SUMMARY
The Cbl family of ubiquitin ligases function as negative regulators of activated receptor tyrosine kinases by facilitating their ubiquitination and subsequent lysosomal targeting. Here, we have investigated the role of Cbl ubiquitin ligase activity in the negative regulation of a non-receptor tyrosine kinase, the Src-family kinase Fyn. Using primary embryonic fibroblasts from Cbl+/+ and Cbl−/− mice, we demonstrate that endogenous Cbl mediates the ubiquitination of Fyn and dictates the rate of Fyn turnover. By analyzing CHO-TS20 cells with a temperature-sensitive ubiquitin activating enzyme, we demonstrate that intact cellular ubiquitin machinery is required for Cbl-induced degradation of Fyn. Analyses of Cbl mutants, with mutations in or near the RING finger domain, in 293T cells revealed that the ubiquitin ligase activity of Cbl is essential for Cbl-induced degradation of Fyn by the proteasome pathway. Finally, use of a SRE-luciferase reporter demonstrated that Cbl-dependent negative regulation of Fyn function requires the region of Cbl that mediates the ubiquitin ligase activity. Given the conservation of structure between various Src-family kinases and the ability of Cbl to interact with multiple members of this family, Cbl-dependent ubiquitination could serve a general role to negatively regulate activated Src-family kinases.
Keywords: tyrosine kinase, ubiquitin, regulation, degradation
INTRODUCTION
Src-family kinases (SFKs) constitute a large family of evolutionarily conserved protein tyrosine kinases (PTKs) that mediate crucial biological functions, including critical roles in tissue and organ development, cell differentiation, adhesion and migration, mitogenesis, and immune responses [1;2]. The ease with which subtle mutations can render SFKs dominantly oncogenic [2] has also made them an important model for understanding the mechanisms of PTK regulation.
All SFKs share a conserved domain structure, consisting of a membrane-anchoring N-terminal myristylation signal, adjacent SH3 and SH2 domains, a kinase domain, and a tyrosine residue near the C-terminal tail whose phosphorylation by the C-terminal Src kinase (CSK) is required for repression [1]. The crystal structures of Src and Hck proteins, together with a large body of mutational data, have established a general model of SFK repression and have suggested potential mechanisms of activation [3;4]. Intra-molecular SH3 domain binding to a type II polyproline-like helix within the SH2-kinase linker region together with SH2 domain binding to the phosphotyrosine residue near the C-terminus force the kinase domain into an inactive conformation [3;4]. Activation signals are hypothesized to displace the SH2 and SH3 domains from their intra-molecular ligands, promoting the open, active conformation of the kinase domain and concurrently releasing the SH2 and SH3 domains for assembly of signaling complexes. Consistent with this model, inactivating point mutations in the SFK SH3 or SH2 domains can significantly enhance the kinase activity [5]. Furthermore, mutations within the SH2-kinase linker that abolish its binding to the SH3 domain, or overexpression of high affinity SH3 domain-binding ligands, result in increased kinase activity of Hck, Src, or Lck [6-8]. Similarly, deletion or substitution of the negative regulatory tyrosine within the carboxyl tail of SFKs results in enhanced kinase activity and oncogenesis [2], and deletion of the CSK gene leads to constitutively activated SFKs [9;10]. Conversely, substitutions that enhance the affinity of the C-terminal phosphotyrosine motif for the SH2 domain decrease the kinase activity [11].
While the above paradigm elegantly accounts for basal repression and provides a plausible scheme for activation of SFKs, it is not clear at present if and how activated SFKs are returned to their basal repressed conformation. Given recent evidence that SFKs require cellular chaperones, such as members of the HSP90 family, for proper folding [12], it is likely that cells utilize additional mechanisms for deactivation of SFKs and, by implication, other PTKs. Without such ancillary mechanisms, activated SFKs could accumulate resulting in deleterious consequences for a cell. Recent studies indicate that the proto-oncoprotein Cbl provides one such mechanism for deactivation of SFKs [13;14].
Cbl is a member of an evolutionarily conserved family of cytoplasmic proteins that have emerged as negative regulators of PTK signaling [15]. The Cbl homologues in Caenorhabditis elegans and Drosophila function as negative regulators of epidermal growth factor receptor (EGFR) signaling [16]. Furthermore, genetic ablation of murine Cbl produced hypercellularity and altered development of several organ systems [17;18], whereas Cbl-b deletion led to hyperproliferation and hyperactivation of immune cells resulting in autoimmunity [19;20].
Recent studies have demonstrated that Cbl functions as a ubiquitin ligase towards activated receptor tyrosine kinases (RTKs), a modification that facilitates sorting of ligand-activated receptors to lysosomes where they are degraded [21-23]. This proposed mechanism is analogous to the genetically well-characterized, ubiquitin-dependent, lysosomal targeting of yeast membrane receptors [24]. It is thought that lysosomal enzymes degrade the extracellular regions of growth factor receptors, while the cytoplasmic portion of these receptors may be targeted for proteasomal degradation [21-23]. Notably, transfection studies have shown that Cbl can target the activated pools of non-receptor PTKs such as Syk, ZAP-70 and the SFK Fyn for degradation [13;25;26]. However, the role of Cbl ubiquitin ligase function in the negative regulation of these non-receptor PTKs has not been addressed. Importantly, if non-receptor PTKs are indeed targeted for Cbl-dependent ubiquitination, their fate is likely to differ from that of ubiquitinated RTKs, as their ubiquitination is likely to target them directly to the proteasome rather than serving as a lysosomal sorting signal.
Defining the role of ubiquitination in Cbl-dependent regulation of SFKs is important not only due to the intrinsic biological significance of SFK regulation, but also because these PTKs interact with Cbl in a manner that is far more complex than the interactions of Cbl with other PTK targets [13]. The evolutionarily conserved N-terminal region tyrosine kinase binding (TKB) domain of Cbl, composed of a four-helical bundle, an EF-hand and an incomplete SH2 domain [27], specifically interacts with negative regulatory phosphorylation sites within Syk/ZAP-70 and EGFR tyrosine kinases, providing a basis for the selective recruitment of Cbl to activated pools of these PTKs [23;25;28]. Mutations (in Cbl or its target PTKs) that abrogate Cbl TKB domain interaction with PTKs block Cbl-dependent negative regulation of EGFR, platelet-derived growth factor (PDGFR) and Syk/ZAP-70 PTKs [23;29;25;30;26]. Furthermore, an intact Cbl RING finger domain, which interacts with E2 ubiquitin conjugating enzymes (UBCs) [31], is also required for ubiquitination and downregulation of the EGFR [21;32]. Notably, the TKB and RING finger domains, without the C-terminal half of Cbl, are sufficient for the negative regulation of Syk or EGFR, as well as the ubiquitination of EGFR [32;29;33].
