Abstract
Motivation: Much of a cell's regulatory response to changing environments occurs at the transcriptional level. Particularly in higher organisms, transcription factors (TFs), microRNAs and epigenetic modifications can combine to form a complex regulatory network. Part of this system can be modeled as a collection of regulatory modules: co-regulated genes, the conditions under which they are co-regulated and sequence-level regulatory motifs.
Results: We present the Combinatorial Algorithm for Expression and Sequence-based Cluster Extraction (COALESCE) system for regulatory module prediction. The algorithm is efficient enough to discover expression biclusters and putative regulatory motifs in metazoan genomes (>20 000 genes) and very large microarray compendia (>10 000 conditions). Using Bayesian data integration, it can also include diverse supporting data types such as evolutionary conservation or nucleosome placement. We validate its performance using a functional evaluation of co-clustered genes, known yeast and Escherichea coli TF targets, synthetic data and various metazoan data compendia. In all cases, COALESCE performs as well or better than current biclustering and motif prediction tools, with high accuracy in functional and TF/target assignments and zero false positives on synthetic data. COALESCE provides an efficient and flexible platform within which large, diverse data collections can be integrated to predict metazoan regulatory networks.
Availability: Source code (C++) is available at http://function.princeton.edu/sleipnir, and supporting data and a web interface are provided at http://function.princeton.edu/coalesce.
Contact: ogt@cs.princeton.edu; hcoller@princeton.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
While the genome sequence of an organism describes its complement of potential proteins, it is the controlled expression, translation and modification of these proteins that allows cells to survive and grow. At the level of transcription and mRNA stability, a complex regulatory network of transcription factors (TFs), RNA binding proteins and microRNAs governs the interactions between components of a cell's internal state and its external environment. Understanding the elements of this regulatory network and the stimuli to which it responds in higher organisms has been of increasing recent interest as a key to metazoan systems biology (Bonneau, 2008; Long et al., 2008), particularly as genetic misregulation is a major cause of human disease.
One means of discovering regulatory modules is the analysis of gene expression data, since a consequence of transcriptional co-regulation is co-expression. While a wealth of assays has also been developed to explore the transcriptional regulatory network under specific experimental conditions, regulatory module prediction from microarray data is a widely studied problem that remains unsurpassed for inference of general regulatory networks, particularly when additional genomic data sources are also integrated (Bussemaker et al., 2007). Prediction of regulatory relationships has been particularly well-studied in unicellular systems, where regulation often occurs based on well-defined TF binding sites and discrete activation or repression of transcription (Beer and Tavazoie, 2004; Roth et al., 1998). These assumptions have led to the current motif discovery paradigm, in which microarray data are clustered, each cluster's promoter sequences tested for enriched motifs, and the resulting consensus sequences matched again known TF binding sites.
In many cases, however, and particularly in more complex organisms, these assumptions no longer hold, and predicting regulatory modules from expression data becomes an increasingly difficult problem. It combines the challenges of biclustering [i.e. grouping together co-expressed genes and the subset of conditions where they are co-expressed (Kloster et al., 2005; Tanay et al., 2004)] with the difficulty of de novo motif discovery from DNA sequences, where regulatory motifs can be short, degenerate and frequently present without being functional (Hannenhalli, 2008). Note that this is distinct from the related tasks of inferring regulatory networks with prior knowledge of potential regulators or regulatory motifs (e.g. Kundaje et al., 2008; Lemmens et al., 2009; Segal et al., 2003) or while omitting the process of motif discovery (e.g. Margolin et al., 2006), both of which have also been intensively studied. Most existing approaches to regulatory module discovery break the biclustering and motif discovery tasks into separate stages: first, expression data is clustered or biclustered, and afterwards, each cluster is analyzed for enrichment of sequence motifs (Elemento et al., 2007). To discover regulatory modules most effectively, though, it would be natural to perform both tasks at the same time, discovering clusters of genes that are both co-expressed and enriched for regulatory motifs. Recent work (Halperin et al., 2009; Reiss et al., 2006) has indeed confirmed the intuition that regulatory module discovery by simultaneous analysis of expression and sequence data can be extremely effective, but this has neither been developed to incorporate heterogeneous data integration, nor has it been scaled for application to complex metazoan genomes.
Here, we describe a Combinatorial Algorithm for Expression and Sequence-based Cluster Extraction (COALESCE), which allows the discovery of regulatory motifs and modules from large collections of genomic data (Fig. 1). COALESCE takes advantage of Bayesian integration of multiple data types (primarily expression data) on a large scale (Huttenhower and Troyanskaya, 2008) to predict co-expressed gene modules, the conditions under which they are co-regulated, and the consensus binding motifs responsible for their regulation. The algorithm is practical for use with complex metazoan genomes (>25 000 genes), analyzes extremely large expression data collections (>15 000 conditions), can explicitly model dependencies between related gene expression conditions (e.g. points in a time course) and can integrate heterogeneous supporting data types in order to improve predictions (nucleosome positioning and evolutionary conservation are specifically demonstrated below). An implementation of COALESCE (including C++ source code) is provided as part of the Sleipnir software package at http://function.princeton.edu/sleipnir, and a web interface is available at http://function.princeton.edu/coalesce. We have validated COALESCE's ability to discover functionally relevant biclusters and transcriptional motifs in synthetic data and in Saccharomyces cerevisiae, demonstrating improvements over previous methods in both expression biclustering and binding site prediction. We provide further evaluation of TF target predictions using the Yeastract (S.cerevisiae; Teixeira et al., 2006) and RegulonDB (E.Coli; Gama-Castro et al., 2008) motif databases and results including regulatory modules for Caenorhabditis elegans, Drosophila melanogaster, Mus musculus and Homo sapiens.
