23 for SLI2). Vernes et al [5]detected CNTNAP2-related associations with an omnibus language assessment phenotype (CELF) as well as NWR in 184 families. Although these phenotypes are clearly promising, other phenotypes are also of interest and could clarify genetic effects.
The condition of SLI is related to speech and reading phenotypes. Speech sound disorder (SSD) is characterized by deficits in articulation, phonological processing, and in the cognitive representation of language. This diagnosis excludes cases of speech dyspraxia, identified as part of the FOXP2 phenotype on chromosome 7 [20, 32], although in practice this distinction is not always made, and study populations may include both SSD and dyspraxia [33]. This heterogeneity can complicate efforts to measure genetic influence and localize genes. For example, speech problems are often included within clinical cases of SLI, and as noted above, there is evidence that heritability estimates are increased when probands are ascertained through clinical referral for speech problems [16]. An epidemiologically ascertained sample [34] yielded a prevalence of SSD as 3.8% of 6-year-old children; of the children with speech delay, an average of 0.51% met the SLI diagnostic criteria. When indexed by SLI, 15% of boys and 11% of girls showed SSD. The overlap is somewhat higher in children who have language impairments with lower levels of nonverbal cognitive performance (15% for boys and 28% of girls). In studies of children ascertained for SSD, findings link this condition to dyslexia-related loci on chromosomes 3, 6 and 15, with suggestive links to chromosome 1 [35–38].
Reading impairments are also related to SLI and SSD. Catts [39] reports about 50% of children with language impairment have subsequent reading impairments. The relationship of reading and language abilities changes over time. The early stages of reading development involve rapid improvement in word recognition skills, which are associated with phonological processing abilities including nonword repetition ability. The later stages involve the development of text comprehension which is associated with language comprehension abilities [40, 41]. Nonword repetition ability is also related to the reading phenotype, interpreted as an index of verbal memory thought to influence the learning processes for reading as well as language acquisition. Genetic studies have also illustrated the overlap between reading and SLI with the finding of linkage of a reading discrepancy phenotype to chromosome 13q21 in families ascertained for SLI [42, 43].
Reading disability has high heritabilities and segregation analyses have estimated that there are several major loci involved [44, 45]. Linkage analyses identified at least eight regions [6, 46, 47], particularly on chromosomes 15q (DYX1; [48]), 6p (DYX2; [49–52], 2p (DYX3; [53], 3p (DYX5; [54], and 1p (DYX8; [55–57]. In addition, SSD has also shown linkage to markers in DYX1 [36], DYX2 [36], and DYX5 regions [35]; [38], suggesting common genetic influences. Candidate genes for reading disability have been proposed for several of these loci: MRPL19/C2ORF3 for chromosome 2 [58], ROBO1 for chromosome 3 [59], DCDC2 and KIAA0319 on chromosome 6 [60–63], and DYX1C1 on chromosome 15 [64]. At least four of these genes have a role in neuronal or axonal migration in the CNS [59, 61, 62, 65].
The findings of multiple and shared linkages for RD and SSD are consistent with multi-gene influences on language phenotypes. These findings in turn have inspired theoretical multi-gene models for complex cognitive traits. Galaburda et al [66] posit that multiple genes contribute to reading disability in a complex interaction of genetics, developmental brain changes, and perceptual and cognitive effects associated with dyslexia. They note that although common genetic factors are expected for dyslexia and language impairment, no overlaps have yet been detected. Similarly, Pennington [67] posits a “probabilistic, multiple cognitive deficit” model with shared cognitive factors and pleiotropic genes and other influences that determine the phenotypic outcome. In contrast, Kovas and Plomin [68] propose a “generalist gene” hypothesis which stipulates that there are very many genes that affect cognitive development, each with small effects, and their interactions with environmental factors determine the resulting phenotype. Under this model, detection of the individual genes would be difficult without very large sample sizes. This hypothesis stands in contrast to the results of segregation analyses cited above, however, which have supported a more oligogenic hypothesis.
To date, one study [7] has examined language and reading phenotypes in the same sample ascertained for SLI. This study reports a multivariate linkage analysis of SLI with the SLI Consortium database, with phenotypes consisting of eight scores from a language omnibus test as multiple linguistic phenotypes, three measures of reading/spelling, and a measure of nonword repetition ability. Multivariate analyses provided further support for SLI1 and SLI2 loci, with additional complexities. The conclusion is that their findings “implied that the effect of SLI1 on non-word repetition was equally strong on reading and spelling phenotypes. In contrast, SLI2 appears to have influences on a selection of expressive and receptive language phenotypes in addition to non-word repetition, but did not show linkage to literacy phenotypes” (p. 660).
The principal aims of this investigation were to explore linkage and association of language, speech and reading phenotypes to previously identified QTLs and genes linked to SSD and RD. We aim to replicate previous linkage and association findings for SSD and RD, determine if the linkages extend to SLI diagnostic phenotypes as well, and, if so, to identify new candidates for linkages and associations for SLI.
Subjects and methods
Subjects
A total of 322 participants, including 86 probands, 134 siblings, and 102 parents and other relatives were drawn from an ongoing longitudinal study of Specific Language Impairment. The study was approved by the institutional review boards at the University of Kansas and at the University of Nebraska Medical Center. Appropriate informed consent was obtained from the subjects. There were 86 probands, mean ages 6;1 to 8;10 across variables, ascertained from school speech pathology caseloads followed by assessment to meet the requirements of the study. There were a total of 134 siblings: 77 males, mean age 8;6; 57 females, mean age 8;5. Previous studies report longitudinal outcomes for part of this sample, documenting that the children’s language impairments persist into adolescence [13, 69–72].
Probands met four entrance screening criteria. The first was nonverbal intelligence above 85. For children ages 3;6 to 6;11 it was measured with the Columbia Mental Maturity Scales [73] and for children ages 7–17, the performance IQ scales from the Wechsler Intelligence Test for Children [74] were utilized. Parents and children ages 17 years and older were evaluated with the performance scales for the Wechsler Intelligence Test for Adults [75]. Probands met exclusionary criteria for nonverbal intelligence; this requirement was not met for parents and siblings whose intellectual status was an outcome of the study. The second criterion for the probands was normal hearing acuity. The third was no history of neurological disorders or diagnosis of autism. The fourth was intelligible speech sufficient for language transcription and production of target phonemes used in word final morphology, as in “goes” and “talks.” Probands were identified as SLI based on language performance one standard deviation or more below the mean on an age appropriate language test. All probands were screened for articulation to ensure they could produce the phonemes needed for morphological measurement and sufficient intelligibility for reliable spontaneous language transcription. Family members received age appropriate speech, language, and reading assessments. Siblings were recruited from age 2 years to adulthood. Within age levels, all participants received the same assessments. The probands and siblings received multiple times of measurement as part of the longitudinal study. For the phenotyping in this study, the lowest value of each variable of interest was selected. This is in keeping with the methods used in the SLI Consortium studies where past or current language performance was used to identify probands [3]. Further, the lowest performance estimate captures the late talker status of siblings.
Measures
The phenotypes assessed the following traits for speech, language, reading, and the related area of nonword repetition. For children ages 2;6 to 9 years, speech was measured by the Goldman Fristoe Test of Articulation (GFTA) standard score [76]. Language was subdivided into three dimensions. The first, general language skills, was measured by an omnibus standardized language test appropriate for the individual’s age (Omnibus): for children at or under age 2;6, Preschool Language Scale-3 [77] Total Language Score; ages 2;6–3;11, the Test of Early Language Development-3rd edition, Spoken Language Standard Score [78]; ages 4–6;11, the Test of Language Development-2: Primary Spoken Language Standard Score[79]; ages 7–17 + , Clinical Evaluation of Language Fundamentals-3rd edition Total Language Standard Score (or Expressive Language Score if that is the only one available) [80]. The second language dimension was Vocabulary: ages 2;6-adults was assessed with the Peabody Picture Vocabulary Test-Revised or 3rd edition (PPVT) [81, 82]. The third language dimension was early spontaneous speech production (mean length of utterance, MLU): for children ages 2;6–10 years of age, the Mean Length of Utterance was computed with the Systematic Analysis of Language Transcripts, with z scores calculated from the norms provided by Leadholm & Miller [83]. Finally, the construct of TNS was evaluated in children ages 3–9 years of age on the Test of Early Grammatical Impairment (TEGI) [26] . An experimental version of two of the subtests of TEGI were used in the twin study of Bishop and colleagues [84].
Reading was subdivided into word level reading and comprehension/text reading. Word level reading for children (beginning with children enrolled in kindergarten) through adulthood was measured by the Woodcock Reading Mastery Tests-Revised [85] Letter Identification (to 9 years only), Word Identification and Word Attack (from kindergarten to adulthood) standard scores. Two quantitative indices were used, one a standard score adjusted for age expectations (WRMT) and one a raw score adjusted to an interval scale benchmarked to fifth grade reading levels (WRMT-w). Beginning at age 7 into adulthood, text reading was assessed with the Gray Oral Reading Test (GORT) [86] standard scores.
Following earlier precedents, a related processing phenotype, nonword repetition, was included. Beginning at age 4 years into adulthood, nonword repetition was assessed with the Comprehensive Test of Phonological Processing subtest (CTOPP) [87] standard score.
In addition to the quantitative phenotypes for these tests, categorical phenotypes were also determined with a criterion of standard score of one standard deviation or more below the mean as cut-offs for affected status for each phenotype.
Preliminary analyses
The means for the full sample per age level per measure and the proportion of affected participants is reported in Table 1. The proportion affected varied by trait and age of the participant. In general the Omnibus assessments identified 32–48% of the family members as affected; vocabulary deficits were detected more in younger children than in adults (46% versus 7%); speech impairments were least likely, at 13%; for children ages 3–9 years the MLU identified 72% of the children as affected, and the level drops to 36% for children somewhat older; the TNS measure, TEGI, identified 95% of the probands as affected and 57% of the siblings in the 3–9 year age range. For reading impairments, word level reading was affected in 43% of younger children, 29% of older children and 14% of parents; text level reading was affected in 58% of children and 26% of the parents. As expected, the proportion of reading impairments in the probands was high, 70–88%. The mean nonverbal IQ score was 102.6 for parents; 96.38 for probands; 98.71 for siblings. With an arbitrary level of nonverbal IQ of 75 or below as an indicator of intellectual limitations, three parents and 10 siblings met this criterion.
Table 1.
