Abstract
The enzyme myo-inositol oxygenase (MIOX) catalyzes conversion of myo-inositol (cyclohexan-1,2,3,5/4,6-hexa-ol or MI) to D-glucuronate (DG), initiating the only known pathway in humans for catabolism of the carbon skeleton of cell-signaling inositol (poly)phosphates and phosphoinositides. Recent kinetic, spectroscopic, and crystallographic studies have shown that the enzyme activates its substrates, MI and O2, at a carboxylate-bridged nonheme diiron(II/III) cluster, making it the first of many known nonheme diiron oxygenases to employ the mixed-valent form of its cofactor. Evidence suggests that (1) the Fe(III) site coordinates MI via its C1 and C6 hydroxyl groups, (2) the Fe(II) site reversibly coordinates O2 to produce a superoxo-diiron(III/III) intermediate, and (3) the pendant oxygen atom of the superoxide ligand abstracts hydrogen from C1 to initiate the unique C-C-bond-cleaving, four-electron oxidation reaction. This review recounts the studies leading to the recognition of the novel cofactor requirement and catalytic mechanism of MIOX and forecasts how remaining gaps in our understanding might be filled by additional experiments.
Bacterial multi-component monooxygenases [BMMs; e.g., soluble methane monooxygenase (sMMO), toluene/o-xylene monooxygenase (ToMO), and phenol hydroxylase], plant fatty acyl desaturases (e.g., stearoyl acyl carrier protein Δ9-desaturase) and the R2 subunits of conventional class I ribonucleotide reductases (RNR-R2s) all use carboxylate-bridged dinuclear iron clusters to activate O2 for cleavage of strong C-H or O-H bonds.1–5 Each of these reaction begins with the reduction of O2 to the peroxide oxidation state by the diiron(II/II) form of the cofactor. In several cases, peroxide-bridged diiron(III/III) intermediates have been directly characterized.6–11 Several of the peroxide complexes are known or believed to undergo O-O-bond cleavage to generate high-valent iron complexes that cleave the target C/O-H bonds.1–5 For example, the diiron(III/IV) complex, X, in the RNR-R2 reaction oxidizes a tyrosine residue by one electron, cleaving the phenolic O-H bond and activating the protein for participation in nucleotide reduction with its partner subunit, RNR-R1.12–21 Similarly, the diiron(IV/IV) complex, Q, in the sMMO reaction cleaves a C-H bond of methane to initiate its hydroxylation.1, 3, 7, 22–24 In each of these reactions, a diiron(III/III) form of the cluster is generated at the end of the oxidation sequence. For the reactions that are catalytic, a complete “turnover” therefore requires reduction of the cluster back to the O2-reactive diiron(II/II) state by additional proteins, with electrons provided ultimately by NAD(P)H.1, 3, 4 Although this mechanistic strategy is both versatile (permitting one-electron oxidation, hydroxylation, and desaturation of the various aromatic and aliphatic targets) and highly potent (permitting cleavage of even the 104 kcal/mol C-H bond of methane), it does have a fundamental chemical limitation: the redox cycling of the cofactor and provision of two electrons by the nicotinamide “co-substrate” dictate that only two electrons, at most, can be extracted from the substrate.
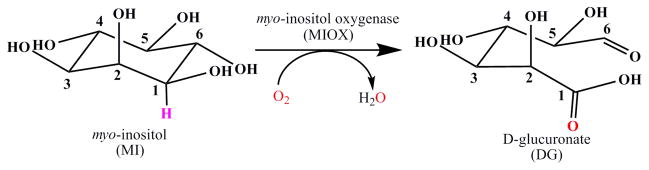
When we recently began to investigate myo-inositol oxygenase (MIOX), the enzyme that catalyzes the ring-opening glycol cleavage of myo-inositol (MI) to D-glucuronate (DG) in higher eukaryotes and some bacteria (Scheme 1),25–28 and discovered that it also contains a non-heme diiron cluster,29 we were soon struck by a conundrum. The MIOX reaction is a four-electron oxidation, converting the C1 alcohol to an acid (which cleaves the C1-H bond and yields four electrons), the C6 alcohol to an aldehyde (which cleaves the O6-H bond and yields two electrons), and the cyclic carbon skeleton to a linear one (which cleaves the C1–C6 bond and formally requires two electrons). It was difficult to imagine how the conventional diiron-oxygenase mechanism could be adapted to yield such an outcome, especially because the obvious possibility of sequential two-electron oxidations by two equivalents of O2 could be ruled out on the basis of the published stoichiometry of one DG per O2.25–28 Indeed, our subsequent mechanistic dissection of the reaction revealed that MIOX uses a decidedly unconventional strategy for O2 and C-H activation by its nonheme diiron cluster.30, 31 In this account, we summarize our current understanding of the structure, evolution, and mechanism of MIOX, point out gaps in this understanding, and forecast how these gaps might be filled.
Scheme 1.
Reaction catalyzed by MIOX.
1. Discovery and Physiology of MIOX
First described in the late 1950s by Charalampous,25–27 the MIOX reaction initiates the only known pathway in humans for catabolism of MI, the sugar backbone of the array of cellular ‘second messengers’ known collectively as inositol (poly)phosphates and phosphoinositides.32 Subsequent transformations convert DG to xylitol. It has been known for some time that diabetes mellitus is associated with aberrant depletion of MI33 and accumulation of polyols, including xylitol.34, 35 Both MI depletion and polyol accumulation have been cited as contributing factors in the devastating complications associated with diabetes (e.g., neuropathy, retinopathy, and nephropathy).34 For these reasons, MIOX has been touted as a potential target for treatment of diabetic complications.36–41
2. Published Mechanistic Clues
The pioneering work by Charalampous25–27 and subsequent studies by Hamilton and co-workers through the 1980s42–45 had revealed certain essential features of the MIOX reaction and important, if cryptic, mechanistic clues. Most importantly, both groups had documented the enzyme’s dependence on iron. Both groups had also shown that only one of the two atoms from O2 is incorporated into the product (into the C1 acid).25, 28 Finally, Hamilton and co-workers had reported complex activation and inactivation behavior.44 For example, incubation with Fe(II) and L-cysteine prior to addition of substrate and inclusion of L-cysteine in the subsequent catalytic reaction were initially found to give optimal activity. Other thiol compounds could substitute for L-cysteine, but with varying efficacy. The trends in efficacy were not explained and seemed inconsistent with a simple, general-reductant effect. Later, a slow-freeze/thaw cycle in the presence of a high concentration of glutathione, a very inefficient activator in the absence of the freeze/thaw, was found to give optimal activity.45 These complexities (especially the requirement for a reducing activator in a charge-balanced reaction) had remained mysterious.
