Abstract
RME-1/EHD1 family proteins are key residents of the recycling endosome required for endosome to plasma membrane transport in C. elegans and mammals. Recent studies suggest parallels of the RME-1/EHD proteins to the Dynamin GTPase superfamily of mechanochemical pinchases that promote membrane fission. Here we show that that endogenous C. elegans AMPH-1, the only C. elegans member of Amphiphysin/BIN1 family of BAR-domain proteins, colocalizes with RME-1 on recycling endosomes in vivo, that amph-1 deletion mutants are defective in recycling endosome morphology and function, and that binding of AMPH-1 NPF (D/E) sequences to the RME-1 EH-domain promotes the recycling of transmembrane cargo. We also show a requirement for human BIN1/Amphyphysin 2 in EHD1-regulated endocytic recycling. In vitro we find that the purified recombinant AMPH-1/RME-1 complexes produce short, coated, membrane tubules that are qualitatively distinct from those produced by either protein alone. Our results indicate that AMPH-1 and RME-1 cooperatively regulate endocytic recycling, likely through functions required for the production of cargo carriers exiting the recycling endosome for the cell surface.
Keywords: Endocytosis, endocytic recycling, RME-1, EHD1, Amphiphysin, Bin1, Dynamin, mechanoenzyme
Previous work has shown that RME-1 and mRme-1/EHD1 are ATPases that require ATP binding for homo-oligomerization and function in vivo1–4. Like RME-1 and mRme-1/EHD1, vertebrate-specific paralogs EHD2, EHD3, and EHD4 also function in endocytic transport but at different steps5–8. Although not apparent in the primary sequence, recent structural analysis of EHD2 revealed that its central ATP binding “G-domain” resembles the GTP binding domain of the large GTPase Dynamin9, a protein that mediates membrane fission through its ability to constrict vesicle necks10, 11. Additional parallels of EHD2 to Dynamin were also found, including the ability of EHD2 to assemble into spiral rings around acidic liposomes when bound to a non-hydrolysable ATP analog9. It was further shown that EHD2 ATPase activity is stimulated upon lipid binding and oligomerization, reminiscent of the assembly stimulated GAP activity characteristic of Dynamin9.
However, outside of the G-domain, RME-1/EHD family proteins are distinct from established Dynamin superfamily members. RME-1/EHD family proteins lack PH-domains. Rather the primary lipid binding region of EHD2 is helical in nature, forming a unique scissor-like interface in the EHD2 dimer9. RME-1/EHD family proteins also lack proline-rich domains. Instead, RME-1/EHD family proteins contain a C-terminal Eps15-homology (EH) domain, a different type of peptide binding interface known to target NPF (Asn-Pro-Phe) containing partner proteins12, 13. Given the high degree of overall similarity among RME-1/EHD family proteins, about 65% sequence identity, and the recently identified similarities to Dynamin, it has been suggested that all RME-1/EHD family proteins could have Dynamin like properties in promoting membrane fission4, 9. In particular, because recycling receptors accumulate in endosomes in the absence of RME-1 or EHD11–3, RME-1/EHD1 could function as the fission machinery for tubules emanating from endosomes, promoting the release of transport carriers during receptor recycling events.
RESULTS
Identification of RME-1 interacting proteins
To gain greater insight into the role of RME-1 at the recycling endosome, we sought functional interactors that might aid RME-1 in the recycling process. Alignment of several known binding partners of the mammalian EHD proteins suggested that RME-1/EHD family EH-domains prefer NPF-target sequences found in multiples and/or followed by acidic residues (reviewed in Ref. 13). The acidic residues following an NPF sequence could potentially neutralize the unique positive surface charge near the NPF binding pocket of RME-1/EHD family EH-domains13, 14.
Using bioinformatic searches of the predicted C. elegans proteome we identified 839 predicted worm proteins containing at least one NPF sequence (Fig.1a and Methods). 74 of these predicted proteins contained multiple NPFs and/or NPFs followed by acidic D/E stretches. To determine which of these 74 candidate RME-1 interactors might be physiologically relevant, we performed RNAi knockdown of each candidate in transgenic animals expressing GFP-tagged RME-1. We reasoned that knockdown of a physiologically relevant RME-1 binding partner could alter RME-1 subcellular localization and/or alter recycling endosome morphology. Among the small number of candidate interactors that affected RME-1 localization after RNAi we noted AMPH-1, the only C. elegans member of the Amphiphysin/Bin1 family of BAR and SH3 domain proteins (Fig.1a). This was especially intriguing given the known interactions of mammalian Amphiphysin with Dynamin in pre-synaptic membranes of the nervous system15, 16.
Figure 1.
AMPH-1 physically interacts with RME-1. (a) A flowchart representation of the steps involved in identifying AMPH-1 as an RME-1 EH-domain interacting protein. Bioinformatic searches of the C. elegans proteome identified multi-NPF and NPF(D/E) containing candidates which were assayed for effects on GFP-RME-1 subcellular localization after RNAi-mediated depletion, leading to identification of AMPH-1, a BAR (Bin1-Amphiphysin-Rvs161p/167p) and SH3 (Src-homology domain 3) domain protein. A diagram of the C. elegans amph-1 gene indicating 5’ and 3’ untranslated regions (dark gray boxes), exons (light gray boxes), introns (connecting lines), and the location of the amph-1(tm1060) deletion. (b) RME-1 (residues 447–555) was expressed as bait in a yeast reporter strain. AMPH-1 (residues 230–394) and its mutant forms were expressed as prey in the same yeast cells. Interaction between bait and prey was assayed by quantitative β-galactosidase (β-gal) assays. Mutation of either NPF motif to NPA is sufficient to significantly disrupt the interaction. The y-axis is labeled in Miller units. n=2, data from independent experiments is indicated above the graphs. (c) Glutathione beads loaded with recombinant GST or GST-RME- 1(442–576) were incubated with in vitro-expressed HA-AMPH-1(+) or HA-AMPH-1(F309A, F363A). Western blot for bound proteins was probed with anti-AMPH-1 antibody. Input lanes contain 50% target protein. The lower panel shows equivalent loading of bait GST fusion proteins. (d) Glutathione beads loaded with recombinant GST or GST-AMPH-1(full length) were incubated with in vitro-expressed HA-RME-1(full length). Bound proteins were analyzed by Western blot probed with anti-HA antibody. Input lanes contain 10% of the target protein HA-RME-1. The lower panel shows equivalent loading of bait GST fusion proteins. (e) Glutathione beads loaded with recombinant GST, GST-RME- 1(442–576), GST-mRme1/EHD1(EH domain), GST-Eps15(EH domain 2) or GST-Intersectin(EH domains a+b) were incubated with in vitro-expressed target protein HA-AMPH-1(+). Bound proteins were analyzed by Western blot with anti-AMPH-1 antibody. Input lane contains 10% target protein. The lower panel shows equivalent loading of bait GST fusion proteins. (f) Glutathione beads loaded with recombinant GST or GST-RME- 1(442–576) were incubated with in vitro-expressed target proteins HA-AMPH-1(+) or HA-AMPH-1(D310-312A, D364-366A). Bound proteins were analyzed by western blot with anti-AMPH-1 antibody. Input lanes contain 50% of the target protein. The lower panel shows equivalent loading of bait GST fusions.
Physical interaction between RME-1 and AMPH-1
In order to determine if AMPH-1 can physically bind to RME-1, we performed yeast two-hybrid based tests and GST pull-down analysis. AMPH-1 interacted with the RME-1 EH domain in the yeast two-hybrid system (Fig.1b). The interaction was abolished when either or both AMPH-1 NPF sequences were altered by alanine substitution at phenylalanine F309 and/or F363 (Fig.1b). In the pull down experiment, full length AMPH-1 from wild-type worm lysates, or in vitro translated AMPH-1(+) was efficiently isolated by immobilized GST-RME-1 EH-domain fusion protein (Fig.1c; Supplemental information S1 b). AMPH-1 also bound strongly to the EH-domain of mammalian mRme-1/EHD1 in this assay (Fig. 1e). Little or no binding of AMPH-1 to the EH-domains of other endocytic proteins Eps15 or Intersectin was observed, indicating the specificity of the interaction (Fig 1e). Furthermore in vitro translated AMPH-1(F309A, F363A), lacking intact NPF motifs, failed to bind to GST-RME-1 EH-domain fusion protein (Fig. 1c), and alanine substitution of the first three aspartic acid residues following each NPF motif reduced binding (Fig. 1f). Full length in vitro translated RME-1 could be isolated by immobilized full length GST-AMPH-1 fusion protein (Fig.1d). We conclude that AMPH-1 binds directly to RME-1, and that this interaction requires the RME-1 EH domain and the two NPF [D/E] motifs present within AMPH-1.
AMPH-1 is enriched on recycling endosomes
RME-1 is ubiquitously expressed in C. elegans, and is highly enriched on tubulovesicular basolateral recycling endosomes in the polarized intestinal epithelium1. In transgenic animals, AMPH-1-GFP under the control of amph-1 genomic DNA regulatory sequences was similarly widely expressed (Supplemental information, Fig. S1c–j). Affinity-purified anti-AMPH-1 antibodies detected a single band of the expected size for AMPH-1 in Western blots of wild-type animals. This band was absent in amph-1(tm1060) deletion mutant animals obtained from the Japanese National Bioresource Project for the Experimental Animal “Nematode C. elegans” (Supplemental information, Fig. S1a and see below), establishing the specificity of the antibody.
