Summary
Autophagy is a physiologically and immunologically controlled intracellular homeostatic pathway that sequesters and degrades cytoplasmic targets including macromolecular aggregates, cellular organelles such as mitochondria, and whole microbes or their products. Recent advances show that autophagy plays a role in innate immunity in several ways: (i) direct elimination of intracellular microbes by digestion in autolysosomes, (ii) delivery of cytosolic microbial products to pattern recognition receptors (PRRs) in a process referred to as topological inversion, and (iii) as an antimicrobial effector of Toll-like receptors and other PRR signaling. Autophagy eliminates pathogens in vitro and in vivo but, when aberrant due to mutations, contributes to human inflammatory disorders such as Crohn's disease. In this review, we examine these relationships and propose that autophagy is one of the most ancient innate immune defenses that has possibly evolved at the time of α-protobacteria-pre-eukaryote relationships, leading up to modern eukaryotic cell-mitochondrial symbiosis, and that during the metazoan evolution, additional layers of immunological regulation have been superimposed and integrated with this primordial innate immunity mechanism.
Keywords: autophagy, inflammatory bowel disease, Toll-like receptors/pattern recognition receptors, viral, bacterial, parasitic-protozoan infectious disease
Introduction
Autophagy is an evolutionarily ancient cytoplasmic homeostasis process conserved in all eukaryotes (1, 2). Autophagy is a collection of biomass quantity and quality control systems targeting a range of cytoplasmic components. Autophagic targets range from individual macromolecules, processed by chaperone-mediated autophagy, to large often toxic protein aggregates, sizeable portions of the cytosol, or even whole organelles, all subject to macroautophagy (sensu stricto autophagy). A distinguishing morphological signature of autophagy is the formation within the cytosol of crescent-shaped slivers of membrane that wrap around cytoplasmic targets to form the double membrane autophagosome, emblematic of autophagy (Fig. 1). The portions of the cytoplasm sequestered into autophagosomes are degraded in autolysosomes, which are the product of autophagic maturation or flux, resulting from fusion between autophagosomes and late endosomal/lysosomal organelles (1, 2).
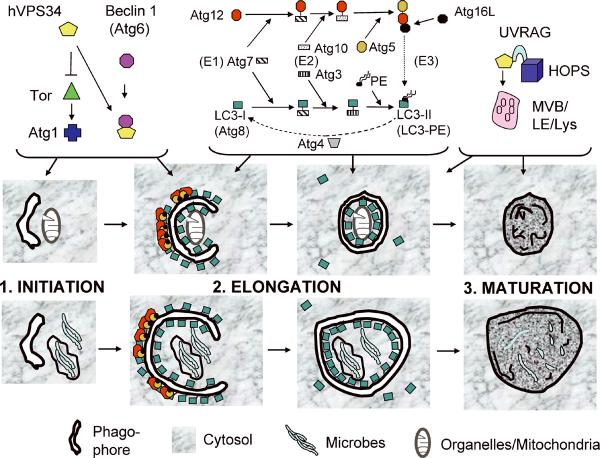
Fig. 1. Autophagy pathway: signaling and execution stages of autophagy.
The Tor signaling cascade controls autophagy execution. During starvation or other physiological conditions, Tor is inhibited. This is possibly dependent on the type III PI3K. When Tor is inhibited, Atg1 and hVPS34 complexed with Beclin 1 (Atg6) lead to activation of downstream Atg factors (Atg1) and set in motion the autophagy execution phases: 1. Initiation, formation of phagophore (isolation membrane). 2. Elongation, growth of the phagophore and its closure to generate a double membrane autophagosome. 3. Maturation of the double membrane autophagosomes into autolysosomes. The membrane elongation and shape are controlled by two protein (and lipid) conjugation systems, akin in activation and conjugation cascade principle to the ubiquitination system but distinct from it. Atg12 is initially conjugated to Atg7, which acts as an E1-activating enzyme, and then gets transferred to the E2-like conjugating enzyme Atg10. This intermediate presents Atg12 to be conjugated to a Lys residue in Atg5. Atg5 is essential for autophagy in the mouse. The Atg5–12 conjugate, stabilized in a noncovalent complex with Atg16, acts as an E3 enzyme for the second conjugation system, and triggers oligomerization on the outside membrane of the growing phagophore, and also enhances conversion of LC3-I (Atg8) into its C-terminally lipidated (with PE) form LC3-II. Upon autophagosome closure, sealing the typical double membrane organelle, Atg5–12/16 and LC3 (delipidated by Atg4) dissociate from the outer autophagosomal membrane and get recycled. The LC3 associated with the lumenal membrane remains trapped in the autophagosome and is degraded during maturation into the autolysosome, which involves fusion of autophagosomes with late endosomal (LE), multivesicular bodies endosomes (MVB) and lysosomal organelles (Lys), dissolution of internal membrane, and conversion of the maturing autophagic organelles into autolysosomes. hVPS34, interacting with UVRAG and HOPS, plays a role in maturation stages of autophagy. Depicted are autophagosomes capturing an intracellular organelle (mitochondria) or microbes such as intraphagosomal bacteria or bacteria released into the cytosol.
Autophagy plays a multitude of physiological roles. A quintessential survival function of autophagy is to reuse cell's own cytoplasm by autodigestion of cytosol, thus maintaining essential anabolic functions during times of nutrient starvation or growth factor deprivation (2, 3). In contrast to its pro-life function, autophagy can, via its interactions with other types of cell death (apoptosis and necrosis) and possibly by itself when excessive, lead to cell death (4), referred to as programmed cell death type II. Since autophagy impacts all living cells, it manifests itself in a broad range of health and disease states, including aging, development, neurodegenerative diseases such as Huntingon's, Alzheimer's, and Parkinson's, myodegeneration, and cancer (1, 2). In the context of immunology (5), autophagy eliminates intracellular microbes (6–9), contributes to major histocompatibility complex class II (MHC II)-restricted endogenous antigen presentation (10–13), is an effector of T-helper 1 (Th1)/Th2 cell polarization (14), affects B and T-cell homeostasis and repertoire selection (15–19), and assists pattern recognition receptors (PRRs) by delivering cytosolic pathogen-associated molecular patterns (PAMP) to endosomal Toll-like receptors (TLRs) (20, 21). It also acts as an effector of TLR sand other PRRs (22–25).
Autophagy in mammalian cells
Autophagy activation and execution can be divided in two phases: (i) signaling, with molecular switches that induce or shut down autophagy, and (ii) the morphologically detectable execution stages subdivided into initiation, elongation, and maturation (Fig. 1). The signaling systems controlling autophagy overlap with the well known regulatory cascade governing cellular growth and biomass centered on Tor (target of rapamycin) (26). In contrast, the execution stages of autophagy depend on a number of unique, specialized autophagy factors (2, 27, 28). Following induction by physiological signals (e.g. starvation) (2), pharmacological agonists (e.g. rapamycin, an inhibitor of Tor) (2), or immunological stimuli [interferon-γ (IFN-γ), tumor necrosis factor-α (TNF-α)] (7, 14, 29–31), autophagy is set in motion. First, it undergoes initiation, whereby nascent autophagosomal structures (termed phagophores) are formed in the cytoplasm from membranes of still undefined origin (Fig. 1). During the elongation stage, the phagophore is enlarged, envelops the target, and finally seals to form an autophagosome. During the final stage (maturation), autophagosomes fuse with lysosomes to form autolysosomes, degradative compartments that loose the inner of the two membranes and expose the captured cargo to the hydrolytic enzymes.