Percent of participants affected by age group: probands, siblings, and parents
| Group/Variable | N | Age | Mean | % Affected |
|---|---|---|---|---|
| Omnibus | ||||
| Toddler | 11 | 1,7 | 86.00 | 45% |
| 2;6–9 years | 61 | 6,1 | 86.57 | 43% |
| 9+ years | 62 | 12,3 | 85.97 | 48% |
| Parent | 100 | 36,7 | 90.24 | 32% |
| Proband | 86 | 7,8 | 71.13 | 100% |
| PPVT | ||||
| 2;6–9 years | 68 | 5,8 | 84.44 | 46% |
| 9+ years | 60 | 12,4 | 95.97 | 18% |
| Parent | 101 | 36,6 | 98.26 | 7% |
| Proband | 86 | 7,1 | 77.05 | 76% |
| GFTA | ||||
| 3–9 | 117 | 7,11 | 46.10 | 13% |
| Proband | 84 | 6,4 | 24.37 | 38% |
| MLU | ||||
| 2;6–9 | 72 | 6,4 | −1.46 | 72% |
| 9–12 | 36 | 11,0 | −0.91 | 36% |
| Proband | 85 | 6,7 | −1.94 | 87% |
| Woodcock | ||||
| 5–9 | 63 | 6,8 | 87.37 | 43% |
| 9+ | 42 | 12,5 | 91.52 | 29% |
| Parent | 56 | 36,4 | 95.9 | 14% |
| Proband | 84 | 6,8 | 78.83 | 70% |
| GORT | ||||
| 7+ | 97 | 10,5 | 7.51 | 58% |
| Parent | 98 | 36,5 | 10.05 | 26% |
| Proband | 73 | 8,7 | 4.58 | 88% |
| CTOPP | ||||
| 4–9 | 45 | 6,5 | 6.5 | 71% |
| 9+ | 51 | 13,0 | 5.7 | 86% |
| Parent | 44 | 35,7 | 5.5 | 93% |
| Proband | 83 | 8,10 | 5.05 | 100% |
| TEGI | ||||
| 3;0–9;0 | 75 | 6,1 | −2.28 | 57% |
| Proband | 85 | 6,1 | −5.59 | 95% |
Zero order correlations were calculated among the variables and reported in Table 2. As expected, there is a moderate and significant level of association among the variables, in the range of .25–.718, accounting for about 6% – 52% of the variance.
Table 2.
Correlations
| MLU z score | GFTA Std | Woodcock Std | GORT Std | Omnibus score | PPVT | TEGI z | CTOPP Std | |
|---|---|---|---|---|---|---|---|---|
| MLU z score | 1 | .367(**) | .382(**) | .416(**) | .490(**) | .375(**) | .472(**) | .241(**) |
| GFTA Std | .367(**) | 1 | .332(**) | .328(**) | .327(**) | .250(**) | .485(**) | .325(**) |
| Woodcock Std | .382(**) | .332(**) | 1 | .657(**) | .701(**) | .564(**) | .446(**) | .414(**) |
| GORT Std | .416(**) | .328(**) | .657(**) | 1 | .700(**) | .718(**) | .475(**) | .314(**) |
| Omnibus score | .490(**) | .327(**) | .701(**) | .700(**) | 1 | .686(**) | .553(**) | .437(**) |
| PPVT | .375(**) | .250(**) | .564(**) | .718(**) | .686(**) | 1 | .364(**) | .376(**) |
| TEGI z | .472(**) | .485(**) | .446(**) | .475(**) | .553(**) | .364(**) | 1 | .431(**) |
| CTOPP Std | .241(**) | .325(**) | .414(**) | .314(**) | .437(**) | .376(**) | .431(**) | 1 |
** Correlation is significant at the 0.01 level (2-tailed)
* Correlation is significant at the 0.05 level (2-tailed)
Genetic analyses
DNA samples were obtained from probands, parents, and siblings using buccal cell samples obtained from buccal swabs or sputum (Oragene; Genotek, Ottawa, Ontario, Canada) and extracted using standard protocols from the manufactures (Gentra, Oragene). Several extended families were included, and some phenotypes were available for relatives besides siblings. The pair counts for each phenotype by the type of relative are shown in Table 3, and these data were included in the linkage analyses. Thus, this is a family based study with mainly sibling and parent-offspring pairs, but including other relative pairs, thereby adding to the power to detect linkage.
Table 3.
Pair counts for each quantitative phenotype
| Sib | Half Sib | Cousin | Parent Child | Grandparent | Avuncular | |
|---|---|---|---|---|---|---|
| GFTASTD | 55 | 13 | 0 | 0 | 0 | 0 |
| Woodcock | 151 | 25 | 26 | 138 | 0 | 5 |
| mlu_z | 101 | 22 | 21 | 0 | 0 | 0 |
| GORTS | 119 | 23 | 33 | 209 | 1 | 20 |
| omnibusscore | 202 | 41 | 33 | 280 | 5 | 21 |
| CTOPP_S | 145 | 23 | 26 | 112 | 0 | 5 |
| PPVT | 193 | 38 | 33 | 270 | 4 | 20 |
| TEGI_Z | 97 | 18 | 2 | 0 | 0 | 0 |
Linkage analysis was used to screen candidate chromosomal regions on chromosomes 1p36, 3p12-q13, 6p22, and 15q21 which have previously been identified as likely to contain genes influencing RD, and 7q31, which contains the FOXP2 gene and surrounding region. Since most of these regions are large, linkage analysis of microsatellite markers was preferred over high density genotyping with single nucleotide polymorphism (SNP) markers, and studies have demonstrated that a density of microsatellite markers at approximately 2 cM distances can give as much information as a more dense map of SNPs, particularly when parental genotypes are included [88]. Well-characterized microsatellite markers in the critical regions of linkage were identified through the NCBI UNISTS website, with intermarker centimorgan distances taken from the Rutgers Combined Linkage-Physical map v2. [89]. Markers were selected to be about 2 cM apart, particularly targeting the candidate genes. The positions and heterogeneity of each marker are shown in Table 4.
Table 4.
Microsatellite marker location and heterogeneity
| Rutgers genetic map (cM) | NCBI physical map (MB) | heterogeneity | |
|---|---|---|---|
| 1p36 | |||
| D1S2667 | 23.99 | 11.41 | 0.82 |
| D1S2740 | 26.20 | 11.84 | 0.62 |
| D1S507 | 31.90 | 14.77 | 0.78 |
| D1S2672 | 32.79 | 15.02 | 0.74 |
| D1S2697 | 36.04 | 16.16 | 0.7 |
| D1S1592 | 38.86 | 17.81 | 0.63 |
| D1S2826 | 39.60 | 18.18 | 0.65 |
| D1S2644 | 42.05 | 18.77 | 0.81 |
| D1S199 | 43.66 | 19.7 | 0.84 |
| D1S478 | 46.05 | 21.35 | 0.74 |
| D1S2698 | 49.56 | 23.01 | 0.74 |
| D1S2885 | 51.97 | 25.82 | 0.87 |
| D1S2749 | 53.45 | 26.98 | 0.8 |
| D1S470 | 55.69 | 29.83 | 0.76 |
| D1S2783 | 61.42 | 34.02 | 0.68 |
| 3p12-q13 | |||
| D3S1566 | 94.20 | 70.38 | 0.84 |
| D3S3568 | 95.95 | 71.63 | 0.68 |
| D3S3551 | 96.29 | 71.86 | 0.87 |
| D3S3614 | 98.99 | 72.45 | 0.75 |
| D3S3581 | 102.58 | 74.16 | 0.59 |
| D3S3653 | 104.14 | 76.67 | 0.65 |
| D3S3507 | 106.60 | 78.64 | 0.6 |
| *ROBO1 | 78.72 | ||
| D3S3049 | 106.76 | 78.99 | 0.66 |
| D3S1604 | 107.05 | 79.65 | 0.41 |
| D3S1595 | 108.51 | 86.25 | |
| D3S1552 | 109.72 | 88.8 | 0.62 |
| D3S1603 | 111.25 | 99.94 | 0.71 |
| D3S3655 | 112.41 | 103.19 | 0.76 |
| D3S1591 | 114.59 | 106.81 | 0.75 |
| D3S3045 | 116.74 | 108.47 | 0.82 |
| D3S1572 | 119.35 | 112.75 | 0.69 |
| D3S3683 | 120.84 | 114.74 | 0.73 |
| D3S1575 | 124.52 | 117.67 | 0.61 |
| 6p22 | |||
| D6S1597 | 45.77 | 21.83 | 0.54 |
| D6S1663 | 47.95 | 22.71 | 0.68 |
| D6S461 | 48.71 | 23.68 | 0.72 |
| *DCDC2 | 24.28 | ||
| *KIAA0319 | 24.65 | ||
| D6S1554 | 51.19 | 24.95 | 0.71 |
| D6S306 | 53.19 | 28.03 | 0.64 |
| D6S1560 | 55.68 | 33.66 | 0.84 |
| D6S291 | 57.66 | 36.27 | 0.7 |
| D6S2427 | 61.86 | 39.58 | 0.77 |
| D6S1549 | 65.8 | 41.49 | 0.6 |
| 7q31 | |||
| D7S2453 | 115.66 | 105.44 | 0.69 |
| D7S2459 | 118.18 | 107.12 | 0.77 |
| D7S799 | 119.61 | 108.39 | 0.88 |
| D7S471 | 122.34 | 111.82 | 0.8 |
| *FOXP2 | 114.09 | ||
| D7S2554 | 123.59 | 114.23 | |
| D7S486 | 124.45 | 115.68 | 0.8 |
| D7S522 | 124.45 | 115.86 | |
| D7S677 | 125.69 | 116.92 | 0.63 |
| *CFTR | 116.99 | ||
| D7S643 | 126.56 | 120.5 | 0.74 |
| 15q21 | |||
| D15S1012 | 37.16 | 36.79 | 0.73 |
| D15S1044 | 38.97 | 37.45 | 0.69 |
| D15S146 | 40.15 | 37.91 | 0.69 |
| D15S132 | 45.29 | 44.98 | 0.75 |
| D15S143 | 45.72 | 45.69 | 0.64 |
| D15S1028 | 46.89 | 46.78 | 0.82 |
| D15S119 | 47.92 | 47.28 | 0.71 |
| *CYP19A1 | 49.29 | ||
| D15S982 | 48.57 | 50.14 | 0.74 |
| D15S1016 | 49.77 | 51.32 | 0.88 |
| *EKN1 | 53.50 | ||
| D15S1049 | 51.55 | 53.54 | 0.74 |
| D15S1033 | 55.77 | 56.54 | 0.68 |
| D15S155 | 58.52 | 58.2 | 0.73 |
Fluorescent labeled primers for the selected markers were obtained from Applied Biosystems (Foster City, CA) or IDT (Coralville, IA) and genotyping was done on an AB 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). Allele calls were reviewed by two experienced technologists and were checked for inheritance and recombination errors using the programs GAS [90] and MERLIN [91]. Any markers with unresolvable genotypes were re-run and re-evaluated or eliminated from the analysis.
Heritability estimates were calculated using the variance components function in MERLIN. Some caveats apply. Reliability is affected by the small, selected samples, and distributional properties of some of the variables. The heritabilities for the standard scores are: GFTA, 96.05%; Woodcock 62.86%; GORT 18.08%; MLU 23.97%; Omnibus score 30.01%; CTOPP 14.83%; PPVT 22.76%; TEGI 19.30%. We note that these are in the range reported for the variables of [7]).
Linkage studies
Linkage was performed with quantitative and categorical measures using the MERLIN package of programs [92]. The MERLIN-regress program was used for the quantitative measures, and the MERLIN nonparametric linkage method was used for affected status for the same measures. These two methods were selected because the quantitative method should have more power to detect linkage across the range of severity, but the categorical measures may highlight genetic differences between clinically affected vs. unaffected individuals. This approach was also applied in previous linkage studies [3]. Interval linkage analysis was used for both methods, with steps of 0.5 cM, and results are expressed as LOD scores as well as p-values.
To verify the results of the MERLIN analyses on a separate platform, the same quantitative phenotypes were analyzed for linkage using the DeFries-Fulker Augmented analysis as implemented in the SAS macro QMS2 [93]. Both of these methods are optimal for families in which probands are selected but siblings are not highly discordant. We performed the two types of analysis as a check for false positive linkages, assuming that true linkages would be detected regardless of analysis platform. For the DeFries-Fulker analysis, linkage was only performed at the marker loci, and the analysis only includes sib-pairs and; the results were reported as p values.