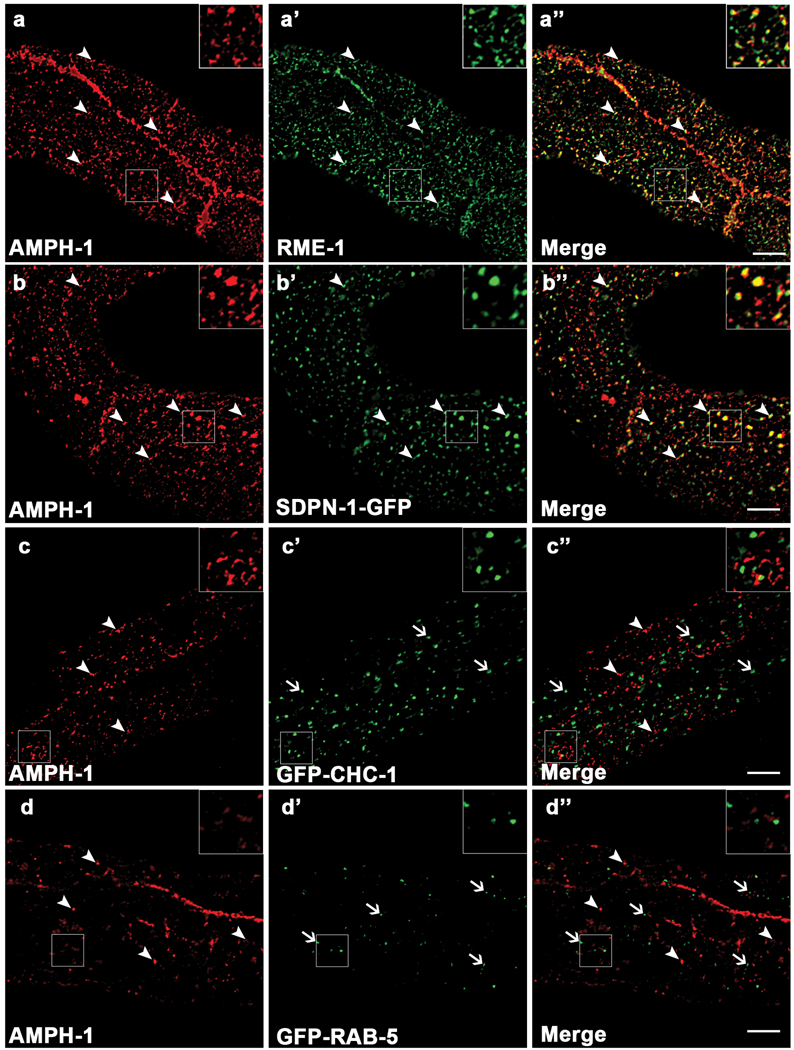
Using immunofluorescence, we found that endogenous AMPH-1 colocalizes extensively with endogenous RME-1 on tubulovesicular basolateral recycling endosomes (Fig. 2a-a’’), and on more weakly labeled sub-apical structures of the worm intestine, possibly the apical recycling endosomes (Supplemental information, Fig. S2c-c’’). While there was significant overlap of RME-1 and AMPH-1 labeling on the recycling endosome, the two proteins do not label the endosome identically. This may reflect differences in the degree of association with specific endosomal sub-domains, or may reflect temporal differences in the assembly or disassembly kinetics of the two proteins. Endogenous AMPH-1 also colocalized with basolateral GFP-tagged SDPN-1/Syndapin/Pacsin, another marker for the C. elegans recycling endosome (Fig. 2b-b’’) 17. Anti-AMPH-1 staining failed to colocalize with markers for the clathrin coated pits (GFP-tagged CHC-1/clathrin)18, early endosome (GFP-RAB-5)19, late endosome (GFP-RAB-7)19 or Golgi (Mannosidase-GFP)19, indicating that AMPH-1 is specifically enriched on recycling endosomes (Fig. 2c-c’’, Fig. 2d-d’’ and Supplemental information, Fig. S3). We also observed extensive colocalization of mCherry-RME-1 and AMPH-1-GFP in living transgenic animals, supporting the data derived from fixed tissue (Supplemental information, Fig. S2a-a’’).
Figure 2.
AMPH-1 co-localizes with RME-1 and SDPN-1/Syndapin on recycling endosomes. Wild type N2 worms or various GFP labeled transgenic worm strains were synchronized to the adult stage and decapitated by microsurgery. The extruded worm intestines were fixed and immunostained with antibodies for visualization of endogenous protein localization. (a-a’’) Endogenous AMPH-1 (red) labeled with rabbit polyclonal anti-AMPH-1 antibody co-localizes extensively with endogenous RME-1 labeled with mouse monoclonal antibody (green). Arrowheads indicate structures labeled by both AMPH-1 and RME-1. (b-b’’) Endogenous AMPH-1 (red) co-localizes significantly with the recycling endosome marker SDPN-1-GFP (green). Arrowheads indicate structures labeled by both AMPH-1 and SDPN-1. (c-c’’) Endogenous AMPH-1 (red) does not co-localize significantly with clathrin (GFP-CHC-1). Arrowheads indicate structures labeled by AMPH-1 and arrows indicate structures labeled by GFP-CHC-1. (d-d’’) Endogenous AMPH-1 (red) does not co-localize with the early endosome marker GFP-RAB-5 (green). Arrowheads indicate structures labeled by AMPH-1 and arrows indicate structures labeled by GFP-RAB-5. Scale bars represent 10 µm.
The lack of association of AMPH-1 with clathrin-coated pits is consistent with the absence of a consensus CLAP (clathrin and adaptor AP2 binding) domain in worm AMPH-1. In fact the CLAP domain is specific for certain isoforms of vertebrate Amphiphysins and is not found in any known invertebrate Amphiphysin family members20. In Drosophila, dAmph has been shown to lack Shibire/Dynamin binding ability, does not colocalize with Shibire/Dynamin, and does not produce shibire-like phenotypes when mutated21–23. While Drosophila Shibire/Dynamin and C. elegans DYN-1/Dynamin do posses proline rich sequences, neither Dynamin ortholog contains the PXRPXR consensus motif that is recognized by Amphiphysin SH3 domains in vertebrates24. Notably, Drosophila Amphiphysin does contain an NPF sequence followed by acidic residues, suggesting potential interaction with an RME-1/EHD family member.
amph-1 null mutants are defective in endocytic recycling
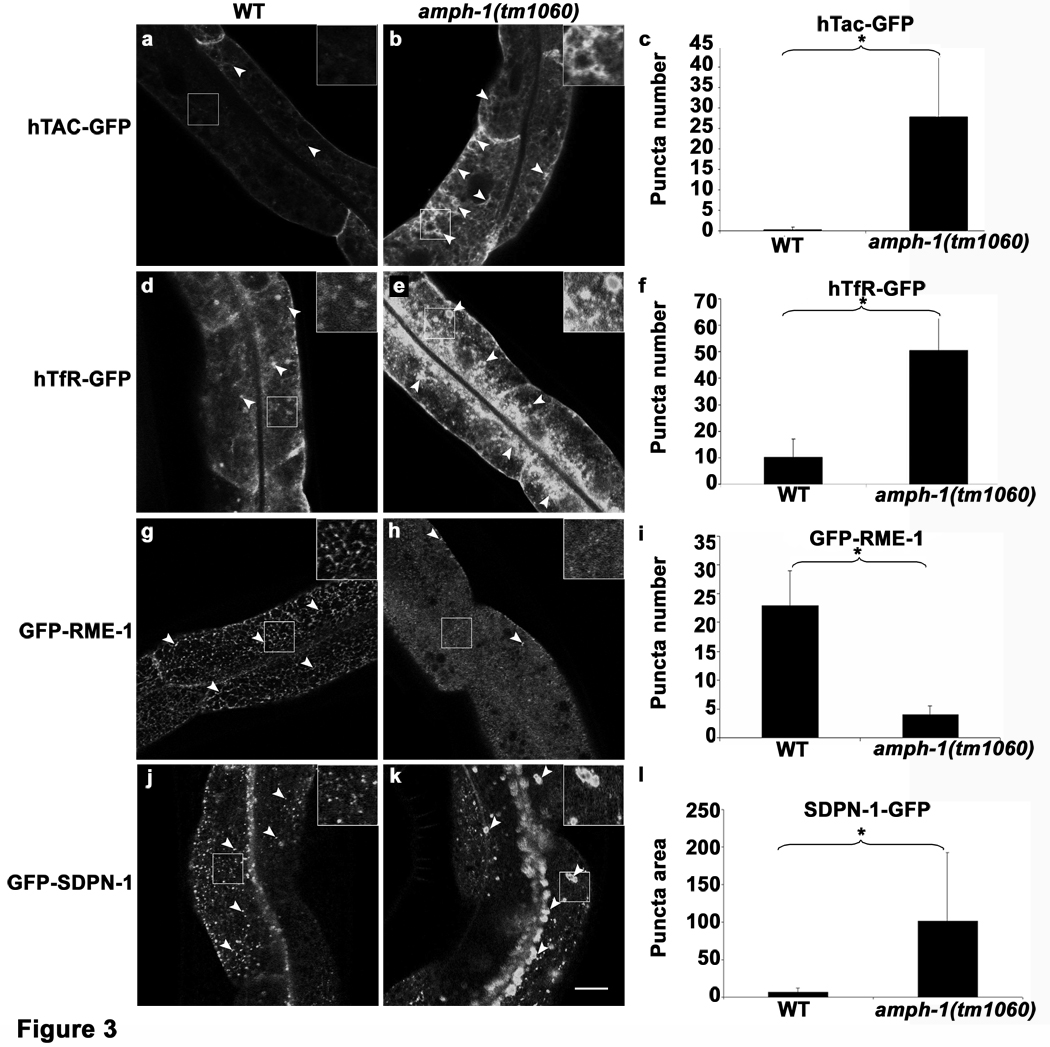
In order to determine the function of AMPH-1 in vivo, we characterized the phenotype of amph-1(tm1060) null mutants. To detect recycling defects, we first focused on previously validated model recycling cargo proteins hTfR-GFP (human transferrin receptor) and hTac-GFP (IL2-receptor alpha chain)25, 26. hTfR is internalized via clathrin-dependent mechanisms, while hTac is internalized by clathrin-independent mechanisms27. Both of these model recycling cargo proteins depend upon RME-1/EHD1 for their recycling in mammalian cells and worm cells, accumulating in recycling endosomes in the absence of RME-1/EHD1 function 2, 3 17, 19. The number of hTac-GFP and hTfR-GFP intracellular puncta number increased by more than 10-fold compared to wild type controls in amph-1 mutants (Fig. 3 a–c, d–f). These results indicate a defect in basolateral recycling in the intestinal epithelia of amph-1 mutants, very similar to that found in rme-1 mutants.
Figure 3.