Autophagy is regulated by a core signaling pathway (Fig. 1), with two marquee players, the Ser/Thr kinase Tor and the phosphatidylinositol 3-kinase (PI3K) hVPS34 along with its interacting partners: (i) Tor is a cellular biomass sentinel governing whether a cell undergoes biomass increase or decrease. Tor integrates growth factor signaling (26), energy status of the cell (32), several types of stressors including genotoxicity (33) and hypoxia (34). (ii) The type III PI3K hVPS34 is an ancient PI3K generating phosphatidylinositol 3-phosphate on endomembranes, evolutionarily predating the familiar type I PI3K acting proximally to growth factor receptors and generating phosphatidylinositol (3,4,5) triphosphate on plasma membrane (35). The hVPS34 lipid kinase is the target for the well-known pharmacological inhibitor of autophagy, 3-methyl adenine (3MA). During the execution phase, hVPS34 plays a positive role when in a complex with Beclin 1 (Atg6), an autophagy-inducing factor (36). Recent studies have identified Bcl-2 (36), controlled by the mitogen-activated protein kinase (MAPK) c-Jun N-terminal kinase (JNK) (37) and UVRAG (38), as two critical Beclin 1-interacting partners, acting as an inhibitor and an activator of autophagy, respectively. During the maturation stages of autophagy, UVRAG is important for activation of the protein complex termed HOPS, specifically its subunit Vps39 that acts as a Rab7 guanine-nucleotide exchange factor, activating this guanosine triphosphatase (GTPase) and allowing recruitment of lysosomal components (39).
The most detailed picture regarding the autophagy-specific proteins (Atgs) is available in the yeast system (27, 28). About a dozen of yeast Atg factors have their orthologs in mammals, emphasizing both the conservation of the basic apparatus and a divergence in its use by metazoans (Fig. 2). Inhibition of Tor activity leads to the initiation of autophagy via Atg1 in yeast, a role likely played by one or both of mammalian homologs, Ulk1 and Ulk2 (40). Autophagosome formation and wrapping around its target during initiation and elongation stages are facilitated by the two highly conserved specialized protein conjugation systems (Fig. 2) The Atg5–12/16 complex acts a an E3 enzyme (borrowing designations from the ubiquitin system) (41) and stimulates a second conjugation system, whereby Atg8 undergoes conversion from its free C-terminus state to its C-terminally lipidated form covalently modified by phosphatidylethanolamine (PE) (Fig. 1). The yeast Atg8 has multiple paralogs in mammals termed LC3A (with two splice isoforms, a and b in humans), LC3B, GABARAP, GABARAPL1, and GABARAPL2 (GATE-16), encoded on different chromosomes, with the exception of LC3B and GABARAPL2, which are linked on chromosome 16 (humans) or chromosome 8 (mouse). The LC3 routinely referred in publications is LC3B (Fig. 1).
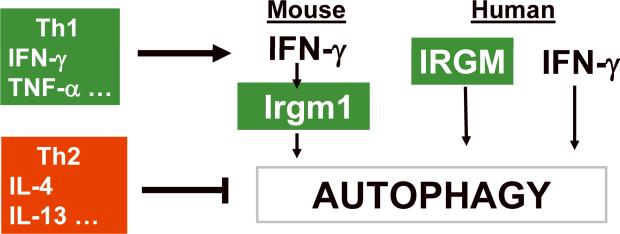
Fig. 2. Th1 and Th2 cytokines activate and inhibit autophagy, respectively.
See text for description. Note that in the mouse, IFN-γ activates expression of immunity related GTPase Irgm1 (LRG-47) to induce autophagy, while in human cells, the IRG factor IRGM is expressed independent of IFN-γ but is required for IFN-γ-induced autophagy.
During its final maturation stage, the double membrane autophagosome fuses with late endosomal/lysosomal organelles. Sometimes an intermediate step is discerned, whereby late endosomes referred to as multivesicular body endosomes fuse with autophagosomes to form an intermediate termed amphisome (42). Eventually, all maturing autophagosomes will lose the inner of the two bilayers and convert into degradative autolysosomal organelles where the digestion of the sequestered cytoplasmic material takes place. The type III PI3K VPS34 plays a role in the formation of late endosomal multivesicular bodies and lysosomal organelles, thus contributing indirectly to the terminal maturation stages of autophagy. A recent study showed that the Beclin 1-interacting partner UVRAG also acts as a factor spurring the late endosomal pathway (39). This occurs via interactions with HOPS containing the Rab7 guanine nucleotide exchange factor Vps39 (39), independently of Beclin 1 and the Beclin 1-UVRAG regulation of autophagic initiation.
Autophagy is a cell autonomous defense mechanism against bacteria, parasites, and viruses
The observations of autophagy in cells containing microbial pathogens hark back to a study in 1984 with rickettsia-laden neutrophils (43). The relationship between autophagy and microbes has remained ill-defined until a recent convergence of studies showing that autophagy is an innate immune defense against bacteria, protozoa, and viral pathogens (5, 9, 12). A group of papers has demonstrated a role of autophagy in elimination of microorganisms such as Mycobacterium tuberculosis residing within phagosomes (7), intracellular pathogens escaping into the cytosol such as Shigella (8), or extracellular pathogens, if and when they manage to invade the interior of host cells, as shown with group A streptococci (6). A stream of these and additional publications has established a role for autophagy in innate immunity against a variety of microbes (6–8, 25, 31, 44–51). Autophagy eliminates intracellular pathogens in a process similar to the capture and digestion of unwanted or damaged intracellular organelles (Fig. 1). Thus, autophagy serves as a mechanism for the removal of intracellular bacteria and protozoans, in keeping with its primary function as a cytoplasmic clean-up process. It may be necessary to underscore that until the recent convergence of studies described above, autophagy was considered to be an epiphenomenon or a process beneficial for intracellular pathogens (52). This can now be explained in the context of evolutionary adaptations of pathogens that have developed protective mechanisms against autophagy (53–55): (i) Levine and colleagues (54) have shown that a virus (HSV-1) can interfere with autophagy using a specific gene product ICP34.5; (ii) Sasakawa and colleagues (8) found that Shigella normally evades autophagy but becomes a substrate for autophagic elimination upon loss of one of its intracellular motility-associated proteins, IscB; (iii) Brumell and colleagues (55) have reported that Listeria can block completion of autophagic maturation (a stage of the autophagy pathway dependent on establishing a low lumenal pH in autophagosomes) using its pore-forming toxins to dissipate protons. This appears to be related to establishment of persistent infection in liver granulomas.