In contrast to Syk/ZAP-70, which interact with Cbl exclusively via its TKB domain, and RTKs, which require a Cbl TKB-mediated interaction for negative regulation, SFK regulation by Cbl is more complex. Previous studies have demonstrated that Cbl-SFK association involves binding between the SFK SH3 domain and the proline-rich sequences in the C-terminal half of Cbl [34]. Furthermore, the SH2 domains of SFKs can interact with phosphopeptide motifs in the C-terminal half of Cbl [35], and an uncharacterized motif in Fyn can interact with the Cbl TKB domain [13]. Consistent with these multiple modes of physical association, a TKB domain mutant of Cbl was fully capable of decreasing the levels and activity of Fyn when analyzed in a 293T cell transfection system; abrogation of Fyn SH3 binding to the proline-rich region of Cbl, in addition to a Cbl TKB mutation, was required to block the effect of Cbl on Fyn [13]. Given these complexities of Cbl-SFK association, and the fact that two of these interactions involve the C-terminal region of Cbl that is dispensable for EGFR and Syk/ZAP-70 regulation, it is critical to determine if Cbl-mediated negative regulation of SFKs indeed involves its activity as a ubiquitin ligase.
Several lines of evidence support the possibility that Cbl-mediated negative regulation of SFKs may be mediated through ubiquitination. We showed that coexpression of Fyn with Cbl resulted in Fyn degradation, and cell lines from Cbl−/− mice showed elevated Fyn levels [13]. Recent studies of other SFKs have revealed them to be targets of ubiquitination [36-39]. For example, Blk was reported to interact with the ubiquitin ligase E6AP and undergo E6AP-dependent ubiquitination and degradation [38]. Similarly, oncogenic v-Src as well as c-Src, the latter in CSK-deficient fibroblasts, were shown to be ubiquitinated; furthermore, treatment with proteasome inhibitors led to increased protein levels [36;37]. While the role of the Cbl proteins in the above situations has not been investigated, these findings are consistent with Cbl regulation of SFKs via ubiquitination.
Here, we have addressed this hypothesis through analyses of Cbl+/+ and Cbl−/− cell lines, Chinese Hamster Ovary (CHO) cells with a temperature-sensitive defect in ubiquitin activating enzyme (E1) and 293T cells co-expressing Cbl and its ubiquitination-deficient mutants. We provide direct evidence that Cbl negatively regulates the SFK Fyn by targeting it for ubiquitination, and that ubiquitination is a critical mechanism to regulate Fyn protein levels and activity. Given the conservation of structure among SFKs, and the ability of Cbl to interact with multiple SFKs, Cbl-dependent ubiquitination may provide a general mechanism to negatively regulate activated SFKs.
MATERIALS AND METHODS
Cells
293T human embryonic epithelial kidney cells and mouse embryonic fibroblasts (MEFs) from wildtype (Cbl+/+) and Cbl knockout (Cbl−/−) mice were maintained as previously described [13]. The CHO cell line CHO-TS20, harboring a temperature-sensitive ubiquitin activating enzyme (E1), was maintained as previously described [40].
Antibodies
The following antibodies were used: monoclonal antibody (mAb) 12CA5 (anti-influenza hemagglutinin [HA] epitope tag; IgG2b) [41]; mAb anti-ubiquitin (IgG1, MMS-258R) from Covance; rabbit polyclonal antibody (pAb) anti-p44/42 MAP kinase (9102) from New England BioLabs; mAb anti-EGFR (IgG2a, sc-120), pAb anti-Fyn (sc-16) and pAb anti-Cbl (sc-170) from Santa Cruz Biotechnology Inc.
Expression plasmids
The HA-ubiquitin, pSRαNeo-CD8-ζ chimera, Cbl and Fyn expression constructs in the pAlterMAX plasmid backbone (Promega) and GFP-Cbl expression constructs in the pCDNA3 vector backbone (Invitrogen) have been previously described [13;29;25;33;42]. The Cbl RING finger mutant C3AHN was previously referred to as Cbl-C3HC4C5 [33].
Transient Transfections
293T cells were transfected as previously described using the calcium phosphate method. Cell lysates were prepared 48 hr post-transfection with Triton X-100 lysis buffer [26] supplemented with 0.1 % SDS and 0.1% DOC. TS20 cells were transfected using the Lipofectamine™ reagent (Life Technologies), according to the manufacturer’s protocol. The cells were cultured at 30°C for 56 hr, then either maintained at 30°C (permissive temperature for E1 function) or shifted to 42°C (non-permissive temperature). Cell lysates were prepared in the lysis buffer described above.
Generation of Fyn-overexpressing MEFs
Cbl+/+ and Cbl−/− MEFs overexpressing Fyn were established by retrovirus-mediated transfection of Cbl+/+ and Cbl−/− MEFs. The retroviral construct MSCVpac-Fyn-T was generated by subcloning murine Fyn-T cDNA fragment from pAlterMAX-Fyn into EcoRI digested MSCVpac. Retroviral supernatants were produced and used to infect target cells as described [43]. Bulk transfectant lines were selected in 5 μg/ml puromycin (Sigma) and used as such.
Immunoprecipitation, gel electrophoresis and immunoblotting
Immunoprecipitations were performed as described [44]. The immunoprecipitated proteins and total cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidine difluoride (PVDF) membranes (NEN), and immunoblotted with the indicated antibodies as described [45]. Blots were visualized as described [26]. Photographs were generated by direct scanning of films using a Hewlett Packard ScanJet 4c™ scanner.
Pulse-Chase Analysis of Fyn Protein Turnover
Fyn-overexpressing Cbl+/+ and Cbl−/− MEFs were grown in 150-mm tissue culture dishes to about 70% confluence, labeled for 1 hr at 37°C with 300μCi/ml EXPRE35S35S labeling mix (NEN), and pulse-chase analysis was performed as previously described [13]. Autoradiography signals were quantified by densitometric analysis of bands using ScionImage software (version beta 3b).
Luciferase Assay
293T cells were transfected by the calcium phosphate method with a serum response element (SRE)-luciferase reporter construct and the appropriate Cbl and Fyn constructs, as previously described [13]. At 48 hr post-transfection, cells were lysed with Cell Culture Lysis Reagent (Promega) and lysate protein concentrations were determined using the Bradford assay. Luciferase activity was determined on equal protein aliquots using a Monolight 3010C luminometer (Analytical Bioluminescence Laboratory Inc.) and Luciferin Reagent (Promega).
RESULTS
Severely reduced ubiquitination of Fyn in MEFs derived from Cbl-deficient mice
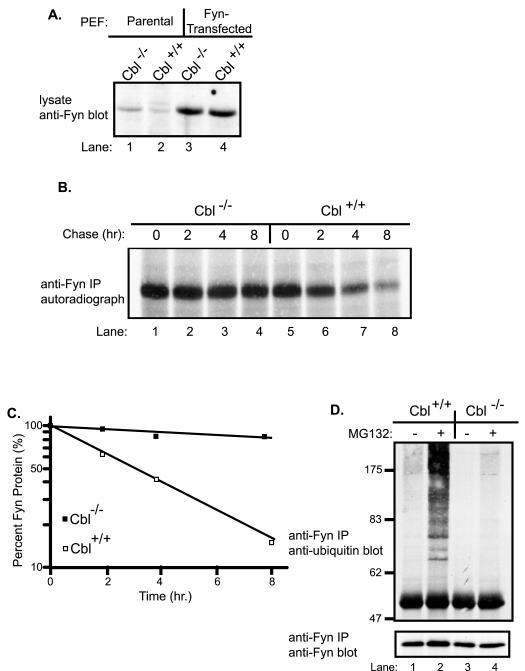
We have previously demonstrated that Cbl targets the Fyn protein for degradation in a 293T cell overexpression system, and that steady-state levels of Fyn were elevated in Cbl−/− MEF and T cell lines [13]. The latter system provided an opportunity to directly assess if endogenous Cbl controls Fyn ubiquitination. In view of the known difficulties in detecting ubiquitinated proteins such as Fyn [46], we established a matched pair of Cbl+/+ and Cbl−/− MEF lines expressing approximately ten-fold higher levels of Fyn compared to parental MEFs (Fig. 1A).