C. elegans amph-1 mutants display abnormal trafficking of recycling cargo and morphologically abnormal recycling endosomes.(a–c) hTAC-GFP, a cargo protein internalized independently of clathrin, accumulates intracellularly in amph-1 mutants. Arrowheads indicate punctate and tubular hTAC-GFP signal in the intestine. (c) Quantification of hTAC-GFP signal in the intestine of living wild-type and amph-1 mutant animals with respect to average number of labeled structures per unit area. The asterisk indicates a significant difference in the one-tailed Student’s T-test, p-value= 1.2×10−12 (d–f) hTfR-GFP, a clathrin-dependent cargo protein, accumulates intracellularly in amph-1 mutants. (f) Quantification of hTfR-GFP signal in the intestine of living wild-type and amph-1 mutant animals with respect to average number of labeled structures per unit area. The asterisk indicates a significant difference in the one-tailed Student’s T-test, p value=9.7×10−22. (g–i) Loss of GFP-RME-1 labeling of basolateral recycling endosomes in the amph-1(tm1060) mutants. Arrowheads indicate GFP-RME-1 signal. (i) Quantification of GFP-RME-1 signal in the intestine of living wild-type and amph-1 mutant animals with respect to average number of labeled structures per unit area. The asterisk indicates a significant difference in the one-tailed Student’s T-test, p-value =1.1424×10−07. (j–l) SDPN-1-GFP labeled recycling endosomes are altered in the amph-1(tm1060) null mutant, appearing fewer and larger. Arrowheads indicate SDPN-1-GFP labeled structures. (l) Quantification of SDPN-1-GFP signal in the intestine of living wild-type and amph-1 mutant animals with respect to average number of labeled structures per unit area. The asterisk indicates a significant difference in the one-tailed Student’s T-test, p-value=2.4×10−06.
For all the presented data, mean values were plotted on the graph and error bars represent ± s.d. from the mean. In all the experiments, n = 30 sampled fields (six animals of each genotype sampled in five different regions of each intestine). Scale bar represents 10 µm.
Specific disruption of recycling endosomes in amph-1 mutants
Further analysis showed that RME-1 labeling of basolateral recycling endosomes was profoundly abnormal in amph-1 mutant animals, with loss of most tubular labeling and an increase in diffuse GFP-RME-1 labeling. Only a small number of RME-1 labeled punctate structures remained (Fig. 3 g–i; Supplemental information, Fig. S5). The loss of RME-1 from recycling endosomes confirmed the amph-1(RNAi) results from the initial screen, suggesting that AMPH-1 functions, at least in part, to recruit RME-1 to the recycling endosome membrane. We also found that loss of amph-1 disrupted another recycling endosome marker SDPN-1-GFP, with greatly reduced SDPN-1-GFP puncta number, and gross enlargement of remaining labeled structures (Fig. 3 j–l). This phenotype is very similar to that we previously found in rme-1 mutants17, further indicating that AMPH-1 functions with RME-1 in vivo. We did note however that some amph-1 phenotypes appear weaker than rme-1 phenotypes, such as in the accumulation of fluid filled vacuoles in the intestine, and the transport of yolk proteins from the intestine to the oocyte (data not shown). This might indicate some functional redundancy between AMPH-1 and other BAR domain adaptor proteins, or residual RME-1 activity in the absence of AMPH-1.
We also examined amph-1 mutants for effects on early endosomes. This was particularly important because mutants known to affect an earlier recycling step, from the early endosome to the recycling endosome, produce a similar set of phenotypes, including loss of RME-1-labeled recycling endosomes and intracellular trapping of recycling cargo proteins such as hTac-GFP19. The key distinguishing characteristic of an earlier block, such as that displayed by rab-10 mutants, is the dramatic over-accumulation of early endosomes, a phenotype not found in rme-1 mutants19. Consistent with a later function for AMPH-1 at the recycling endosome to plasma membrane step, the morphology and distribution of early endosome markers GFP-RAB-5 and GFP-RAB-10 was unperturbed in amph-1 mutants (Supplemental information, Fig. S4 a–c, d–f).
Finally, because mammalian Amphiphysin has been associated with plasma membrane clathrin-coated pits16, 28–30, we investigated whether amph-1 mutants alter the morphology or distribution of functional GFP-tagged Dynamin or Clathrin. We found that DYN-1-GFP and GFP-CHC-1 localization was unaltered in amph-1 mutants, consistent with the proposal that C. elegans AMPH-1 does not interact with Dynamin, similar to previous findings in Drosophila (Supplemental information Fig. S4 g–i, j–l) 21–23. Thus, we do not find any evidence that worm AMPH-1 is involved in recruiting DYN-1/dynamin to the clathrin coated pit during endocytic uptake at the plasma membrane. Instead, C. elegans AMPH-1 appears to be specific for RME-1 mediated recycling processes.
AMPH-1 requires its interaction with RME-1 for recycling function
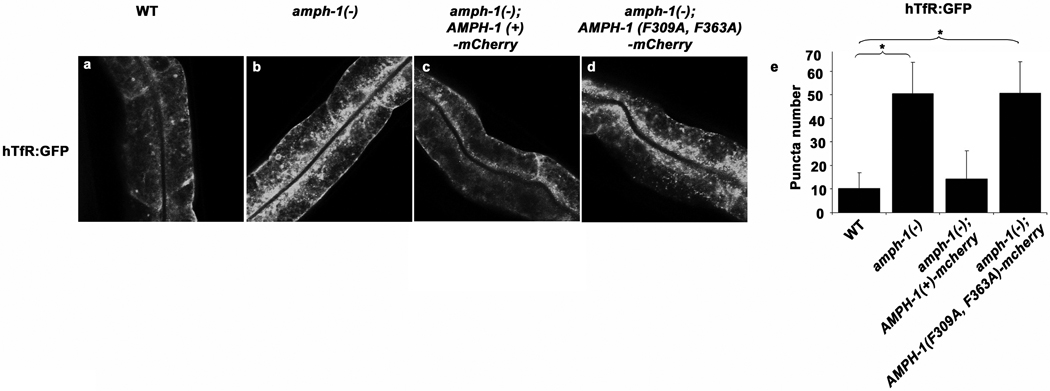
Having established the effects of loss of AMPH-1 on endocytic traffic, and the close association of AMPH-1 with RME-1, we sought to determine the importance of the AMPH-1/RME-1 physical association in vivo. Thus we tested the ability of transgenically expressed AMPH-1, with or without intact RME-1 interaction motifs, to rescue amph-1 mutant phenotypes. We found that although AMPH-1(F309A, F363A) expressed as well as AMPH-1(+) (Supplemental information Fig. S5 a–b), only AMPH-1(+) rescued the amph-1 mutant phenotypes with respect to hTfR accumulation and RME-1 localization (Fig. 4 and Supplemental information Fig. S5c–g), indicating that AMPH-1 requires its interaction with RME-1 to perform its role as an endocytic recycling regulator. We also found that AMPH-1 protein is mislocalized in rme-1(0) mutants. Instead of an extended tubulovesicular pattern, we observed a more concentrated localization of endogenous AMPH-1 to enlarged structures, similar to the abnormal localization of SDPN-1 in rme-1 mutants (Supplemental information, Fig. S5h–l). However AMPH-1 still appeared to be membrane associated in the absence of RME-1, suggesting that AMPH-1 does not require RME-1 for membrane recruitment.
Figure 4.
AMPH-1 function in endocytic recycling requires interaction with RME-1. AMPH-1(+) and interaction deficient AMPH-1(F309A, F363A) were expressed as mCherry fusions in amph-1(tm1060) mutant intestines. AMPH-1(+) ( c) but not AMPH-1 (F309A, F363A) (d) could rescue the abnormal accumulation of recycling cargo hTfR-GFP in amph-1(tm1060) mutants. Synchronized adult worms were imaged by confocal microscopy. (e) Quantification of the hTfR-GFP labeled structures in the worms with respect to average number of labeled structures per unit area. The asterisk indicates a significant difference in the one-tailed Student’s T-test, p value for WT compared against amph-1(tm1060) = 9.7×10−22 and for WT compared with amph-1(tm1060); AMPH-1(F309A, F363A)-mCherry=9.4×10−22. For all the presented data mean values were plotted on the graph and error bars represent ± s.d. from the mean. In all the experiments, n = 30 sampled fields (six animals of each genotype sampled in five different regions of each intestine). Scale bar represents 10 µm.
Mammalian Bin1/Amphiphysin2 functions in recycling
Unlike C. elegans, which appears to express only one Amphiphysin isoform, the mammalian genome contains two distinct Amphiphysin genes denoted Amphiphysin 1 and Bin1/Amphiphysin 2. Each of these genes encodes multiple protein isoforms. Amph1 is brain specific and Amph2a is brain enriched. Both of these isoforms contain a CLAP domain for Clathrin and AP2 adaptor binding. Amph1 and Amph2a form heterodimers and are thought to aid endocytic uptake at the synaptic clathrin coated pit, at least in part through interaction with Dynamin15, 16, 28, 29, 31–33. Other forms of BIN1/Amph2 lack clathrin/AP2 interaction modules. One specific isoform functions in muscle development34, while other isoforms, such as BIN1 isoform 10 discussed below, have no known function. In fact, it has been shown that BIN1/Amph2 knockout MEF cells are not defective in endocytic uptake of transferrin, rather showing an approximately two-fold increase in transferrin uptake35.