3. Isolation of the Gene and Heterologous Expression of MIOX: A Crucial Advance
Earlier this decade, Reddy and co-workers isolated the gene encoding Mus musculus (mouse) MIOX, determined its amino acid sequence, and created Escherichia coli strains for over-expression of the MIOX orthologues from mouse and several other mammals.46 These achievements set the stage for structural and mechanistic studies on the enzyme. The amino acid sequences did not, however, by themselves provide any obvious structural or mechanistic clues, as the orthologues are too highly similar to one another (> 45 % identity) to infer functional importance of residues that are conserved but are insufficiently globally similar to any other protein of known structure and function to provide hints as to either the nature of the iron cofactor or the mechanistic pathway. Attracted by the possible biomedical significance and likely mechanistic novelty of the reaction, we began to study MIOX with our collaborators in the Reddy group.
4. Discovery that MIOX Contains a Coupled Dinuclear Iron Cluster
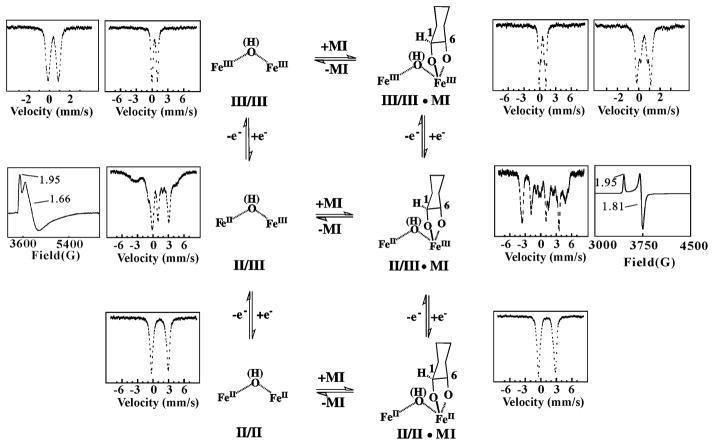
At the outset, there was no reason to suspect that MIOX would prove to be a di-iron oxygenase, much less one that employs a novel mechanism. A very simple Mössbauer spectroscopic experiment immediately revealed the binuclear nature of the cofactor.29 Exposure of a solution of pure apo MIOX to just less than 2 equiv Fe(II) followed by excess O2 or H2O2 yielded a form that exhibited a Mössbauer quadrupole doublet (at 4.2 K and 53 mT) with isomer shift and quadrupole splitting parameters characteristic of high-spin Fe(III) ions with nitrogen and oxygen coordination (Scheme 2, top left). The absence of magnetic hyperfine splitting suggested that the complex might have an integer-spin ground state. Indeed, the 4.2-K spectrum acquired in a strong external magnetic field (6 T) showed that the complex has a diamagnetic (S = 0) ground state, which could most simply be rationalized by antiferromagnetic coupling of two high-spin Fe(III) ions in a diiron(III/III) cluster. Perturbations to the Mössbauer spectrum upon addition of MI suggested that the substrate binds either directly to or in the vicinity of the diiron cluster (Scheme 2, top right).
Scheme 2.
Interconversion of three different oxidation states of the MIOX diiron cluster [(II/II), (II/III), and (III/III)] in the absence and presence of substrate, MI. Selected X-band EPR and 4.2K/53-mT Mossbauer reference spectra are shown next to the respective structures.29
5. Initial Indications of a New Pathway
Familiarity with the conventional diiron proteins conditioned us to expect reaction of O2 with the MI complex of MIOX having its cluster in the II/II oxidation state. An early experiment, in which a solution containing enzyme, 2 equiv Fe(II), and saturating MI (100 mM, compared to a KM of ~ 10 mM) was mixed with limiting O2, was carried out under this presumption.30 Expecting that one DG per O2 would be produced and, importantly, that the reduced form of the enzyme would be regenerated after consumption of the limiting reactant, we instead observed production of much less than one DG per O2 and conversion of the enzyme to the mixed-valent, diiron(II/III) state with a stoichiometry of 1.1 ± 0.2 per O2. The accumulation of the mixed-valent form under turnover conditions hinted, remarkably, that the II/III state might be the active enzyme form.30 When the fully-oxidized enzyme was found to be reduced to the II/III state by the activator, L-cysteine, it seemed even more likely that MIOX might operate in this uncharted territory.
6. Preparation of MIOX(II/III) and Characterization of MI Binding to it
Slow diffusion of O2(g) into a solution containing MIOX, 2 equiv Fe(II), and either L-cysteine or the alternative reductant, ascorbate permitted preparation of the mixed-valent state in ~ 60% yield, sufficient to test the hypothesis that it is the active state. Reduction of MIOX(III/III) by L-cysteine can give a similar yield. This yield could be near the thermodynamic upper limit if the standard reduction potential for the III/III→II/III step is only modestly (~ 52 mV) more positive than that for the II/III→II/II step (Scheme 2). These potentials remain to be determined.
Mixing of MIOX(II/III) with MI gave rich absorption changes (albeit with relatively small effective molar absorptivities of <1000 M−1cm−1), with the difference spectrum for substrate binding (−MI – +MI) exhibiting a positive feature at ~ 390 nm and a negative feature at ~ 490 nm (Scheme 3, second spectrum from left).30 Kinetic measurements monitoring these changes showed that binding of the substrate to mixed-valent MIOX is kinetically competent to be on the catalytic pathway, with the rate constant for association (260 ± 20 M−1s−1) exceeding kcat/KM (70 M−1s−1) by almost four-fold. Dissociation rate and equilibrium constants of ~ 0.1 s−1 and ~ 0.4 mM, respectively, were estimated from the kinetic data. MI binding also markedly perturbs the Mossbauer and electron paramagnetic resonance (EPR) spectra of the mixed-valent diiron cluster (Scheme 2, middle row). The 4.2-K/53-mT Mössbauer spectrum of MIOX(II/III) exhibits broad, poorly resolved absorption from −6 mm/s to + 6 mm/s, whereas the MI complex exhibits well-resolved peaks at −1.9 mm/s and 3.7 mm/s arising from the Fe(II) site and at −3.8 mm/s and +4.5 mm/s arising from the Fe(III) site (inner spectra).29 Each spectrum collapses to a pair of quadrupole doublets with parameters indicative of a high-spin Fe(II) and a high-spin Fe(III) at higher temperature (> 120 K) and zero field. The broad, axial EPR spectrum of the MI-free form has g-values of 1.95, 1.66, and 1.66 and is readily detected only at relatively low temperature (< 8 K) and high microwave power (20 mW), whereas the spectrum of the substrate complex is much sharper, has g-values of 1.95, 1.81, and 1.81, and is readily observable under less stringent conditions (e.g., 20 K and 0.1 mW) (outer spectra). The EPR perturbation has also been used to estimate KD for the complex as 0.7 ± 0.3 mM (unpublished data), in agreement with the estimate from the stopped-flow absorption kinetic data.