Fig. 1. Stabilization of Fyn protein and impaired Fyn ubiquitination in Cbl−/− primary embryonic fibroblasts.
A, Fyn protein levels in parental versus Fyn-transfected primary embryonic fibroblasts (MEFs). Equal amounts (50 μg) of protein lysates from the parental and Fyn-transfected Cbl−/− and Cbl+/+ MEFs were resolved by SDS-PAGE and immunoblotted with anti-Fyn antibody. B, metabolic pulse-chase analysis of Fyn protein in Fyn-transfected Cbl−/− and Cbl+/+ MEFs. Cbl−/− and Cbl+/+ MEFs were methionine-starved for 1 hr and pulse-labeled with 35S-methionine for 1 hr, as described in materials and methods. The cells were incubated in methionine-supplemented, unlabeled medium (chase) for the indicated times (hr, hours), and cell lysates were prepared. Anti-Fyn immunoprecipitates (IP) of cell lysates (1 mg) were resolved by SDS-PAGE, and labeled Fyn signals were detected by autoradiography. C, the radioactive Fyn signals in B were quantified using densitometry, expressed as a percentage of the maximal signal intensity and plotted as a function of chase times. D, impaired Fyn ubiquitination in Cbl−/− MEFs. Fyn-transfected Cbl−/− and Cbl+/+ MEFs were incubated with 50 μM MG132 (+) or DMSO control (−) for 5 hr and then lysed. Anti-Fyn immunoprecipitates of 1 mg aliquots of lysate were immunoblotted with anti-ubiquitin antibody (top panel), followed by anti-Fyn antibody (bottom panel).
To determine the impact of the presence or absence of endogenous Cbl on the stability of Fyn protein, we carried out a metabolic pulse-chase analysis of Fyn in the Fyn-transfected Cbl+/+ and Cbl−/− MEFs. Equal aliquots of cell lysates were subjected to anti-Fyn immunoprecipitation and radiolabeled Fyn was detected by autoradiography (Fig. 1B). Comparable 35S-Fyn signals were observed in the two cell lines prior to chase (time zero) (Fig. 1B, compare lane 1 with lane 5). Whereas the radiolabeled Fyn signal in Cbl+/+ MEFs showed a substantial time-dependent reduction of nearly 80% over the chase period, with a half-life of about 3 hr, Fyn protein in Cbl−/− cells was substantially more stable with only a small decrease in signal during the chase period (Fig. 1B and C). These results established that endogenous Cbl controls the stability of the Fyn protein, and provided crucial reagents to directly assess if Cbl regulates the ubiquitination of Fyn.
To assess Fyn ubiquitination in Fyn-transfected Cbl+/+ and Cbl−/− MEFs, the cells were incubated for 5 hr with (+) or without (−) the proteasome inhibitor MG132, and their lysates were subjected to anti-Fyn immunoprecipitations followed by anti-ubiquitin immunoblotting. A low but detectable ubiquitin signal, seen as a smear similar to ubiquitinated species of other SFKs [37;39], was observed in anti-Fyn immunoprecipitates of Cbl+/+ MEFs incubated without MG132; this signal dramatically increased upon MG132 treatment (Fig. 1D, top panel, compare lanes 1 with lane 2). In contrast, the ubiquitin signal was essentially undetectable in anti-Fyn immunoprecipitates of Cbl−/− MEFs and remained very low even after MG132 treatment (Fig. 1D, top panel, compare lane 2 with lane 4). Anti-Fyn immunoblotting showed that MG132 treatment increased the Fyn protein level in Cbl+/+ but not Cbl−/− MEFs (Fig. 1D, bottom panel). These findings demonstrate that Fyn protein undergoes ubiquitination, and that the level of endogenous Cbl protein controls the extent of Fyn ubiquitination.
Intact cellular ubiquitination machinery is essential for Cbl-mediated Fyn degradation
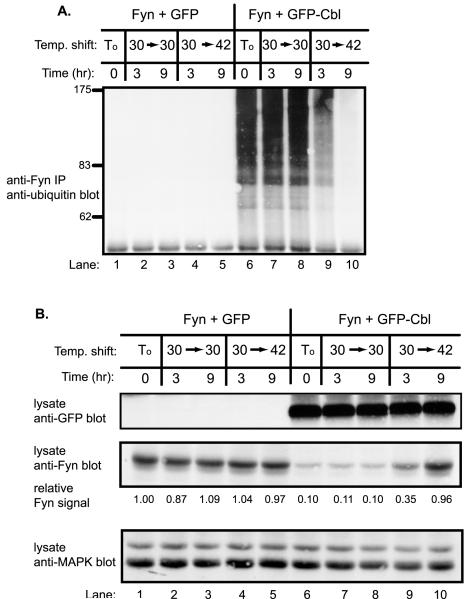
Stabilization of Fyn protein in Cbl−/− cells, together with accumulation of ubiquitinated Fyn in MG132-treated Cbl+/+ cells, strongly suggested that Cbl-induced ubiquitination serves as a signal for proteasome-mediated degradation. To directly assess the requirement of Fyn ubiquitination for its Cbl-induced degradation, we utilized CHO-TS20 cells. In these cells, the ubiquitin activating enzyme (E1) is fully active at 30°C but nonfunctional at 42°C [40], allowing manipulation of Fyn ubiquitination by using a temperature shift.
Very little Fyn ubiquitination (anti-Fyn immunoprecipitations immunoblotted with anti-ubiquitin) was observed in the absence of co-transfected Cbl at either temperature (Fig. 2A, lanes 1–5). In contrast, co-expression of Cbl resulted in a marked increase in the levels of ubiquitinated Fyn when cells were maintained at 30°C (Fig. 2A, lane 6). When these cells were shifted to 42°C, Fyn ubiquitination decreased rapidly with nearly undetectable signals after 9 hr (Fig. 2A, compare lane 6 with lanes 9–10). Thus, the level of Cbl-induced ubiquitination of Fyn could be precisely regulated in CHO-TS20 cells upon temperature shift.
Fig. 2. Cbl-mediated loss of Fyn protein requires intact cellular ubiquitination machinery.