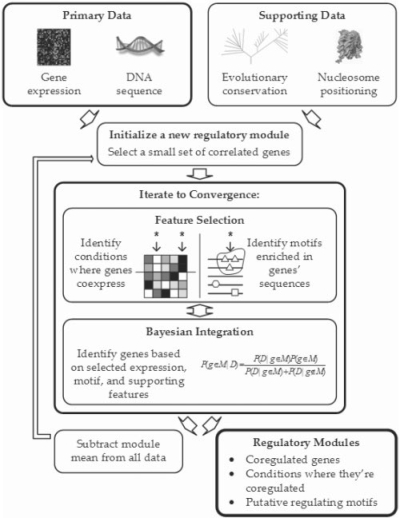
Fig. 1.
A schematic overview of the COALESCE algorithm for regulatory module discovery. COALESCE predicts regulatory modules, each consisting of a gene expression bicluster (co-regulated genes and the conditions under which they are co-regulated) plus zero or more putative regulating motifs. Its primary input data are gene expression microarrays (to form biclusters) and flanking sequences (to predict motifs, although these can be omitted to output only expression biclusters). Additional supporting data types can be flexibly integrated using a Bayesian framework; for example, highly conserved sequence locations may be more likely to contain motifs, and sites occluded by nucleosomes may be less likely. COALESCE is efficient enough to integrate thousands of expression conditions and supporting data for large (> 20 000 genes) metazoan genomes.
2 METHODS
The COALESCE algorithm provides an efficient, iterative framework for predicting regulatory modules (co-regulated genes, the conditions under which they are co-regulated, and putative regulatory motifs) from very large collections of gene expression data. Supporting data types (e.g. nucleosome positioning or evolutionary conservation) can be integrated in a Bayesian framework, and the algorithm scales sufficiently to handle very large genomes (>25 000 genes) and gene expression collections compendia (>15 000 conditions). We validate COALESCE's performance on gold standards from S.cerevisiae and E.coli, provide additional results from C.elegans, D.melanogaster, M.musculus and H.sapiens, evaluate results on synthetic data, and compare COALESCE to existing algorithms for biclustering (Kloster et al., 2005; Reiss et al., 2006; Tanay et al., 2004) and de novo motif prediction (Elemento et al., 2007; Pavesi et al., 2004). Example inputs for yeast are provided in Supplementary Dataset 1, and predicted regulatory modules for all datasets and organisms are available in Supplementary Dataset 2. Methods are presented here in summary, with additional details provided in Supplementary Text 1.
2.1 The COALESCE algorithm
The basic COALESCE algorithm consumes gene expression and DNA sequence data as input to produce putative co-regulated modules as output; extensions allowing supporting data types and in-depth sequence analysis are discussed below. Each resulting module consists of a set of co-regulated genes, one or more expression conditions under which they are co-expressed and zero or more motifs predicted to drive the co-regulation. The algorithm finds modules in a serial manner by seeding each new module with a set of co-expressed genes and iteratively refining the module to convergence. Each iteration begins with a process of feature extraction, in which expression conditions and sequence motifs showing differential expression/enrichment are associated with the developing module. This is followed by a Bayesian integration step, in which each gene's values for the selected features are combined probabilistically to determine whether the gene should be included in the module, with priors proportional to the fraction of features actually selected. After these two stages are alternated to convergence, the module's centroid is subtracted from the selected genes and features and the process begins again for the next cluster. This algorithm is presented schematically in Figure 1; it is summarized in pseudocode in Supplementary Text 1 and described in more detail below.
COALESCE receives as input a standard genes-by-conditions expression matrix, DNA sequences for each gene in the regions of interest (e.g. upstream and/or downstream of the coding region), and four parameters: a k-mer length k, maximum P-value cutoffs pe and pm for expression condition and motif significance, respectively, and a minimum probability cutoff pg for inclusion of genes in modules. Each module is then computed beginning with an initial seed of the two genes maximally correlated across all expression conditions. Three steps are then iterated to modify the module until it converges: selection of significant expression conditions, selection of significant motifs and inclusion of probable genes. An expression condition is considered to be significant (and thus included in the module) if the distribution of expression values for genes currently in the module differs below threshhold pe from the genomic background (based on a standard Z-test). Similarly, a motif is significant if its frequency in gene sequences currently in the module differs below threshhold pm from background (based on a Z-test modified to use Cohen's d; Supplementary Text 1). Based on these selected features (significant conditions and motifs), each gene's probability P(g∈G|C,M) of inclusion in the developing module is calculated using Bayesian data integration of P(C|g∈G), observed from the expression data, P(M|g∈G), observed from the sequence data and P(g∈G), a prior used to stabilize module convergence. Genes above probability pg are included and those below are excluded. When the module converges to a final set of conditions C, motifs M and genes G, its mean expression values and motif frequencies are subtracted from the underlying data and the process is begun again with a new pair of seed genes. C++ source code for the algorithm is available at http://function.princeton.edu/sleipnir, and a detailed description including pseudocode can be found in Supplementary Text 1.