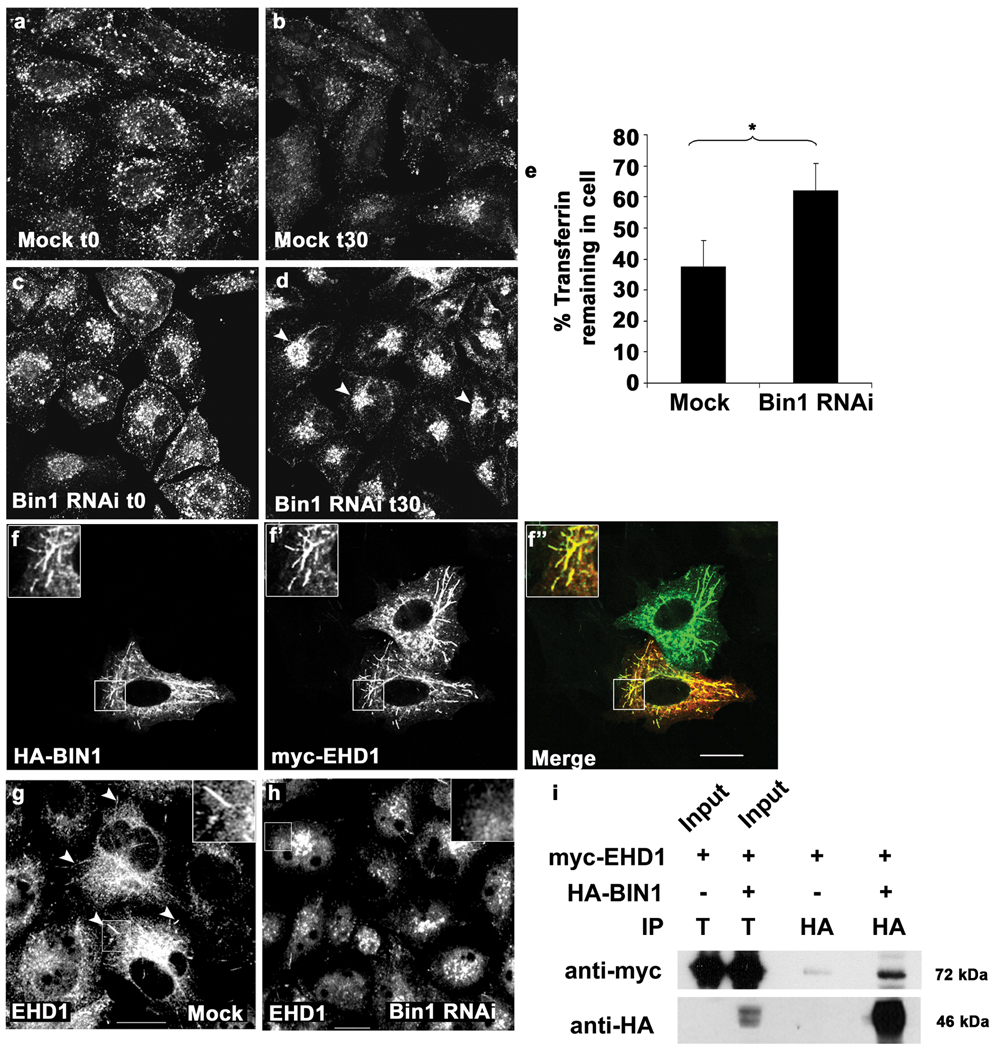
Although the known BIN1 isoforms lack NPF sequences, and so may not bind to mammalian RME-1/EHD proteins directly, we considered the possibility that as in C. elegans, mammalian BIN1/Amph2 might play a role in endocytic recycling. In fact, we observed weak co-immunoprecipitation of myc-EHD1 with HA-Bin1 isoform 10 in transfected cells (Fig. 5 i). As a direct measure of the importance of BIN1 in endocytic transport, we followed fluorescently labeled transferrin transport in BIN1 depleted HeLa cells using previously described pulse-chase assays27. BIN1 knockdown strongly reduced expression of all forms of BIN1/Amph2 in HeLa cells as determined by Western blot (Supplemental information Fig. S6a). Consistent with the previously published results in knockout MEFs, we found that HeLa cells depleted of BIN1/Amph2 are not defective in transferrin uptake. Rather uptake was increased in BIN1 knockdown cells after 7 min of Tf uptake as measured by FACS (mock treated 348±118 a.u., BIN1 RNAi 662±325 a.u., averaged from three independent experiments) or visualized by microscopy (Fig. 5a, c). Consistent with a role for BIN1 in recycling from the ERC, we found that transferrin accumulated in the juxtanuclear compartment in BIN1/Amph2 depleted cells (Fig. 5b, d), and recycling was delayed as measured by image quantification (Fig. 5e) as well as by FACS analysis (Supplemental information Fig. S6b).
Figure 5.
RNAi-mediated depletion of human Bin1 impairs Transferrin recycling and disrupts mRme-1/EHD1 positive recycling endosome tubules. Representative fields are shown for mock treated or Bin1 RNAi treated HeLa cells after 5 min Alexa-488 transferrin uptake (a, c) and 30 min chase (recycling) (b, d). The average total cell fluorescence was measured from confocal micrographs of 80 HeLa cells, mock treated or Bin1 RNAi treated, after Alexa-488 transferrin uptake followed by chase in complete medium for 30 min. (e) The data shown represents the mean of three independent experiments with n=80 cells per experimental condition. The mean values were plotted and error bars indicate ± s.e. from the mean value. The asterisk indicates a significant difference in the one-tailed Student’s T-test p value= 1.06 × 10−35. (f-f’) Bin1 and mRme-1/EHD1 colocalize on intracellular tubules. HeLa cells transfected with 2×-HA-BIN1 isoform 10 and myc-EHD1 were visualized by confocal microscopy after immunostaining with primary antibodies (f) mouse anti-HA and (f’) rabbit anti-EHD1 followed by appropriate fluorochrome-labeled secondary antibodies. The insets and merged image (in panel f’’) demonstrate colocalization of HA-BIN1 isoform 10 with myc-EHD1 positive tubules. (g–h) mRme-1/EHD1 positive tubules are disrupted by Bin1 depletion. HeLa cells grown on coverslips were mock treated or subjected to Bin1 RNAi. Endogenous EHD1 was visualized by confocal microscopy after immunostaining with affinity-purified polyclonal rabbit anti-EHD1 antibody, followed by Alexa Fluor 488 anti-rabbit secondary antibodies. Note the loss of tubular EHD1 labeling after Bin1 depletion. Scale bar represents 10 µm. (i) mRme1/EHD1 physically associates with Bin1 isoform 10. HeLa cells were transfected with myc-EHD1 or co-transfected with myc-EHD1 and 2×-HA-Bin1 isoform 10. Cells were lysed and lysates were immunoprecipitated with goat anti-HA antibody-conjugated agarose beads. Unbound protein was removed by successive washes. Bound proteins were eluted with Laemmli sample buffer, separated by SDS-PAGE, and analyzed by western blot with anti-myc antibody. Input lane contains 1% of the lysate used in the binding assay.
Consistent with the idea that BIN1/Amph2 functions directly in transport from the ERC, we found that BIN1 was enriched on distinctive mRME-1/EHD1-positive tubules. This could be visualized in cells overexpressing myc-EHD1 and HA-Bin1 isoform 10 (Fig.5 f-f’’), and in untransfected cells visualizing the endogenous proteins using EHD1 and BIN1 (isoform non-specific) antibodies (Supplemental information, Fig. S6c-c’’). The EHD1-positive tubules have been previously shown to be endogenous features of HeLa cells, emanating from the endocytic recycling compartment (ERC) 3. These tubular endosomes are involved in the recycling of cargo proteins from the ERC to the plasma membrane 3, and the absence of EHD1 from these structures leads to impaired recycling36. We found that depletion of BIN1/Amph2 disrupted the EHD1-positive recycling tubules (Fig. 5g–h). After depletion of BIN1/Amph2, EHD1 localization was limited to a compact juxtanuclear localization, consistent with the main body of the ERC (Fig. 5h)37. Taken together these results indicate the BIN1 is important for the formation and/ or maintenance of ERC derived tubules. Furthermore these results indicate that the requirement for an amphiphysin family protein in the endocytic recycling pathway is a conserved evolutionary feature from worms to mammals.