A, impaired Fyn ubiquitination upon E1 inactivation in Cbl-transfected CHO-TS20 cells. CHO-TS20 cells were transfected with Fyn (0.2 μg) expression plasmid together with 4 μg of GFP or GFP-Cbl plasmids and incubated at 30°C for 56 hr. At this point (To), cells were either maintained at 30°C (30−>30) or shifted to 42°C (30−>42) for the indicated times. Anti-Fyn immunoprecipitates from aliquots of lysate protein (1 mg) were immunoblotted with anti-ubiquitin antibody. B, stabilization of Fyn protein upon E1 inactivation in Cbl-transfected CHO-TS20 cells. Equal amounts (30 μg) of the same cell lysate used in A were immunoblotted with anti-GFP antibody (top panel), anti-Fyn antibody (middle panel), and anti p42/44 MAPK antibody (bottom panel). The levels of Fyn protein were quantified by densitometry, and the values at various times are expressed as a function of the initial Fyn protein level (lane 1) that was assigned a value of 1.0.
The cell lysates used above were directly immunoblotted with anti-GFP and anti-Fyn antibodies to assess the levels of transfected GFP-Cbl and Fyn proteins, respectively. As anticipated, cells co-transfected with GFP-Cbl and Fyn showed a marked reduction in Fyn protein levels when compared to cells cotransfected with GFP vector (Fig. 2B, middle panel, compare lane 1 with lane 6; densitometric units of 1.0 versus 0.1). When transfected cells were maintained at the permissive temperature (30°C), no substantial changes in the steady-state levels of Fyn protein were observed. In contrast, when Fyn plus GFP-Cbl transfected cells were shifted to 42°C, a marked time-dependent increase in Fyn protein levels was observed (Fig. 2B, middle panel, lanes 6–10; densitometric units of 0.35 and 0.96 at 3 hr and 9 hr at 42°C versus 0.11 and 0.10 at 30°C, respectively). Relatively little change in Fyn protein level was observed when GFP and Fyn transfected cells were shifted to 42°C (Fig. 2B, middle panel, lanes1–5; densitometric units of 1.04 and 0.97 at 3 hr and 9 hr at 42°C versus 0.87 and 1.09 at 30°C, respectively). Anti-MAP kinase immunoblotting of cell lysates revealed no substantial changes in the levels of MAP kinase protein (Fig. 2B, bottom panel). These results establish that Cbl-mediated degradation of Fyn requires intact cellular ubiquitination machinery.
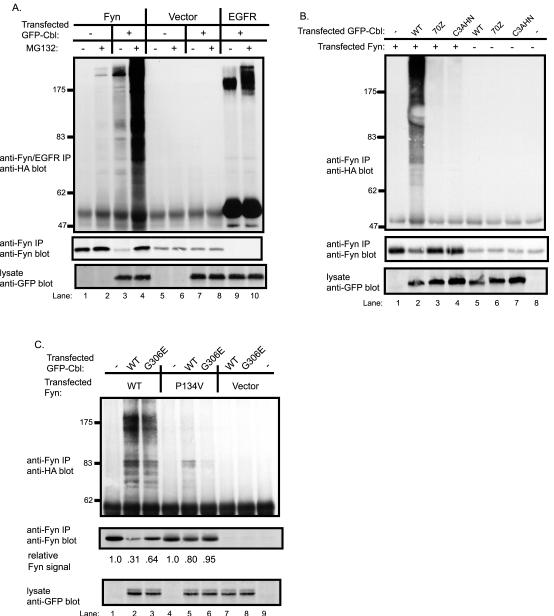
Cbl-mediated ubiquitination and degradation of Fyn requires an intact Cbl RING finger domain and Fyn SH3 domain
Given the ability of Cbl to control Fyn ubiquitination, we wished to determine if this activity is mediated by the Cbl RING finger-domain encoded ubiquitin ligase activity. For this purpose, we compared the ability of wildtype Cbl protein with its RING finger domain mutants to target Fyn for ubiquitination. To assess Cbl-dependent Fyn ubiquitination in vivo, 293T cells were co-transfected with Fyn together with GFP or GFP-Cbl, and a plasmid encoding HA-tagged ubiquitin to facilitate detection of ubiquitinated Fyn.
As expected [32], transfection with GFP-Cbl led to easily detectable ubiquitination of EGFR and this signal was markedly enhanced by MG132 treatment of cells (Fig. 3A, top panel, compare lane 9 with lane 10). Relatively little ubiquitin signal was observed on Fyn in the absence of co-transfected Cbl. In contrast, co-expression of GFP-Cbl led to easily detectable ubiquitination of Fyn, which was accompanied by an expected decrease in the level of Fyn protein (Fig. 3A, compare lane 1 with lane 3). MG132 treatment of cells prior to lysis resulted in marked accumulation of ubiquitinated Fyn and an increase in the level of Fyn protein (Fig. 3A, compare lane 3 with lane 4). Equivalent expression of GFP-tagged Cbl protein in the appropriate lysates was confirmed by anti-GFP immunoblotting of whole cell lysates (Fig. 3A, bottom panel).
Fig. 3. Cbl-dependent ubiquitination of Fyn in 293T cells and an essential role for the Cbl RING finger domain.
A, Cbl-dependent Fyn ubiquitination is enhanced by treatment with a proteasome inhibitor. 293T cells were transfected with plasmids encoding HA-ubiquitin (7 μg), Fyn (0.15 μg), EGFR (0.15 μg), GFP-Cbl (+) (3 μg) or a GFP (−) control (3 μg). Five hr prior to cell lysate preparation, cells were treated with 50μM MG132 (+) or DMSO control (−). Anti-Fyn or anti-EGFR immunoprecipitates from aliquots of lysate protein (800 μg) were immunoblotted with anti-HA antibody (top panel) followed by anti-Fyn antibody (middle panel). Equal aliquots (30 μg) of the same cell lysates used above were immunoblotted with anti-GFP antibody (bottom panel). B, an intact RING finger domain is required for Cbl-dependent Fyn ubiquitination. 293T cells were transfected with the indicated expression plasmids, lysed and anti-Fyn immunoprecipitations were carried out as in A and immunoblotted with anti-HA antibody (top panel) and with anti-Fyn antibody (middle panel). The lysate proteins (30 μg) were immunoblotted with anti-GFP antibody (bottom panel). C, the role of Fyn SH3 domain and Cbl TKB domain-mediated interaction for Cbl-dependent Fyn ubiquitination. 293T cells were transfected with the vector, wildtype Fyn or Fyn-SH3 domain mutant (P134V), lysed and anti-Fyn immunoprecipitations were carried out as in A and immunoblotted with anti-HA antibody (top panel) and with anti-Fyn antibody (middle panel). The lysate proteins (10 μg) were immunoblotted with anti-GFP antibody (bottom panel). The levels of Fyn protein were quantified by densitometry, and the values are expressed as a function of the initial Fyn protein level for each Fyn construct (lane 1 and 4) that was assigned a value of 1.0.