Integration of additional data types modifies the algorithm only minimally and is discussed below. All significance tests involving P-values are Bonferroni corrected for multiple hypotheses. For all experiments in this manuscript, P-value threshholds were fixed at 0.05, probability threshholds at 0.95 and k = 7.
2.1.1 Motifs and DNA sequences
COALESCE considers three types of motifs. The pseudocode above describes simple k-mers, each a string of k characters drawn from the alphabet {A, C, G, T}. Our implementation also considers reverse complement pairs (RCs) and probabilistic suffix trees (PSTs) in an equivalent manner. An RC is the equally weighted union of a k-mer and its reverse complement. A PST is the union of two or more arbitrary k-mers and RCs in a weighted (probabilistic) manner; such a structure can be constructed and matched against DNA sequence rapidly at runtime (Pavesi et al., 2004). Briefly, just as a Position Weight Matrix (PWM) or Position Specific Score Matrix (PSSM) contains a single column per base to be matched, a PST contains a single node in a tree for each base. A PST thus has some depth equivalent to the length of a PWM/PSSM, and the maximal length match against some sequence is thus the depth or the length of the sequence, whichever is shorter.
Initially, all possible k-mers and RCs are considered, but no PSTs. During each runtime iteration, a new PST is constructed for any pair of existing motifs m and m′ for which (i) Z-score (Mm, Mm′) is small and (ii) the minimum edit distance between m and m′ is small (experiments here used gap penalty 1, mismatch penalty 2.1 and threshhold 2.5). Each PST so constructed is treated identically to k-mers and RCs with respect to frequency calculations etc. in all future iterations, subsequent to calculation of gene-specific scores Mg,m. For a PST p with depth |p|, maximum length match |p[s, i]| for some sequence s beginning at offset i and probability p[s, i] of specifically matching position i, these are calculated as:
| (1) |
That is, each base is treated as a simple independent probability, and normalized by the likelihood of matching the observed number of bases by chance; this is the normalized product over each base of the probability of a match, just as would be calculated for a PWM/PSSM. Note that, as defined below, scores Mg,m are calculated in a simpler manner for static k-mers and for k-mers weighed by supporting datasets. Using this definition, PSTs can thus capture degeneracy (similarly to PWMs), reverse complementation and motifs longer than any given k.
See Supplementary Text 1 for details on discretization of motif frequencies, Z-tests for enrichment and decomposition of sequence data into partitioned subareas (e.g. upstream versus downstream and flank versus UTR).
2.1.2 Gene expression data and prior structural knowledge
COALESCE is robust to missing values in the input expression data and to differences in microarray platform and processing across conditions. The input expression matrix E is initially Z-scored per condition (column) to have mean=0, SD=1. Since all subsequent tests occur within single conditions, differences in platform and preprocessing do not affect the remainder of the algorithm, and missing values are excluded from distribution comparisons. See Supplementary Text 1 for details on the prior β for different data types and the incorporation of dataset covariance (i.e. block structure such as a time course).
2.1.3 Supporting data types: nucleosome occupancy and conservation
Additional data types can be incorporated into the COALESCE algorithm in one of two ways. First, since the data integration step uses a flexible Bayesian framework, any dataset for which P(D|g∈G) can be calculated can be included. This will effectively treat the dataset D in a ‘microarray-like’ manner, in that its distribution of values will drive the inclusion and exclusion of genes in a module similarly to a single microarray condition; priors can, of course, be applied to any dataset or condition to up- or down-weight its contribution to the integration process. This might include any dataset quantifying a gene product's behavior under some environmental condition: degree of localization to a particular compartment, quantitative phenotypes from gene deletions or physical interactions with particular prey.
However, other data types are more appropriately analyzed in a ‘sequence-like’ way, in that they associate values with individual genomic bases rather than with genes as a whole. Two such data types are nucleosome occupancy, which describes the probability with which each base is occluded by a nucleosome under some condition, and evolutionary conservation, which describes how conserved each base is over some set of organisms of interest. Data that scores individual bases can be used by COALESCE as weights to transform instances of each motif prior to the assembly of the matrix M. For example, let sg[i, j] represent the substring of sequence sg at position i (zero-indexed) with length j. Then the calculation of Mg,m in the unweighted case as described above can be written as:
| (2) |
for δ (the Kronecker delta function)=1, when its inputs are identical and 0 otherwise. Each supporting dataset w can be represented as a function w(sg, i) mapping the i-th base of sequence sg to some continuous value. Given a set of such supporting datasets W, this allows each entry of Mg,m to be weighted appropriately:
| (3) |
The specific nucleosome occupancy and evolutionary conservation data used in our experiments are described below and available in Supplementary Dataset 3.