We looked at eight different phenotypes in these studies, two which largely measure reading (Woodcock and GORT), one which measures articulation (GFTA), and five which examine facets of language (MLU, TEGI, CTOPP, PPVT and the Omnibus language score). The reading and articulation phenotypes were used for replication of the linkages of dyslexia and speech sound disorder in our population. The language phenotypes, which were correlated with the other phenotypes in this population (see Table 2), were selected to determine if the linkages extended to SLI diagnostic phenotypes as well. While this gives us a comprehensive view of the phenotypes that may be linked to these regions, we must acknowledge that the multiple tests make it difficult to interpret our overall significance levels. Except where noted, all p-values reported in this study are nominal p-values, not corrected for multiple testing. Because the phenotypes analyzed are all correlated, and the linkage or association tests should be consistent, a Bonferroni correction would be too conservative. Therefore, for the MERLIN analyses we have reported LOD scores, and for the largest LOD scores we have provided nominal p-values, as well as empirical p-values, based on simulations under the null hypothesis.
To determine the empirical significance of the p-values, repeated simulations were performed for all markers and phenotypes across each chromosome using the simulation function in MERLIN and MERLIN-regress. This procedure uses permutations of genotypes simulated under the null hypothesis, while maintaining phenotypes and family structure. The number of simulations for each chromosome was adjusted to obtain at least 500 representations of the highest LOD score for that chromosome. Based on these calculations, between 1000 and 4000 simulations were performed for each chromosomal region, generating more than 400,000 observations for each phenotype.
SNP association analyses
Based on the results of the linkage, we decided to test three known candidate genes for association with a battery of SNP markers. We genotyped 53 SNPs covering the candidate genes DCDC2 and KIAA0319 on chromosome 6p22 and the FOXP2 region of chromosome 7. SNPs were selected which tag regions of linkage disequilibrium using the Tagger function on HapMap (URL), along with SNPs selected to replicate previously reported associations and haplotypes with RD. In all, 36 SNPs were genotyped on chromosome six spanning the genes DCDC2, KIAA0319, and TTRAP. On chromosome 7, we genotyped 17 SNPs spanning FOXP2, including the region upstream of the gene. Although we found minimal linkage to this gene in our sample, the linkage of this region with SLI [21] and identification of mutations in dyspraxia [20]; [32] made it a candidate. Only quantitative traits were used in this analysis, and analysis was again done by two methods: QTDT [94] and FBAT [95]. The same quantitative measures were used as in the linkage analyses. Genotyping was done on a Sequenom MassArray iPlex system. While replication of associated SNPs would verify a relationship between disorders at an etiologic level, it is possible that the disorder that is manifested is due to different allelic mutations which would have different associated SNPs. In this case, the patterns of association among individuals selected for SLI, SSD, or RD could serve as a method of “triangulating” on the causal genes.
Results: microsatellite linkage analysis
Chromosome 1
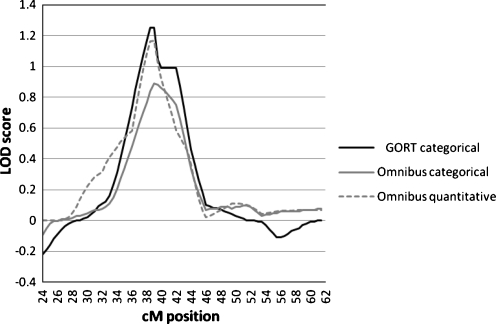
Table 5 and Fig. 1 include only those phenotypes which reached a LOD score of at least 0.60 (equivalent to p < 0.05) for markers on chromosome 1. Two phenotypes showed LOD scores greater than 1.0, the GORT categorical phenotype (LOD 1.25 at 38.49–38.99 cM) and the Omnibus language test quantitative phenotype (LOD 1.165 at 38.99 cM), The Omnibus categorical phenotype also showed a peak in the same area, but with a LOD less than 1.0 (0.890 at 38.99 cM). The peak of linkage spans the marker D1S1592 and is between the two candidate markers, D1S507 (31.9 cM) and D1S199 (43.66 cM), and is precisely within the region defined by de Koval et al. [57] in studies of reading disability.
Table 5.
Chromosome 1 LOD Scores: MERLIN and MERLIN-regress, LOD scores > 0.6 only
| Position (cM) | GORT categorical | Omnibus categorical | Omnibus quantitative |
|---|---|---|---|
| 23.99 | −0.22 | −0.09 | 0.00 |
| 24.49 | −0.19 | −0.05 | 0.00 |
| 24.99 | −0.16 | −0.02 | 0.00 |
| 25.49 | −0.12 | 0.00 | 0.00 |
| 25.99 | −0.09 | 0.00 | 0.00 |
| 26.49 | −0.06 | 0.01 | 0.001 |
| 26.99 | −0.04 | 0.01 | 0.007 |
| 27.49 | −0.02 | 0.01 | 0.023 |
| 27.99 | −0.01 | 0.02 | 0.05 |
| 28.49 | 0.00 | 0.03 | 0.088 |
| 28.99 | 0.00 | 0.03 | 0.135 |
| 29.49 | 0.01 | 0.04 | 0.182 |
| 29.99 | 0.02 | 0.05 | 0.224 |
| 30.49 | 0.04 | 0.06 | 0.258 |
| 30.99 | 0.06 | 0.07 | 0.282 |
| 31.49 | 0.09 | 0.07 | 0.298 |
| 31.99 | 0.11 | 0.08 | 0.317 |
| 32.49 | 0.12 | 0.09 | 0.367 |
| 32.99 | 0.16 | 0.11 | 0.41 |
| 33.49 | 0.23 | 0.15 | 0.443 |
| 33.99 | 0.31 | 0.21 | 0.476 |
| 34.49 | 0.40 | 0.27 | 0.507 |
| 34.99 | 0.51 | 0.34 | 0.535 |
| 35.49 | 0.62 | 0.41 | 0.559 |
| 35.99 | 0.73 | 0.48 | 0.58 |
| 36.49 | 0.85 | 0.55 | 0.706 |
| 36.99 | 0.96 | 0.63 | 0.846 |
| 37.49 | 1.07 | 0.70 | 0.976 |
| 37.99 | 1.17 | 0.77 | 1.083 |
| 38.49 | 1.25 | 0.84 | 1.16 |
| 38.99 | 1.25 | 0.89 | 1.165 |
| 39.49 | 1.04 | 0.88 | 1.015 |
| 39.99 | 0.99 | 0.86 | 0.916 |
| 40.49 | 0.99 | 0.84 | 0.833 |
| 40.99 | 0.99 | 0.81 | 0.749 |
| 41.49 | 0.99 | 0.78 | 0.665 |
| 41.99 | 0.99 | 0.75 | 0.583 |
| 42.49 | 0.87 | 0.66 | 0.538 |
| 42.99 | 0.73 | 0.55 | 0.492 |
| 43.49 | 0.58 | 0.43 | 0.444 |
| 43.99 | 0.46 | 0.34 | 0.362 |
| 44.49 | 0.36 | 0.25 | 0.244 |
| 44.99 | 0.26 | 0.18 | 0.138 |
| 45.49 | 0.18 | 0.12 | 0.064 |
| 45.99 | 0.10 | 0.07 | 0.022 |
| 46.49 | 0.09 | 0.08 | 0.028 |
| 46.99 | 0.08 | 0.08 | 0.04 |
| 47.49 | 0.08 | 0.08 | 0.053 |
| 47.99 | 0.07 | 0.09 | 0.067 |
| 48.49 | 0.06 | 0.09 | 0.081 |
| 48.99 | 0.05 | 0.09 | 0.095 |
| 49.49 | 0.04 | 0.08 | 0.109 |
| 49.99 | 0.03 | 0.09 | 0.111 |
| 50.49 | 0.02 | 0.09 | 0.109 |
| 50.99 | 0.01 | 0.10 | 0.106 |
| 51.49 | 0.00 | 0.10 | 0.101 |
| 51.99 | 0.00 | 0.09 | 0.094 |
| 52.49 | 0.00 | 0.07 | 0.075 |
| 52.99 | −0.01 | 0.05 | 0.057 |
| 53.49 | −0.01 | 0.03 | 0.043 |
| 53.99 | −0.03 | 0.04 | 0.047 |
| 54.49 | −0.06 | 0.04 | 0.051 |
| 54.99 | −0.08 | 0.05 | 0.055 |
| 55.49 | −0.11 | 0.05 | 0.058 |
| 55.99 | −0.11 | 0.06 | 0.061 |
| 56.49 | −0.10 | 0.06 | 0.062 |
| 56.99 | −0.09 | 0.06 | 0.064 |
| 57.49 | −0.07 | 0.06 | 0.065 |
| 57.99 | −0.06 | 0.06 | 0.066 |
| 58.49 | −0.05 | 0.06 | 0.067 |
| 58.99 | −0.03 | 0.07 | 0.068 |
| 59.49 | −0.02 | 0.07 | 0.068 |
| 59.99 | −0.01 | 0.07 | 0.068 |
| 60.49 | −0.01 | 0.07 | 0.068 |
| 60.99 | 0.00 | 0.08 | 0.068 |
| 61.49 | 0.00 | 0.08 | 0.067 |
Fig. 1.
Chromosome 1 MERLIN linkage outcomes
To determine the empirical significance of the results we obtained, random simulations were performed using MERLIN and MERLIN-regress, for the categorical and quantitative phenotypes respectively. These analyses resulted in an empirical p-value of 0.0179 for the Omnibus quantitative maximum LOD score of 1.165, very close to the nominal p-value of 0.01 obtained in the original analysis. Likewise, the statistical significance of the GORT categorical linkage (LOD = 1.25) was changed only minimally by the simulations (nominal p-value = 0.008; empirical p-value = 0.009).
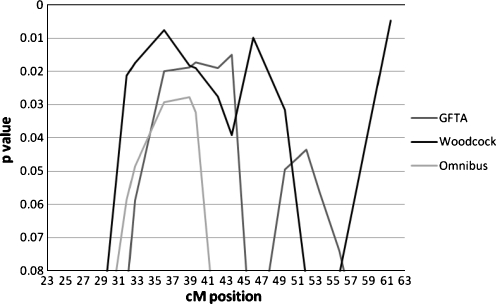
The DeFries-Fulker augmented quantitative linkage analyses mirrored the MERLIN results with major peaks between 36–39 cM for the Woodcock, Omnibus, and GFTA phenotypes (Fig. 2). With the DeFries-Fulker analyses, the maximum significance for the GFTA was at 39.6 cM with p = 0.017, for the Woodcock at 36.04 cM with p = 0.007, and the Omnibus phenotype at 36.04–39.6 cM with p = 0.03. These results involve measures of all three of the clinical disorders, language, reading and speech-sound disorder, within our language-impaired population.
Fig. 2.