Autophagy is an effector of Th1/Th2 polarization
Autophagy is regulated by immunologically relevant cytokines and ligands (7, 10, 14, 15, 29–31, 56–59). TNF-α activates autophagy (associated with increased Beclin 1 levels) but only under conditions when NF-κB is blocked, as NF-κB activates Tor and counteracts autophagy induction (30). Cell-mediated immunity regulatory systems can induce autophagy, e.g. CD40L-CD40 stimulation, in the context of protection against the parasite Toxoplasma gondii (31) in conjunction with TNFα secreted downstream of the CD40-TNF receptor-associated factor 6 (TRAF6) stimulation (60). The prototypical key Th1 cytokine IFN-γ can stimulate autophagy (7, 29, 56) (Fig. 2). In one report (13), the authors could not observe an increase over the basal levels of autophagy upon IFN-γ addition. Autophagy induction may depend on IFN-γ responsiveness of a given cell type, receptor expression status, in vitro conditions, and pre-existing background levels of autophagy that depend on cell type and vary considerably; for example, some phagocytic cells have high basal levels of autophagy, as noted by Schmidt et al. (13). Furthermore, induction of autophagy varies considerably from cell to cell in asynchronous cultures (61), so some caution might be needed in interpreting negative results.
In contrast to Th1 cytokines, which induce autophagy, the Th2 cytokines interleukin-4 (IL-4) and IL-13 are antagonists of autophagy (14) (Fig. 2). This is in part based on the activation of the Akt-Tor cascade by IL-4 and IL-13 (62). In early biochemical studies and out of any immunological context, IL-13 has been used as an antagonist of autophagy (57, 58) and, along with IL-4, has been shown to inhibit autophagy in immunologically relevant cells such as macrophages (14). The IL-4 and IL-13 treatment of macrophages inhibits starvation- or IFN-γ-induced autophagic delivery of mycobacteria into degradative compartments and enhances mycobacterial survival in infected macrophages stimulated for autophagy (14). IL-4 and IL-13 signal through the shared IL-4Rα receptor, which complexes with the γ-common chain (for IL-4) or with IL-13Rα1 (for IL-13) (63). IL-13 can also signal through the high affinity IL-13Rα2. Once IL-4 and IL-13 receptors are engaged, this brings about activation of not only the signal transducer and activator of transcription 6 (STAT-6) pathway but also signaling via the insulin receptor substrate-1 (IRS-1) and IRS-2 (63). The signaling via IRS stimulates the Akt pathway, which provides the basis for IL-4 and IL-13 inhibition of autophagy induced by starvation (14). However, a different signaling pathway, independent of Akt and dependent on STAT-6, is required to suppress IFN-γ-induced autophagy. The inhibitory action of IL-4 and IL-13 translated into the inhibition of autophagic control of intracellular M. tuberculosis (14). The induction of autophagy by IFN-γ and autophagic control of M. tuberculosis upon stimulation with Th1 cytokines versus inhibition by IL-4 and IL-13 of autophagy and suppression of autophagic killing of M. tuberculosis by Th2 cytokines demonstrate that autophagy is an effector of Th1/Th2 polarization, explaining why Th1 cytokines are protective and Th2 cytokines permissive when it comes to controlling intracellular pathogens. Moreover, studies by Harris et al. showed that even in the presence of Th1 cytokine (IFN-γ), Th2 cytokines (IL-4, IL-13) block the protective role of autophagy. This finding indicates that Th1–Th2 polarization does not necessarily need to be sharply defined, as it rarely happens in infection sites, and that the presence of Th2 cytokines may override IFN-γ when mixed cytokine responses are dominant.
Autophagy is regulated by immunity-related GTPases
How does IFN-γ induce autophagy? At least one pathway connecting IFN-γ and autophagy has been described (7, 49): IFN-γ-sponsored autophagy activation depends on the members (Irgm1/LRG47 in the mouse and IRGM in humans) of the family of p47 immunity-related GTPases (IRGs) (Fig. 2). In the mouse, induction of IRG expression is controlled by IFNs [type I IFN through IFN-sensitive response elements (ISRE) or IFN-γ via γ-IFN activation site (GAS) elements] (64). The mechanisms by which IRG factors control and eliminate intracellular pathogens have remained a mystery but now are known to include autophagy (48, 49). The work on M. tuberculosis has led to an initial connection between IRG and autophagy in murine cells (7) and has been recently expanded to the control of mycobacteria in human cells (49) and to elimination of intracellular T. gondii (48). IFN-γ is a major correlate of immunity against tuberculosis, but the exact nature of IFN-γ antimycobacterial action remained an unsolved issue, as neither reactive oxygen nor reactive nitrogen intermediates could explain its potent antimycobacterial action (65). It has been demonstrated that IFN-γ acts through an IRG family member, Irgm1 (LRG-47), to control M. tuberculosis in murine macrophages and in the mouse model of tuberculosis (65). How Irgm1 (and potentially other IRG proteins) in turn act to restrict intracellular pathogens has remained obscure until a recent connection to autophagy (48, 49). Additional roles have been proposed for IRG (66), such as proper phagosome acidification and maturation (65), direct destruction of the parasitophorous vacuole membrane (67), and aberrant function of hemopoietic stem cells during infection (68). It is worth noting that the majority of the above mechanisms are compatible with autophagy or can even be explained by autophagy.
The IRG family (69) in the mouse is represented by a total of 23 Irg genes: (i) 19 IFN-controlled complete Irg genes, Irgm1–Irgm3, Irgb1–Irg6, Irgb8–Irgb10 on the mouse chromosome 11, and Irga1–Irga4, Irg6–Irga8 on the mouse chromosome 18. They are driven by a combination of IRES and GAS elements, with the exception of Irgb5 and Irgb9, where only IRES sites have been identified in the putative promoter regions. (ii) Two Irg pseudogenes (Irga5ψ and Irgb7ψ) intercalated with the rest of the Irga and Irgb loci. (iii) Two IFN-independent (Irgc) or quasi-GTPase (Irgq) genes on the mouse chromosome 7, that have not been implicated or considered in immune functions (69). The murine IRG, Irgm1, Irgb6, Irgd, Irgm3, Irgm2, and Irga6, are also known by their old names (given when they were sporadically identified and studied): LRG-47, TGTP, IRG-47, IGTP, GTPI, and IIGP1, respectively. These, and more recently additional murine IRG, e.g. Irgb10 (70, 71), have been implicated in resistance to specific intracellular pathogens (64, 66).
The extraordinary complexity of the murine irg complement of genes contrasts sharply with the paucity of IRG in the human genome. with only three IRG candidate genes: (i) IRGC and IRGQ on human chromosome 19, syntenic with the mouse Irgc and Irgq not implicated in immunity, and (ii) the sole IRGM gene on human chromosome 5, surrounded by chromosomal segments syntenic with murine Irgm and Irga chromosomal loci attesting to its bona fide genomic relationship to the immunologically defined murine IRG. Moreover, human IRGM has been characterized functionally as playing a role in immune processes (21, 49), even though it is a shortened (N-terminally and C-terminally truncated) version of the murine Irgm proteins.
Human IRGM is required for elimination of M. tuberculosis by autophagy in human macrophages (49). Moreover, IRGM has been identified (72–74), along with another autophagy factor Atg16L1(74–76), in genome-wide association studies as a genetic risk locus for Crohn's disease, a common form of inflammatory bowel disease. Curiously, human IRGM is not regulated by IFN-γ and is instead constitutively expressed from the long terminal repeat (LTR) of a human endogenous retrovirus repetitive element, ERV9. However, IRGM is still required for full autophagy activation in cells stimulated with IFN-γ, starvation, or rapamycin (Fig. 2). The exact mechanisms by which IRGM in human cells and Irgm1 (LRG-47) in murine cells promote autophagy are not known and are presently being investigated.