Next, we examined if the RING finger domain is required for Cbl-mediated ubiquitination of Fyn. The Cbl mutant C3AHN contains four point mutations predicted to abrogate coordination of both zinc atoms that stabilize the RING finger domain [33], whereas the naturally occurring Cbl-70Z mutant, which is unable to induce Fyn degradation [13], has a deletion of the critical linker region that provides additional essential contacts for UBC binding [31]. In contrast to wildtype Cbl, both 70Z and C3AHN Cbl RING finger mutants were unable to mediate Fyn ubiquitination (Fig. 3B, top panel) despite their equivalent or higher expression levels compared to wildtype Cbl (Fig. 3B, bottom panel). Thus, the RING finger domain-mediated ubiquitin ligase activity of Cbl is necessary for Cbl-dependent ubiquitination of Fyn.
Previous studies have demonstrated that the Fyn SH3 domain provides the primary mode of association with Cbl by interacting with its proline-rich region [13]. Furthermore, mutation of the Cbl TKB domain alone had no significant effect on Cbl-mediated Fyn degradation. However, the Cbl TKB domain was capable of associating with Fyn [13], and recent studies using Src have suggested that the Cbl TKB domain binds to the SFK activation loop phosphorylation site [47], which is conserved among SFKs. In order to extend our structure-function analyses and establish which domains of Cbl and Fyn are required for Fyn ubiquitination, we tested the ability of a Fyn SH3 domain mutant (P134V) [13] to be ubiquitinated by wildtype Cbl or its TKB domain mutant (G306E). Compared to wildtype Cbl, the TKB domain mutant was still able to mediate Fyn ubiquitination and degradation although to a lesser extent (Fig. 3C, top and middle panel, compare lanes 1–3) when expressed at a level equivalent to wildtype Cbl (Fig. 3C, bottom panel). In contrast, the Fyn SH3 domain mutant showed only minor ubiquitination and degradation when coexpressed with wildtype Cbl. Essentially no ubiquitination or degradation of the Fyn SH3 mutant was observed when coexpressed with the Cbl G306E mutant, and Fyn protein levels were unchanged (Fig. 3C). Thus, the Fyn SH3 domain mediates the predominant physical interaction required for Cbl-mediated Fyn ubiquitination, while the Cbl TKB domain appears to mediate a less dominant mode of interaction.
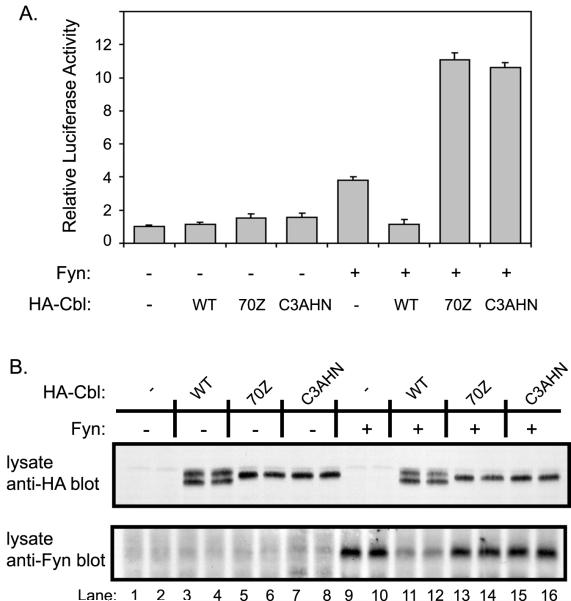
The RING finger domain plays an essential role in Cbl-mediated negative regulation of Fyn-dependent cellular activation
In order to determine the effect of Cbl-dependent ubiquitination on Fyn-mediated cellular activation, we compared the effects of wildtype Cbl and its RING finger domain mutants on Fyn kinase-dependent transactivation of the serum response element (SRE) linked to a luciferase reporter [48]. 293T cells were transfected with the SRE-luciferase reporter plasmid and either Fyn alone or Fyn in combination with wildtype Cbl or its RING finger domain mutants. As expected [13], the expression of Fyn protein led to a modest increase in SRE-luciferase activity compared to mock-transfected cells (Fig. 4A), and this increase was suppressed upon co-expression of wild-type Cbl. In contrast, co-expression of the Cbl RING finger mutant C3AHN as well as the 70Z mutant [13] resulted in a marked enhancement of Fyn-dependent SRE-luciferase reporter activity (Fig. 4A). Expression of Cbl proteins without Fyn had no effect on the SRE luciferase activity. Analysis of cell lysates demonstrated the expected effects of Cbl proteins on Fyn protein levels and confirmed the equivalent expression of various Cbl constructs (Fig. 4B). Overall, these data demonstrate that the RING finger domain, which is required for Fyn ubiquitination and degradation, is also critical for functional negative regulation of Fyn by Cbl.
Fig. 4. The RING finger domain is required for Cbl-dependent negative regulation of Fyn-induced transcriptional activation of a SRE-luciferase reporter.
A, mutations in the RING finger domain of Cbl blocks the negative regulation of SRE-luciferase activation. 293T cells were transfected with plasmids encoding the SRE-luciferase reporter (5 μg), CD8-ζ (0.5 μg) and the indicated combinations of Fyn (0.1 μg), HA-Cbl, HA-Cbl-70Z and HA-Cbl-C3AHN (1 μg) or pAlterMAX vector (−). Cells were lysed 48 hr after transfection and equal aliquots of lysate protein were used to assay the luciferase activity. The luciferase activity was expressed relative to the activity of lysates transfected with the reporter in the absence of Fyn or Cbl. Results represent the mean +/− one standard deviation of five replicate transfections. B, analysis of Fyn protein levels in transfected cells used for SRE-luciferase assay. Aliquots of lysate protein (10 μg) from two of the five replicate samples analyzed in A were resolved by SDS-PAGE and immunoblotted with anti-HA (top panel) and anti-Fyn (bottom panel) antibodies.
DISCUSSION
The recently identified function of Cbl as a ubiquitin ligase [21;32] and our earlier results that Cbl functions as a negative regulator of SFKs [13] led us to hypothesize that Cbl ubiquitin ligase activity provides a physiological mechanism to control the levels of activated SFKs. Here we provide several lines of evidence in support of this hypothesis by examining the regulation of SFK Fyn.
Analyses of multiple cell types, including mouse embryonic fibroblasts, CHO-TS20 cells and 293T human embryonic kidney cells provide evidence for Cbl-dependent ubiquitination of Fyn. An accumulation of ubiquitinated Fyn was also observed upon MG132 treatment of a Jurkat T cell line stably overexpressing Cbl [26] (NR and HB, unpublished results). Importantly, we show that lack of endogenous Cbl leads to a drastic deficiency in Fyn ubiquitination in Cbl−/− MEFs. The reduction in Fyn ubiquitination in Cbl−/− cells is accompanied by a substantial increase in endogenous Fyn levels [13] and a marked increase in the half-life of the Fyn protein, indicating that Cbl-dependent ubiquitination is a critical determinant of Fyn turnover. It is notable that there are two other mammalian Cbl family members [15]. Whether drastically reduced Fyn ubiquitination in Cbl−/− MEFs reflects a lack of expression of other Cbl family members or a lesser role for these proteins in Fyn ubiquitination will require further investigation.