Chromosome 1 DeFries Fulker augmented
Chromosome 3
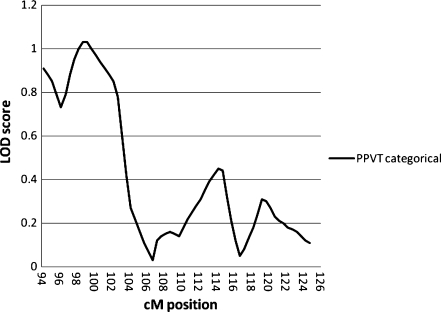
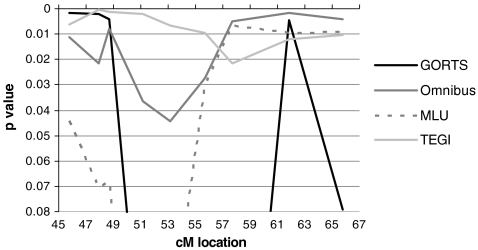
For Chromosome 3, as shown in Table 6 and Fig. 3, the PPVT categorical phenotype had a LOD score of 1.03 with a peak at 98.7 – 99.2 cM, around D3S3614. This is telomeric to the candidate gene ROBO1, which is between 106.60 and 106.76 cM, and the more centromeric region of linkage defined for RD by [96] and SSD by [97], between 106 and 116 cM. No other phenotypes had LOD scores greater than 1.0 with either the categorical or quantitative measures. Random simulations with 1000 replications gave an empirical p-value of 0.015, which is the same as the nominal p value. As shown in Fig. 4, the De-Fries-Fulker augmented analysis showed a similar peak between 94 and 96 cM for the GORT (p = 0.0091), Woodcock (p = 0.015), and Omnibus (p = 0.035) measures. Although this does not appear to overlap the previously reported linkage regions for RD and SSD, these results indicate that this region requires further investigation to determine if this actually defines a separate locus on this chromosome.
Table 6.
Chromosome 3 LOD scores: MERLIN, LOD scores > 0.60 only
| Position (cM) | PPVT categorical |
|---|---|
| 94.2 | 0.91 |
| 94.7 | 0.88 |
| 95.2 | 0.85 |
| 95.7 | 0.79 |
| 96.2 | 0.73 |
| 96.7 | 0.79 |
| 97.2 | 0.88 |
| 97.7 | 0.95 |
| 98.2 | 1 |
| 98.7 | 1.03 |
| 99.2 | 1.03 |
| 99.7 | 1 |
| 100.2 | 0.97 |
| 100.7 | 0.94 |
| 101.2 | 0.91 |
| 101.7 | 0.88 |
| 102.2 | 0.85 |
| 102.7 | 0.78 |
| 103.2 | 0.6 |
| 103.7 | 0.42 |
| 104.2 | 0.27 |
| 104.7 | 0.22 |
| 105.2 | 0.16 |
| 105.7 | 0.11 |
| 106.2 | 0.07 |
| 106.7 | 0.03 |
| 107.2 | 0.12 |
| 107.7 | 0.14 |
| 108.2 | 0.15 |
| 108.7 | 0.16 |
| 109.2 | 0.15 |
| 109.7 | 0.14 |
| 110.2 | 0.18 |
| 110.7 | 0.22 |
| 111.2 | 0.25 |
| 111.7 | 0.28 |
| 112.2 | 0.31 |
| 112.7 | 0.35 |
| 113.2 | 0.39 |
| 113.7 | 0.42 |
| 114.2 | 0.45 |
| 114.7 | 0.44 |
| 115.2 | 0.32 |
| 115.7 | 0.21 |
| 116.2 | 0.12 |
| 116.7 | 0.05 |
| 117.2 | 0.08 |
| 117.7 | 0.13 |
| 118.2 | 0.18 |
| 118.7 | 0.24 |
| 119.2 | 0.31 |
| 119.7 | 0.3 |
| 120.2 | 0.27 |
| 120.7 | 0.23 |
| 121.2 | 0.21 |
| 121.7 | 0.2 |
| 122.2 | 0.18 |
| 122.7 | 0.17 |
| 123.2 | 0.16 |
| 123.7 | 0.14 |
| 124.2 | 0.12 |
| 124.7 | 0.11 |
Fig. 3.
Chromosome 3 MERLIN linkage outcomes
Fig. 4.
Chromosome 3 DeFries Fulker augmented
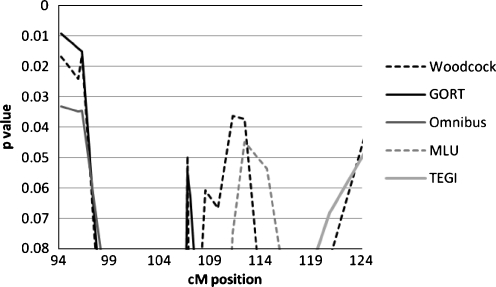
Somewhat weaker linkage results were seen in the previously-described RD/SSD region. The Woodcock and MLU quantitative measures showed marginally significant p-values for replication in the 112–114 cM region (p = 0.036 and 0.045, respectively). The GORT and PPVT categorical scores showed an increase in that area as well with the MERLIN analysis, but were not significant (p = 0.06 and 0.07 respectively). This corresponds to D3S3655 at 113 cM, which was the marker showing maximal linkage for reading disability in a large family reported by [96] and is in the region of linkage for speech sound disorder identified by [97] so it may indicate that reading phenotypes are marginally influenced by a gene or genes in that region in SLI families. We are cautious in this interpretation, however, since strength of linkage may not reliably reflect differential genetic influences on closely related phenotypes [98]; at the same time, these present interesting hypotheses to be investigated further when they involve separate clinically-defined disorders.
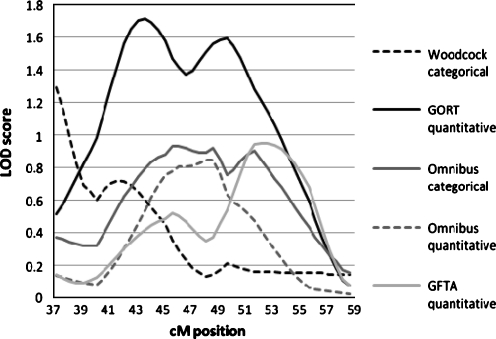
Chromosome 6
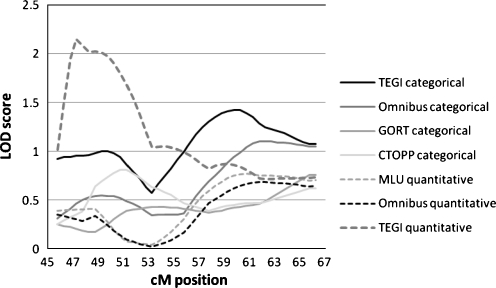
For Chromosome 6, only phenotypes showing a maximum LOD score greater than 0.60 are shown in Table 7 and Figs. 5 and 6. The TEGI quantitative measure had a peak LOD score of 2.145 at 47.27 cm, and the TEGI categorical variable reached a LOD score of 1.0 at 49.77 cM. These peaks are between markers D6S461 and D6S1554 which flank the Reading Disability candidate genes DCDC2 and KIAA0319. Other phenotypes show suggestive peaks in the same region. The TEGI categorical measure also shows a peak of 1.42 at 59.77 and 60.27 cM, between D6S291 and D6S2427. The Omnibus categorical variable also has a peak at a LOD of 1.10 between 61.77–62.77, with trends in that same region for the Omnibus quantitative (LOD 0.684 at 61.77 cM) and MLU quantitative (LOD 0.769 at 60.77 cM) measures. This could correspond to the second peak seen in some previous studies of Reading Disability, although it appears to be slightly centromeric. These differences could be due to variations in the estimates of map distances in the last 10 years, however. Overall, we show strong support for linkage of language phenotypes to the reading disability candidate genes, as well as linkage to a region more centromeric.
Table 7.
Chromosome 6 LOD Scores: MERLIN and MERLIN-regress, LOD scores > 0.60 only
| Position (cM) | TEGI categorical | Omnibus categorical | GORT categorical | CTOPP categorical | MLU quantitative | Omnibus quantitative | TEGI quantitative |
|---|---|---|---|---|---|---|---|
| 45.77 | 0.92 | 0.31 | 0.25 | 0.25 | 0.388 | 0.35 | 1.014 |
| 46.27 | 0.94 | 0.36 | 0.23 | 0.29 | 0.393 | 0.334 | 1.506 |
| 46.77 | 0.94 | 0.41 | 0.22 | 0.32 | 0.397 | 0.318 | 1.947 |
| 47.27 | 0.95 | 0.46 | 0.2 | 0.35 | 0.40 | 0.302 | 2.145 |
| 47.77 | 0.95 | 0.5 | 0.18 | 0.38 | 0.403 | 0.286 | 2.076 |
| 48.27 | 0.96 | 0.53 | 0.17 | 0.5 | 0.41 | 0.308 | 2.015 |
| 48.77 | 0.98 | 0.54 | 0.17 | 0.64 | 0.41 | 0.337 | 2.018 |
| 49.27 | 1.00 | 0.55 | 0.21 | 0.69 | 0.342 | 0.29 | 2.007 |
| 49.77 | 1.00 | 0.54 | 0.25 | 0.74 | 0.268 | 0.238 | 1.965 |
| 50.27 | 0.98 | 0.54 | 0.3 | 0.78 | 0.193 | 0.186 | 1.894 |
| 50.77 | 0.94 | 0.52 | 0.35 | 0.81 | 0.125 | 0.136 | 1.793 |
| 51.27 | 0.86 | 0.49 | 0.39 | 0.81 | 0.077 | 0.094 | 1.67 |
| 51.77 | 0.79 | 0.46 | 0.4 | 0.77 | 0.068 | 0.07 | 1.531 |
| 52.27 | 0.72 | 0.43 | 0.41 | 0.73 | 0.058 | 0.049 | 1.368 |
| 52.77 | 0.63 | 0.38 | 0.42 | 0.68 | 0.047 | 0.03 | 1.188 |
| 53.27 | 0.57 | 0.34 | 0.43 | 0.63 | 0.042 | 0.02 | 1.033 |
| 53.77 | 0.66 | 0.35 | 0.43 | 0.61 | 0.076 | 0.037 | 1.043 |
| 54.27 | 0.74 | 0.35 | 0.43 | 0.58 | 0.121 | 0.059 | 1.044 |
| 54.77 | 0.82 | 0.35 | 0.42 | 0.55 | 0.176 | 0.087 | 1.035 |
| 55.27 | 0.91 | 0.35 | 0.42 | 0.51 | 0.239 | 0.122 | 1.016 |
| 55.77 | 0.99 | 0.36 | 0.41 | 0.47 | 0.307 | 0.165 | 0.988 |
| 56.27 | 1.08 | 0.45 | 0.4 | 0.45 | 0.391 | 0.231 | 0.953 |
| 56.77 | 1.17 | 0.54 | 0.39 | 0.43 | 0.47 | 0.306 | 0.908 |
| 57.27 | 1.24 | 0.62 | 0.38 | 0.41 | 0.538 | 0.387 | 0.856 |
| 57.77 | 1.3 | 0.69 | 0.37 | 0.39 | 0.592 | 0.464 | 0.819 |
| 58.27 | 1.35 | 0.75 | 0.38 | 0.41 | 0.642 | 0.508 | 0.849 |
| 58.77 | 1.39 | 0.82 | 0.39 | 0.43 | 0.685 | 0.549 | 0.865 |
| 59.27 | 1.41 | 0.88 | 0.41 | 0.44 | 0.721 | 0.585 | 0.869 |
| 59.77 | 1.42 | 0.94 | 0.42 | 0.45 | 0.747 | 0.616 | 0.859 |
| 60.27 | 1.42 | 0.99 | 0.43 | 0.46 | 0.763 | 0.642 | 0.838 |
| 60.77 | 1.39 | 1.04 | 0.44 | 0.47 | 0.769 | 0.661 | 0.806 |
| 61.27 | 1.35 | 1.08 | 0.45 | 0.47 | 0.765 | 0.675 | 0.765 |
| 61.77 | 1.28 | 1.10 | 0.46 | 0.47 | 0.752 | 0.684 | 0.719 |
| 62.27 | 1.24 | 1.10 | 0.49 | 0.48 | 0.75 | 0.682 | 0.712 |
| 62.77 | 1.22 | 1.10 | 0.53 | 0.5 | 0.749 | 0.678 | 0.714 |
| 63.27 | 1.2 | 1.09 | 0.57 | 0.52 | 0.746 | 0.674 | 0.716 |
| 63.77 | 1.18 | 1.09 | 0.61 | 0.54 | 0.74 | 0.668 | 0.718 |
| 64.27 | 1.15 | 1.08 | 0.65 | 0.56 | 0.733 | 0.662 | 0.719 |
| 64.77 | 1.12 | 1.07 | 0.69 | 0.58 | 0.723 | 0.655 | 0.721 |
| 65.27 | 1.09 | 1.06 | 0.72 | 0.6 | 0.711 | 0.647 | 0.722 |
| 65.77 | 1.07 | 1.05 | 0.76 | 0.62 | 0.697 | 0.639 | 0.723 |
| 66.27 | 1.07 | 1.05 | 0.76 | 0.62 | 0.703 | 0.644 | 0.729 |
Fig. 5.