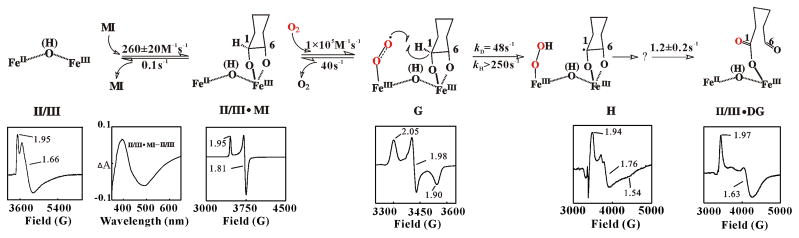
Scheme 3.
Proposed mechanism of conversion of MI to DG initiated by the formally (superoxo)diiron(III/III) intermediate, G, via the abstraction of hydrogen atom, from C1. X-band EPR of the various states and the optical spectroscopy change upon addition of MI to II/III are shown underneath the structures.30, 31 The mechanism of decay of the (hydroperoxo)diiron(III/III) intermediate and subsequent steps, as well as the nature of intermediate H are not well understood at present.
More recent studies on human MIOX revealed similar, but not identical, EPR properties for its mixed-valent state and MI complex thereof.47 Interestingly, this study provided evidence for the presence of two resolvable EPR resonances in the substrate free enzyme. Because only one of these two signals was observed in a partially (18 %) active variant protein lacking the N-terminal 37 residues, which contribute to a “lid” that covers the active site, the authors attributed the two signals to “open” and “closed” conformations of the enzyme. Detecting a more homogeneous signal upon addition of MI to the wild-type protein, they concluded that substrate binding favors lid closure to prepare the enzyme for O2 activation and turnover. These authors also used the temperature and microwave-power dependencies of the EPR signals of the substrate-free and -bound forms to estimate the magnitude of the (antiferromagnetic) exchange coupling (J) between the Fe(II) and Fe(III) sites, concluding that substrate binding weakens coupling from −20 cm−1 to 10 cm−1.47
From our spectroscopic characterization of mouse MIOX(II/III)(• MI), we speculated that MI coordinates directly to the diiron cluster, possibly via an alkoxide bridge. The C1 oxygen was deemed the most likely to bridge the irons, because the C1-H bond is cleaved during the reaction and coordination with deprotonation of O1 might appropriately position and also activate the C1-H for abstraction. To test for direct coordination and probe the coordination mode, Q-band 2H-ENDOR spectra of the complexes prepared either with MI substituted with deuterium at all six carbons [MIOX(II/III) • D6-MI] or with unlabeled MI [MIOX(II/III) • H6-MI] were acquired at magnetic fields spanning the axial EPR signal.48 The magnitudes of the observed deuterium hyperfine couplings were sufficiently large to establish that MI does indeed coordinate directly to the cluster. Further analysis of the spectra seemed consistent with the proposed μ-O1 coordination mode. However, the crystal structure published soon afterward suggested a different coordination mode (see section 12).49
7. Demonstration of Cyclic and Productive Reaction of MIOX(II/III) • MI with O2
Absorption and EPR spectral changes occurring upon mixing of the MIOX(II/III) • MI complex with limiting O2 at 5°C were unequivocally diagnostic of turnover.30 Kinetic-difference absorption spectra were essentially the opposite of the difference spectrum for MI binding. The negative feature at ~ 390 nm, indicating loss of the substrate complex, developed and decayed ahead of the positive feature at 490 nm, corresponding to accumulation of substrate-free enzyme. The temporal separation implied the accumulation of an intermediate. Within ~ 8 s after mixing, the difference absorption had decayed, making the cycle fast enough to account for the steady-state turnover number (~ 0.3 s−1) under similar conditions. Decay of the positive 490-nm peak was accelerated by increased [MI], proving that this decay reflects re-binding of MI to the substrate-free form. The obvious implication that the substrate-free enzyme accumulates as a result of turnover was verified by direct radiometric and mass spectrometric detection of DG (0.8 ± 0.1 DG/O2) after completion.
EPR spectra of samples freeze-quenched through the course of the single-turnover corroborated the cyclic decay and regeneration of MIOX(II/III) • MI and provided direct evidence for the inferred intermediate state.30 Development of a new, rhombic g = (1.92, 1.76, 1.54) signal accompanying the transient decay of the g = (1.95, 1.81, 1.81) signal of MIOX(II/III) • MI was observed. Formation and decay of the new signal were shown to track precisely with decay and re-development of the MIOX(II/III) • MI signal. The possibility that the associated intermediate state, designated H, is merely a product complex was disfavored on the basis that complexes formed by addition of either the product, DG, or its C6-aldehyde-reduced analogue, L-gulonate (which cannot cyclize, and thus should mimic well the acyclic form of DG produced in the MIOX active site), directly to MIOX(II/III) exhibit rhombic EPR signals distinct from that of H (Scheme 3). This tentative conclusion required inclusion of an additional state, the MIOX(II/III) DG product complex, in the minimal kinetic mechanism for the reaction. According to this scenario, only a lower-limit of ~ 8 s−1 could be set for the rate constant for release of DG, due to the fact that conversion of H to the product is sufficiently slow (1.2 ± 0.2 s−1) to prevent the MIOX(II/III) • DG complex from accumulating to a high level. A still-viable alternative scenario is that H is the MIOX(II/III) • product complex, but with the DG and/or cofactor in an altered state that can be accessed only through turnover. Indeed, in previous studies on taurine:α-ketoglutarate dioxygenase, a mononuclear nonheme-iron enzyme, we were unable to generate a known product complex by addition of products to the resting enzme.50 The nature of H and the kinetics of DG release are being investigated further by kinetic and spectroscopic approaches.