Autophagy is a topological inversion device feeding immune processes
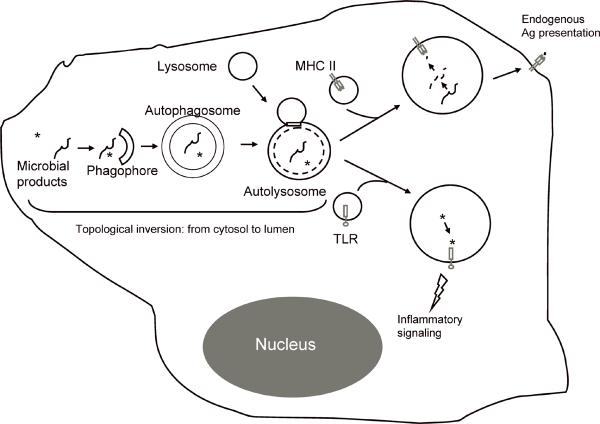
The MHC II-driven T-cell selection for self-tolerant repertoire in the thymus has been recently linked to autophagy in thymic epithelial cells (19). Interference with autophagy in thymic epithelial cells results in multi-organ inflammation in mice (19). These findings link autophagy to thymic selection and place it left, right, front, and center as a safeguard against autoimmunity. For the elimination of self-reactive T cells, MHC II molecules of thymic epithelial cells need to be loaded with cytoplasmic antigens. How do MHC II molecules, facing the lumen of the antigen-processing and loading organelles, become loaded with cytosolic antigens to carry out this and perhaps other processes such as endogenous antigen presentation in general? A favorite view until now has been that bits and pieces of dead cells or otherwise released antigens are endocytosed by antigen-presenting cells and delivered to MHC II. However, it turns out that autophagy can accomplish this in a far more elegant way without any cells having to die in the process. Autophagy works as a topological inversion device (77). It has been documented that close to 50% of autophagosomes merge with MHC II antigen-loading compartments (13). Bulk cytosolic proteins scooped by autophagosomes are delivered to antigen-processing and MHC II-loading compartments following fusion between autophagosomes and lysosomes (78–81). Individual proteins can also be imported one by one directly into the lysosomes by chaperone-mediated autophagy. Hence, autophagy is utilized by immune cells as a topological inversion device, bringing cytosolic antigens into the lumen of MHC II compartments (Fig. 3).
Fig. 3. Autophagy as a topological inversion device feeding immune processes.
For MHC class II-restricted endogenous antigen presentation, cytosolic proteins are captured by autophagosomes and are delivered to intracellular antigen-processing and loading compartments to meet the lumenally-oriented MHC II molecules. Note that the topological inversion occurs by sequestration of cytosolic proteins into the autophagosome, where they now are facing the lumen of endomembranous compartments, which puts them on the same side of the membrane as MHC II groove. For delivery of microbial products, e.g. PAMPs, a similar process sequesters them into autophagosomal lumen and follows the steps as above to deliver PAMP to lumenally oriented TLRs.
In addition to thymic selection and endogenous antigen presentation, autophagy plays a role in adaptive immunological responses against microbial (especially viral) antigens, as in the case of a CD4+ T-cell response to autophagically processed EBNA1 protein of Epstein-Barr virus (10). Of significance is to note the following: (i) constitutive macroautophagy is high in dendritic cells (DCs) and other antigen=presenting cells such as epithelial cells, B cells, and primary monocyte-derived cells (13); (ii) the process shows selectivity and some antigens are favored over others (e.g. different EBNA proteins show differential dependence on autophagy for MHC II presentation) (82); (iii) by coupling antigens to LC3, one can augment MHC II presentation of a desired antigen (13) of potential use in vaccine development.
Other immunity–relevant processes depend on autophagy as a topological inverter device. Delivery of cytosolic PAMPs to endosomally located and lumenally oriented PRRs can be accomplished via autophagy, in a process akin to delivering cytosolic antigens to MHC II molecules (Fig. 3). For example, cytosolic intermediates of replicating vesicular stomatitis virus (VSV), recognized by PRRs (83), are delivered by autophagy into the endosomal lumen, where TLR7 can recognize viral single-stranded RNA (20). This topological inversion is critical for efficient viral recognition in plasmacytoid DCs (pDCs), since this cell type depends exclusively on endosomal TLR7 and TLR9 to detect viral replication intermediates. Again, if a PAMP is located in the cytosol and the corresponding PRR (e.g. TLR7) is facing endosomal lumen, the PAMP and the PRR can meet only after autophagy captures cytosol along with the PAMP and delivers the PAMP to the endosomal compartments (20), in a topological inversion process equivalent to endogenous antigen presentation described above. Another related observation is that autophagy facilitates synergistic B-cell receptor (BCR) and TLR9 signaling from an intracellular autophagosome-like compartment, where BCR and TLR9 cooperate for optimal MAPK signaling (21). This is of significance for production of autoantibodies to DNA-containing antigens in systemic autoimmune diseases.
In the cases described above, autophagy assists TLRs in meeting their cognate ligands (20, 21). However, the relationship between autophagy and PRR signaling in innate immunity does not stop with delivery of PAMPs to endosomal TLRs and will be covered in the next section. Furthermore, autophagy (presumably following PAMP-PRR or other type of stimulation), can deliver antimicrobial products uniquely generated through autophagy by capture of cytosolic substrates into the lumen of autophagosomes. This has been elegantly documented for ubiquitin (102), captured from the cytosol and proteolytically processed into ubiquitin fragments within autophagic organelles, contributing to killing and autophagic elimination of M. tuberculosis in macrophages (102). It is a lesser known fact that ubiquitin fragments (84), ubiqucidin (an unusual, processed ribosomal protein S30 with homology to ubiquitin) and the ribosomal polypeptides S19, and L30 (isolated from colonic epithelial cells enteric bacteria) have antimicrobial activity (85). In the context of autophagy, the antimicrobial properties of digested ubiquitin can be put to work within autophagolysosomes (102). It will be of interest to explore whether ribosomal proteins [ribosomes are common autophagic substrates (86)] may play a role in innate immune defenses in intestinal epithelia, now that connections between two autophagy factors (Atg16L and IRGM) and predisposition to Crohn's disease have been uncovered (72–76).
Autophagy is an effector of TLR signaling
The innate immune system is responsible for early detection and elimination of invading microbes and modulates adaptive immunity responses (87, 88). It senses microbial presence via specific PRRs, which in principle consist of a recognition domain and a protein-protein-interacting domain for downstream signaling. There are three major classes of PRRs: TLRs, retinoic acid-inducible gene I (RIG-I)-like helicase receptors (RLRs), and nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) (89). To initiate immune responses, PRRs recognize PAMPs and induce a number of pro-inflammatory cytokines as a widely known output. A new addition to the repertoire of PRR stimulation outputs is the recently described induction of autophagy downstream of TLR stimulation (20, 22–24) (Fig. 4). In this section we review the links thus far established between PRR signaling and autophagy.