Chromosome 6 MERLIN linkage outcomes
Fig. 6.
Chromosome 6 DeFries Fulker augmented
Simulations were performed to obtain empirical significance values of the LOD scores. The LOD score of 2.145 for the TEGI phenotype had an empirical p-value of 0.0013, compared to the nominal p-value of 0.0008. These simulations also showed that a LOD of greater than 0.701 would be required to meet the significance requirement of p < 0.05 for the Omnibus trait, and a LOD greater than 0.621 would be required for the MLU trait. Thus, the MLU results could be accepted as significant at the 0.05 level. Similarly, simulations with the categorical TEGI phenotype gave an empirical p value of 0.007, similar to the nominal p value of 0.005 for the peak LOD score of 1.42.
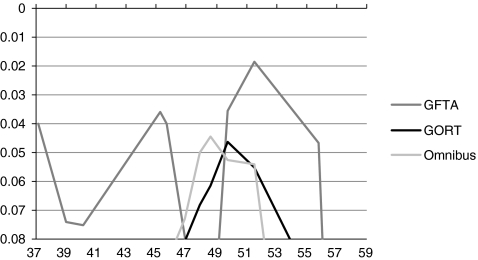
The results of the MERLIN and MERLIN-regress analyses were corroborated by the DeFries-Fulker Augmented analyses. Peaks were seen between 47.95 and 48.71 cM for TEGI (p = 0.00057), GORT (p = 0.0019) and Omnibus score (p = 0.0077), reflecting the linkage to the candidate genes DCDC2 and KIAA0319. A second broad peak of linkage was seen between 58 and 65 cM, with the maximum at 61.88 cm for the Omnibus measure (p = 0.0018, the GORT (p = 0.0047), MLU (p = 0.0094), and TEGI (p = 0.012).
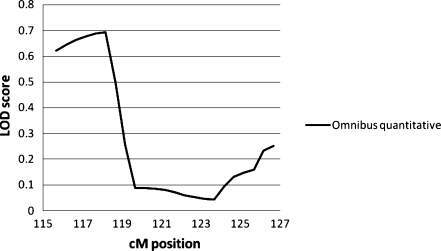
Chromosome 7
For chromosome 7 (see Table 8, Fig. 7), the only phenotype which gave a LOD score over 0.6 (p < 0.05) is the Omnibus measure as a quantitative trait, with a maximum LOD of 0.692 (nominal p = 0.04; empirical p = 0.0493) at 118.16 cM, around D7S2459. This would be upstream of the FOXP2 gene which is between D7S471 and D7S2554, corresponding to 122.34 and 123.59 cM.
Table 8.
Chromosome 7 LOD scores: MERLIN-regress, LOD scores > 0.60 only
| Position (cM) | Omnibus quantitative |
|---|---|
| 115.66 | 0.621 |
| 116.16 | 0.644 |
| 116.66 | 0.663 |
| 117.16 | 0.677 |
| 117.66 | 0.687 |
| 118.16 | 0.692 |
| 118.66 | 0.495 |
| 119.16 | 0.257 |
| 119.66 | 0.087 |
| 120.16 | 0.088 |
| 120.66 | 0.086 |
| 121.16 | 0.081 |
| 121.66 | 0.072 |
| 122.16 | 0.061 |
| 122.66 | 0.054 |
| 123.16 | 0.046 |
| 123.66 | 0.044 |
| 124.16 | 0.095 |
| 124.66 | 0.131 |
| 125.16 | 0.148 |
| 125.66 | 0.159 |
| 126.16 | 0.234 |
| 126.66 | 0.251 |
Fig. 7.
Chromosome 7 MERLIN linkage outcomes
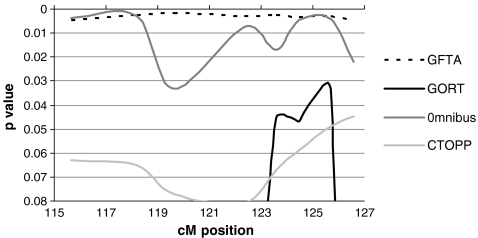
The results for the DeFries-Fulker analysis (Fig. 8) were inconclusive. The GFTA quantitative score gave p values between 0.005 and 0.0018 across the entire region, which may be an artifact. However, this was mirrored somewhat by the Omnibus score, which showed p values of 0.002 between 115 and 118.18 cM, similar to the results of the MERLIN-regress analysis, and also had p values of 0.004 between 124.45 and 126.69 cM. This region is between FOXP2 and CFTR. Overall, while the results of linkage analysis are unclear and thus cannot be considered supportive, the region around FOXP2 is still of interest for language disorders.
Fig. 8.
Chromosome 7 DeFries Fulker augmented
Chromosome 15
Markers on chromosome 15 showed a fairly broad pattern over several phenotypes, as shown in Table 9 and Fig. 9. For simplicity, only those phenotypes showing a LOD score greater than 0.80 are shown in the figure. Two phenotypes had LOD scores greater than 1.0. The Woodcock categorical phenotype had a maximum LOD score of 1.29 at the most centromeric marker, D15S1012 (37.16 cM). The CTOPP quantitative trait had a similar pattern with a maximum LOD of 0.798 (p = 0.03) at the same marker. This is within the region of linkage for SSD previously reported [99], which went from D15S118 (32.39 cM) to D15S209 (50.02 cM), with a peak at D15S214 (40.63 cM). Interestingly, their linkage was found using oral motor variables and Nonword repetition; the latter is equivalent to our CTOPP measure. The second peak of linkage was with the GORT quantitative phenotype, with a maximum LOD of 1.712 at 43.66 cM, with a second peak of 1.594 at 49.66 cM. This region includes the candidate region around D15S119 (47.92 cM) and DYX1C1, between 49.77–51.55. Additional phenotypes had results suggestive of replication of linkage in this region; the Omnibus categorical and quantitative measures (maximum LODs 0.9 and 0.843, respectively), the GFTA quantitative measure (LOD 0.949), and the CTOPP quantitative measure (LOD 0.757). These LODs correspond to nominal p values between 0.05 and 0.02. This region also corresponds to the region of linkage for GFTA and Nonword Repetition on chromosome 15 found in a sample selected for Speech Sound Disorder [100].
Table 9.
Chromosome 15 LOD scores: MERLIN and MERLIN-regress, LOD scores > 0.60 only
| Position (cM) | Woodcock categorical | Omnibus categorical | CTOPP categorical | GORT quantitative | GFTA quantitative | Omnibus quantitative | Woodcock quantitative | CTOPP quantitative | PPVT quantitative |
|---|---|---|---|---|---|---|---|---|---|
| 37.16 | 1.29 | 0.37 | 0.73 | 0.513 | 0.142 | 0.136 | 0.576 | 0.798 | 0.021 |
| 37.66 | 1.15 | 0.36 | 0.6 | 0.577 | 0.118 | 0.126 | 0.58 | 0.709 | 0.031 |
| 38.16 | 0.99 | 0.34 | 0.48 | 0.651 | 0.10 | 0.113 | 0.56 | 0.614 | 0.038 |
| 38.66 | 0.82 | 0.33 | 0.38 | 0.734 | 0.088 | 0.10 | 0.516 | 0.525 | 0.039 |
| 39.16 | 0.69 | 0.32 | 0.32 | 0.82 | 0.09 | 0.089 | 0.487 | 0.476 | 0.034 |
| 39.66 | 0.65 | 0.32 | 0.31 | 0.9 | 0.107 | 0.084 | 0.508 | 0.477 | 0.029 |
| 40.16 | 0.60 | 0.32 | 0.30 | 0.979 | 0.125 | 0.079 | 0.527 | 0.477 | 0.025 |
| 40.66 | 0.66 | 0.39 | 0.30 | 1.134 | 0.165 | 0.12 | 0.583 | 0.491 | 0.054 |
| 41.16 | 0.70 | 0.47 | 0.30 | 1.289 | 0.209 | 0.171 | 0.636 | 0.5 | 0.099 |
| 41.66 | 0.72 | 0.55 | 0.29 | 1.435 | 0.254 | 0.232 | 0.684 | 0.502 | 0.162 |
| 42.16 | 0.71 | 0.62 | 0.28 | 1.558 | 0.3 | 0.301 | 0.724 | 0.497 | 0.243 |
| 42.66 | 0.69 | 0.68 | 0.26 | 1.648 | 0.343 | 0.377 | 0.752 | 0.483 | 0.336 |
| 43.16 | 0.65 | 0.74 | 0.24 | 1.7 | 0.383 | 0.457 | 0.768 | 0.462 | 0.433 |
| 43.66 | 0.6 | 0.79 | 0.22 | 1.712 | 0.417 | 0.537 | 0.77 | 0.434 | 0.523 |
| 44.16 | 0.55 | 0.83 | 0.20 | 1.69 | 0.446 | 0.615 | 0.761 | 0.402 | 0.597 |
| 44.66 | 0.5 | 0.86 | 0.17 | 1.641 | 0.469 | 0.687 | 0.742 | 0.367 | 0.651 |
| 45.16 | 0.45 | 0.88 | 0.15 | 1.574 | 0.486 | 0.751 | 0.715 | 0.332 | 0.686 |
| 45.66 | 0.35 | 0.93 | 0.12 | 1.465 | 0.519 | 0.777 | 0.604 | 0.291 | 0.679 |
| 46.16 | 0.29 | 0.93 | 0.13 | 1.415 | 0.501 | 0.802 | 0.553 | 0.285 | 0.653 |
| 46.66 | 0.23 | 0.92 | 0.15 | 1.372 | 0.465 | 0.806 | 0.511 | 0.281 | 0.619 |
| 47.16 | 0.18 | 0.90 | 0.18 | 1.39 | 0.42 | 0.811 | 0.506 | 0.268 | 0.613 |
| 47.66 | 0.15 | 0.89 | 0.21 | 1.453 | 0.373 | 0.828 | 0.531 | 0.249 | 0.629 |
| 48.16 | 0.13 | 0.89 | 0.27 | 1.507 | 0.349 | 0.842 | 0.557 | 0.247 | 0.64 |
| 48.66 | 0.14 | 0.92 | 0.35 | 1.556 | 0.368 | 0.843 | 0.603 | 0.267 | 0.626 |
| 49.16 | 0.17 | 0.85 | 0.39 | 1.582 | 0.457 | 0.756 | 0.705 | 0.295 | 0.511 |
| 49.66 | 0.21 | 0.75 | 0.41 | 1.594 | 0.533 | 0.634 | 0.789 | 0.323 | 0.38 |
| 50.16 | 0.20 | 0.78 | 0.46 | 1.549 | 0.637 | 0.587 | 0.799 | 0.3 | 0.355 |
| 50.66 | 0.18 | 0.84 | 0.5 | 1.472 | 0.76 | 0.557 | 0.789 | 0.262 | 0.359 |
| 51.16 | 0.17 | 0.88 | 0.54 | 1.376 | 0.872 | 0.52 | 0.775 | 0.225 | 0.361 |
| 51.66 | 0.16 | 0.9 | 0.56 | 1.277 | 0.941 | 0.474 | 0.747 | 0.191 | 0.355 |
| 52.16 | 0.16 | 0.85 | 0.53 | 1.213 | 0.949 | 0.414 | 0.679 | 0.165 | 0.321 |
| 52.66 | 0.16 | 0.79 | 0.50 | 1.14 | 0.949 | 0.353 | 0.608 | 0.141 | 0.286 |
| 53.16 | 0.16 | 0.73 | 0.47 | 1.059 | 0.936 | 0.294 | 0.535 | 0.117 | 0.252 |
| 53.66 | 0.15 | 0.68 | 0.43 | 0.971 | 0.911 | 0.237 | 0.462 | 0.096 | 0.218 |
| 54.16 | 0.15 | 0.62 | 0.4 | 0.878 | 0.871 | 0.184 | 0.39 | 0.076 | 0.186 |
| 54.66 | 0.15 | 0.55 | 0.37 | 0.782 | 0.818 | 0.137 | 0.321 | 0.059 | 0.155 |
| 55.16 | 0.15 | 0.49 | 0.34 | 0.685 | 0.752 | 0.096 | 0.258 | 0.043 | 0.127 |
| 55.66 | 0.15 | 0.43 | 0.30 | 0.59 | 0.678 | 0.062 | 0.20 | 0.03 | 0.101 |
| 56.16 | 0.15 | 0.37 | 0.27 | 0.479 | 0.564 | 0.051 | 0.152 | 0.019 | 0.077 |
| 56.66 | 0.15 | 0.32 | 0.25 | 0.366 | 0.426 | 0.045 | 0.109 | 0.009 | 0.055 |
| 57.16 | 0.14 | 0.26 | 0.22 | 0.264 | 0.295 | 0.039 | 0.073 | 0.003 | 0.037 |
| 57.66 | 0.14 | 0.22 | 0.19 | 0.177 | 0.19 | 0.033 | 0.044 | 0 | 0.022 |
| 58.16 | 0.14 | 0.17 | 0.17 | 0.109 | 0.113 | 0.027 | 0.022 | 0 | 0.011 |
| 58.66 | 0.14 | 0.15 | 0.15 | 0.071 | 0.075 | 0.023 | 0.012 | 0 | 0.005 |
Fig. 9.