2.2 Data collection and processing
COALESCE consumes two primary data types, gene expression and genomic sequence data, as well as arbitrary supporting data of other experimental types; for the latter, we use probabilities of nucleosome occupancy and a per-base measure of evolutionary conservation. The experiments in this manuscript were run on S.cerevisiae expression data drawn from a large (>2200 condition) compendium (Huttenhower and Troyanskaya, 2008), E.coli expression data from GEO (Barrett et al., 2009), three synthetic datasets, and a variety of metazoan (H.sapiens, M.musculus, D.melanogaster and C.elegans) expression data from GEO. Sequence data were obtained from BioMart (Durinck et al., 2005) and RSAT (Thomas-Chollier et al., 2008) using 2 kb upstream and downstream flanks for each organism. 5′ and 3′ UTR annotations and repeat masking were available for all organisms except yeast, and sequences overlapping adjacent ORFs were excluded. See Supplementary Text 1 for details on the specific expression data, nucleosome placements, evolutionary conservation and synthetic data used in this study.
2.3 Testing and evaluation
We evaluated COALESCE using four different metrics: ability to recover functionally informative modules in S.cerevisiae; correspondence of predicted motifs in S.cerevisiae and E.coli with known TF sites from the Yeastract (Teixeira et al., 2006) and RegulonDB (Gama-Castro et al., 2008) databases; comparison with previous methods [biclusters from SAMBA (Tanay et al., 2004) and PISA (Kloster et al., 2005), motifs from Weeder (Pavesi et al., 2004) and FIRE (Elemento et al., 2007), and total results from cMonkey (Reiss et al., 2006)]; and recovery of biclusters and motifs from three synthetic datasets. For all analyses, default parameters were used as described above. Functional evaluation was performed as in (Myers et al., 2006); see Supplementary Text 1 for details and for information on functional evaluation of COALESCE's predicted human modules and TF/target evaluations in yeast and E.coli.
3 RESULTS
We have evaluated COALESCE's performance in four broad areas: its ability to construct functionally informative biclusters from S.cerevisiae data; its ability to correctly predict known TF targets in yeast and E.coli; its efficiency and accuracy on synthetic data; and its ability to discover regulatory modules in large metazoan datasets, including ∼15 000 human gene expression conditions. Example yeast input files are provided in Supplementary Dataset 1, and predicted modules for all datasets and organisms are available in Supplementary Dataset 2.
3.1 Predicted S.cerevisiae regulatory modules are functionally cohesive
We ran COALESCE on a compendium of ∼2200 S.cerevisiae expression conditions comprising ∼125 datasets (Huttenhower and Troyanskaya, 2008), using 2 kb of upstream and downstream sequence for regulatory motif prediction. An evaluation of the functional accuracy of the resulting putative regulatory modules appears in Figure 2, which describes the ability of these gene/condition biclusters to recapitulate known associations between functionally related genes as annotated in the Gene Ontology. In comparison to standard k-means clustering over the entire compendium or the representative PISA (Kloster et al., 2005) and SAMBA (Tanay et al., 2004) biclustering methods, COALESCE (both with and without information from gene sequences) succeeds in recovering regulatory modules strongly enriched for specific functional activities and functionally related genes.
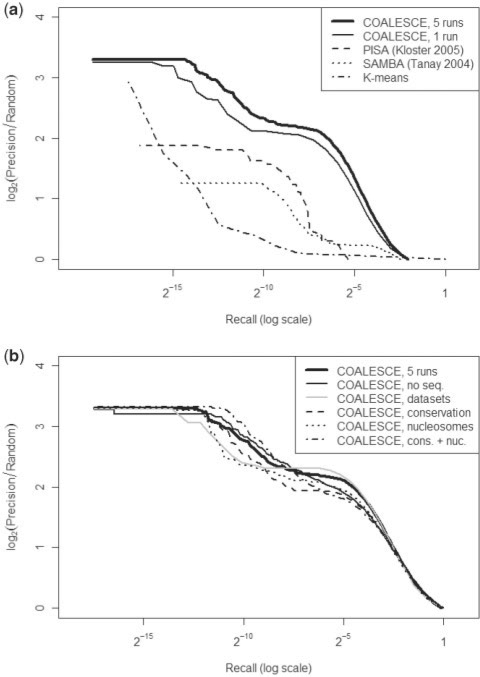
Fig. 2.
Evaluation of the functional consistency of S.cerevisiae expression biclusters predicted by COALESCE. Precision and recall are over gene pairs co-annotated in the Gene Ontology as described in (Myers et al., 2006). Unless noted, COALESCE was executed on ∼2200 yeast expression conditions using 2 kb of up- and downstream flanking sequence. See Supplementary Figure 1 for a plot with standard scale axes and Supplementary Figure 2 for a comparable evaluation using human data. (a) A comparison of COALESCE with the PISA and SAMBA expression-only biclustering systems. This comprises 1870 modules integrating five runs of COALESCE, 428 modules from one run of COALESCE, ∼1000 modules integrating ∼20 runs of PISA, 492 modules from one run of SAMBA (lower recall results are not available from PISA or SAMBA), and k-means clusters with k ranging from 10 to 5000 for comparison. (b) Effects of supporting data types (evolutionary conservation and nucleosome placement) and of dataset correlation structure on COALESCE predictions. While neither supporting data nor prior knowledge of dataset correlation structure (e.g. sets of related conditions such as time courses) significantly influence overall performance, accounting for correlation structure greatly improves conciseness, achieving comparable functional accuracy using <1/3 as many modules.