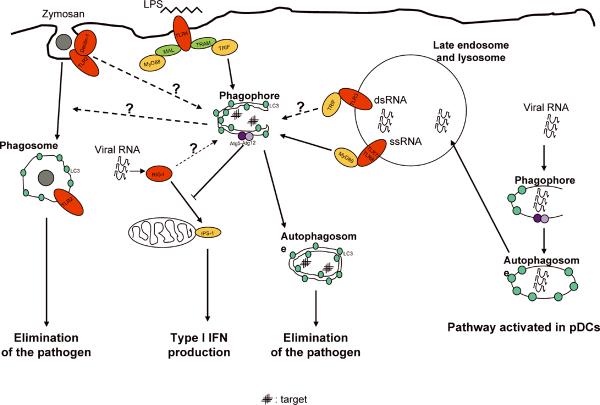
Fig. 4. Autophagy is a downstream effector of TLR signaling.
LPS induces autophagy in macrophages by activating the TLR4/TRIF pathway. Zymosan induces autophagy and additionally may induce LC3 translocation to the phagosome (directly or mediated by rapid fusion with LC3-positive autophagic organelles). TLR3 (or RIG-I/MDA5) may activate autophagy upon recognition of double-stranded RNA (dsRNA). TLR7 (and TLR8, not shown) induce autophagy. In a viral infection, viral RNA is recognized by TLR7 (endocytic ssRNA) or by RIG-I (short dsRNA or 3′-triphosphate ssRNA). In macrophages, TLR7 ligands induce autophagy, activating the TLR7/MyD88 pathway. In most cells, but not in pDCs, the viral ligands detected by RIG-I induce the IPS-1 pathway (IPS-1 is associated with mitochondria). As a potential negative feedback, Atg5–Atg12 conjugate inhibits this pathway. In pDCs, constitutive autophagy delivers TLR7 ligands from the cytosol to the compartments containing TLR7.
TLRs are among the best-characterized PRRs. TLR1, TLR2, TLR4, TLR5, and TLR6 are located mainly on the cell surface and primarily recognize bacterial components, and TLR3, TLR7, TLR8, and TLR9 are mostly in the endocytic compartments and mainly recognize viral products (90). After recognition of pathogen-derived components, individual TLRs trigger distinctive responses by recruiting a different combination of four Toll-IL-1 receptor (TIR) domain-containing adapter molecules: myeloid differentiation primary response protein 88 (MyD88), used by all TLRs except TLR3; TIR domain-containing adapter protein (TIRAP) or MyD88 adapter-like (MAL), used by TLR2 and TLR4 as a bridge to recruit MyD88; TIR domain-containing adapter-inducing IFN-β (TRIF) or TIR domain-containing adapter molecule 1 (TICAM-1), used by TLR3 and TLR4; and TRIF-related adapter molecule (TRAM) or TICAM-2, used only by TLR4 for interactions with TRIF (20, 91, 92). Some TLRs signal exclusively in a MyD88-dependent manner (TLR1, TLR2, TLR5, TLR6, TLR7, TLR8, and TLR 9), while TLR3 signals exclusively in a MyD88-independent manner (by using TRIF). TLR4 signals by both MyD88-dependent and -independent pathways (by using TRIF/TRAM); the reasons for this duality have recently been revealed by showing that TLR4 does so in a sequential manner, with the TIRAP-MyD88 pathway being engaged by TLR4 on the plasma membrane, while the TRAM-TRIF pathway is engaged from early endosomes (93). These signaling cascades activate the transcription factors nuclear factor-κB (NF-κB) and activator protein-1 (AP-1), which are common to all TLRs, leading to the production of inflammatory cytokines and chemokines (90). TLR3, TLR4, TLR7, TLR8, and TLR9 also activate IFN regulatory factor 3 (IRF3) and/or IRF7, leading to the production of type I IFN (IFN-α and IFN-β) (90). Type I IFN can induce antiviral state in most cells (90). Other cytokines and chemokines initiate and amplify inflammatory responses by recruiting and activating various innate immune cells such as monocytes, neutrophils, and natural killer cells (90).
It has been recently demonstrated that stimulation of a number of TLRs with their cognate ligands activates autophagy as a defense mechanism capable of directly eliminating intracellular pathogens (22–24). Since autophagy as an effector mechanism has direct antimicrobial action (22, 24), it differs from other PRR outputs that often require multiple downstream events to occur. Thus, autophagy can be viewed as a true blue collar worker, executing elimination of microbes, with an apparent simplicity rarely observed with other complex inflammatory processes. The TLR4 ligand lipopolysaccharide (LPS) is able to induce autophagy in murine RAW264.7 macrophage-like cells (RAW). This has been reported by three different groups showing that cytoplasmic LC3 changes from diffuse to punctuate distribution (indicative of autophagosome formation) upon LPS stimulation (22–24). LPS also increased the amount of the lipidated form of LC3 (LC3-II), detected by immunoblotting, in RAW macrophages and in murine primary bone marrow macrophages (BMMs) (22, 24). An increase in double-membrane vacuoles monitored by electron microscopy (EM) was reported in RAW cells after LPS treatment (22). The punctate distribution of LC3, a sign of autophagosome formation, was also observed by immunofluorescence in human alveolar macrophages upon LPS stimulation (22). Induction of autophagy with LPS was reported by Xu et al. (22) to be dependent on TLR4, TRIF, RIP1, and p38 MAPK, but independent on MyD88. Xu and colleagues also showed that LPS increased the association of hVPS34 with membranes and the induced expression of LRG47 (Irgm1), both known to be involved in autophagy induction (7, 49). The same group reported that LPS induced localization of M. tuberculosis in autophagosomes.
Two different ligands for mouse TLR7 induce autophagy in RAW, J774 macrophage-like cells, and BMMs, albeit less vigorously in the latter cells (24). The classical TLR7 ligand single-stranded RNA (ssRNA) induces LC3 puncta formation, LC3-I-to-LC3-II conversion, and formation of late stage autophagosomal profiles (autolysosomes) detected and quantified by EM (24). LC3-II conversion can be detected as early as 30 min upon ssRNA stimulation, in the presence of Bafilomycin A1 (to inhibit LC3-II degradation by the autophagic flux) (24). Imiquimod, another mouse TLR7 ligand, induces LC3 puncta formation (23, 24) and increases the proteolysis of long-lived proteins in RAW macrophages (24). Both TLR7 ligands were able to induce green fluorescence protein (GFP)-LC3 puncta formation in murine primary macrophages from transgenic knockin GFP-LC3 mice (24). The autophagy induced by TLR7 ligands depends on Beclin 1, TLR7, and MyD88 (24). Artificial activation of autophagy by TLR7 ligands (TLR7 has not been implicated in mycobacterial infections) can induce killing of intracellular mycobacteria (24), indistinguishable from autophagy induced by starvation, rapamycin, or overexpression of LRG47 (Irgm1) (7, 49, 94). The mycobacterial killing induced by TLR7 stimulation was dependent on MyD88, Beclin 1, and Atg5, shown in small interfering RNA (siRNA) knockdown experiments (24). Induction of autophagy in human cells is detectable after infection with human immunodeficiency virus (HIV), a virus normally causing TLR7/8 activation. This has been detected by LC3-II conversion in HeLa cells infected with HIV and was abrogated when TLR8 was knocked down with siRNA (24). The above studies demonstrate that autophagy is a physiological, potentially highly relevant output for TLR signaling. TLR signaling in the context of autophagy may be cell-type dependent. For example, in pDCs there is no detectable autophagic increase upon infection with VSV (20). This can however be attributed to high baseline autophagy levels reported for DCs (13). While this issue needs to be experimentally addressed, autophagy does play a role in TLR signaling in pDCs, as it serves to deliver viral TLR7 ligand to endosomal TLR7 (in the process of topological inversion) (20).
