Abstract
Summary: G-compass is designed for efficient comparative genome analysis between human and other vertebrate genomes. The current version of G-compass allows us to browse two corresponding genomic regions between human and another species in parallel. One-to-one evolutionarily conserved regions (i.e. orthologous regions) between species are highlighted along the genomes. Information such as locations of duplicated regions, copy number variations and mammalian ultra-conserved elements is also provided. These features of G-compass enable us to easily determine patterns of genomic rearrangements and changes in gene orders through evolutionary time. Since G-compass is a satellite database of H-InvDB, which is a comprehensive annotation resource for human genes and transcripts, users can easily refer to manually curated functional annotations and other abundant biological information for each human transcript. G-compass is expected to be a valuable tool for comparing human and model organisms and promoting the exchange of functional information.
Availability: G-compass is freely available at http://www.h-invitational.jp/g-compass/.
Contact: t.imanishi@aist.go.jp
1 INTRODUCTION
Information regarding evolutionary sequence conservation enables us to exchange biological knowledge between human and model organisms, since conserved sequences generally have important and similar functions across species. There are various genome browsers that show gene structures with patterns of genomic conservation, such as the UCSC Genome Browser (Karolchik et al., 2008), the Ensembl Genome Browser (Hubbard et al., 2009) and the VISTA Genome Browser (Visel et al., 2007). However, these browsers were designed to consider one genome as a reference, showing the patterns of genomic conservation of other genomes along the reference genome. The previous version of G-compass (Fujii et al., 2005), which only provided human–mouse genome comparisons, also considered the human genome as a reference. These browsers do not provide a quick overview of the genomic conservations and synteny information simultaneously in parallel. We found that only Combo (Engels et al., 2006) displays two genomic regions for different species in parallel and allows us to easily determine genomic rearrangements and changes in gene orders through evolutionary time. However, because Combo is provided as a Java program, it requires installation and configuration of a Java client. Furthermore, users have to prepare data in an appropriate format to be shown, making it difficult to use Combo, especially for bench biologists who are not familiar with computer-based analysis.
Here we present a new version of G-compass, which is freely available for all researchers via commonly used web browsers in any operating system, such as Windows, Mac or Linux, and has greatly improved user-friendly interfaces. Information on genomic conservations between human and another species is visually displayed simultaneously in parallel. G-compass also allow in silico biologists to download all information of one-to-one conserved genomic regions between human and other species for further analyses.
2 DATA CONSTRUCTION AND USAGE
Whole genome sequences were obtained from the UCSC web site (http://genome.ucsc.edu/): human (hg18), chimpanzee (panTro2), rhesus monkey (rheMac2), mouse (mm8), rat (rn4), dog (canFam2), cow (bosTau3), horse (equCab1), opossum (monDom4), chicken (galGal3), zebrafish (danRer4), medaka (oryLat1) and tetraodon (tetNig1). To identify evolutionary-conserved regions with duplicated regions, BLASTZ sequence similarity searches (Schwartz et al., 2003) were performed. Whole genome sequences were cut into overlapping fragments (‘10 Mb + 10 kb overlap’ for the reference genome and ‘50 Mb + 10 Mb overlap’ for the query genome). Round-robin BLASTZ searches were performed for genome fragments with the following parameters: ‘C=0 H=2000 Y=3400’ for primates, ‘C=0 H=2000’ for other mammals and ‘C=0 H=2000 Q=HoxD55.q’ for non-mammalian species. Subsequently, ‘reversed’ round-robin BLASTZ searches were performed by replacing the reference with the query and vice versa. Results of these searches were merged to produce genome alignments. Because the BLASTZ parameters used in these searches were more relaxed than those used for comparative genomics tracks in the UCSC Genome Browser, more conserved-regions are provided in G-compass; for example, numbers of conserved regions between the human and mouse genomes were 4 462 310 and 1 769 239, respectively, and coverage of the human genome was 36.3% and 35.9% for G-compass and axtNet data of UCSC, respectively. In addition to the conserved regions with duplicated regions, one-to-one (1:1) orthologous genomic regions were identified by removing redundancy in the genome alignments.