COALESCE allows a variety of supporting information and data types to be incorporated into the process of regulatory module prediction: information about the dependency structure within datasets, supporting data such as individual nucleotides' evolutionary conservation or occlusion by nucleosomes, or even the simple aggregation of multiple predictions into a single set of putative regulatory modules. A single execution of COALESCE on this data typically produces ∼450 regulatory modules, while aggregation of five executions produces ∼1900 predicted modules with diminishing returns as additional executions are included (Supplementary Figs 3 and 4, Supplementary Table 3). As indicated in Figure 2B, incorporation of information on dataset covariance introduces only minor changes in the precision and recall of these functional modules, but with a great improvement in their conciseness: each execution of COALESCE with dataset covariance produces ∼150 modules, with ∼550 predicted by the aggregation of five executions. Inclusion of additional data types has no substantial impact on the functional cohesiveness of predicted modules, which may be indicative that (i) individual binding sites are not sufficiently conserved at the genome-wide level to enhance functional specificity; (ii) conservation and/or nucleosome placement may be sufficiently condition-specific that global effects are not reflected in our single-condition datasets; and/or (iii) nucleosome placement is sufficiently dynamic that our single dataset does not reflect substantial global effects. Interestingly, when individual TFs are inspected, supporting data types do influence motif predictions as discussed below.
3.2 Predicted motifs match known S.cerevisiae and E.coli TFs and targets
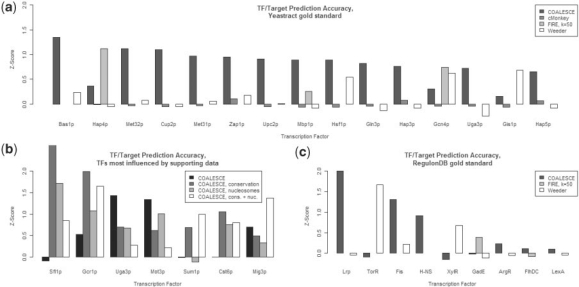
In order to compare COALESCE to other algorithms that explicitly predict regulatory modules (instead of solely gene expression biclusters), we evaluated its TF/target assignments based on the curated Yeastract (Teixeira et al., 2006) database (Fig. 3A). Yeastract includes one or more experimentally determined consensus binding sequences for ∼100 yeast TFs, as well as known regulatory targets for an additional ∼75 TFs. Using this information, we evaluated the cMonkey (Reiss et al., 2006) system in a similar manner, in addition to the FIRE (Elemento et al., 2007) and Weeder (Pavesi et al., 2004) motif predictors. While running cMonkey on the 2200-condition yeast compendium proved to be computationally impractical, we were able to run COALESCE on the 667 conditions used by Reiss et al. to validate cMonkey. Likewise, since FIRE and Weeder require data to be pre-clustered before motif prediction, we evaluated these two algorithms on these 667 conditions as clustered by k-means with k∈{50, 100, 500} and on the clusters produced by COALESCE itself. FIRE's best results were produced for k=50 (likely due to the relatively large size of the resulting clusters) and Weeder's for COALESCE's clusters (likely due to their smaller size). The resulting predictive performance of all four algorithms (COALESCE, cMonkey, FIRE and Weeder) is shown in Figure 3A. This figure displays a representative sample of predictions for individual TFs in order of decreasing algorithm-independent performance; COALESCE consistently provides accurate predictions for a variety of TFs, while cMonkey, FIRE and Weeder produce much sparser results and generally focus on common motifs with many strongly co-regulated targets (e.g. Gcn4p, Hap4p). It is worth noting that COALESCE runs over four times faster for S.cerevisiae data than comparable algorithms such as cMonkey (for which additional results are shown in Supplementary Fig. 5), and many times faster for higher organisms, enabling it to perform more in-depth analyses of larger, complex datasets; this is particularly important when exploring large compendia of metazoan (e.g. human) expression data as discussed below.
Fig. 3.
Accuracy of predicted S.cerevisiae and E.coli TF targets using the Yeastract and RegulonDB databases. COALESCE and cMonkey (Reiss et al., 2006) both predict expression biclusters and putative motifs simultaneously; FIRE (Elemento et al., 2007) and Weeder (Pavesi et al., 2004) require data to be pre-clustered and have been provided here with clusters from k-means and COALESCE, respectively (Section 2). TF/target prediction accuracies were assessed by assigning predicted motifs to known TF consensus binding sites, ranking predicted targets by frequency and confidence, and Z-scoring the resulting separation of known targets [from Yeastract (Teixeira et al., 2006) or RegulonDB (Gama-Castro et al., 2008)] from the background distribution. (a) The 15 yeast TFs predicted most accurately by any of the four systems. (b) Seven yeast TFs most influenced by integration of supporting data using COALESCE. This indicates that, for example, functional Sum1p binding sites may be particularly well-conserved among the Saccharomyces. (c) The nine E.coli TFs predicted most accurately by the three applicable systems; results are sparse due to the smaller coverage of RegulonDB.
Surprisingly, the incorporation of supporting data types did not substantially increase the overall accuracy of motif prediction, with data on evolutionary conservation or nucleosome positioning introducing only modest gains in accuracy with at best borderline statistical significance (paired t-test P < 0.05 and >0.5, respectively). However, the accuracies with which several individual TFs were assigned to known target genes were strongly affected by supporting data (Fig. 3B), indicating that these TFs may (i) have unusually conserved binding sites and/or (ii) strongly interact with nucleosome positioning in order to carry out their regulation. As seen in Figure 3B, examples include Sfl1p, Gcr1p and Cst6p, for which predictive performance is increased by the inclusion of supporting data. Interestingly, predicted targets of Uga3p and Mot3p both disagree with Yeastract more when supporting data types are included, suggesting that these TFs may have unusually weakly conserved binding sites or that the predicted nucleosome placements used in our analysis may differ from those in the conditions recorded by Yeastract. Finally, predicted targets of Sum1p are specifically improved by the inclusion of evolutionary conservation scores, raising the possibility that functional Sum1p target sites are consistently well-conserved within the (Kellis et al., 2003) Saccharomyces clade.