Reconstitution of RME-1/AMPH-1 interactions in a liposome system
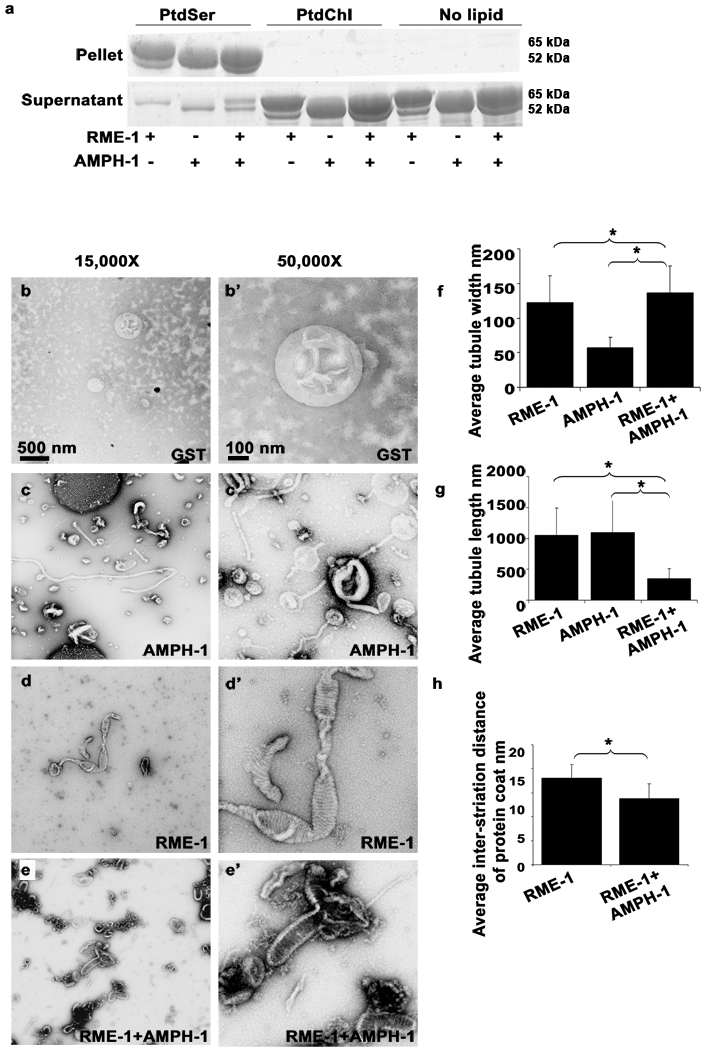
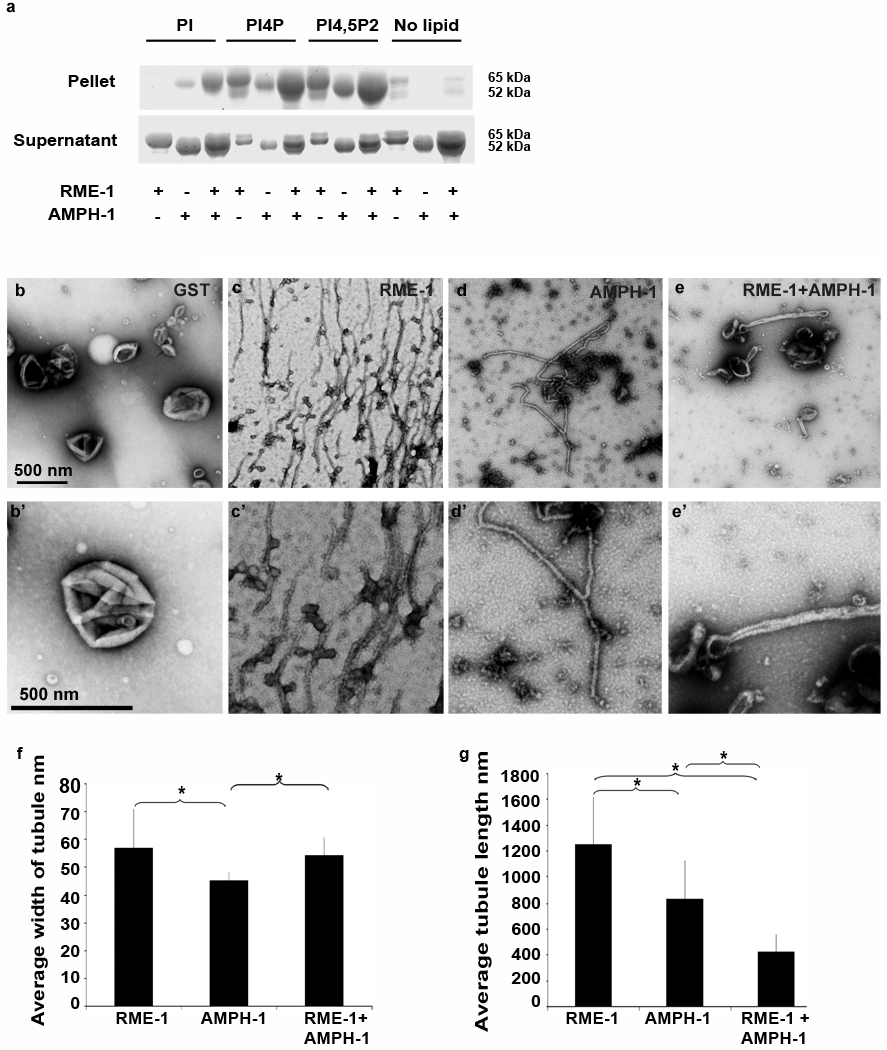
Previous reports have shown that mammalian Amphiphysin38, mammalian Dynamin15, 33, 38, and mammalian EHD29 have the capacity to bind to negatively charged liposomes and deform the membrane to produce tubules15, 38,9. Mammalian HeLa cell recycling endosomes are enriched in PIP2 due to the activation of PI5K by Arf6 on these endosomes39 and PI4P has also been recently identified as a component of these endosomes36. As a first step toward determining the membrane binding capacity of worm RME-1 and AMPH-1, we tested the interaction of purified recombinant RME-1 and AMPH-1 with liposomes in a sedimentation assay. We found that both RME-1 and AMPH-1 pelleted with acidic Phosphatidylserine (PtdSer) liposomes, or with a more physiological liposome composition containing 10% PS and 10% PIP2 (phosphatidylinositol-[4,5]-bisphosphate) or 10% PI4P (phosphatidylinositol-[4]-phosphate) (Fig 6a, Fig. 7a). Interestingly the binding to PIP2, PI4P, and PI-containing liposomes appeared enhanced when both proteins were added to the reaction, compared to adding the proteins individually (Fig. 7a). We observed no binding of either protein to neutral Phosphatidylcholine (PtdChl) liposomes (Fig. 6a, Fig. 7a).
Figure 6.
Membrane binding and tubulation by AMPH-1 and RME-1 in vitro. (a) Coomassie stained gels of supernatant and pellet fractions from liposome co-sedimentation assays are shown. Binding reactions were performed in the absence or presence of 0.33 mg/ml, 0.4 µm (average diameter) 100% Phosphatidylserine (PtdSer), or 100% Phosphatidylcholine (PtdChl) liposomes. Liposomes were incubated with 1mM ATP-γ-S and 1 µM of full length AMPH-1 or RME-1 proteins, or equimolar quantities of both proteins, as indicated. (b-e’) Electron micrographs of negatively stained PtdSer liposomes, prepared as above, but used at 0.05 mg/ml in the presence of 1mM ATP-γ-S with 2.5 µM proteins, GST (b-b’), AMPH-1 (c-c’), RME-1 (d-d’), or equimolar quantities of AMPH-1 and RME-1 (e-e’). (f) Quantification of tubule widths. For each experimental condition, width was measured for every tubule on each of 15 electron micrographs. For tubules demonstrating uneven width, an average measurement was made after taking measures at several representative points along the tubule. For n=15 representative tubules per experimental condition, the mean value for tubule width was plotted and error bars represent ± s.d. from the mean. The asterisk indicates a significant difference in the one-tailed Student’s T-test: RME-1 vs. RME-1+AMPH-1, p value= 0.0074 and for AMPH-1 vs. RME-1+AMPH-1, p value= 1.29 × 10−13. (g) Quantification of tubule lengths. The length from the edge of the liposome body to end of tubule was measured for every tubule on each of 15 electron micrographs per experimental condition. For n=15 representative tubules for each condition, the mean value for tubule length was plotted and error bars represent ± s.d. from the mean. The asterisk indicates a significant difference in the one-tailed Student’s T-test for tubule length: RME-1 vs. RME-1+AMPH-1, p value=1.2 × 10−07 and for AMPH-1 vs. RME-1+AMPH-1, p value= 3.2 × 10−04. (h) Quantification of inter-striation distance. For n=15 representative tubules, the average distance between every successive ring-like striation was counted for each experimental condition. Mean values were plotted and error bars represent ± standard deviation from the mean. The asterisk indicates a significant difference in the one-tailed Student’s T-test, p value=0.0031.
Figure 7.
Membrane binding and tubulation by AMPH-1 and RME-1 in vitro with PI-[4,5]P2 liposomes. (a) Coomassie stained gels of supernatant and pellet fractions from liposome co-sedimentation assays are shown. Binding reactions were performed in the absence or presence of 0.33 mg/ml, 0.4 µm (average diameter) 80% Phosphatidylcholine (PtdChl), 10% Phosphatidylserine (PtdSer) and 10% of either unphosphorylated phosphatidylinositol (PI) or phosphatidylinositol-[4]-phosphate (PI-[4]-P) or phosphatidylinositol-[4,5]-bisphosphate (PI-[4, 5] P2) containing liposomes. Liposomes were incubated with 1mM ATP-γ-S and 1 µM of full length AMPH-1 or RME-1 proteins, or equimolar quantities of both proteins, as indicated. (b-e’) Electron micrographs of negatively stained PI-[4, 5]-P2 liposomes, prepared as above, but used at 0.05 mg/ml in the presence of 1mM ATP-γ-S with 2.5 µM proteins, GST (b-b’), RME-1-1 (c-c’), AMPH-1 (d-d’), or equimolar quantities of AMPH-1 and RME-1 (e-e’). Panels b–e represent 50, 000× magnification images while panels b’-e’ are enlarged regions from panels b–e respectively. (f) Quantification of tubule widths. For each experimental condition, width was measured for every tubule on each of 10 electron micrographs. For tubules demonstrating uneven width, an average measurement was made after taking measures at several representative points along the tubule. For n=10 representative tubules, the mean value for tubule width was plotted and error bars represent ± s.d. from the mean. The asterisk indicates a significant difference in the one-tailed Student’s T-test: RME-1 vs. AMPH-1, p value=0.01, RME-1 vs. RME-1+AMPH-1, p value= 0.29 and for AMPH-1 vs. RME-1+AMPH-1, p value= 0.004. (g) Quantification of tubule lengths. The length from the edge of the liposome body to end of tubule was measured for every tubule on each of 10 electron micrographs per experimental condition. For n=10 representative tubules for each experimental condition, the mean value of tubule length was plotted and error bars represent ± s.d. from the mean. The asterisk indicates a significant difference in the one-tailed Student’s T-test for tubule length: RME-1 vs. AMPH-1, p value= 0.005, RME-1 vs. RME-1+AMPH-1, p value=1.8 × 10−06 and for AMPH-1 vs. RME-1+AMPH-1, p value= 3.8 × 10−04.