Chromosome 15 MERLIN linkage outcomes
To determine the empiric p values for these results, 2000 simulations were performed for the quantitative and categorical analyses respectively. These showed that the maximum LOD score of 1.712 with a nominal p value of 0.002 corresponded to an empirical p value of 0.005. The maximum LOD scores for the Omnibus, GFTA, and CTOPP quantitative phenotypes all meet the empirical criteria for p < 0.05. For the Omnibus measure, the simulated LOD score for a p value of 0.05 was 0.701, for the GFTA it was 0.741, and for the CTOPP it was 0.637. With the categorical Woodcock phenotype, the empirical p value for the LOD of 1.29 was 0.008, similar to the nominal p value of 0.007.
The DeFries-Fulker augmented analyses (Fig. 10) show some corroboration of the second peak of linkage that was seen with the MERLIN and MERLIN-regress analyses, although p values are low. Peaks were seen between 47 and 55 cM for GFTA (p = 0.019 at 51.55 cM, in the DYX1C1 gene), Omnibus score (p = 0.044 at 48.57 cM), and GORT (p = 0.046 at 49.77 cM). The GFTA score also had a p value of 0.04 at the most centromeric marker, which may reflect the SSD linkage reported earlier [99].
Fig. 10.
Chromosome 15 DeFries Fulker augmented
The current results and those from the literature results suggest there may be two loci on chromosome 15 that are linked to language disorders, one on proximal 15q and perhaps associated with the Prader Willi/Angelman syndrome region [99] and at least one more distal locus associated with the candidate RD regions D15S143 and DYX1C1 which may affect RD, SSD, and SLI.
The results of the linkage analyses are summarized in Table 10. Overall, we find the best evidence for replication of linkage to our candidate regions on chromosomes 1, 6, and 15, with suggestive evidence on chromosomes 3 and 7. As in other studies, our sample sizes are small, and some of the phenotypes have been evaluated in only a subset of subjects because they weren’t old enough.
Table 10.
Summary of Linkage Outcomes
| Chr 1 | Chr 3 | Chr 6 | Chr7 | Chr 15 | |
|---|---|---|---|---|---|
| Woodcock | *DF | *DF | **c, *q | ||
| GORT | *c | **DF | *c, **DF | *DF | **q, *DF |
| Omnibus | *c, *q, *DF | *DF | **c, *q, **DF | *q, **DF | *c, *q, *DF |
| CTOPP | *c | *DF | *c, *q | ||
| TEGI | *DF | **c, **DF | |||
| MLU | *DF | *q, **DF | |||
| PPVT | *c | *q | |||
| GFTA | *DF | **DF | *q, *DF |
* LOD > 0.6 (p < 0.05)
**LOD > 1.0 p < 0.01)
c = categorical measure
q = quantitative measure
DF = DeFries-Fulker augmented analysis, quantitative measure
Results: SNP association analysis
Detailed outcomes for the SNP analyses can be found in the “Supplemental Information”. Table 11 summarizes the results for SNPs on chromosome 6 which had p values less than 0.05 for the QTDT and FBAT analyses. The significant results cluster in the 5’ region of KIAA0319 (the gene is read on the “negative” strand), which is the same region of the gene that has shown association in studies of reading disability. In particular, we replicate the associated alleles for rs4504469 (allele C); rs761100 (allele G); rs6935076 (allele T) and rs3756821 (allele A) from previous studies of reading disability [101–104]. It is particularly notable that reading, SSD, and language phenotypes show association to the same alleles, with the exception of the PPVT test, which showed marginal association to the opposite allele. This may be due in part to the small number of informative subjects with the T allele with data for this measure. It is also somewhat surprising that the TEGI phenotype did not show significant association.
Table 11.
Chromosome 6 SNP associations
| QTDT | FBAT | |||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Location (bp) | gene | phenotype | p value | phenotype | allele | p value | |
| rs6456605 | 24444995 | DCDC2 | GFTASTD | 0.0181 | ||||
| rs807530 | 24653918 | KIAA | GFTASTD | 0.0343 | ||||
| rs807533 | 24657885 | KIAA | GFTASTD | C | 0.0187 | |||
| rs2760179 | 24658972 | KIAA | GFTASTD | 0.0141 | ||||
| rs6901322 | 24691783 | KIAA | GORTS GFTASTD | 0.0470 0.0203 | GFTASTD PPVT | T A | 0.0124 0.0413 | |
| rs4504469 | 24696863 | KIAA | GORTS | 0.0400 | ||||
| rs761100 | 24740621 | KIAA | GORTS | G | 0.0412 | |||
| rs6935076 | 24752301 | KIAA | GORTS Omnibus | T T | 0.0167 0.0263 | |||
| rs3756821 | 24754800 | KIAA | GORTS Omnibus | A A | 0.0106 0.0426 | |||
For chromosome 7, summarized in Table 12, the greatest evidence for association was found with the 2 most proximal SNPs, rs7785744 and rs1852638. These reflect the small linkage peak that was observed, and together suggest a localization in a possible regulatory region of FOXP2. Two SNPs located within FOXP2 also showed marginal association.
Table 12.
Chromosome 7 SNP associations
| QTDT | FBAT | ||||||
|---|---|---|---|---|---|---|---|
| SNP | Location (bp) | gene | Phenotype | p value | Phenotype | allele | p value |
| rs7785744 | 113531068 | Woodcockw | 0.0460 | ||||
| GORTS | 0.0130 | ||||||
| Omnibusscore | 0.0240 | ||||||
| rs1852638 | 113632185 | GFTASTD Omnibusscore | T T* | 0.0440 0.0397 | |||
| rs1358278 | 113750570 | GFTASTD | A | 0.0465 | |||
| rs17137004 | 113816487 | Omnibusscore | 0.0430 | ||||
| rs17137124 | 113998050 | FOXP2 | Omnibusscore | T | 0.0408 | ||
| rs12705970 | 114094386 | FOXP2 | GFTASTD | C | 0.0295 | ||
Discussion
This study considers the question of whether regions known to influence RD or SSD also affect related language phenotypes. The results of the linkage and association analyses indicate that it is highly likely that loci exist in the candidate regions that influence language ability, and not just RD or SSD. Linkage analysis does not have the precision to confirm that the same genes in these regions are involved, however. For that reason, association analysis of SNP markers was subsequently done. The SNP association analyses, in an unprecedented finding, point to KIAA0319 as a gene of interest for pleiotropic effects on omnibus language ability, speech impairments, and text comprehension. This common genetic influence is consistent with the pattern of correlations reported in Table 2. The correlation of the omnibus language score and the text comprehension measure (GORT) is high, r = .70, p < .01; the correlation of the speech (GFTA) and reading measure (GORT) is also high, r = .657, p < .01. It should be noted that a vocabulary measure (PPVT) also yielded high correlations with GORT, r = .718, p < .01, with a significant association with one SNP location on KIAA0319. Although the vocabulary association is a weak signal, it is of interest because vocabulary level is a likely mediator of a language effect on text comprehension. Overall, these findings are congruent with investigations of children identified as “poor comprehenders” that report a strong relationship of language impairments and text comprehension performance [105, 106]. In short, the role of KIAA0319 in contributing to the observed overlap of SSD, language impairments and text comprehension warrants further investigation. Other genes or variants in this or other chromosomal regions, not tested in the current work, may also contribute to the shared genetic factors among these speech, reading and language skills.
This is the first evidence of KIAA0319’s possible effect on general language impairment. This finding adds to the earlier reports from the SLI consortium for linkage of chromosomes 16 and 19 to performance on the CELF instrument. It may be that some genes are more influential, in the strength of their effect, in the language domain and others in the overlapping variance shared by reading and language. The findings here suggest that clarification of multi-gene effects can be achieved from focusing on the genes linked to reading as well as the sites associated with language impairments.
The findings here were less clear on the more specific measures of TNS and NWR. Increased sample size will be important in determining if we can differentiate linkages for the more specific measures as suggested by the outcomes of TEGI with chromosome 6 and the correspondence of reading and Omnibus language measures on chromosomes 1 and 15. Yet the sample size of Falcaro et al [3] was also small and yielded significant linkage for chromosomes 16 (NWR) and 19 (CELF/TNS). It may be that the effects are stronger for chromosomes 16 and 19 than for the loci/genes studied here, which would explain why these loci were missed in the original genome screen.
Differences in outcomes, or power to detect linkages, could also be attributable to differences in phenotype measurement. The measures selected for study in this investigation are standardized test instruments, normed on epidemiologically stratified population-based samples of children external to this study. The TNS and non-word repetition tasks in the previous studies have been internally normed on the sample used for genetics investigation, or normed on selected experimental samples available from investigators’ labs. The import of the differences in measurement instruments is whether the binary variables of affectedness are benchmarked to broader population-based samples of children or to more selected samples. Stronger effects may be apparent in binary classifications based on the low end of the ascertained sample versus the low end of an externally-derived sample. As it now stands, the comparison across studies is confounded by differences in the genes/loci of interest, the instruments used for determination of affectedness, and the methods of analyses. Although it appears that multiple genes contribute in different ways to TNS and NWR, further investigation is needed to sort out the number of genes involved, relative robustness of possible effects across measures, and whether these are separate functions that must both be impaired for severe language impairment [4].