To further quantify COALESCE's ability to recover accurate regulatory modules, we performed a similar performance analysis in E.coli using the RegulonDB database of known TF consensus binding sequences and target genes (Gama-Castro et al., 2008) (Fig. 3C). Part of the goal of COALESCE is to enable large-scale data integration for regulatory module discovery in complex metazoans; however, while examples of predicted mammalian modules are discussed below, it is difficult to evaluate such predictions quantitatively due to the sparsity of transcriptional regulatory gold standards in higher organisms. RegulonDB itself is much smaller than Yeastract, even taking E.coli's smaller genome into account, and this sparsity is reflected in the relative performance shown in Figure 3C. Appropriate E.coli results were not available for cMonkey, but results from FIRE and Weeder were comparable to those in yeast: particularly for TFs with relatively general, low information content motifs (e.g. Lrp, Fis, H-NS), Of the three methods evaluated, only COALESCE consistently predicts more than 1–2 TFs' known target genes with high accuracy, as well as providing a variety of predictions that could represent novel direct targets or downstream effects of targeted pathways.
3.3 High accuracy with zero false positives in synthetic data
To assess COALESCE's accuracy more precisely than is possible with currently available biological databases, we analyzed three sets of synthetic data designed to resemble biological systems of interest, detailed in Table 1. These included a ‘yeast-like’ dataset Y1 comprising 5000 synthetic genes and 100 conditions, a ‘human-like’ dataset H1 comprising 25 000 genes and 100 conditions, and a ‘human-like’ compendium H2 of 25 000 genes and 10 000 conditions, comparable to the total amount of human microarray datasets currently available from GEO (Barrett et al., 2009). As shown in Table 1, these synthetic datasets were spiked with 10, 100 and 500 synthetic ‘TFs’, respectively, and each was analyzed with five different random seeds. Average runtimes for the three datasets were 4.1 min, 11.2 h and >24 h, respectively, resulting in average motif, gene and condition and F-scores of 0.92, 0.94 and 0.96 (module F-scores are not calculated since the H2 dataset analysis was terminated after 24 h, making recall inapplicable). Zero false positives were predicted for any dataset in the absence of spiked transcriptional modules.
Table 1.
COALESCE performance on synthetic data
| DS | Genes | Conditions | TFs | Prec./Rec. modules | Prec./Rec. motifs | Prec./Rec. genes | Prec./Rec. conditions |
|---|---|---|---|---|---|---|---|
| Y1 | 5000 | 100 | 10 | 1.00±0.00 0.68±0.17 | 1.00±0.00 0.91±0.28 | 1.00±0.01 0.94±0.08 | 0.99±0.02 1.00±0.00 |
| H1 | 25 000 | 100 | 100 | 0.99±0.01 0.75±0.04 | 0.98±0.10 0.92±0.27 | 0.98±0.05 0.94±0.11 | 0.99±0.08 1.00±0.01 |
| H2 | 25 000 | 10 000 | 500 | 0.90±0.02 NA | 0.83±0.30 0.87±0.33 | 0.80±0.28 0.99±0.02 | 0.83±0.23 0.98±0.05 |
COALESCE was a run using default parameters on three synthetic datasets constructed to resemble biological systems of interest: a ‘yeast-like’ dataset Y1 of 5000 genes, a ‘human-like’ dataset H1 of 25 000 genes, and a ‘human-like’ compendium H2 of 25 000 genes and 10 000 conditions. Each dataset was generated five times using different random seeds, with the average and SD of the resulting analyses shown here for module, motif, gene and condition precision and recall; see Supplementary Dataset 4 for details. When the datasets were generated without spiked TFs, zero false positives were predicted for any dataset. Prec., Precision; Rec., Recall.
3.4 Recovery of biologically relevant regulatory modules from large metazoan datasets
COALESCE is efficient enough to process substantial collections of metazoan data, which can comprise both large genomes (≥20 000 genes) and thousands of gene expression conditions. In addition to the quantitative evaluations, we have described here for yeast and E.coli, Figure 4 shows sample modules predicted by COALESCE from four metazoan datasets as detailed in Supplementary Table 4: tissue- and development-specific data from C.elegans, diverse conditions from D.melanogaster, neural data from M.musculus and ∼15 000 diverse conditions from H.sapiens. Full results for these datasets are available in Supplementary Dataset 2.