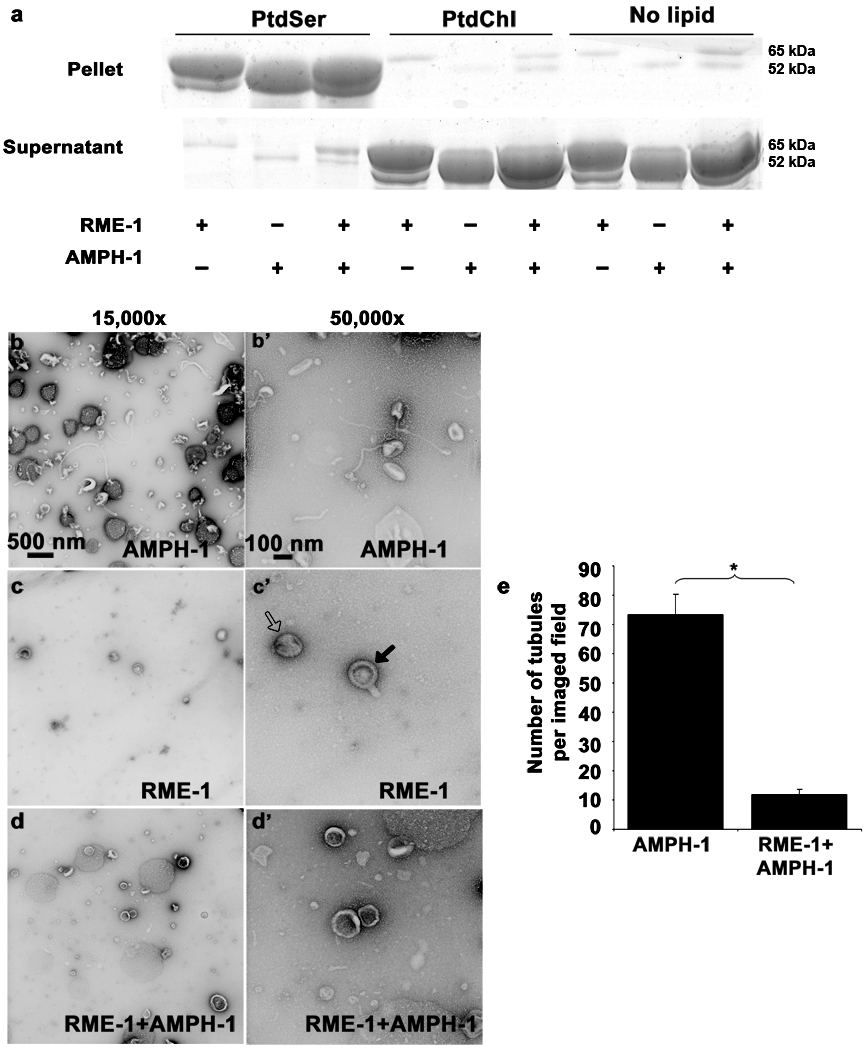
We observed equivalent binding of RME-1 to PS liposomes in the presence of ADP or non-hydrolysable ATP analog ATP-γ-S, indicating that under these binding conditions, RME-1 association with liposomes was independent of its nucleotide state (Fig. 6a, Fig. 8a).
Figure 8.
Nucleotide effects on RME-1 and AMPH-1 mediated liposome tubulation. (a) Coomassie stained gels of supernatant and pellet fractions from liposome co-sedimentation assays are shown. Binding reactions were performed in the absence or presence of 0.33 mg/ml, 0.4 µm (average diameter) 100% Phosphatidylserine (PtdSer), or 100% Phosphatidylcholine (PtdChl) liposomes. Liposomes were incubated with 1mM ADP and 1 µM of full length AMPH-1 or RME-1 proteins, or equimolar quantities of both proteins, as indicated. Note that RME-1 can bind to PtdSer liposomes in the presence of ADP. Tubulation experiments were performed with 2.5µM of each protein and 1mM ADP incubated with 0.05mg/ml 100% PtdSer liposomes. (b-b’) AMPH-1 incubated with PtdSer liposomes in the presence of ADP (compare with ATP-γ-S, Fig. 5 c-c’). (c-c’) RME-1 incubated with PtdSer liposomes in the presence of ADP (compare with ATP-γ-S, Fig. 5 d-d’). RME-1(ADP) lacks tubulation capacity and most liposomes remain spherical (open arrow) in the presence of RME-1(ADP). Striations are often visible and rare short protrusions (closed arrows) are present on occasional liposomes. (d-d’) RME-1 in its ADP bound state can affect the tubulation ability of AMPH-1 as observed in an experiment containing equimolar concentrations of both proteins in the presence of ADP. (e) Quantification reveals an approximately 7 fold decrease in number of tubules in conditions where RME-1 is present with AMPH-1, as compared to tubulation produced by AMPH-1 alone. n=10 fields for each experimental condition (imaged at 3,000× magnification), the mean value for number of observed tubules was plotted and error bars represent ± s.d. from the mean. The asterisk indicates a significant difference in the one-tailed Student’s T-test (p value=1.48×10−15).
We also examined the morphology of liposomes incubated with RME-1 and/or AMPH-1 by electron microscopy. In the presence of ATP-γ-S, RME-1 tubulates 400 nm average diameter PS liposomes into approximately 120 nm wide tubules with an average length of 1 µm (Fig. 6d-d’, 6f–g). The tubules were decorated with ring-like striations reminiscent of structures formed by mammalian Dynamin and EHD2 on liposomes. The rings likely represent highly ordered RME-1(ATP-γ-S) oligomers wrapping around the tubule, with regions of close ring packing and regions of looser packing. We found that in the presence of ATPγS, RME-1 tubulated PIP2/PS/PC containing liposomes into long but narrower tubules than those produced from PS-only liposomes (Fig 7c-c’, f–g). In profile, these tubules displayed an irregular spike like coated appearance. Frequently, we also observed structures resembling constrictions along the length of the tubule, imparting a beads-on-string appearance to the tubulating liposomes.
In the absence of RME-1, AMPH-1 tubulated 400 nm average diameter PS liposomes into 50 nm wide tubules with an average length of 1.1 µm (Fig. 6c-c’, 6f–g), much narrower than the tubules formed in the presence of RME-1 alone. Similar results were obtained for AMPH-1 incubated with PIP2/PS/PC liposomes (Fig. 7d-d’).
The physical and functional interaction of AMPH-1 and RME-1 suggests that the two proteins work together to promote recycling through endosomal tubules. We therefore investigated the biochemical interaction of AMPH-1 and RME-1 in membrane tubulation. Tubules generated from PS liposomes incubated with AMPH-1 and RME-1, in the presence of ATP-γ-S, were qualitatively different from tubules generated by the individual proteins (Fig. 6e-e’). The most striking difference is reduction in length of the tubules produced by AMPH-1 and RME-1(ATP-γ-S) together. Such tubules are about one-third the length of tubules produced by the individual proteins (Fig. 6f–g). In addition, in the presence of AMPH-1, the apparent RME-1(ATP-γ-S) spiral coat on the tubules is more regular and closely spaced than in RME-1(ATP-γ-S) tubules (Fig. 6h). The tubules produced by AMPH-1 and RME-1(ATP-γ-S) together are more rigid and the decoration is more pronounced in profile than that produced by the individual proteins (Fig. 6e’). Similar to our observation with 100% PS liposomes, upon incubation with 10% PIP2 liposomes we found that a combination of AMPH-1 and RME-1 generated tubules that were dramatically shorter than those produced by the individual proteins (Fig. 7e-e’, g).
The physical interaction of AMPH-1 and RME-1 through the AMPH-1 NPF motifs appears critical for the formation of hybrid tubules. AMPH-1(F309A, F363A) was unable to form short tubules from PS liposomes when co-incubated with RME-1(ATP-γ-S). Rather long thin tubules equivalent to AMPH-1(F309A, F363A) alone dominated the reaction (Supplemental information, Fig. S7). A few wide RME-1 type tubules were also observed but were relatively rare.
The ratio of AMPH-1 to RME-1 in the reaction mixture does not appear to control tubule length, since reducing AMPH-1 by 10-fold to 250nM, while keeping RME-1 levels constant (2.5µM), produced many short PS tubules that were similar in length to those produced by the proteins at 1:1 stoichiometry. This suggests that AMPH-1 and RME-1 tend to assemble into a coat of a particular length, rather than one of varying length dependent upon their relative available protein concentrations. (See Supplemental information Fig. S7f–g).
RME-1 nucleotide state is critical for tubulation
Finally we investigated effects of nucleotide state on the in vitro reaction. We found that RME-1(ADP) lacks most tubulation activity in this assay, even though it still binds to PS liposomes (Fig. 8). Furthermore, we found that RME-1(ADP) together with AMPH-1 added to PS liposome preparations led to an approximately 7-fold decrease in tubulation capacity, including loss of most of the characteristically thin tubules produced by AMPH-1 alone. This suggests that RME-1 in the inactive ADP-bound state interacts with AMPH-1 and prevents AMPH-1 mediated tubulation. Our RME-1 tubulation results are somewhat different from those reported for mammalian EHD2, the most diverged member of this protein family. EHD2 was found to be insensitive to nucleotide condition in tubulating PS liposomes9. Although under more stringent binding conditions, with liposomes of lower PS composition, EHD2 liposome tubulation was also dependent on nucleotide state 9.
DISCUSSION
The work of Daumke et.al.9, and the work presented here, suggest that RME-1/EHD family proteins could function in membrane tubulation and potentially membrane fission, using a self-assembly mechanism with overall similarity to Dynamin. AMPH-1, through it’s BAR domain, could function to initiate tubule formation from endosomes, recruiting and activating RME-1 ATPase activity to drive tubule fission. It is also quite interesting to note that AMPH-1 appears to limit the assembly of RME-1 rings on membrane tubules. Recent work on the mechanism of Dynamin-mediated membrane fission indicates that short Dynamin spirals are the active agents of fission40, 41. In those studies, long Dynamin assemblies were found to constrict tubules, but failed to promote fission upon GTP hydrolysis, while GTP hydrolysis and membrane release by short Dynamin assemblies frequently produced full fission. Thus by limiting RME-1 spiral assembly, AMPH-1 may help to generate productive RME-1 assemblies. The AMPH-1 SH3 domain might interact with other endocytic regulatory proteins and help create a hub for protein interactions relevant for the RME-1 recycling function.
Another similarity of RME-1/EHD family proteins and Dynamin is that they interact with Syndapin/Pacsin, a known activator of Wasp and Arp2/3 mediated actin polymerization, suggesting that RME-1 family proteins may promote actin dynamics on endosomes to promote membrane fission42. Membrane associated actin dynamics is thought to promote membrane fission by increasing membrane tension during Dynamin-mediated membrane constriction43. Further analysis of RME-1/EHD function is likely to provide important insights into membrane fission mechanisms that operate on endosomal membranes.
METHODS
Bioinformatic Screen
We searched the C. elegans proteome (WormBase release WS136) for all predicted ORFs containing the Asparagine-Proline-Phenylalanine(NPF) motif using the program myHits version 2.0 (myhits.isb-sib.ch). A total of 839 proteins were identified. These NPF containing sequences were further selected for those containing multiple NPF tripeptides (57 proteins) followed by either aspartic acid or glutamic acid residues within 5 amino acids. We found 17 C. elegans NPF containing proteins with varying degrees of D and/or E enrichment at these positions. C. elegans F58G6.1/Amphiphysin-like /Amphiphysin contained two NPF [D/E] sequences.