Another TLR ligand, double-stranded RNA (dsRNA) can induce autophagy (24). TLR3 can be activated with natural dsRNA or with its synthetic analog polyinosinic-polycytidylic acid (polyI:C). PolyI:C can induce autophagy, as detected by GFP-LC3 puncta formation and increased proteolysis of long-lived proteins in RAW cells and by LC3-II conversion in BMMs (24). However, an exclusive role for TLR3 was not demonstrated in these experiments, since polyI:C can also activate melanoma differentiation-associated gene 5 (MDA5) (89). There is some controversy regarding TLR9 and CpG. One study reported that CpG was not able to induce autophagy in RAW cells (24). CpG did not induce GFP-LC3 puncta formation (in the presence or absence of the autophagic protease inhibitors E-64d and PepstatinA), there was no LC3-II conversion, there was no increased proteolysis of stable proteins, and furthermore there was no killing of intracellular bacille Calmette-Guérin (24). Sanjuan and colleagues (23) reported an opposite result, showing GFP-LC3 puncta formation in RAW264.7 cells upon 3 h of incubation with CpG. Additional work is needed to resolve this discrepancy. It is furthermore possible that some TLRs require a combinatorial stimulation. For example, autophagy was not induced by Pam3CSK4 or by Pam2CSK4 (ligands for TLR1/TLR2 and TLR2/TLR6, respectively), but it was strongly induced by yeast cell wall particles (zymosan) (24), engaging TLR2/TLR6 and dectin-1, a lectin receptor for β-glucan (a fungal cell wall component) (90, 95). This was demonstrated by the GFP-LC3 puncta formation (24) and by LC3-I to LC3-II conversion (23). The GFP-LC3 translocation in response to zymosan was shown to be TLR2 dependent but MyD88 independent, making it difficult to interpret this as a TLR2-dependent process (23).
Of further interest, Sanjuan et al. (23) reported that the autophagic marker LC3-II was recruited to phagosomal membranes, noting that these were surprisingly not double membranes as expected from conventional autophagosomes (23). The point that LC3-II was not on double membranes, perhaps intriguing, is less of significance, since even on bona fide autophagosomes LC3-II is recruited only to a single membrane bilayer. Indeed, it is just the overall microscopic morphology of autophagic organelles that makes them appear (very transiently) as `double membranes'. It is nevertheless of some interest whether LC3-II can be recruited in a physiologically relevant manner to membranes other than autophagosomes, or whether phagosomes [which may be topologically viewed as autophagosomal equivalents ingesting the external space (96)] possess a special relationship with the autophagic machinery. Another explanation is that phagocytosis of particulate material that can stimulate TLRs may send danger signals to the cell to accelerate its degradative functions, and thus perhaps compress phagocytosis and autophagy in a single process or in a rapidly linked sequence difficult to resolve by methodologies used.
Signaling downstream of TLRs and induction of autophagy
The signaling pathways leading to autophagy activation downstream of TLR stimulation remain to be determined. The present information is based on a single very recent study (97) showing that TLR-dependent induction of autophagy may involve TRIF or MyD88-dependent recruitment of Beclin 1. Shi and colleagues (97) have shown that MyD88 and TRIF co-immunoprecipitated with Beclin 1, a key autophagy regulator. TLR signaling led to increased Beclin 1 presence in protein complexes containing MyD88 and TRIF, and, most importantly, reduced the binding of Beclin 1 to Bcl-2 (97). Given that disruption of Bcl-2-Beclin 1 interaction is key to autophagy induction (98), the observations by Shi et al., appear very promising in terms of deciphering how TLRs induce autophagy. The above findings suggest a common signaling pathway leading to autophagy induction downstream of TLR4 and TLR7, however unlikely it appeared from the initial reports showing that TRIF or MyD88, but not both, may be engaged in autophagy stimulation: Induction by LPS has been shown to require TRIF downstream of TLR4, while ssRNA and imiquimod acting via TLR7 use MyD88 to induce autophagy (22, 24).
Both TLR4 and TLR7 stimulate induction of type I IFN. However, after 4 h of stimulation in the study by Delgado et al. (24), IFN-β was secreted by RAW264.7 macrophages treated with LPS but not with ssRNA, suggesting that type I IFNs are unlikely to be key inducers of autophagy. In keeping with this finding, it has been reported that type I IFN does not induce autophagy in RAW264.7 macrophages (7). The majority of TLR signaling pathways do activate NF-κB and MAPK, and both of these outputs affect autophagy: NFκB is a negative regulator of autophagy (30), while different MAPK have varying effects autophagy. Extracellular signal-regulated kinase (ERK) and p38 activation affect the maturation step of autophagy, playing a positive and a negative role, respectively (99, 100). p38 can activate NADPH oxidase to generate reactive oxygen species (ROS) (101), and ROS are known to play a role in autophagy induction (30, 102). Another MAK, JNK, has a clearly positive role in activating autophagy by phosphorylating Bcl-2 thus releasing Beclin 1 from inhibitory complexes with Bcl-2 (37). Since all TLRs induce MAPK and NF-κB, the autophagic response will most likely depend on the nature, strength, timing, and duration of the pathways activated.
Autophagy and other PRRs
RIG-I (Fig. 4) and MDA5 are cytoplasmic PRRs collectively referred to as RLRs. RLRs can detect cytosolic viral RNA to induce IFN-α/β production by infected cells (68). Signaling by RLRs occurs through homotypic caspase activation and recruitment domain (CARD) interactions with the IFN promoter stimulator-1 (IPS-1) adapter protein [also known as mitochondrial antiviral signaling protein (MAVS), virus-induced signaling adapter (VISA), or CARD adapter inducing IFN-γ (Cardif)] (103–105). Interestingly, IPS-1 recruits RIG-I and MDA5 to the outer membrane of the mitochondria to form a macromolecular complex that serves to activate downstream signaling (106). At present, there are no reports addressing induction of autophagy by RIG-I or MDA5 activation, although Jounai and colleagues (107) reported that infection with VSV (a virus that generates 5′-triphosphate RNA) induces autophagy in mouse embryonic fibroblasts, raising the possibility of a direct participation of RIG-I engagement in autophagy induction. The same group also reported a non-canonical role for autophagic proteins as suppressors of the innate antiviral immune response: the Atg5–Atg12 conjugate, a key regulator of autophagy, directly associates with RIG-I and IPS-1 through the CARD domains, negatively regulating the signaling pathway mediated by IPS-1 and suppressing type I IFN production (107).