In sum, this investigation replicated previous reports of linkages of SSD and RD to QTLs on chromosomes 1, 3, 6, 7, and 15. We identified new suggestive linkages to SLI diagnostic phenotypes, as well, and identified new and promising indications of association of SNPs on chromosome 6 to language impairment, SSD and RD. In particular, KIAA0319 appears to play a role in the shared variance in speech, language, and reading phenotypes. The outcomes add to the growing evidence of the likelihood of multiple gene effects on language and related abilities, and the need for studies of participants with concurrent measurements across the domains of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by NIDCD DC01803. We wish to thank the following research staff for assistance with genotyping: Judith Kenyon, James Askew, Denise Hoover. Behavioral data management and analyses were carried out by Denise Perpich, Allen Richman, and Hiromi Morikawa. Behavioral data collection was carried out by Jennifer Francois, Amy Kepler, Travis Thompson, Andrea Ash, Karla Barnhill, Amy Chadwell, Pat Cleave, Sean Redmond, Tim Brackenbury, Billie Higheagle, Tracy Hirata-Edds, Stacy Betz, April Matthews, Alyson Abel, Anita Alsop, Michelle Knoll, and Julie Hudson. We thank Ken Wexler for his theoretical insights about possible genetic contributions to grammatical impairments of children with SLI. We especially appreciate the participation of the children and families who contributed time and effort to this longitudinal study.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.SLIConsortium A genomewide scan identifies two novel loci involved in specific language impairment. Am J Hum Genet. 2002;70:384–98. doi: 10.1086/338649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.SLIConsortium Highly significant linkage to the SLI1 locus in an expanded sample of individuals affected by specific language impairment. Am J Hum Genet. 2004;74:1225–38. doi: 10.1086/421529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falcaro M, Pickles A, Newbury DF. Genetics and phenotypic effects of phonological short-term memory and grammatical morphology in specific language impairment. Genes Brain Behav. 2008;7:393–402. doi: 10.1111/j.1601-183X.2007.00364.x. [DOI] [PubMed] [Google Scholar]
- 4.Pennington BF, Bishop DVM. Relations among speech, language, and reading disorders, Annu Rev Psychol. 2009;60:283–306. doi: 10.1146/annurev.psych.60.110707.163548. [DOI] [PubMed] [Google Scholar]
- 5.Vernes SC, Newbury DF, Abrahams BS. A functional genetic link between distinct developmental language disorders. N Eng J Med. 2008;359:2337–45. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGrath L, Smith SD, Pennington BF. Breakthroughs in the Search for Dyslexia Candidate Genes, Trends Mol Med. 2006;12:333–41. doi: 10.1016/j.molmed.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Monaco PM. Multivariate linkage analysis of specific language impairment (SLI) Ann Hum Genet. 2007;71:660–73. doi: 10.1111/j.1469-1809.2007.00361.x. [DOI] [PubMed] [Google Scholar]
- 8.Tomblin JB, Records N, Buckwater P. Prevalence of specific language impairment in kindergarten children. J Speech Lang Hear Res. 1997;40:1245–60. doi: 10.1044/jslhr.4006.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leonard L. Children with Specific Language Impairment. Cambridge: MIT Press; 1998. [Google Scholar]
- 10.Rice ML, Taylor CL, Zubrick SR. Language outcomes of 7-year-old children with or without a history of late language emergence at 24 months. JSLHR. 2008;51:394–407. doi: 10.1044/1092-4388(2008/029). [DOI] [PubMed] [Google Scholar]
- 11.Johnson C, Beitchman JH, Young A, et al. Fourteen-year follow-up of children with and without speech/language impairments: Speech/language stability and outcomes. J Speech Lang Hear Res. 1999;42:744–60. doi: 10.1044/jslhr.4203.744. [DOI] [PubMed] [Google Scholar]
- 12.Tomblin JB, Zhang X. The dimensionality of language ability in school-aged children. J Speech Lang Hear Res. 2006;49:1193–208. doi: 10.1044/1092-4388(2006/086). [DOI] [PubMed] [Google Scholar]
- 13.Rice ML, Hoffman L, Wexler K. "Judgments of omitted BE and DO in questions as extended finiteness clinical markers of SLI to fifteen years. ," Journal of Speech , Language, and Hearing Research, in press. [DOI] [PMC free article] [PubMed]
- 14.Bishop DVM. Motor Immaturity and Specific Speech and Language Impairment: Evidence for a common genetic basis, Am J Med Genet. 2002;114:56–63. doi: 10.1002/ajmg.1630. [DOI] [PubMed] [Google Scholar]
- 15.DeThorne L, Hart S, Petrill S, et al. Children’s History of Speech-Language Difficulties: Genetic Influences and Associations with Reading-Related Measures. J Speech Lang Hear Res. 2006;49:1280–93. doi: 10.1044/1092-4388(2006/092). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop DVM, Hayiou-Thomas ME. Heritability of specific language impairment depends on diagnostic criteria. Genes Brain Behav. 2008;7:365–72. doi: 10.1111/j.1601-183X.2007.00360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice ML, Haney KR, Wexler K. Family histories of children with SLI who show extended optional infinitives. J Speech Lang Hear Res. 1998;41:419–32. doi: 10.1044/jslhr.4102.419. [DOI] [PubMed] [Google Scholar]
- 18.Tallal P, Hirsch LS, Raelpe-Bonilla T, et al. Familial aggregation in specific language impairment. J Speech Lang Hear Res. 2001;44:1172–82. doi: 10.1044/1092-4388(2001/091). [DOI] [PubMed] [Google Scholar]
- 19.Tomblin JB. Familial concentration of developmental language impairment, Journal of Speech and Hearing Disorders. 1989;54:287–95. doi: 10.1044/jshd.5402.287. [DOI] [PubMed] [Google Scholar]
- 20.Lai CSL, Fisher SE, Hurst JA, et al. A forkhead-domain gene is mutated in a severe speech and language disorder, Nature. 2001;413:519–23. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 21.O’Brien EK, Zhang X, Nishimura C, et al. Association of specific language impairment (SLI) to the region of 7q31, Am J Hum Genet. 2003;72:1536–43. doi: 10.1086/375403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alarcón M, Abrahams BS, Stone JL. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–9. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semel E, Wiig E, Secord W. Clinical evaluation of language fundamentals, 3rd edition., 3rd ed. San Antonio, TX: Psychological Corporation, 1995
- 24.Tager-Flusberg H, Cooper J. Present and future possibilities for defining a phenotype for specific language impairment. J Speech Lang Hear Res. 1999;42:1275–8. doi: 10.1044/jslhr.4205.1275. [DOI] [PubMed] [Google Scholar]
- 25.Bishop DVM, Adams CV, Norbury CF. Distinct genetic influences on grammar and phonological short-term memory deficits: evidence from 6-year-old twins. Genes Brain Behav. 2006;5:158–69. doi: 10.1111/j.1601-183X.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 26.Rice ML, Wexler K. Rice/Wexler Test of Early Grammatical Impairment. San Antonio: The Psychological Corporation; 2001. [Google Scholar]
- 27. Marchman VA, Wulfeck B, Ellis Weismer S. “Morphological productivity in children with normal language and SLI: A study of the English past tense,” Journal of Speech, Language, and Hearing Research, pp. 206–219, 1999 [DOI] [PubMed]
- 28.Newbury DF, Bishop DVM, Monaco AP. Genetic influences on language impairment and phonological short-term memory, Trends Cogn Sci. 2005;9:528–34. doi: 10.1016/j.tics.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Conti-Ramsden G, Botting N, Faragher B. Psycholinguistic markers for specific language impairment (SLI). J Child Psychol Psychiatry. 2001;42:741–8. doi: 10.1111/1469-7610.00770. [DOI] [PubMed] [Google Scholar]
- 30.Gathercole SE, Willis CS, Baddeley AD, et al. The children’s test of nonword repetition: a test of phonological working memory. Memory. 1994;2:103–27. doi: 10.1080/09658219408258940. [DOI] [PubMed] [Google Scholar]
- 31.Bishop DVM. “Beyond words: Phonological short-term memory and syntactic impairment in specific language impairment.,” Applied Psycholinguistics, pp. 545–547, 2006.