RNAi Screen
Strain RT33, expressing GFP-RME-1 under rme-1 promoter control in all somatic tissues, was examined by epifluorescence microscopy at 630X magnification for abnormal RME-1 subcellular localization or recycling endosome morphology after RNAi of each of the 74 multi-NPF and NPF(D/E) candidates. RNAi clones were obtained from the Ahringer library44 or were constructed by cloning cDNAs into RNAi vector L4440 and transforming HT115 cells. RNAi screening was performed by the feeding method as previously described45. Animals were scored as young adults in the P0 generation and after 4 more days of growth on fresh RNAi plates, animals of the next (F1) generation were also scored. All trials included a positive control (L4440-rab-10) and a negative control (L4440 empty vector). The screen was performed twice under identical experimental conditions.
Immunofluorescence
Immunofluorescence on dissected intestine preparations was performed as previously described1. Briefly, dissected tissues were fixed in 1.25% Paraformaldehyde (Electron Microscopy Sciences, Fort Washington, PA) for 10 minutes at room temperature. Rabbit anti-AMPH-1 antibody G2 was used at 1:100 dilution overnight at 4°C. Mouse monoclonal anti-RME-1 5G11 was used at 1:10 dilution. Secondary antibodies conjugated to Alexa-488 or Alexa-568 were purchased from Molecular Probes and were used at a dilution of 1:1000.
HeLa cells were grown on cover-glasses and fixed with 4% (v/v) paraformaldehyde in PBS. Fixed cells were incubated with primary antibodies prepared in staining solution [0.2% saponin (w/v) and 0.5% (w/v) BSA in PBS] for 1 hr at room temperature. After washes in PBS, the cells were incubated with the appropriate fluorochrome-conjugated secondary antibody mixture in staining solution for 30 min at room temperature. All images were acquired using a Zeiss LSM 5 Pascal confocal microscope (Carl Zeiss, Thornwood, NY) using a 63X objective with a numerical aperture of 1.4.
Transferrin (Tf) Recycling assay
Tf-568 was purchased from Molecular Probes (Invitrogen, Carlsbad, CA). HeLa cells on plates were mock treated or treated with Bin-1-RNAi oligonucleotides. After 72 h, cells starved for 30 minutes in DMEM lacking serum (but containing 0.5% BSA) were either pulsed with 16.7 µg/ml transferrin-Alexa Fluor 568 (Tf-568; Invitrogen) for 15 min and fixed immediately (t=0, uptake measurement), or then 'chased' in complete media for the indicated times prior to fixation. Images were acquired using a Zeiss LSM 5 Pascal confocal microscope (Carl Zeiss, Thornwood, NY) by using a 63× 1.4 numerical aperture objective with appropriate filters. All micrographs shown are representative images from experiments that have been repeated at least three times. Quantification of mean fluorescence was done using LSM Pascal software. Bars represent the mean ± standard error calculated from counting 70–80 cells each from three independent experiments. For Flow cytometry analysis, the remaining intracellular Tf-568 levels for 10,000 cells were determined at indicated time points after the start of chase. Error bars indicate standard error from three independent experiments.
Protein expression and purification of full length tagless proteins
C. elegans AMPH-1, AMPH-1(F309A, F363A) and RME-1 full-length proteins were expressed as N-terminal GST-fusions followed by a PreScission cleavage site in Escherichia coli Arctic Express cells (Stratagene, Agilent La Jolla, CA). Bacterial cultures in LB medium were induced at an OD600 of 0.5 with 1 mM IPTG, and grown overnight at 15°C. Bacteria were lysed in 50 mM HEPES (pH 7.5), 400 mM NaCl, 1 mM DTT, 1 mM Phenyl-methyl-sulfonyl fluoride (PMSF) (Sigma Aldrich, St. Louis, MO) using a microfluidizer M-110Y (Microfluidics, Newton, MA). The bacterial lysate was centrifuged at 10,000 × g for 30 minutes in a Sorvall SS-34 rotor (Sorvall, Guelph, ON, Canada). The soluble supernatant fraction was centrifuged for another 40 minutes at 4 °C at 100,000 × g in a Beckman Ti-70 ultracentrifuge rotor (Beckman, Fullerton, CA). The soluble extract was applied to a Glutathione sepharose 4B column (GE Amersham, Piscataway, NJ) equilibrated with lysis buffer. The column with bound protein was washed thoroughly with wash buffer (20 mM HEPES (pH 7.5), 300 mM NaCl, 1mM MgCl2, 1 mM DTT). To remove the GST tag, column-bound GST tagged protein was treated overnight at 4 °C with 225 µg PreScission protease (GE Amersham, Piscataway, NJ). The protein was eluted with several volumes of wash buffer. Eluted protein was re-applied to a Glutathione sepharose 4B column to bind any residual GST tag. The eluate containing purified tagless protein was analyzed by SDS-PAGE followed by Coomassie staining to check the purity of the proteins and Western blot with anti-RME-1 or anti-AMPH-1 antibodies. The proteins were extensively dialyzed against 20 mM HEPES (pH 7.5), 300 mM NaCl, 1mM MgCl2, 1 mM DTT and snap frozen in liquid Nitrogen and stored in aliquots at −80 °C. For use in all the biochemical experiments, the frozen aliquots were rapidly thawed, then spun at 20,000 g at 4°C to remove any aggregated protein. Protein concentration was determined under denaturing conditions by absorbance at 280 nm 46.
Liposome preparation
Lipids were obtained in chloroform solution from Avanti Polar Lipids Inc. (Alabaster, AL). For experiments utilizing PI, PI4P or PI4, 5 P2 lipids, the liposome composition comprised molar ratio of 80% Phosphatidylcholine(PC), 10% Phosphatidylserine(PS) and 10% of the respective phosphatidylinositol derivative. All glassware was thoroughly washed with chloroform (Extra dry Chloroform, Acros Organics, Geel, Belgium) and flushed with argon gas. Lipids were dried in a stream of argon gas. Residual chloroform was removed by vacuum over several hours or overnight. The dried lipid was resuspended at 1 mg/ml concentration in argon-purged liposome buffer (20 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM MgCl2). 0.4 µm average diameter liposomes were formed by using 0.4 µm polycarbonate track-etched membrane filters (Whatman Ltd, Kent, UK) with a Lipex Extruder, as per manufacturer’s instructions (Northern Lipids Inc. Burnaby, BC Canada).
Liposome binding
For liposome binding experiments, 1 µM final concentration of RME-1 or AMPH-1 proteins were incubated with 0.33 mg/ml final concentration of either 100% Phosphatidylserine (PtdSer), 100% Phosphatidylcholine (PtdChl) liposomes or 80% Phosphatidylcholine (PtdChl), 10% Phosphatidylserine (PtdSer) and 10% of either unphosphorylated phosphatidylinositol(PI) or phosphatidylinositol-[4]- phosphate (PI4P) or phosphatidylinositol-[4, 5]- bisphosphate (PI4,5P2) for 15 minutes at room temperature in the liposome buffer. The binding reaction was centrifuged at 90,000 × g in a TLA100.2 rotor (Beckman Coulter, Fullerton CA) at 25°C for 15 minutes using an Optima TLX100 centrifuge (Beckman Coulter). Supernatants were precipitated with trichloro acetic acid (TCA) and washed. Supernatant precipitates and pellets from the same experiment were suspended in equal volumes of 2X Laemmli sample loading buffer and equal volumes of pellet versus supernatant were analyzed by Coomassie gel staining.
Liposome tabulation
For liposomes tubulation experiments, 2.5 µM RME-1, AMPH-1 or AMPH-1(F309A, F363A) were incubated with 0.05 mg/ml final concentration of 100% Phosphatidylserine (PtdSer) liposomes in liposome buffer with 1 mM final concentration of either ATP-γ-S, or ADP (Sigma Aldrich, St. Lois, MO). For another experiment, 2.5 µM RME-1 and/or AMPH-1 were incubated with 0.05 mg/ml final concentration of 80% Phosphatidylcholine (PtdChl), 10% Phosphatidylserine (PtdSer) and 10% Phosphatidylinositol-[4, 5]-bisphosphate(PI4, 5P2) liposomes. The experiments were performed in the liposome buffer and in the presence of 1 mM final concentration of non-hydrolyzable ATP analog ATPγS. For experiments to determine stoichiometry of protein action, 2.5µM RME-1 andAMPH-1 or 2.5 µM RME-1 and 250 nM AMPH-1 were incubated with 0.05 mg/ml final concentration of 100% PS liposomes and 1mM ATP-γ-S. For experiments to determine the contribution of AMPH-1 NPF sequences to the tubulation reaction, experiments were performed as described in this section and using 2.5 µM RME-1 and/or AMPH-1(F309A, F363A) incubated with 0.05 mg/ml final concentration of 100%PS liposomes and 1mM ATP-γ-S. All samples were incubated on ice for 4 minutes and 12 minutes after the start of protein addition the samples were spotted on formvar carbon coated 300 mesh Copper grid. After negative staining with 1% uranyl acetate, electron microscopy was performed using a JEOL 1200 EX or JEOL 100 CX transmission electron microscope at 80 KV. Control experiments included no-liposome, protein incubations under identical experimental conditions, but with 2.5µM each of GST alone, GST with RME-1 or GST with AMPH-1. Images were obtained at 15,000× and 50,000× magnification as indicated. For quantification of AMPH-1 tubulation in the presence and absence of RME-1 in an ADP nucleotide condition, micrographs were imaged at 3,000×, for large field in which tubules could be clearly distinguished. The electron micrographs were scanned using a CanoScan scanner. For the selected images included in Figure 6–Figure 8 and Figures S7, the Adobe Photoshop CS Levels function was used to alter contrast to best represent the observed features. Quantification of liposome tubule length and diameter were performed using Adobe Photoshop CS Ruler function. Individual tubule lengths, diameters or inter striation distances were measured and converted to nm measures utilizing the measured length of the scale bar at that EM magnification. Mean values were calculated and plotted as graphs. Standard deviations were used for Y-error bars on graphs. Statistical analysis was performed utilizing a Student’s paired T-test. Significance was determined at p-values less than 0.01 and is noted by asterisks on figures and in figure legends.