8. Detection of the C1-H-Cleaving Intermediate, G, and Kinetic Description of the Catalytic Cycle
Use of D6-MI allowed the formation of H from MIOX(II/III) • MI and O2 to be resolved into two steps, (1) formation of a preceding intermediate, G, in a reversible bimolecular reaction between O2 and MIOX(II/III) • MI and (2) conversion of G to H in a reaction involving abstraction of hydrogen from the substrate (almost certainly from C1, as this is the only hydrogen lost in conversion to DG; Scheme 1).31 G is characterized by a very sharp rhombic EPR signal with g = (2.05, 1.98, 1.90) (Scheme 3). That G abstracts hydrogen was implied by the observations that (1) it accumulates to a much lesser extent with H6-MI and (2) the lesser accumulation of G results from a greater accumulation of H. In other words, there is a significant substrate deuterium kinetic isotope (2H-KIE) effect on decay of G to H. Only a rough estimate could be obtained for the magnitude of the intrinsic 2H-KIE (8–16), because the modest accumulation of G in the H6-MI reaction made it difficult to determine accurately the rate constant for the G → H step. This estimate was facilitated by the photolytic lability of G to ambient laboratory light at cryogenic temperatures (77 K), which was used to quantify G in H6-MI and D6-MI samples from difference spectra. Kinetic simulations constrained by experimentally determined parameters were used to estimate the magnitude of 2H-KIE necessary to account for the [G](H6-MI)/[G](D6-MI) ratios at two reaction times. The propagation of this substrate 2H-KIE on the G → H step back into the decay of the reactant complex (i.e., the appearance of a slow phase in decay of MIOX(II/III) • MI only in the reaction with D6-MI) established the reversibility of G formation. The freeze-quench EPR kinetic data were accounted for by a mechanism comprising four states, G, H, MIOX(II/III) • DG, and MIOX(II/III), in addition to the reactant complex, and this mechanism accurately rationalized both the steady-state kcat (0.7 s−1) and absence of 2H-KIE thereupon (Scheme 3). Notable features of the mechanism are: (1) a kon for O2 (1 × 105 M−1s−1) very similar to those that we have seen in a number of oxidase/oxygenase systems50–54 and a KD for the O2-addition step of ~ 400 μM; (2) rate-limiting decay of H, which leaves knowledge of the chemical nature of the rate-limiting step dependent upon the aforementioned further characterization of H.
9. Basis for Assignment of G as a (Superoxo)diiron(III/III) Complex with Terminal Superoxide
As for H, insight into the nature of G is scant. What is clear is that it has an S = 1/2 ground state and, therefore, an odd number of electrons. This fact is established by the g ~ 2 EPR signal. Its S = 1/2 ground state is most simply interpreted to mean that G is iso-electronic with the formally (superoxo)diiron(III/III) complex formed by simple addition of O2 to the Fe(II) site of the reactant complex. In principle, it might be a more advanced state of the transformation, in electrons have already been extracted from the substrate, but there is no obvious, chemically reasonable pathway for extraction of electrons from the cyclohexan-hexa-ol prior to C-H cleavage, and the 2H-KIE on its decay establishes that G precedes C-H cleavage (see above). Moreover, formulations with high-valent oxidation states and/or a cleaved O-O bond [e.g., (peroxo)diiron(III/IV) or di-(μ-oxo)diiron(IV/V)] seem unlikely. Oxidation of the cofactor to high-valency should be endergonic without O-O cleavage, and formulations with a cleaved O-O bond would require that the bond reform at ≥ 40 s−1 to account for the dissociation of O2 from G implied by the kinetic data. We are unaware of any precedent for this rapid, reversible O-O cleavage in a diiron complex, although a noteworthy example has been reported for a dicopper complex.55 Thus, the (superoxo)diiron(III/III) formulation is most consistent with the limited available data.
Formulations with either terminal or bridging superoxide coordination can be envisaged for G. The hyperfine coupling of the two I = 1/2 57Fe nuclei to the electron spin, which is expected to be different for the two modes, was examined in an attempt to clarify this issue.31 The photolysis procedure was used to resolve the spectra of G prepared from 56Fe- and 57Fe-containing MIOX(II/III) • MI. Simulations of the EPR spectra, starting with that of the 56Fe-labeled complex and applying isotropic hyperfine couplings of 13 G and 25 G for the two (presumed) high-spin Fe(III) sites to this theoretical spectrum to reproduce the experimental spectrum of the 57Fe-labeled complex, were carried out. The relatively large couplings necessary to produce the experimentally observed broadening were judged on the basis of the magnitude of the spin projection factors predicted for the two models to be more consistent with the terminal coordination mode. The anisotropy of the g-tensor of G is unexpected, because the g-tensors of the constituent spins [two high-spin Fe(III) and a superoxide radical] are nearly isotropic and close to the free-electron g-value. However, mixing of S > 1/2 excited spin states into the S = 1/2 ground state can account for the observed anisotropy. More detailed insight into the atomic and electronic structure of G should be obtainable by multi-nuclear, multi-frequency electron-nuclear double resonance and electron spin echo envelope modulation (ENDOR/ESEEM) spectroscopy, as in previous studies by the Hoffman and Stubbe groups on the diiron(III/IV) intermediate X from E. coli RNR-R2. 16, 17, 19
10. Nature of C1-H Cleavage by G
The substrate 2H-KIE on decay of G showed that it abstracts hydrogen from MI. The C1-H of MI is the only (non-exchangeable) hydrogen obviously removed in conversion to DG (Scheme 1). Because G is a radical species (i.e., it has an S = 1/2 ground state), we favor the possibility that the C1-H is abstracted as a hydrogen atom (coupled one-proton/one-electron transfer) by the pendant oxygen of the coordinated superoxide. This step would generate an as-yet-undetected C1-radical/hydroperoxo-diiron(III/III) complex (Scheme 3). A coupled one-proton/two-electron transfer (e.g., hydride transfer) step to yield a 1-ketone intermediate is also possible but seems less likely.