A complimentary line of evidence for a critical role of Cbl-dependent ubiquitination in regulating Fyn protein levels was provided by analyses of CHO-TS20 cells, which express a thermolabile ubiquitin activating (E1) enzyme. This genetic approach provided further evidence that ubiquitin machinery is essential for Cbl to induce the degradation of Fyn. The results obtained in MEFs and CHO-TS20 cells clearly implicate the ubiquitin ligase activity of Cbl in the negative regulation of Fyn. In vivo analysis in 293T cells, using Cbl RING finger domain mutants, established that this indeed was the case. Taken together, our results establish Cbl-dependent ubiquitination as an important mechanism of negative regulation for Fyn, a prototype SFK. Given the ability of Cbl to interact with multiple SFKs and the conservation of structure among members of the SFK family, we propose that Cbl-dependent ubiquitination may provide a general mechanism to negatively regulate activated SFKs. The relatively intense ubiquitin signal on higher molecular weight species, as compared to Fyn signal, is likely due to multi-ubiquitinated Fyn providing increased numbers of epitopes reactive with anti-ubiquitin antibody. We consider it unlikely that these higher molecular weight species represent a Fyn-associated protein, as lysates were prepared in RIPA lysis buffer containing SDS and deoxycholate in order to disrupt protein-protein interactions. Indeed, under such conditions, Fyn-Cbl association was disrupted (data not shown). Moreover, these experiments were performed under conditions that had been optimized to detect Cbl-mediated Fyn ubiquitination and subsequent degradation.
Demonstration of a non-receptor PTK as a target of Cbl-mediated ubiquitination is of considerable significance since all of the targets identified previously are RTKs. Ubiquitin modification of RTKs facilitates their sorting to lysosomes where their extracellular regions are degraded by lysosomal enzymes accounting for receptor downregulation. In contrast, ubiquitination of non-receptor PTKs, such as Fyn, is likely to serve as a direct targeting signal for proteasomal degradation, as supported by our results using proteasome inhibitors. While in vivo studies and the known direct association between Cbl and SFKs are consistent with Cbl-mediated ubiquitination of SFKs, further analyses using purified SFK, Cbl and ubiquitination enzymes in in vitro reconstitution assays will be needed to establish this definitively.
It is likely that Cbl-mediated degradation functions in concert with other mechanisms for deactivation of SFKs, such as the return of activated SFKs to their repressed state through CSK-mediated phosphorylation of the C-terminal tyrosine and potential chaperone-mediated folding into a closed, inactive conformation. The ability of Cbl to target SFKs for ubiquitination and degradation also provides a likely explanation for why Cbl, unlike other SH3 domain ligands such as Sin and HIV NEF [48;6;8], does not activate SFKs. This proposal is supported by the ability of ubiquitin ligase-deficient Cbl mutants, such as 70Z and C3AHN, to activate rather than downregulate SFK activity.
The proposed role of ubiquitin in Cbl-mediated SFK regulation is consistent with recent findings that other SFKs, such as v-Src, c-Src, Lyn and Blk undergo ubiquitination [36-39]. A recent report published while the present paper was under review indicates that Cbl can indeed function as a ubiquitin ligase towards v-Src and c-Src [49]. Interestingly, Blk was shown to interact with and serve as a target of the HECT domain-containing ubiquitin ligase E6AP, which has been previously implicated in ubiquitin-dependent degradation of the nuclear tumor suppressor protein p53 by the human papilloma-virus oncoprotein E6 [50]. Whether E6AP is a physiological ubiquitin ligase for Blk or other SFKs, and whether Cbl and E6AP might work in concert are obvious questions that will require further examination.
A number of observations suggest that Cbl-dependent ubiquitination and degradation primarily target the activated pool of SFKs. The SFK SH3 and SH2 domains, which are primarily responsible for association with Cbl [13], are predicted to be intra-molecularly sequestered in repressed SFKs but available for inter-molecular interactions after activation. The additional interaction between Cbl and Fyn, mediated via Cbl’s TKB domain, is also likely to involve an activation-dependent autophosphorylation site on Fyn, very likely the activation loop phosphorylation site [47]. Consistent with our proposal, mutation of the Fyn SH3 domain drastically reduced Cbl-mediated Fyn ubiquitination and degradation. However, mutation of the Cbl TKB domain also reduced its ability to induce Fyn ubiquitination and degradation quite significantly. Together, these results suggest that both the Fyn SH3 domain and Cbl-TKB domain-mediated interactions, expected only upon activation of Fyn, contribute to Cbl-dependent Fyn ubiquitination. The more dominant effect of abrogating the Fyn SH3 domain-mediated interaction with Cbl may reflect a requirement for this primary association for the secondary Cbl TKB-domain-mediated Cbl-Fyn interaction to occur. Further support for selective regulation of the activated pool of SFKs by Cbl is provided by the observation that the level of autophosphorylated Fyn was markedly increased in Cbl−/− MEFs and T cell lines when compared to their Cbl+/+ counterparts [13]. We also show here that wild-type Cbl reduces whereas ubiquitination-defective Cbl mutants increase the Fyn-dependent SRE luciferase reporter activity, a readout of the kinase activity of SFKs. Notably, co-expression of Cbl was also shown to reduce the Src-dependent induction of DNA synthesis in NIH 3T3 cells, and the inhibitory effect of Cbl was abrogated by deletion of the RING finger domain [14]. Finally, ubiquitination of Src and Blk also correlated with their kinase activity [36-38], and CSK-deficient cells were shown to have elevated kinase activity but reduced protein levels of Src, Fyn and Lyn [9;10]. It will be important to determine if reduction in SFK protein levels in these situations is Cbl-dependent.
While our results support a model that the major function of Cbl is to downregulate the level of activated SFKs by ubiquitin-mediated degradation, other studies have suggested that Cbl transduces signals downstream of SFKs. For example, several SFK-mediated cellular functions, such as integrin-induced macrophage spreading and bone resorption by osteoclasts, were severely reduced when cells were treated with Cbl antisense oligonucleotides [51;52]. Furthermore, introduction of Cbl into v-abl transformed NIH 3T3 cells restored cell adhesion [43;53], and a truncated Cbl protein (Cbl 1–480) lacking the C-terminal region, enhanced lamellipodia formation in transfected NIH 3T3 cells [54]. It is therefore possible that Cbl can downregulate SFKs by targeting them for ubiquitination while simultaneously serving as an adapter for SH2 domain-containing proteins, thereby positively regulating signal transduction. In this regard, it is notable that the C-terminal phosphorylation sites of Cbl interact with the p85 subunit of PI3 kinase, the Rac/Rho exchange factor Vav and Crk adapter proteins [15], all of which are known to be involved in cytoskeletal remodeling, cell spreading and cell migration.
In conclusion, our results demonstrate that Cbl functions as a key regulator of the SFK Fyn by enhancing its ubiquitination and subsequent degradation via the proteasome. The negative regulatory role of Cbl is dependent on intact cellular ubiquitin machinery as well as the Cbl RING finger domain, which recruits the ubiquitin machinery. Given the ability of Cbl to interact with multiple SFKs and the conservation of structure among various SFKs, we propose that Cbl-dependent ubiquitination may provide a general mechanism to negatively regulate activated SFKs.