- 32.MacDermot KD, Bonora E, Sykes N, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits, Am J Hum Genet. 2005;76:1074–80. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stromswold K. The genetics of speech and language impairments, N Eng J Med. 2008;359:2381–3. doi: 10.1056/NEJMe0807813. [DOI] [PubMed] [Google Scholar]
- 34.Shriberg LD, Tomblin JB, McSweeny JL. “Prevalence of speech delay in 6-year-old children and comorbidity with language impairment,” Journal of Speech, Language, and Hearing Research, vol. 42, 1999 [DOI] [PubMed]
- 35.Stein CM, Schick J, Taylor H, et al. Pleiotropic effects of a chromosome 3 locus on speech-sound disorder and reading. Am J Hum Genet. 2004;74:283–97. doi: 10.1086/381562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith SD, Pennington BF, Boada R, et al. Linkage of speech sound disorder to reading disability loci. J Child Psychol Psychiatry. 2005;46:1057–66. doi: 10.1111/j.1469-7610.2005.01534.x. [DOI] [PubMed] [Google Scholar]
- 37.Stein CM, Millard C, Kluge A, et al. Speech Sound Disorder Influenced by a Locus in 15q14 Region. Behav Genet. 2006;36:858–68. doi: 10.1007/s10519-006-9090-7. [DOI] [PubMed] [Google Scholar]
- 38.Miscimarra L, Stein CM, Millard C, et al. Further evidence of pleiotropy influencing speech and language: Analysis of the DYX8 region. Hum Hered. 2007;63:47–58. doi: 10.1159/000098727. [DOI] [PubMed] [Google Scholar]
- 39.Catts HW. Language impairment and reading disability. In: Kent R, editor. The MIT Encyclopedia of Communication Disorders. Cambridge: Bradford Books; 2004. [Google Scholar]
- 40.Hoover WA, Gough PB. The simple view of reading. Read Writ. 1990;2:127–60. doi: 10.1007/BF00401799. [DOI] [Google Scholar]
- 41.Catts HW, Adlof SM, Ellis Weismer S. Language deficits in poor comprehenders: A case for the simple view of reading. J Speech Lang Hear Res. 2006;49:278–93. doi: 10.1044/1092-4388(2006/023). [DOI] [PubMed] [Google Scholar]
- 42.Bartlett CW, Flax JF, Logue M, et al. A major susceptibility locus for specific language impairment is located on 13q21, Am J Hum Genet. 2002;71:45–55. doi: 10.1086/341095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartlett CW, Flax JF, Logue MW, et al. Examination of potential overlap in autism and language loci on chromosomes 2, 7, and 13 in two independent samples ascertained for specific language impairment. Hum Hered. 2004;57:10–20. doi: 10.1159/000077385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wijsman E, Peterson D, Leuteneggar A, et al. Segregation analysis of phenotypic components of learning disabilities. I. Nonword memory and digit span. Am J Hum Genet. 2000;67:631–46. doi: 10.1086/303044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chapman N, Raskind W, Thomson J, et al. Segregation analysis of phenotypic components of learning disabilities. II. Phonological decoding. Am J Med Genet. 2003;121:60–70. doi: 10.1002/ajmg.b.20068. [DOI] [PubMed] [Google Scholar]
- 46.Williams J, O’Donovan M. The genetics of developmental dyslexia, Eur J Hum Genet. 2006;14:681–9. doi: 10.1038/sj.ejhg.5201575. [DOI] [PubMed] [Google Scholar]
- 47.Schumacher J, Hoffmann P, Schmal C, et al. Genetics of Dyslexia: The evolving landscape, J Med Genet. 2007;44:289–97. doi: 10.1136/jmg.2006.046516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith SD, Kimberling WJ, Pennington B. Screening for multiple genes influencing dyslexia, Reading and Writing: An Interdisciplinary Journal. 1991;3:285–98. doi: 10.1007/BF00354963. [DOI] [Google Scholar]
- 49.Cardon LR, Smith SD, Fulker DW, et al. Quantitative train locus for reading disability on chromosome 6, Science. 1994;266:276–9. doi: 10.1126/science.7939663. [DOI] [PubMed] [Google Scholar]
- 50.Gayán J, Smith SD, Cherny SS, et al. Quantitative trait locus for specific language and reading deficits on Chromosome 6p. Am J Hum Genet. 1999;64:157–64. doi: 10.1086/302191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fisher S, Marlow A, Lamb J, et al. “A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia.,” Am J Hum Genet, vol. 64, 1999. [DOI] [PMC free article] [PubMed]
- 52.Deffenbacher K, Kenyon J, Hoover D, et al. Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: linkage and association analyses. Hum Genet. 2004;115:128–38. doi: 10.1007/s00439-004-1126-6. [DOI] [PubMed] [Google Scholar]
- 53.Fagerheim T, Raeymaekers P, Tonnessen FE, et al. A new gene (DYX3) for dyslexia is located on chromosome 2, J Med Genet. 1999;36:664–9. [PMC free article] [PubMed] [Google Scholar]
- 54.Nopola-Hemmi J, Myllyluoma B, Haltia T, et al. A dominant gene for developmental dyslexia on chromosome 3. J Med Genet. 2001;38:658–64. doi: 10.1136/jmg.38.10.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rabin M, Wen XL, Hepburn M, et al. Suggestive linkage of developmental dyslexia to chromosome 1p34–p36. Lancet. 1993;342:178. doi: 10.1016/0140-6736(93)91384-X. [DOI] [PubMed] [Google Scholar]
- 56.Grigorenko EL, Wood FB, Meyer MS, et al. Linkage studies suggest a possible locus for developmental dyslexia on chromosome 1p. Am J Hum Genet. 2001;105:120–9. [PubMed] [Google Scholar]
- 57.de Koval CGF, Franke B, Hol FA, et al. confirmation of dyslexia susceptibility loci on chromosomes 1p and 2p, but not 6p in a Dutch sib-pair collection. American Journal of Medical Genetics.Part B Neuropsychiatry Genetics. 2008;147:294–300. doi: 10.1002/ajmg.b.30598. [DOI] [PubMed] [Google Scholar]
- 58.Anthoni H, Zucchelli M, Matsson H, et al. A locus on 2p12 containing the co-regulated MRPL19 and C2ORF3 genes is associated to dyslexia. Hum Mol Genet. 2007;16:667–77. doi: 10.1093/hmg/ddm009. [DOI] [PubMed] [Google Scholar]
- 59.Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, et al. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexic. PLoS Genet. 2005;1:50. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cope N, Harold D, Hill G, et al. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet. 2005;76:581–91. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paracchini S, Thomas A, Castro S, et al. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15:1659–66. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- 62.Meng H, Smith SD, Hager K, et al. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci U S A. 2005;102:17053–8. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schumacher J, Anthoni H, Dahdouh F, et al. Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am J Hum Genet. 2006;78:52–62. doi: 10.1086/498992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taipale M, Kaminen N, Nopola-Hemmi J, et al. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci U S A. 2003;100:11553–8. doi: 10.1073/pnas.1833911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Paramasivam M, Thomas A, et al. DYX1C1 functions in neuronal migration in developing neocortex. Neuroscience. 2006;143:515–22. doi: 10.1016/j.neuroscience.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 66.Galaburda AM, Loturco J, Ramus F, et al. From genes to behavior in developmental dyslexia. Nat Neurosci. 2006;9:1213–7. doi: 10.1038/nn1772. [DOI] [PubMed] [Google Scholar]
- 67.Pennington BF. From single to multiple deficit models of developmental disorders. Cognition. 2006;101:385–413. doi: 10.1016/j.cognition.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 68.Kovas Y, Plomin R. Generalist genes: Implications for the cognitive sciences. Trends Cogn. Sci. 2006;10:198–203. doi: 10.1016/j.tics.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 69.Rice ML, Wexler K, Hershberger S. Tense over time: The longitudinal course of tense acquisition in children with specific language impairment. J Speech Lang Hear Res. 1998;41:1412–31. doi: 10.1044/jslhr.4106.1412. [DOI] [PubMed] [Google Scholar]
- 70.Rice ML, Wexler K, Redmond SM. Grammaticality judgments of an extended optional infinitive grammar: Evidence from English-speaking children with specific language impairment. J Speech Lang Hear Res. 1999;42:943–61. doi: 10.1044/jslhr.4204.943. [DOI] [PubMed] [Google Scholar]
- 71.Rice ML, Wexler K, Marquis J, et al. Acquisition of irregular past tense by children with specific language impairment. J Speech Lang Hear Res. 2000;43:1126–45. doi: 10.1044/jslhr.4305.1126. [DOI] [PubMed] [Google Scholar]
- 72.Rice ML, Redmond SM, Hoffman L. “MLU in children with SLI and young control children shows concurrent validity, stable and parallel growth trajectories,” Journal of Speech, Language, and Hearing Research, pp. 793–808, 2006. [DOI] [PubMed]
- 73.Burgemeister BB, Blum LH, Lorge I. The Columbia Mental Maturity Scale. San Antonio: The Psychological Corporation; 1972. [Google Scholar]
- 74.Wechsler D. Wechsler Intelligence Scale for Children-Third edition. San Antonio, TX:: The Psychological Corporation., 1991.
- 75.Wechsler D. Wechsler Adult Intelligence Scale-Third edition. San Antonio, TX:: The Psychological Corporation., 1997.
- 76.Goldman R, Fristoe M. Goldman Fristoe Test of Articulation-2 (2 ed.) Circle Pines: American Guidance Service; 2000. [Google Scholar]
- 77.Zimmerman IL, Steiner VG, Pond RE. Preschool Language Scale-3. San Antonio, Texas.: The Psychological Corporation, 1992.
- 78.Hresko WR, Kim Reid D, Hammill DD. Test of Early Language Development (TELD-3) Austin: Pro-Ed; 1999. [Google Scholar]
- 79.Newcomer PL, Hammill DD. Test of Language Development — Primary. 3rd Edition. Austin, TX: Pro-Ed., 1999.
- 80.Semel E, Wiig E, Secord W. Clinical Evaluation of Language Fundamentals (3rd ed.). San Antonio,TX:: The Psychological Corporation., 1995.
- 81.Dunn A, Dunn A. Peabody Picture Vocabulary Test-Revised. Circle Pines: American Guidance Service; 1981. [Google Scholar]
- 82.Dunn LM, Dunn LM, Williams KT. Peabody Picture Vocabulary Test III. Circle Pines, MN:: American Guidance Service., 1997.
- 83.Leadholm B, Miller J. Language sample analysis: The Wisconsin guide. Milwaukee: Wisconsin Department of Public Instruction; 1992. [Google Scholar]
- 84.Bishop DVM, Adams CV, Norbury CF. Distinct genetic influences on grammar and phonological short-term memory deficits: Evidence from 6-year-old twins. Genes, Brain, and Behavior. 2005;5:158–69. doi: 10.1111/j.1601-183X.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 85.Woodcock R. Woodcock Reading Mastery Tests — Revised. Circle Pines, MN:: American Guidance Service., 1998.
- 86.Wiederholt JL, Bryant BR. Gray Oral Reading Tests-Third Edition. Austin, TX:: Pro-Ed., 1992.
- 87.Wagner R, Rashotte C, Torgesen J. Comprehensive Test of Phonological Processing. Austin: Pro-Ed; 1999. [Google Scholar]
- 88.Evans DM, Cardon LR. “Guidelines for genotyping in genomewide linkage studies: Single-nucleotide-polymorphism maps versus microsatellite maps.,” Am. J Hum Genet. 2004;75:687–92. doi: 10.1086/424696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matise TC CF, Chen W, De La Vega FM, Hansen M, He C, Hyland FC, Kennedy GC, Kong X, Murray SS, Ziegle JS, Stewart WC, Buyske S. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–6. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Young A. “Genetic Analysis System,” “Genetic Analysis System”http://users.ox.ac.uk/~ayoung/gas.html, 1995.
- 91.Abecasis G, Cherny S, Cookson W, et al. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees, Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 92.Abecasis GR, SS C, WO C, et al. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 93.Lessem JM, Cherny SS. DeFries-Fulker multiple regression of Sibship QTR data: a SAS macro, Bioinformatics. 2001;17:371–2. doi: 10.1093/bioinformatics/17.4.371. [DOI] [PubMed] [Google Scholar]
- 94.Abecasis G, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19:S36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 96.Nopola-Hemmi J. Myllyluoma, et al, “A dominant gene for developmental dyslexia on chromosome 3,”. J Med Genet. 2001;38:658–64. doi: 10.1136/jmg.38.10.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stein CM, Schick JH, Taylor HG, et al. Pleiotropic effects of a chromosome 3 locus on speech-sound disorder and reading. Am J Hum Genet. 2004;74:283–97. doi: 10.1086/381562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pennington BF. Using genetics to dissect cognition, Am J Hum Genet. 1997;60:13–6. [PMC free article] [PubMed] [Google Scholar]
- 99.Stein CM, Millard C, Kluge A, et al. “Speech sound disorder influenced by locus in 15q14 region.,” Behavioral Genetics, vol. 36, 2006. [DOI] [PubMed]
- 100.Smith SD, Pennington BF, Boada R, et al. Linkage of speech and sound disorder to reading disability loci, J Child Psychol Psychiatry. 2005;46:1057–66. doi: 10.1111/j.1469-7610.2005.01534.x. [DOI] [PubMed] [Google Scholar]
- 101.Francks C, Paracchini S, Smith S, et al. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and the United States. Am J Hum Genet. 2004;75:1046–58. doi: 10.1086/426404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cope N, Harold D, Hil G, et al. Strong evidence that KIAA0319 on chromosome 6p is a suceptibility gene for developmental dyslexia. Am J Hum Genet. 2005;76:581–91. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harold D, Paracchini S, Scerri T, et al. Further evidence that the KIAA0319 gene confers susceptibility to developmental dyslexia. Mol Psychiatry. 2006;11:1085–91. doi: 10.1038/sj.mp.4001904. [DOI] [PubMed] [Google Scholar]
- 104.Luciano M, Lind P, Duffy D, et al. A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. Biol Psychiatry. 2007;62:811–7. doi: 10.1016/j.biopsych.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 105.Hoover WaGP. The simple view of reading. Read Writ. 1990;2:127–60. doi: 10.1007/BF00401799. [DOI] [Google Scholar]
- 106.Catts HW, Adlof SM, Weismer SE. Language deficits in poor comprehenders: A case for the simple view of reading. J Speech Lang Hear Res. 2006;49:278–93. doi: 10.1044/1092-4388(2006/023). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.