We initially viewed the proposed abstraction of the aliphatic hydrogen atom before O-O-bond cleavage as potentially thermodynamically unfeasible: the majority of the decrease in free energy associated with the four-electron reduction of dioxygen is realized in the last two steps, as the peroxide equivalent is reductively cleaved to water/hydroxo/oxo and hydroxyl radical equivalents and the latter is then reduced to a second water/hydroxide equivalent. Indeed, in the best-understood mechanism for cleavage of strong C-H bonds by iron enzymes, O2 is (formally) fully reduced to two water/hydroxo/oxo equivalents prior to C-Hcleavage. One or both of the O-atoms remains coordinated to the iron cofactor in a ferryl [Fe(IV)=O2−] complex. The ferryl unit has sufficient potency to cleave strong C-H bonds,54 including, for the case of the aforementioned formally bis-ferryl complex Q in soluble methane monooxygenase, the 104 kcal/mol C-H bond of methane. Much less is known about the potency of the preceding, mid-valent superoxide complexes for H • abstraction.56 However, a recent study showed that a (superoxo)diiron(II/III) complex, one electron less oxidized than G in MIOX, is capable of formal hydrogen abstraction from the hydroxyl of a phenol.57 Morevover, the mid-valent-metal-superoxide manifold is thought to be operant in the uncoupled dicopper oxygenases, dopamine β-monooxygenase and peptidylglycine α-hydroxylating monooxygenase,58, 59 and in the mononuclear non-heme-iron oxidase, isopenicillin N synthase (IPNS).60 The four-electron oxidation of its substrate by a single equivalent of dioxygen in IPNS is particularly reminiscent of the situation in MIOX and provides a second example of the special utility of the alternative manifold. Its limitations, especially with respect to the strength of C-H bonds that can be cleaved, remain to be established.56
11. Possible Pathways for DG Production after C1-H Abstraction by G
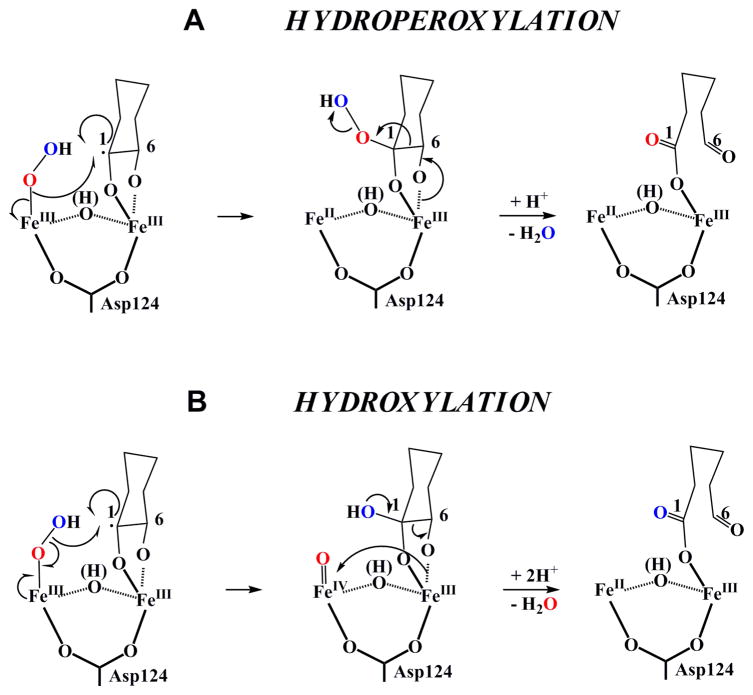
After abstraction of the C1-bonded H-atom by the uncoordinated O-atom of G, subsequent oxidative steps lead to formation of a new C1-O bond and cleavage of the O-O and substrate O6-H and C1-C6 bonds. For these steps, two classes of mechanisms were envisaged (Scheme 4).31 In the first, which we designate the “hydroperoxylation” path, formation of the new C1-O bond precedes the bond-cleavage steps. The intermediate in this pathway, (1-hydroperoxy)-myo-inositol, was first envisaged by Hamilton and co-workers in the 1980s, who cited chemical precedent that it would break down to DG.61 In the second class, designated the “hydroxylation” path, the (hydroperoxo)diiron(III/III) intermediate decays by attack of the C1 radical on the distal O-atom of the hydroperoxide, leading simultaneously to hydroxylation of C1 and cleavage of the O-O bond. The resultant substrate species would be a coordinated gem-diol(ate), and the cofactor would be in the III/IV oxidation state. The electron-deficient cofactor would then serve as a “sink” to permit cleavage of the C1–C6 bond.
Scheme 4.
Two possible reaction pathways for breakdown of the (hydroperoxo)diiron(III/III) intermediate. The upper pathway involves O-O homolysis and rebound of the hydroxyl radical equivalent with the C1-centered substrate radical. The lower pathway involves Fe-O homolysis and rebound of the hydroperoxyl radical with the substrate radical.
12. The Three-Dimensional Structure of MIOX and its Mechanistic Implications
A major breakthrough in the understanding of MIOX came when the Baker group reported the three-dimensional structure of the mouse enzyme determined by X-ray crystallography.49 The structure revealed the overall protein fold, identified the iron ligands, and confirmed the direct coordination of MI to the cofactor (albeit in a mode distinct from the μ-O1 mode that we had speculated).48 Before summarizing the structure and how it impacts our thinking about the mechanism, we recall that Baker and co-workers speculated that their structure probably represents the inactive MIOX(III/III) • MI form. Precedent indicates that cluster reduction could be associated with ligand rearrangements (e.g., carboxylate shifts62–67) affecting cofactor and MI-coordination geometries in ways that would impact mechanistic analysis. However, a higher-resolution structure of the aforementioned partially active, N-terminally truncated form of human MIOX revealed very similar geometries (see Figure 1D).47 Crystals used in this study were obtained in the presence of 1 mM L-cysteine and 2 mM tris-(carboxyethyl)phospine and subsequently “swept through” a solution containing 1 mM L-cysteine. Given the ability of L-cysteine to reduce inactive MIOX(III/III) to MIOX(II/III), it is conceivable that this structure represents the mixed-valent form of the partially active, truncated human MIOX. If so, then the similarity of the active site geometries would suggest that the details deduced by Baker are essentially correct. In addition, our preliminary (unpublished) assessments of the binding of MI epimers to MIOX(II/III) have shown that their relative affinities are qualitatively consistent with the more stringent steric constraints on the C1, C2, C4 and C6 positions and the relatively less constrained environments of C3 and C5 observed by Baker and co-workers. Thus, the evidence weighs in favor of the accuracy of the Baker model for the active, mixed-valent form.
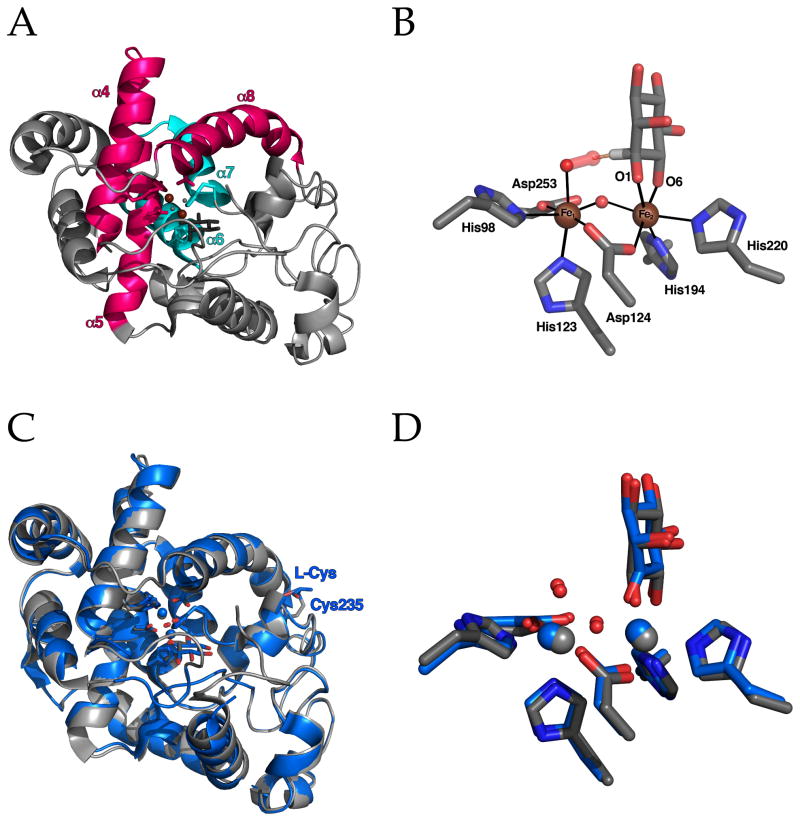
Figure 1.