ACKNOWLEDGEMENTS
We thank Dr. Ger Strous (University Medical Center Utrecht, Netherlands) for providing the CHO-TS20 cell line, Dr. Dirk Bohmann (EMBO, Heidelberg, Germany) for the HA-ubiquitin expression construct, Dr. Alexander Tysgankov (Temple University, Philadelphia, USA) for the pHIT60 and pMD.G packaging plasmids and Dr. Robert Hawley (University of Toronto, Toronto) for the MSCVpac retroviral vector.
This work was supported by grants to HB from the National Institutes of Health (CA87986, CA75075 and CA76118) and the American Cancer Society (066-04-CIM). NR is a Howard Hughes Medical Institute Predoctoral Fellow. CEA, AG and PD are US Department of Defense Breast Cancer Research Program Postdoctoral Fellows (CEA and AG by DAMD 17-98-1-8032 and PD by DAMD17-99-1-9085).
Abbreviations
- CHO
Chinese hamster ovary
- CSK
C-terminal Src kinase
- EGFR
epidermal growth factor receptor
- GFP
green fluorescent protein
- HA
influenza hemagglutinin
- IP
immunoprecipitate
- mAb
monoclonal antibody
- MEF
mouse embryonic fibroblast
- pAb
rabbit polyclonal antibody
- PDGFR
platelet-derived growth factor
- PTK
protein tyrosine kinase
- RTK
receptor tyrosine kinase
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SFK
Src-family kinase
- SRE
serum response element
- TKB
tyrosine kinase binding
- UBC
ubiquitin conjugating enzyme
- WT
wild-type
REFERENCES
- [1].Chow LM, Veillette A. The Src and Csk families of tyrosine protein kinases in hemopoietic cells. Semin Immunol. 1995;7:207–226. doi: 10.1006/smim.1995.0026. [DOI] [PubMed] [Google Scholar]
- [2].Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- [3].Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- [4].Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- [5].Seidel-Dugan C, Meyer BE, Thomas SM, Brugge JS. Effects of SH2 and SH3 deletions on the functional activities of wildtype and transforming variants of c-Src. Mol Cell Biol. 1992;12:1835–1845. doi: 10.1128/mcb.12.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Briggs SD, Sharkey M, Stevenson M, Smithgall TE. SH3-mediated Hck tyrosine kinase activation and fibroblast transformation by the Nef protein of HIV-1. J Biol Chem. 1997;272:17899–17902. doi: 10.1074/jbc.272.29.17899. [DOI] [PubMed] [Google Scholar]
- [7].Gonfloni S, Williams JC, Hattula K, Weijland A, Wierenga RK, Superti-Furga G. The role of the linker between the SH2 domain and catalytic domain in the regulation and function of Src. Embo J. 1997;16:7261–7271. doi: 10.1093/emboj/16.24.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- [9].Imamoto A, Soriano P. Disruption of the csk gene, encoding a negative regulator of Src family tyrosine kinases, leads to neural tube defects and embryonic lethality in mice. Cell. 1993;73:1117–1124. doi: 10.1016/0092-8674(93)90641-3. [DOI] [PubMed] [Google Scholar]
- [10].Nada S, Yagi T, Takeda H, Tokunaga T, Nakagawa H, Ikawa Y, Okada M, Aizawa S. Constitutive activation of Src family kinases in mouse embryos that lack Csk. Cell. 1993;73:1125–1135. doi: 10.1016/0092-8674(93)90642-4. [DOI] [PubMed] [Google Scholar]
- [11].Porter M, Schindler T, Kuriyan J, Miller WT. Reciprocal regulation of Hck activity by phosphorylation of Tyr(527) and Tyr(416). Effect of introducing a high affinity intramolecular SH2 ligand. J Biol Chem. 2000;275:2721–2726. doi: 10.1074/jbc.275.4.2721. [DOI] [PubMed] [Google Scholar]
- [12].Bijlmakers MJ, Marsh M. Hsp90 is essential for the synthesis and subsequent membrane association, but not the maintenance, of the Src-kinase p56(lck) Mol Biol Cell. 2000;11:1585–1595. doi: 10.1091/mbc.11.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Andoniou CE, Lill NL, Thien CB, Lupher ML, Jr., Ota S, Bowtell DD, Scaife RM, Langdon WY, Band H. The Cbl proto-oncogene product negatively regulates the Src-family tyrosine kinase Fyn by enhancing its degradation. Mol Cell Biol. 2000;20:851–867. doi: 10.1128/mcb.20.3.851-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Broome MA, Galisteo ML, Schlessinger J, Courtneidge SA. The proto-oncogene c-Cbl is a negative regulator of DNA synthesis initiated by both receptor and cytoplasmic tyrosine kinases. Oncogene. 1999;18:2908–2912. doi: 10.1038/sj.onc.1202873. [DOI] [PubMed] [Google Scholar]
- [15].Lupher ML, Jr., Rao N, Eck MJ, Band H. The Cbl protooncoprotein: a negative regulator of immune receptor signal transduction. Immunol Today. 1999;20:375–382. doi: 10.1016/s0167-5699(99)01484-x. [DOI] [PubMed] [Google Scholar]
- [16].Thien CB, Langdon WY. Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- [17].Murphy MA, Schnall RG, Venter DJ, Barnett L, Bertoncello I, Thien CB, Langdon WY, Bowtell DD. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol Cell Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Naramura M, Kole HK, Hu RJ, Gu H. Altered thymic positive selection and intracellular signals in Cbl- deficient mice. Proc Natl Acad Sci U S A. 1998;95:15547–15552. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chiang YJ, Kole HK, Brown K, Naramura M, Fukuhara S, Hu RJ, Jang IK, Gutkind JS, Shevach E, Gu H. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- [20].Krawczyk C, Bachmaier K, Sasaki T, Jones RG, Snapper SB, Bouchard D, Kozieradzki I, Ohashi PS, Alt FW, Penninger JM. Cbl-b Is a Negative Regulator of Receptor Clustering and Raft Aggregation in T Cells. Immunity. 2000;13:463–473. doi: 10.1016/s1074-7613(00)00046-7. [DOI] [PubMed] [Google Scholar]
- [21].Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2- dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- [22].Lee PS, Wang Y, Dominguez MG, Yeung YG, Murphy MA, Bowtell DD, Stanley ER. The Cbl protooncoprotein stimulates CSF-1 receptor multiubiquitination and endocytosis, and attenuates macrophage proliferation. Embo J. 1999;18:3616–3628. doi: 10.1093/emboj/18.13.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hicke L, Zanolari B, Riezman H. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J Cell Biol. 1998;141:349–358. doi: 10.1083/jcb.141.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lupher ML, Jr., Rao N, Lill NL, Andoniou CE, Miyake S, Clark EA, Druker B, Band H. Cbl-mediated negative regulation of the Syk tyrosine kinase. A critical role for Cbl phosphotyrosine-binding domain binding to Syk phosphotyrosine 323. J Biol Chem. 1998;273:35273–35281. doi: 10.1074/jbc.273.52.35273. [DOI] [PubMed] [Google Scholar]