Another class of intracellular PRRs referred to as NLRs is characterized by the presence of a conserved NOD domain, represent the largest family of PRRs (108). NLRs often have three distinct domains: a C-terminal domain, mediating autorepression and ligand sensing; an intermediary NOD domain that is required for nucleotide binding and self-oligomerization; and an N-terminal effector domain, mediating protein-protein interactions for initiating downstream signaling (108). Although the ligands and functions of many of these receptors are not known, one of their major roles is to recognize cytoplasmic microbial PAMPs and/or endogenous danger signals, initiating immunological responses (108). There are at present no reports showing NLRs affecting autophagy, although NLRs are ideally poised for recognition of cytosolic microbial products. This positioning and the association between Crohn's disease with both NOD2 (108) and autophagy regulators Atg16 (74–76) and IRGM (72–74) suggest that we may see some connections in the future. Furthermore, a recent study in Drosophila (25) has shown that a cytosolic PRR, PGRP-LE, recognizing diaminopimelic acid-type peptidoglycan, induces autophagy and that autophagy protects the fly from Listeria monocytogenes infection. On the flip side of the autophagy-PRR relationships, Takeshita and colleagues (109) have announced in their recent paper that Atg5 interacts with caspase-1 and NOD1 CARD domains. If this can be demonstrated experimentally, it would mirror the relationships between Atg5–Atg12, IPS-1 and RIG-I. It is anticipated that more information will be emerging on the relationships between RLRs, NLRs, and autophagy.
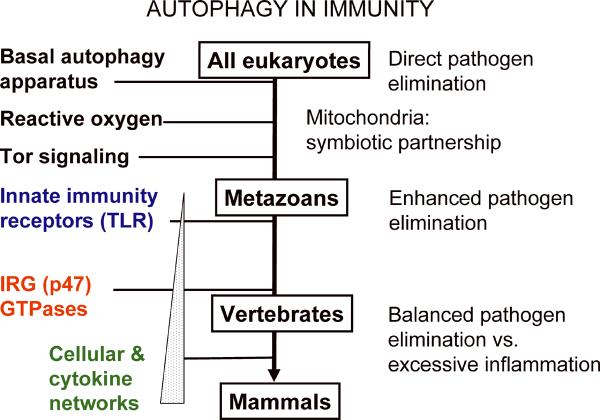
A model linking autophagy, mitochondria, and innate immunity regulators
The fast pace at which the connections between immunity and autophagy are being unraveled beckons to attempt to understand the overall position of autophagy vis-à-vis other innate and adaptive immunity systems. Is autophagy just a peripheral process recently recruited into the realm of innate immunity? Or is it perhaps centrally positioned, at the root of innate immunity? We propose a model (Fig. 5) consistent with the latter possibility. In this model, we postulate that autophagy was possibly the very initial cell-autonomous defense of the early eukaryotic cells against microbes. We envision that a rickettsia-like α-protobacterium, believed to be mitochondrial ancestor, was initially subject to elimination by autophagy along with other microbes that managed to invade the cytoplasm of the early pre-eukaryotic cell. This ancient relationship may have remained evolutionarily preserved and is reflected today in the cell-autonomous elimination of intracellular pathogens by autophagy (6–8, 25, 31, 44–51). After a symbiotic relationship was established with mitochondria, these organelles remained a substrate for autophagic elimination (mitophagy) (110, 111) to the extent that can lead to mitochondrial elimination (112, 113), sometimes referred to as mitoptosis (114). Mitochondria are prime autophagic targets, and this process can become so excessive to result in cell death referred to as mitophagic cell death (115). More routinely, eliminating damaged or dysfunctional mitochondria occurs at low level in all cells at all times (110), lest leaky mitochondria induce unscheduled cell death or otherwise damage (e.g. via excessive reactive oxygen generation) the host cell (116). We further envision that upon this backdrop of primordial relationships, layers of immune regulation may have become superimposed during evolution. This is perhaps reflected in the recently uncovered TLR-dependent induction of autophagy (22–25) and the mitochondrial localization of RLR signaling networks (88). In vertebrates, p47 GTPases (117) were added to the system as (in one of their roles) additional regulators of autophagy (7, 48, 49), followed by advanced cytokine networks produced by T-cell subsets (7, 14). We believe that, albeit speculative, this model can serve as a starting point for useful discussions and generation of testable hypotheses.
Fig. 5. Model of interactions between autophagy and immune regulatory systems presented as an evolutionary timeline.
Acknowledgments
This work was supported by grants AI069345, AI45148, AI42999 from National Institutes of Health, 107160-44-RGRL from amfAR, and a grant from Crohn's & Colitis Foundation of America
References
- 1.Levine B, Kroemer G. Autophagy in the Pathogenesis of Disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 4.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakagawa I, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 8.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 9.Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005;26:523–528. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Paludan C, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 11.Dengjel J, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris J, et al. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–517. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, Lu B. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 16.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pua HH, He YW. Maintaining T lymphocyte homeostasis: another duty of autophagy. Autophagy. 2007;3:266–267. doi: 10.4161/auto.3908. [DOI] [PubMed] [Google Scholar]
- 18.Miller BC, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 19.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 20.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 21.Chaturvedi A, Dorward D, Pierce SK. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity. 2008;28:799–809. doi: 10.1016/j.immuni.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 24.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yano T, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS letters. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 29.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002;157:455–468. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Djavaheri-Mergny M, et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem. 2006;281:30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 31.Andrade RM, Wessendarp M, Gubbels MJ, Striepen B, Subauste CS. CD40 induces macrophage anti-Toxoplasma gondii activity by triggering autophagy-dependent fusion of pathogen-containing vacuoles and lysosomes. J Clin Invest. 2006;116:2366–2377. doi: 10.1172/JCI28796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr Opin Cell Biol. 2006;18:589–597. doi: 10.1016/j.ceb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nobukuni T, Kozma SC, Thomas G. hvps34, an ancient player, enters a growing game: mTOR Complex1/S6K1 signaling. Curr Opin Cell Biol. 2007;19:135–141. doi: 10.1016/j.ceb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 36.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 30:678–688. doi: 10.1016/j.molcel.2008.06.001. 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 39.Liang C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanada T, et al. The Atg12-Atg5 Conjugate Has a Novel E3-like Activity for Protein Lipidation in Autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 42.Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rikihisa Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced by rickettsiae. Anat Rec. 1984;208:319–327. doi: 10.1002/ar.1092080302. [DOI] [PubMed] [Google Scholar]
- 44.Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 2003;5:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- 45.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci USA. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling YM, et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 50.Py BF, Lipinski MM, Yuan J. Autophagy limits Listeria monocytogenes intracellular growth in the early phase of primary infection. Autophagy. 2007;3:117–125. doi: 10.4161/auto.3618. [DOI] [PubMed] [Google Scholar]