(A) Ribbon diagram of the mouse MIOX fold showing helices 4, 5 and 8 (in pink) comprising the conserved HD domain structure that contributes four of the six Fe ligands and helices 7 and 8 (in blue) containing the remaining two unique histidine ligands that complete the Fe2 site. (B) Active site of mouse MIOX adapted from the structure solved by Baker and coworkers49 with superoxide modeled into the Fe1 site representing the superoxo-diiron(III/III) intermediate, G. The distance from the terminal oxygen atom of superoxide to C1 of MI is 1.92 Å. (C and D) Superposition of the structures of mouse and human enzymes (in grey and blue, respectively). Note that in the structure of human MIOX by Thorsell et al. the protein has a known inhibitor, myo-inosose-1, rather than MI bound and the active site and one molecule of L-cysteine, a known activator that reduces MIOX(III/III) state to MIOX(II/III), bound to the periphery of the enzyme in a disulfide with Cys235.47
12.1 Fold and Evolution
As was deduced from sequence analysis, MIOX is structurally unrelated to the conventional diiron oxygenase/oxidase proteins. Like these proteins, the MIOX diiron site is buried within a bundle of four anti-parallel α-helices (α4-α7) (Figure 1A), but the arrangement of the helices and the fact that a fifth helix contributes a ligand make the overall folds dissimilar. Indeed, the MIOX fold is not globally similar to that of any other structurally characterized protein. However, one of the two iron sub-sites (Fe1) is contained within an “HD domain,” a conserved divalent-metal-binding structure first recognized by Aravind and Koonin in metal-dependent phosphohydrolases.68 HD domains conform to the HX20–55HDX47–152D motif, which contributes the strictly conserved metal ligands, two histidines (H) and two aspartates (D), for a single metal site. In MIOX, these ligands are contained in α4, α5 and α8. Within an unusually long (127-residue) variable region between the two D ligands (X47–152) in MIOX are helices α6 and α7, which contribute the two histidine residues (H194 in α6 and H220 in α7) for the second iron sub-site (Fe2) that coordinates the substrate. Although helices corresponding to α6 and α7 are present in other HD domains, their orientations are more variable than those of α4, α5 and α8, and they do not generally provide metal ligands. Thus, it appears that MIOX evolved by elaboration of an HD cassette with two additional ligands that provide a second iron sub-site (Fe2) to form the coupled cluster.
12.2 The Diiron Site
In the conventional diiron oxygenase/oxidase proteins, the cluster site is constructed, in part, from the carboxylate and imidazole groups of tandem EXXH motifs.63, 67, 69–71 In crystal structures of the diiron(II/II) forms, the glutamates from these tandem motifs bridge the Fe ions, and each His imidazole coordinates a single iron.64, 65, 71 Each sub-site has an additional carboxylate ligand, resulting in a relatively symmetrical (His)2(Asp/Glu)4 overall architecture. The much greater asymmetry in the MIOX diiron site reflects its different mechanism for O2 and C-H activation (especially its use of the mixed-valent state of the cofactor). The Fe1 sub-site formed by the HD domain has a (His)2(Asp)2 composition, with Asp124 of the HD dyad for which the domain is named providing the only protein-derived bridge to the Fe2 sub-site (Figure 1B). The remaining ligands in the nearly octahedral Fe1 sphere are one terminal and one bridging solvent molecule. The Baker group assigned the terminal ligand as H2O and the bridging ligand tentatively as hydroxide. The Fe2 sub-site has a His2Asp composition that is reminiscent of the sites in several mononuclear nonheme-iron oxygenases (e.g., the α-ketoglutarate-dependent dioxygenases), which share what has been called the “(His)2(Asp/Glu) facial triad.72” The coordination sphere of Fe2 is completed by O1 and O6 of MI (see below) and the bridging hydroxide.
12.3 Substrate Binding and Coordination by the Diiron Cluster
By contrast to the μ-O1 substrate-binding mode that we had proposed on the basis of chemical considerations and the ENDOR data, the Baker structure showed MI coordinated only to Fe2 in an η2-(O1,O6) chelating mode with the substrate in its most stable chair conformation (O2 axial, O1 and O3-6 equatorial). The asymmetric disposition of the substrate suggested a satisfying division of labor between the two iron sub-sites: substrate activation by Fe2 and oxygen activation by Fe1. Baker and co-workers proposed just that, arguing that Fe1 should be the ferrous site in the active, mixed-valent form (recall that they speculated that the structure represents the inactive III/III state) and that the site trans to His123 of the HD sequence, occupied by the terminal solvent molecule in their structure, is the site to which O2 adds in formation of G. Their assignment seems reasonable also on the grounds that the Fe(III) ion should be the stronger Lewis acid and thus better suited to promote ionization of O1 to activate for C1-H cleavage (and possibly ionization of O6 to activate for C1–C6 cleavage).
The structure of the diiron site in the truncated human enzyme is very similar, but subtle differences from the earlier mouse MIOX structure could potentially reflect different oxidation states [Fe2(III/III) in the mouse MIOX versus Fe2(II/III) in the human MIOX]. Notably, the Fe Fe distance is 0.2 Å greater in the human MIOX structure, and both bridging ligands (Asp124 and the water/hydroxo) are more asymmetrically disposed toward Fe2, which is expected to be the Fe(III) site in the mixed-valent form. The most striking difference, however, is the assignment by the authors of the bound species as myo-inosose-1 (the 2-electron-oxidized 1-ketone form of MI), despite the fact that only MI was added in the crystallization. The authors suggested that this known MIOX inhibitor61 could have been formed during the long (~ 2-month) incubation required for crystal growth or could have been a contaminant in the commercial MI stock that they employed. An interesting possibility is that the inhibitor could have formed in the active site of the truncated protein as a consequence of derangement of the catalytic cycle by removal of the active-site-enclosing N-terminal loop. If so, the bound myo-inosose-1 could be an intermediate in the normal catalytic pathway (as originally proposed by Hamilton and co-workers61) or a breakdown product thereof. Mechanistic studies on the truncated protein could, therefore, potentially be informative. Regardless of how it formed, the presence of the ketone ligand, which should be capable of forming only a mono-anion and not the dianion that MI most likely forms in binding to Fe2, could be partly if not wholly responsible for the shorter bonds of the bridging ligands to Fe2 and consequent longer Fe Fe vector in the structure of the human variant. Thus, the oxidation state of the cofactor in the truncated human MIOX structure remains in doubt and the structure of a verifiably active, mixed-valent MIOX from any species remains an important objective.