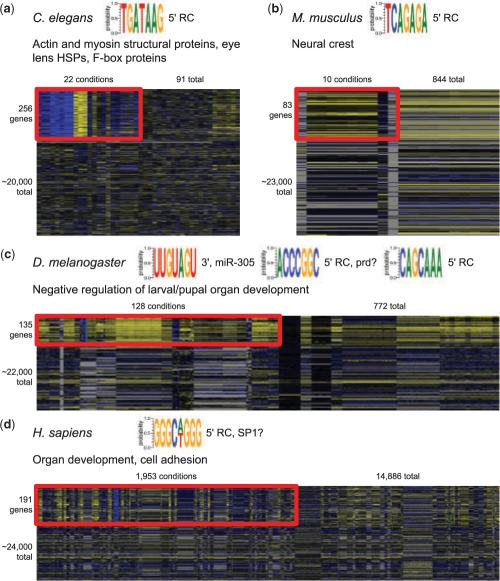
Fig. 4.
Metazoan regulatory modules predicted by COALESCE from worm, mouse, fly and human data. Each module shows associated functional enrichments of the contained genes, predicted motifs and their locations (5′ upstream or 3′ downstream, RC if predicted as a reverse complement), and a trimmed subset of the resulting normalized expression heatmap. (a) C.elegans module enriched for structural proteins co-expressing primarily in neural tissues with a predicted GATA motif. (b) M.musculus module co-expressing in neural crest samples, enriched for an uncharacterized reverse complement motif. (c) D.melanogaster module enriched for regulation of larval and pupal organ development, predicted to have two upstream and one downstream motif, the latter corresponding to miR-305; module genes also overlap significantly with known miR-305 targets (P < 0.005). (d) H.sapiens module enriched for organ development and adhesion proteins, with a predicted SP1-like upstream motif.
Briefly, the worm module in Figure 4A comprises ∼250 proteins functionally enriched for various structural functions (actins, myosins and the extracellular matrix), as well as known components of the eye lens (hsp-12.3, hsp-16.1, hsp-16.2 and others) and several F-box proteins. These co-express primarily in the neural tissues of (Colosimo et al., 2004) and (Von Stetina et al., 2007), and they are enriched for an upstream GATA motif. In Figure 4B, 83 mouse genes have been predicted to co-express under 10 conditions, seven of which are neural crest tissue samples (of 30 such samples in the compendium, hypergeometric P < 10−9); these are predicted to contain an uncharacterized upstream reverse-complement motif. Figure 4C shows a fly module functionally enriched for larval and pupal organ development and regulation. Of its three predicted motifs, two are upstream [one with strong similarity to the developmental TF paired consensus binding sequence (Underhill, 2000)] and one in the 3′ flank resembles the known miR-305 seed (Ruby et al., 2007); the 135 genes in the module also significantly overlap known miR-305 targets (hypergeometric P < 0.005). Finally, the human module in Figure 4D contains 191 genes co-regulated over almost a 10th of the expression data compendium, functionally enriched for organ development and with the consensus sequence of the known developmental TF SP1 (Zhao and Meng, 2005) predicted in the upstream region.
4 DISCUSSION AND CONCLUSIONS
The COALESCE algorithm is a biclustering and de novo motif prediction system capable of extracting regulatory modules from very large expression data compendia. It can also integrate supporting data such as sequence-level evolutionary conservation, nucleosome positioning or dataset-level correlation structure in order to improve its predicted regulatory networks. Here, we have detailed the algorithm and shown its effectiveness in predicting functionally enriched S.cerevisiae biclusters, in finding known yeast and E.coli TF targets, in analyzing synthetic data with zero false positives, and in predicting regulatory modules from metazoan data collections of up to tens of thousands of expression conditions.
Of course, COALESCE can also be used to analyze single datasets; an example of this is provided in Supplementary Dataset 2, which contains modules predicted from the S.cerevisiae controlled growth conditions of (Brauer et al., 2008). In combination with condition-specific supporting data (e.g. ChIP-chip for a TF of interest), this can provide a powerful means of querying an organism's regulatory network in a particular environment, although more focused systems also exist for this type of analysis (e.g. Lerman et al., 2007; Toedling and Huber, 2008). COALESCE's Bayesian integration step could easily be adapted, however, to incorporate data type-specific probabilities (e.g. based on a ChIP-chip or ChIP-seq physical binding model), which is a potential avenue for future development. Similarly, a query-based system could be developed to explore individual datasets by seeding modules with specific genes or known TFBS motifs of interest.
While COALESCE as an algorithm can scale efficiently to the complexities of metazoan genomes and datasets, it is less clear whether a simple computational model of regulatory modules—genes, conditions and short, independent sequence motifs—is completely appropriate for the biology of higher organisms. As demonstrated above, this model certainly captures some fraction of the regulatory information inherent in complex datasets (e.g. the ∼15 000 condition human compendium), and if sequence-level regulatory interactions are ignored, COALESCE remains an accurate and efficient biclustering algorithm. However, transcriptional regulation in multi-cellular organisms is substantially more complex than in prokaryotes or unicellular eukaryotes: individual binding motifs can be more degenerate, distal or both (Maston et al., 2006); combinatorial regulation is more prevalent (Smale, 2001); regulatory systems can buffer the effects of copy number changes (Stranger et al., 2007); and epigenetic and post-transcriptional/translational effects are more common and more complex (Reik et al., 2001; Wu and Belasco, 2008). A richer computational model will be necessary to more completely unravel the complexities of metazoan regulatory networks; this should both integrate additional experimental data types and include specific knowledge of the multiple ways in which gene products' activities can be modulated.