- [26].Rao N, Lupher ML, Jr., Ota S, Reedquist KA, Druker BJ, Band H. The linker phosphorylation site Tyr292 mediates the negative regulatory effect of Cbl on ZAP-70 in T cells. J Immunol. 2000;164:4616–4626. doi: 10.4049/jimmunol.164.9.4616. [DOI] [PubMed] [Google Scholar]
- [27].Meng W, Sawasdikosol S, Burakoff SJ, Eck MJ. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature. 1999;398:84–90. doi: 10.1038/18050. [DOI] [PubMed] [Google Scholar]
- [28].Lupher ML, Jr., Songyang Z, Shoelson SE, Cantley LC, Band H. The Cbl phosphotyrosine-binding domain selects a D(N/D)XpY motif and binds to the Tyr292 negative regulatory phosphorylation site of ZAP-70. J Biol Chem. 1997;272:33140–33144. doi: 10.1074/jbc.272.52.33140. [DOI] [PubMed] [Google Scholar]
- [29].Lill NL, Douillard P, Awwad RA, Ota S, Lupher ML, Jr., Miyake S, Meissner-Lula N, Hsu VW, Band H. The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J Biol Chem. 2000;275:367–377. doi: 10.1074/jbc.275.1.367. [DOI] [PubMed] [Google Scholar]
- [30].Miyake S, Lupher ML, Jr., Druker B, Band H. The tyrosine kinase regulator Cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor alpha. Proc Natl Acad Sci U S A. 1998;95:7927–7932. doi: 10.1073/pnas.95.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin- protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- [32].Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- [33].Ota S, Hazeki K, Rao N, Lupher ML, Jr., Andoniou CE, Druker B, Band H. The RING finger domain of Cbl is essential for negative regulation of the Syk tyrosine kinase. J Biol Chem. 2000;275:414–422. doi: 10.1074/jbc.275.1.414. [DOI] [PubMed] [Google Scholar]
- [34].Miyake S, Lupher ML, Jr., Andoniou CE, Lill NL, Ota S, Douillard P, Rao N, Band H. The Cbl protooncogene product: from an enigmatic oncogene to center stage of signal transduction. Crit Rev Oncog. 1997;8:189–218. doi: 10.1615/critrevoncog.v8.i2-3.30. [DOI] [PubMed] [Google Scholar]
- [35].Tsygankov AY, Mahajan S, Fincke JE, Bolen JB. Specific association of tyrosine-phosphorylated c-Cbl with Fyn tyrosine kinase in T cells. J Biol Chem. 1996;271:27130–27137. doi: 10.1074/jbc.271.43.27130. [DOI] [PubMed] [Google Scholar]
- [36].Hakak Y, Martin GS. Ubiquitin-dependent degradation of active Src. Curr Biol. 1999;9:1039–1042. doi: 10.1016/s0960-9822(99)80453-9. [DOI] [PubMed] [Google Scholar]
- [37].Harris KF, Shoji I, Cooper EM, Kumar S, Oda H, Howley PM. Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc Natl Acad Sci U S A. 1999;96:13738–13743. doi: 10.1073/pnas.96.24.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Oda H, Kumar S, Howley PM. Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination. Proc Natl Acad Sci U S A. 1999;96:9557–9562. doi: 10.1073/pnas.96.17.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Winberg G, Matskova L, Chen F, Plant P, Rotin D, Gish G, Ingham R, Ernberg I, Pawson T. Latent membrane protein 2A of epstein-barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol Cell Biol. 2000;20:8526–8535. doi: 10.1128/mcb.20.22.8526-8535.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kulka RG, Raboy B, Schuster R, Parag HA, Diamond G, Ciechanover A, Marcus M. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J Biol Chem. 1988;263:15726–15731. [PubMed] [Google Scholar]
- [41].Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- [42].Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- [43].Feshchenko EA, Shore SK, Tsygankov AY. Tyrosine phosphorylation of C-Cbl facilitates adhesion and spreading while suppressing anchorage-independent growth of V-Abl-transformed NIH3T3 fibroblasts. Oncogene. 1999;18:3703–3715. doi: 10.1038/sj.onc.1202672. [DOI] [PubMed] [Google Scholar]
- [44].Lupher ML, Jr., Reedquist KA, Miyake S, Langdon WY, Band H. A novel phosphotyrosine-binding domain in the N-terminal transforming region of Cbl interacts directly and selectively with ZAP-70 in T cells. J Biol Chem. 1996;271:24063–24068. doi: 10.1074/jbc.271.39.24063. [DOI] [PubMed] [Google Scholar]
- [45].Miyake S, Mullane-Robinson KP, Lill NL, Douillard P, Band H. Cbl-mediated negative regulation of platelet-derived growth factor receptor-dependent cell proliferation. A critical role for Cbl tyrosine kinase-binding domain. J Biol Chem. 1999;274:16619–16628. doi: 10.1074/jbc.274.23.16619. [DOI] [PubMed] [Google Scholar]
- [46].Mimnaugh EG, Bonvini P, Neckers L. The measurement of ubiquitin and ubiquitinated proteins. Electrophoresis. 1999;20:418–428. doi: 10.1002/(SICI)1522-2683(19990201)20:2<418::AID-ELPS418>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- [47].Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, Antoine E, Levy J, Gailit J, Bowtell D, Horne WC, Baron R. Cbl associates with Pyk2 and Src to regulate Src kinase activity, alpha(v)beta(3) integrin-mediated signaling, cell adhesion, and osteoclast motility. J Cell Biol. 2001;152:181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Alexandropoulos K, Baltimore D. Coordinate activation of c-Src by SH3- and SH2-binding sites on a novel p130Cas-related protein, Sin. Genes Dev. 1996;10:1341–1355. doi: 10.1101/gad.10.11.1341. [DOI] [PubMed] [Google Scholar]
- [49].Yokouchi M, Kondo T, Sanjay A, Houghton A, Yoshimura A, Komiya S, Zhang H, Baron R. Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J Biol Chem. 2001;276:35185–35193. doi: 10.1074/jbc.M102219200. [DOI] [PubMed] [Google Scholar]
- [50].Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- [51].Meng F, Lowell CA. A beta 1 integrin signaling pathway involving Src-family kinases, Cbl and PI-3 kinase is required for macrophage spreading and migration. Embo J. 1998;17:4391–4403. doi: 10.1093/emboj/17.15.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tanaka S, Amling M, Neff L, Peyman A, Uhlmann E, Levy JB, Baron R. c-Cbl is downstream of c-Src in a signalling pathway necessary for bone resorption. Nature. 1996;383:528–531. doi: 10.1038/383528a0. [DOI] [PubMed] [Google Scholar]
- [53].Ribon V, Printen JA, Hoffman NG, Kay BK, Saltiel AR. A novel, multifuntional c-Cbl binding protein in insulin receptor signaling in 3T3-L1 adipocytes. Mol Cell Biol. 1998;18:872–879. doi: 10.1128/mcb.18.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Scaife RM, Langdon WY. c-Cbl localizes to actin lamellae and regulates lamellipodia formation and cell morphology. J Cell Sci. 2000;113(Pt 2):215–226. doi: 10.1242/jcs.113.2.215. [DOI] [PubMed] [Google Scholar]