- 51.Cullinane M, et al. Stimulation of autophagy suppresses the intracellular survival of Burkholderia pseudomallei in mammalian cell lines. Autophagy. 2008;4:744–753. doi: 10.4161/auto.6246. [DOI] [PubMed] [Google Scholar]
- 52.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson WT, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orvedahl A, et al. HSV-1 ICP34.5 Confers Neurovirulence by Targeting the Beclin 1 Autophagy Protein. Cell Host and Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature. 2005;451:350–354. doi: 10.1038/nature06479. [DOI] [PubMed] [Google Scholar]
- 56.Pyo JO, et al. Essential Roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 57.Arico S, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 58.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno O. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 59.Schlottmann S, et al. Prolonged classical NF-kappaB activation prevents autophagy upon E. coli stimulation in vitro: a potential resolving mechanism of inflammation. Mediators Inflamm. 2008:725854. doi: 10.1155/2008/725854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabauste CS, Andrade RM, Wessendarp M. CD40-TRAF6 and autophagy-dependent anti-microbial activity in macrophages. Autophagy. 2007;3:245–248. doi: 10.4161/auto.3717. [DOI] [PubMed] [Google Scholar]
- 61.Tasdemir E, et al. Cell cycle-dependent induction of autophagy, mitophagy and reticulophagy. Cell Cycle. 2007;6:2263–2267. doi: 10.4161/cc.6.18.4681. [DOI] [PubMed] [Google Scholar]
- 62.Wright K, Ward SG, Kolios G, Westwick J. Activation of phosphatidylinositol 3-kinase by interleukin-13. An inhibitory signal for inducible nitric-oxide synthase expression in epithelial cell line HT-29. J Biol Chem. 1997;272:12626–12633. doi: 10.1074/jbc.272.19.12626. [DOI] [PubMed] [Google Scholar]
- 63.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 64.Taylor GA, Feng CG, Sher A. p47 GTPases: regulators of immunity to intracellular pathogens. Nat Rev Immunol. 2004;4:100–109. doi: 10.1038/nri1270. [DOI] [PubMed] [Google Scholar]
- 65.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- 66.Howard J. The IRG proteins: a function in search of a mechanism. Immunobiology. 2008;213:367–375. doi: 10.1016/j.imbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 67.Martens S, et al. Disruption of Toxoplasma gondii Parasitophorous Vacuoles by the Mouse p47-Resistance GTPases. PLoS Pathog. 2005;1:e24. doi: 10.1371/journal.ppat.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng CG, Weksberg DC, Taylor GA, Sher A, Goodell MA. The p47 GTPase Lrg-47 (Irgm1) links host defense and hematopoietic stem cell proliferation. Cell Stem Cell. 2008;2:83–89. doi: 10.1016/j.stem.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bekpen C, et al. The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol. 2005;6:R92. doi: 10.1186/gb-2005-6-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miyairi I, et al. The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. J Immunol. 2007;179:1814–1824. doi: 10.4049/jimmunol.179.3.1814. [DOI] [PubMed] [Google Scholar]
- 71.Coers J, et al. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. J Immunol. 2008;180:6237–6245. doi: 10.4049/jimmunol.180.9.6237. [DOI] [PubMed] [Google Scholar]
- 72.Massey D, Parkes M. Common pathways in Crohn's disease and other inflammatory diseases revealed by genomics. Gut. 2007;56:1489–1492. doi: 10.1136/gut.2007.127928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 76.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deretic V, Klionsky DJ. How cells clean house. Sci Am. 2008;298:74–81. doi: 10.1038/scientificamerican0508-74. [DOI] [PubMed] [Google Scholar]
- 78.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 79.Zhou D, et al. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 80.Lunemann JD, Munz C. Autophagy in CD4(+) T-cell immunity and tolerance. Cell Death Differ. 2008 doi: 10.1038/cdd.2008.113. in press. [DOI] [PubMed] [Google Scholar]
- 81.Crotzer VL, Blum JS. Cytosol to lysosome transport of intracellular antigens during immune surveillance. Traffic. 2008;9:10–16. doi: 10.1111/j.1600-0854.2007.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taylor GS, Long HM, Haigh TA, Larsen M, Brooks J, Rickinson AB. A role for intercellular antigen transfer in the recognition of EBV-transformed B cell lines by EBV nuclear antigen-specific CD4+ T cells. J Immunol. 2006;177:3746–3756. doi: 10.4049/jimmunol.177.6.3746. [DOI] [PubMed] [Google Scholar]
- 83.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kieffer AE, et al. The N- and C-terminal fragments of ubiquitin are important for the antimicrobial activities. FASEB J. 2003;17:776–778. doi: 10.1096/fj.02-0699fje. [DOI] [PubMed] [Google Scholar]
- 85.Howell SJ, Wilk D, Yadav SP, Bevins CL. Antimicrobial polypeptides of the human colonic epithelium. Peptides. 2003;24:1763–1770. doi: 10.1016/j.peptides.2003.07.028. [DOI] [PubMed] [Google Scholar]
- 86.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nature cell biology. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 87.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 88.Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3:352–363. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 89.Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr Opinion Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 90.Lee MS, Kim YJ. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]
- 91.Kawai T, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 92.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 93.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci USA. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 96.Deretic V. Autophagosome and phagosome. Methods Mol Biol. 2008;445:1–10. doi: 10.1007/978-1-59745-157-4_1. [DOI] [PubMed] [Google Scholar]
- 97.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008 doi: 10.1074/jbc.M804478200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maiuri MC, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Corcelle E, et al. Control of the autophagy maturation step by the MAPK ERK and p38: lessons from environmental carcinogens. Autophagy. 2007;3:57–59. doi: 10.4161/auto.3424. [DOI] [PubMed] [Google Scholar]
- 100.Corcelle E, et al. Disruption of autophagy at the maturation step by the carcinogen lindane is associated with the sustained mitogen-activated protein kinase/extracellular signal-regulated kinase activity. Cancer Res. 2006;66:6861–6870. doi: 10.1158/0008-5472.CAN-05-3557. [DOI] [PubMed] [Google Scholar]
- 101.Laroux FS, Romero S, Wetzler L, Engel P, Terhorst C. Cutting edge: MyD88 controls phagocyte NADPH oxidase function and killing of gram-negative bacteria. J Immunol. 2005;175:5596–5600. doi: 10.4049/jimmunol.175.9.5596. [DOI] [PubMed] [Google Scholar]
- 102.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 104.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 105.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 106.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jounai N, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 109.Takeshita F, Kobiyama K, Miyawaki A, Jounai N, Okuda K. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy. 2008;4:67–69. doi: 10.4161/auto.5055. [DOI] [PubMed] [Google Scholar]
- 110.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 112.Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lyamzaev KG, et al. Novel mechanism of elimination of malfunctioning mitochondria (mitoptosis): formation of mitoptotic bodies and extrusion of mitochondrial material from the cell. Biochim Biophysica Acta. 2008;1777:817–825. doi: 10.1016/j.bbabio.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 115.Kim EH, et al. Sodium selenite induces superoxide-mediated mitochondrial damage and subsequent autophagic cell death in malignant glioma cells. Cancer Res. 2007;67:6314–6324. doi: 10.1158/0008-5472.CAN-06-4217. [DOI] [PubMed] [Google Scholar]
- 116.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Martens S, Howard J. The interferon-inducible GTPases. Annu Rev Cell Dev Biol. 2006;22:559–589. doi: 10.1146/annurev.cellbio.22.010305.104619. [DOI] [PubMed] [Google Scholar]