12.4 Possible Mechanistic Implications
The mode of coordination of MI to the cofactor has important mechanistic implications. For example, the proposed transfer of the C1-H to the coordinated superoxide of G requires that the two be held in close mutual proximity. In addition, the hydroperoxylation pathway has the rather stringent geometric requirement for the Fe-coordinated O-atom of the O2 unit to come into sufficient proximity to C1 to be transferred after the other O-atom abstracts the C1-H. The hydroxylation path is, by comparison, less demanding because the O-atom that abstracts the C1-H itself “rebounds” to C1. If it is assumed that the structure of MIOX(II/III) • MI is not drastically different from the published crystal structures47, 49 and that the site-valence assignments of Baker and co-workers are correct, then several mechanistic inferences may be drawn. As noted by the Baker group, the pendant oxygen of the G superoxide coordinated end-on to Fe1 at the site occupied in their model by the terminal water/hydroxo would (with proper Fe-O-O angle and O-O vector) be ideally positioned to abstract the C1-H (Figure 1B).49 They proposed subsequent transfer of the resultant hydroperoxide from Fe1 to the C1 radical, but we view this step as improbable, because the distance from C1 to the Fe1-coordinated peroxide oxygen would (without significant structural rearrangement) be too great for direct hydroperoxyl-radical rebound. A two-step transfer initiated by Fe1-O homolysis or heterolysis seems extremely unlikely on the grounds that it would probably be associated with unproductive (and possibly deleterious) oxidative chemistry. Thus, the hydroxylation pathway would seem to be more likely than the hydroperoxylation pathway, if the active complex is not significantly different from the published structures. However, several other pathways from either the coordinated gem-diolate or a ketone intermediate61 can also be envisaged.
13. Conclusions and Outlook
Detailed kinetic and spectroscopic studies on MIOX,29–31, 47 made possible by the isolation of the gene by the Reddy group,73 have revealed that the enzyme employs an unprecedented chemical mechanism involving both substrate and O2 activation by a mixed-valent nonheme diiron cluster. A large substrate deuterium kinetic isotope effect on decay of the first detectable intermediate, G, implies that this formally mid-valent superoxo-diiron(III/III) complex abstracts hydrogen from the substrate to initiate its oxidative ring-cleavage.31 Details of the hydrogen-abstraction, ring-cleavage, and rate-determining steps are currently being elucidated. Understanding of the chemical mechanism will be greatly facilitated by insight into the structures of G and its successor, H. Such insight should be obtainable by multi-nuclear electron-nuclear double resonance (ENDOR and ESEEM) spectroscopies. As in studies on high-valent iron-oxo intermediates in other enzyme systems,74 inorganic models are expected to play important roles in defining the structures of the MIOX intermediates and the nature of C-H cleavage by the superoxo complex, G. The mechanisms of the enzyme’s univalent, oxidative and reductive activation and inactivation reactions remain completely unexplored. The relatively rapid activation of the fully reduced form by the co-substrate, O2, is an especially intriguing processes, as it is a one-electron oxidation by the four-electron oxidant and could potentially produce reactive oxygen intermediates (e.g., superoxide) as by-products. The mechanisms of these activation and inactivation reactions, including the impact of substrate and product binding thereupon, are under investigation.
References
- 1.Wallar BJ, Lipscomb JD. Chem Rev. 1996;96:2625–2657. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- 2.Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee SK, Lehnert N, Neese F, Skulan AJ, Yang YS, Zhou J. Chem Rev. 2000;100:235–349. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 3.Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Müller J, Lippard SJ. Angew Chem, Int Ed Engl. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 4.Fox BG, Lyle KS, Rogge CE. Acc Chem Res. 2004;37:421–429. doi: 10.1021/ar030186h. [DOI] [PubMed] [Google Scholar]
- 5.Krebs C, Price JC, Baldwin J, Saleh L, Green MT, Bollinger JM., Jr Inorg Chem. 2005;44:742–757. doi: 10.1021/ic048523l. [DOI] [PubMed] [Google Scholar]
- 6.Liu KE, Wang D, Huynh BH, Edmondson DE, Salifoglou A, Lippard SJ. J Am Chem Soc. 1994;116:7465–7466. [Google Scholar]
- 7.Liu KE, Valentine AM, Wang D, Huynh BH, Edmondson DE, Salifoglou A, Lippard SJ. J Am Chem Soc. 1995;117:10174–10185. [Google Scholar]
- 8.Bollinger JM, Jr, Krebs C, Vicol A, Chen S, Ley BA, Edmondson DE, Huynh BH. J Am Chem Soc. 1998;120:1094–1095. [Google Scholar]
- 9.Saleh L, Krebs C, Ley BA, Naik S, Huynh BH, Bollinger JM., Jr Biochemistry. 2004;43:5953–5964. doi: 10.1021/bi036099e. [DOI] [PubMed] [Google Scholar]
- 10.Murray LJ, Garcia-Serres R, Naik S, Huynh BH, Lippard SJ. J Am Chem Soc. 2006;128:7458–7459. doi: 10.1021/ja062762l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yun D, Garcia-Serres R, Chicalese BM, An YH, Huynh BH, Bollinger JM., Jr Biochemistry. 2007;46:1925–1932. doi: 10.1021/bi061717n. [DOI] [PubMed] [Google Scholar]
- 12.Bollinger JM, Jr, Stubbe J, Huynh BH, Edmondson DE. J Am Chem Soc. 1991;113:6289–6291. [Google Scholar]