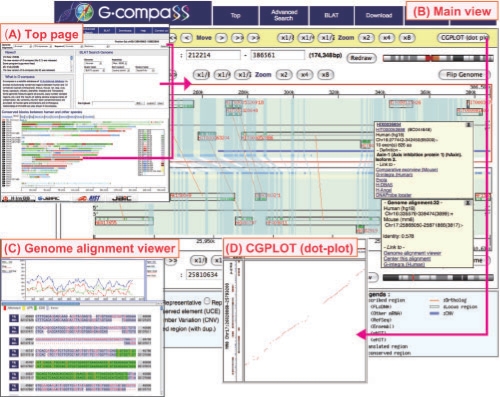
In the top page of G-compass, three entrances to the Main view are available: (i) keyword search for all transcripts and one-to-one orthologous genomic regions; (ii) the blast-like alignment tool (BLAT; Kent, 2002) for sequence similarity searches against the human, chimpanzee and mouse genomes; and (iii) clickable human chromosome maps with patterns of conserved regions with other species painted (Fig. 1A). In the Main view, two corresponding genomic regions in different species are displayed in parallel. For both genomes, mapped transcripts are shown and one-to-one orthologous genomic regions are highlighted as bands (light blue) (Fig. 1B). Copy number variations (CNVs) among human individuals and ultra-conserved elements (UCEs), which are genomic regions of at least 200 bp with 100% nucleotide identity between human and other mammalian organisms, are provided in genomic feature tracks. Clicking on each transcript or one-to-one orthologous genomic region, a pop-up window will display information. Users can get more detailed functional annotation and orthologous relationships from the links to H-InvDB (Genome Information Integration Project and H-Invitational 2, 2008) and Evola (Matsuya et al., 2008), respectively, in the pop-up window. The genome alignment viewer, also available from the link in the pop-up windows, provides detailed nucleotide sequence alignments and a user-interactive sliding window analysis tool for calculating the rate of nucleotide substitutions, gaps and GC contents (Fig. 1C). CGPLOT is a dot-plot viewer used to effectively investigate genome rearrangements between two genomic regions by comparing mapped gene structures (Fig. 1D).
Fig. 1.
Screenshots of G-compass. (A) Top page providing three entrances for users. (B) Main view showing conserved genomic regions between human and mouse in parallel. (C) Genome alignment viewer showing the levels of sequence conservation by sliding window analysis and the nucleotide sequence alignment with gene structures. (D) CGPLOT showing a dot-plot graph of two genomic regions with gene structures.
3 CONCLUSION
G-compass has been designed to serve as a comparative genome browser aiding general biologists in obtaining information by intuitive manipulation. It displays evolutionary conserved regions for human and another species in parallel. It also displays genomic features, such as UCEs, CNVs and conserved cis-regulatory elements. Close relationships with the annotated human gene database H-InvDB and its ortholog database Evola form the background of G-compass by adding standardized annotation resource of genes and transcript. Therefore, G-compass provides valuable information needed to investigate the patterns of vertebrate genome evolution and the function of conserved genomic regions.
ACKNOWLEDGEMENTS
We thank the members of the Integrated Database and Systems Biology Team, Biomedicinal Information Research Center, National Institute of Advanced Industrial Science and Technology (AIST) for valuable comments and suggestions. We also thank Masaru Watanabe for constructing cis-regulatory elements data.
Funding: Ministry of Economy, Trade and Industry of Japan, AIST; Japan Biological Informatics Consortium.
Conflict of Interest: none declared.
REFERENCES
- Engels R, et al. Combo: a whole genome comparative browser. Bioinformatics. 2006;22:1782–1783. doi: 10.1093/bioinformatics/btl193. [DOI] [PubMed] [Google Scholar]
- Fujii Y, et al. A web tool for comparative genomics: G-compass. Gene. 2005;364:45–52. doi: 10.1016/j.gene.2005.05.043. [DOI] [PubMed] [Google Scholar]
- Genome Information Integration Project H-Invitational 2. The H-Invitational database (H-InvDB), a comprehensive annotation resource for human genes and transcripts. Nucleic Acids Res. 2008;36:D793–D799. doi: 10.1093/nar/gkm999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuya A, et al. Evola: ortholog database of all human genes in H-InvDB with manual curation of phylogenetic trees. Nucleic Acids Res. 2008;36:D787–D792. doi: 10.1093/nar/gkm878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, et al. VISTA enhancer browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]