Statistical analysis
For all data sets the standard deviation value from data sets was utilized as Y-error bars on bar graphs plotted for mean value of the data. Data sets were subjected to Student’s one-tailed paired T-test analysis. Data were considered significantly different if p-values were lower than 0.01. p-values for various experiments are included in the figure legends. In all experiments presented, p-values less than 0.05 were considered statistically significant and are marked by asterisks in the Figures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank P. McPherson, Z. Zhou, B. Kay, G. Prandergast and S. Mitani for important reagents. The monoclonal against RME-1 was made at Washington University School of Medicine and funded by Grant R24RR22234 (PI Michael L. Nonet). We also thank C. Martin, P. Yurchenco, D. Winkelman, F. Matsumura, S. Yamashiro and S.H. deGregori for their generous sharing of instruments, reagents and protocols. We thank A. Shi for the construction and generous gift of the GFP-SDPN-1 strain. We thank R. Patel and V. Starovoytov for expert assistance with EM protocols and Z. Pan for usage of the confocal microscopy facility. We thank O. Daumke and T. Pucadyil for generous discussions and advice on liposome tubulation experiments. We thank P. Schweinsberg for technical assistance. This work was supported by NIH Grants GM67237 to B.D.G and GM074876 to S.C., NJCSCR Graduate Fellowship 06-2915-SCR-E-0, Anne B. and James B. Leathem Fellowship, McCallum Fund Fellowship Grant to S.P., an American Heart Association student fellowship to M.S. and Rutgers Summer Undergraduate Research Fellowship (SURF) to K.P.
Footnotes
AUTHOR CONTRIBUTIONS.
S.P. participated in the experimental design, performed the C. elegans related experiments, biochemical and liposome experiments (Figure 1–Figure 4, Figure 6–Figure 8 and Supplemental figures 1–5 and 7) and wrote the paper. M.S. performed the mammalian cell experiments(Figure 5 and Supplemental figure 6) and contributed to writing about those experiments. K.P. contributed to developing some strains used in the study. S.C. designed the mammalian cell experiments. C.M.C. designed the experiments related to liposome biochemistry and trained S.P. for the same. B.D.G. designed the experiments, trained S.P. for all C. elegans experiments and wrote the paper.
REFERENCES
- 1.Grant B, et al. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nature cell biology. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- 2.Lin SX, Grant B, Hirsh D, Maxfield FR. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nature cell biology. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- 3.Caplan S, et al. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. The EMBO journal. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee DW, et al. ATP binding regulates oligomerization and endosome association of RME-1 family proteins. The Journal of biological chemistry. 2005;280:17213–17220. doi: 10.1074/jbc.M412751200. [DOI] [PubMed] [Google Scholar]
- 5.Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Molecular biology of the cell. 2006;17:163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma M, Naslavsky N, Caplan S. A role for EHD4 in the regulation of early endosomal transport. Traffic (Copenhagen, Denmark) 2008;9:995–1018. doi: 10.1111/j.1600-0854.2008.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao Y, et al. Pincher, a pinocytic chaperone for nerve growth factor/TrkA signaling endosomes. The Journal of cell biology. 2002;157:679–691. doi: 10.1083/jcb.200201063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilherme A, et al. EHD2 and the novel EH domain binding protein EHBP1 couple endocytosis to the actin cytoskeleton. The Journal of biological chemistry. 2004;279:10593–10605. doi: 10.1074/jbc.M307702200. [DOI] [PubMed] [Google Scholar]
- 9.Daumke O, et al. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- 10.Sever S. Dynamin and endocytosis. Current opinion in cell biology. 2002;14:463–467. doi: 10.1016/s0955-0674(02)00347-2. [DOI] [PubMed] [Google Scholar]
- 11.Roux A, Antonny B. The long and short of membrane fission. Cell. 2008;135:1163–1165. doi: 10.1016/j.cell.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Salcini AE, et al. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes & development. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant BD, Caplan S. Mechanisms of EHD/RME-1 Protein Function in Endocytic Transport. Traffic (Copenhagen, Denmark) 2008 doi: 10.1111/j.1600-0854.2008.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieken F, Jovic M, Naslavsky N, Caplan S, Sorgen PL. EH domain of EHD1. Journal of biomolecular NMR. 2007;39:323–329. doi: 10.1007/s10858-007-9196-0. [DOI] [PubMed] [Google Scholar]
- 15.Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nature cell biology. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 16.David C, McPherson PS, Mundigl O, de Camilli P. A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:331–335. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi A, et al. A novel requirement for C. elegans Alix/ALX-1 in RME-1-mediated membrane transport. Curr Biol. 2007;17:1913–1924. doi: 10.1016/j.cub.2007.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato K, et al. Differential requirements for clathrin in receptor-mediated endocytosis and maintenance of synaptic vesicle pools. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1139–1144. doi: 10.1073/pnas.0809541106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CC, et al. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Molecular biology of the cell. 2006;17:1286–1297. doi: 10.1091/mbc.E05-08-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Zelhof AC. Amphiphysins: raising the BAR for synaptic vesicle recycling and membrane dynamics. Bin-Amphiphysin-Rvsp. Traffic (Copenhagen, Denmark) 2002;3:452–460. doi: 10.1034/j.1600-0854.2002.30702.x. [DOI] [PubMed] [Google Scholar]
- 21.Razzaq A, et al. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes & development. 2001;15:2967–2979. doi: 10.1101/gad.207801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zelhof AC, et al. Drosophila Amphiphysin is implicated in protein localization and membrane morphogenesis but not in synaptic vesicle endocytosis. Development (Cambridge, England) 2001;128:5005–5015. doi: 10.1242/dev.128.24.5005. [DOI] [PubMed] [Google Scholar]
- 23.Leventis PA, et al. Drosophila Amphiphysin is a post-synaptic protein required for normal locomotion but not endocytosis. Traffic (Copenhagen, Denmark) 2001;2:839–850. doi: 10.1034/j.1600-0854.2001.21113.x. [DOI] [PubMed] [Google Scholar]
- 24.Grabs D, et al. The SH3 domain of amphiphysin binds the proline-rich domain of dynamin at a single site that defines a new SH3 binding consensus sequence. The Journal of biological chemistry. 1997;272:13419–13425. doi: 10.1074/jbc.272.20.13419. [DOI] [PubMed] [Google Scholar]
- 25.Burack MA, Silverman MA, Banker G. The role of selective transport in neuronal protein sorting. Neuron. 2000;26:465–472. doi: 10.1016/s0896-6273(00)81178-2. [DOI] [PubMed] [Google Scholar]
- 26.Naslavsky N, Weigert R, Donaldson JG. Characterization of a nonclathrin endocytic pathway: membrane cargo and lipid requirements. Molecular biology of the cell. 2004;15:3542–3552. doi: 10.1091/mbc.E04-02-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naslavsky N, Weigert R, Donaldson JG. Convergence of non-clathrin-and clathrin-derived endosomes involves Arf6 inactivation and changes in phosphoinositides. Molecular biology of the cell. 2003;14:417–431. doi: 10.1091/mbc.02-04-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farsad K, et al. A putative role for intramolecular regulatory mechanisms in the adaptor function of amphiphysin in endocytosis. Neuropharmacology. 2003;45:787–796. doi: 10.1016/s0028-3908(03)00306-x. [DOI] [PubMed] [Google Scholar]
- 29.McMahon HT, Wigge P, Smith C. Clathrin interacts specifically with amphiphysin and is displaced by dynamin. FEBS letters. 1997;413:319–322. doi: 10.1016/s0014-5793(97)00928-9. [DOI] [PubMed] [Google Scholar]
- 30.Olesen LE, et al. Solitary and repetitive binding motifs for the AP2 complex alpha-appendage in amphiphysin and other accessory proteins. The Journal of biological chemistry. 2008;283:5099–5109. doi: 10.1074/jbc.M708621200. [DOI] [PubMed] [Google Scholar]
- 31.Owen DJ, et al. Crystal structure of the amphiphysin-2 SH3 domain and its role in the prevention of dynamin ring formation. The EMBO journal. 1998;17:5273–5285. doi: 10.1093/emboj/17.18.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slepnev VI, De Camilli P. Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nature reviews. 2000;1:161–172. doi: 10.1038/35044540. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida Y, et al. The stimulatory action of amphiphysin on dynamin function is dependent on lipid bilayer curvature. The EMBO journal. 2004;23:3483–3491. doi: 10.1038/sj.emboj.7600355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee E, et al. Amphiphysin 2 (Bin1) and T-tubule biogenesis in muscle. Science (New York, N.Y. 2002;297:1193–1196. doi: 10.1126/science.1071362. [DOI] [PubMed] [Google Scholar]
- 35.Muller AJ, et al. Targeted disruption of the murine Bin1/Amphiphysin II gene does not disable endocytosis but results in embryonic cardiomyopathy with aberrant myofibril formation. Molecular and cellular biology. 2003;23:4295–4306. doi: 10.1128/MCB.23.12.4295-4306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jovic M, Kieken F, Naslavsky N, Sorgen PL, Caplan S. Eps15 homology domain 1-associated tubules contain phosphatidylinositol-4-phosphate and phosphatidylinositol-(4,5)-bisphosphate and are required for efficient recycling. Molecular biology of the cell. 2009;20:2731–2743. doi: 10.1091/mbc.E08-11-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashiro DJ, Tycko B, Fluss SR, Maxfield FR. Segregation of transferrin to a mildly acidic (pH 6.5) para-Golgi compartment in the recycling pathway. Cell. 1984;37:789–800. doi: 10.1016/0092-8674(84)90414-8. [DOI] [PubMed] [Google Scholar]
- 38.Takei K, Slepnev VI, De Camilli P. Interactions of dynamin and amphiphysin with liposomes. Methods in enzymology. 2001;329:478–486. doi: 10.1016/s0076-6879(01)29109-5. [DOI] [PubMed] [Google Scholar]
- 39.Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. The Journal of cell biology. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bashkirov PV, et al. GTPase Cycle of Dynamin Is Coupled to Membrane Squeeze and Release, Leading to Spontaneous Fission. Cell. 2008 doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braun A, et al. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Molecular biology of the cell. 2005;16:3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- 44.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods (San Diego, Calif. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 45.Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nature cell biology. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 46.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.