By providing a general framework for rapidly integrating large, diverse metazoan data collections, COALESCE represents a platform with which richer regulatory models can be built. As demonstrated by our evaluations in yeast and E.coli, the current algorithm can recover substantial portions of unicellular regulatory networks; incorporation of information on combinatorial regulation, alternate splicing, non-coding regulatory elements and additional experimental data types are all possibilities for expanding the breadth of predictions in multi-cellular organisms. A C++ implementation of COALESCE is available as part of the Sleipnir library (Huttenhower et al., 2008) at http://function.princeton.edu/sleipnir, and a web interface is provided at http://function.princeton.edu/coalesce, both of which allow data from any organism to be mined for new regulatory modules.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all of the members of the Coller and Troyanskaya labs for their suggestions and input, particularly Matthew Hibbs, Chris Park, Ana Pop and Erin Haley.
Funding: PhRMA Foundation (grant 2007RSGl9572); New Jersey Commission on Cancer Research fellowship; National Institutes of Health (grants T32 HG003284 and R01 GM071966); National Science Foundation CAREER Award (DBI-0546275); National Science Foundation (grant IIS-0513552); National Institute of General Medical Sciences Center of Excellence (grant P50 GM071508). Alfred P. Sloan Research Fellowship (to O.G.T.); Milton E. Cassel Scholarship of the Rita Allen Foundation (to H.A.C.).
Conflict of Interest: none declared.
REFERENCES
- Barrett T, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- Bonneau R. Learning biological networks: from modules to dynamics. Nat. Chem. Biol. 2008;4:658–664. doi: 10.1038/nchembio.122. [DOI] [PubMed] [Google Scholar]
- Brauer MJ, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemaker HJ, et al. Predictive modeling of genome-wide mRNA expression: from modules to molecules. Ann. Rev. Biophys. Biomol. Struct. 2007;36:329–347. doi: 10.1146/annurev.biophys.36.040306.132725. [DOI] [PubMed] [Google Scholar]
- Colosimo ME, et al. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr. Biol. 2004;14:2245–2251. doi: 10.1016/j.cub.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Durinck S, et al. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Elemento O, et al. A universal framework for regulatory element discovery across all genomes and data types. Mol. Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Castro S, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin Y, et al. Allegro: analyzing expression and sequence in concert to discover regulatory programs. Nucleic Acids Res. 2009;37:1566–1579. doi: 10.1093/nar/gkn1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S. Eukaryotic transcription factor binding sites–modeling and integrative search methods. Bioinformatics. 2008;24:1325–1331. doi: 10.1093/bioinformatics/btn198. [DOI] [PubMed] [Google Scholar]
- Huttenhower C, Troyanskaya OG. Assessing the functional structure of genomic data. Bioinformatics. 2008;24:i330–i338. doi: 10.1093/bioinformatics/btn160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, et al. The Sleipnir library for computational functional genomics. Bioinformatics. 2008;24:1559–1561. doi: 10.1093/bioinformatics/btn237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, et al. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kloster M, et al. Finding regulatory modules through large-scale gene-expression data analysis. Bioinformatics. 2005;21:1172–1179. doi: 10.1093/bioinformatics/bti096. [DOI] [PubMed] [Google Scholar]
- Kundaje A, et al. A predictive model of the oxygen and heme regulatory network in yeast. PLoS Comput. Biol. 2008;4:e1000224. doi: 10.1371/journal.pcbi.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens K, et al. DISTILLER: a data integration framework to reveal condition dependency of complex regulons in Escherichia coli. Genome Biol. 2009;10:R27. doi: 10.1186/gb-2009-10-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman G, et al. Functional genomics via multiscale analysis: application to gene expression and ChIP-on-chip data. Bioinformatics. 2007;23:314–320. doi: 10.1093/bioinformatics/btl606. [DOI] [PubMed] [Google Scholar]
- Long TA, et al. Systems approaches to identifying gene regulatory networks in plants. Ann. Rev. Cell Dev. Biol. 2008;24:81–103. doi: 10.1146/annurev.cellbio.24.110707.175408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl. 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston GA, et al. Transcriptional regulatory elements in the human genome. Ann. Rev. Genomics Hum. Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- Myers CL, et al. Finding function: evaluation methods for functional genomic data. BMC Genomics. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, et al. Weeder web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, et al. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Reiss DJ, et al. Integrated biclustering of heterogeneous genome-wide datasets for the inference of global regulatory networks. BMC Bioinformatics. 2006;7:280. doi: 10.1186/1471-2105-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth FP, et al. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, et al. Revealing modularity and organization in the yeast molecular network by integrated analysis of highly heterogeneous genomewide data. Proc. Natl Acad. Sci. USA. 2004;101:2981–2986. doi: 10.1073/pnas.0308661100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MC, et al. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, et al. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toedling J, Huber W. Analyzing ChIP-chip data using bioconductor. PLoS Comput. Biol. 2008;4:e1000227. doi: 10.1371/journal.pcbi.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill DA. Genetic and biochemical diversity in the Pax gene family. Biochem. Cell Biol. 2000;78:629–638. [PubMed] [Google Scholar]
- Von Stetina SE, et al. Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol. 2007;8:R135. doi: 10.1186/gb-2007-8-7-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Zhao C, Meng A. Sp1-like transcription factors are regulators of embryonic development in vertebrates. Dev. Growth Differ. 2005;47:201–211. doi: 10.1111/j.1440-169X.2005.00797.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.