- 13.Bollinger JM, Jr, Edmondson DE, Huynh BH, Filley J, Norton JR, Stubbe J. Science. 1991;253:292–298. doi: 10.1126/science.1650033. [DOI] [PubMed] [Google Scholar]
- 14.Sturgeon BE, Burdi D, Chen S, Huynh BH, Edmondson DE, Stubbe J, Hoffman BM. J Am Chem Soc. 1996;118:7551–7557. [Google Scholar]
- 15.Bollinger JM, Jr, Tong WH, Ravi N, Huynh BH, Edmondson DE, Stubbe J. J Am Chem Soc. 1994;116:8015–8023. [Google Scholar]
- 16.Burdi D, Sturgeon BE, Tong WH, Stubbe JA, Hoffman BM. J Am Chem Soc. 1996;118:281–282. [Google Scholar]
- 17.Willems JP, Lee HI, Burdi D, Doan PE, Stubbe J, Hoffman BM. J Am Chem Soc. 1997;119:9816–9824. [Google Scholar]
- 18.Riggs-Gelasco PJ, Shu LJ, Chen SX, Burdi D, Huynh BH, Que L, Stubbe J. J Am Chem Soc. 1998;120:849–860. [Google Scholar]
- 19.Burdi D, Willems JP, Riggs-Gelasco P, Antholine WE, Stubbe J, Hoffman BM. J Am Chem Soc. 1998;120:12910–12919. [Google Scholar]
- 20.Stubbe J. Curr Opin Chem Biol. 2003;7:183–188. doi: 10.1016/s1367-5931(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 21.Stubbe J, Nocera DG, Yee CS, Chang MCY. Chem Rev. 2003;103:2167–2202. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 22.Lee SK, Fox BG, Froland WA, Lipscomb JD, Munck E. J Am Chem Soc. 1993;115:6450–6451. [Google Scholar]
- 23.Shu LJ, Nesheim JC, Kauffmann K, Munck E, Lipscomb JD, Que L. Science. 1997;275:515–518. doi: 10.1126/science.275.5299.515. [DOI] [PubMed] [Google Scholar]
- 24.Lee SK, Nesheim JC, Lipscomb JD. J Biol Chem. 1993;268:21569–21577. [PubMed] [Google Scholar]
- 25.Charalampous FC, Lyras C. J Biol Chem. 1957;228:1–13. [PubMed] [Google Scholar]
- 26.Charalampous FC. J Biol Chem. 1959;234:220–227. [PubMed] [Google Scholar]
- 27.Charalampous FC. J Biol Chem. 1960;235:1286–1291. [PubMed] [Google Scholar]
- 28.Moskala R, Reddy CC, Minard RD, Hamilton GA. Biochem Biophys Res Commun. 1981;99:107–113. doi: 10.1016/0006-291x(81)91719-8. [DOI] [PubMed] [Google Scholar]
- 29.Xing G, Hoffart LM, Diao Y, Prabhu KS, Arner RJ, Reddy CC, Krebs C, Bollinger JM., Jr Biochemistry. 2006;45:5393–5401. doi: 10.1021/bi0519607. [DOI] [PubMed] [Google Scholar]
- 30.Xing G, Barr EW, Diao Y, Hoffart LM, Prabhu KS, Arner RJ, Reddy CC, Krebs C, Bollinger JM., Jr Biochemistry. 2006;45:5402–5412. doi: 10.1021/bi0526276. [DOI] [PubMed] [Google Scholar]
- 31.Xing G, Diao Y, Hoffart LM, Barr EW, Prabhu KS, Arner RJ, Reddy CC, Krebs C, Bollinger JM., Jr Proc Natl Acad Sci, USA. 2006;103:6130–6135. doi: 10.1073/pnas.0508473103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hankes LV, Politzer WM, Touster O, Anderson L. Ann NY Acad Sci. 1969;165:564–576. [PubMed] [Google Scholar]
- 33.Winegrad AI. Diabetes. 1987;36:396–406. doi: 10.2337/diab.36.3.396. [DOI] [PubMed] [Google Scholar]
- 34.Cohen RA, MacGregor LC, Spokes KC, Silva P, Epstein FH. Metabolism: clinical and experimental. 1990;39:1026–1032. doi: 10.1016/0026-0495(90)90161-5. [DOI] [PubMed] [Google Scholar]
- 35.Zhu X, Eichberg J. Proc Natl Acad Sci U S A. 1990;87:9818–9822. doi: 10.1073/pnas.87.24.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stribling D, Armstrong FM, Harrison HE. J Diabeties Complicat. 1989;3:70–76. doi: 10.1016/0891-6632(89)90015-9. [DOI] [PubMed] [Google Scholar]
- 37.Cohen AM, Wald H, Popovtzer M, Rosenmann E. Diabetologia. 1995;38:899–905. doi: 10.1007/BF00400577. [DOI] [PubMed] [Google Scholar]
- 38.Crabbe MJ, Goode D. Prog Retinal Eye Res. 1998;17:313–383. doi: 10.1016/s1350-9462(97)00013-x. [DOI] [PubMed] [Google Scholar]
- 39.Khandelwal M, Reece EA, Wu YK, Borenstein M. Teratology. 1998;57:79–84. doi: 10.1002/(SICI)1096-9926(199802)57:2<79::AID-TERA6>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 40.Sundkvist G, Dahlin LB, Nilsson H, Eriksson KF, Lindgarde F, Rosen I, Lattimer SA, Sima AA, Sullivan K, Greene DA. Diabetic Med. 2000;17:259–268. doi: 10.1046/j.1464-5491.2000.00261.x. [DOI] [PubMed] [Google Scholar]
- 41.Prabhu KS, Arner RJ, Vunta H, Reddy CC. J Biol Chem. 2005;280:19895–19901. doi: 10.1074/jbc.M502621200. [DOI] [PubMed] [Google Scholar]
- 42.Fraser MS, Hamilton GA. J Am Chem Soc. 1982;104:4203–4211. [Google Scholar]
- 43.Hamilton GA. In: Molecular Mechanisms of Oxygen Activation. Hayaishi O, editor. Academic; New York: 1974. pp. 405–451. [Google Scholar]
- 44.Reddy CC, Pierzchala PA, Hamilton GA. J Biol Chem. 1981;256:8519–8524. [PubMed] [Google Scholar]
- 45.Reddy CC, Swan JS, Hamilton GA. J Biol Chem. 1981;256:8510–8518. [PubMed] [Google Scholar]
- 46.Arner RJ, Prabhu KS, Thompson JT, Hildenbrandt GR, Liken AD, Reddy CC. Biochem J. 2001;360:313–320. doi: 10.1042/0264-6021:3600313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorsell AG, Persson C, Voevodskaya N, Busam RD, Hammarström M, Gräslund S, Graslund A, Hallberg BM. J Biol Chem. 2008;283:15209–15216. doi: 10.1074/jbc.M800348200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SH, Xing G, Bollinger JM, Jr, Krebs C, Hoffman BM. J Am Chem Soc. 2006;128:10374–10375. doi: 10.1021/ja063602c. [DOI] [PubMed] [Google Scholar]
- 49.Brown PM, Caradoc-Davies TT, Dickson JMJ, Cooper GJS, Loomes KM, Baker EN. Proc Natl Acad Sci, U S A. 2006;103:15032–15037. doi: 10.1073/pnas.0605143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price JC, Barr EW, Hoffart LM, Krebs C, Bollinger JM., Jr Biochemistry. 2005;44:8138–8147. doi: 10.1021/bi050227c. [DOI] [PubMed] [Google Scholar]
- 51.Baldwin J, Krebs C, Ley BA, Edmondson DE, Huynh BH, Bollinger JM., Jr J Am Chem Soc. 2000;122:12195–12206. [Google Scholar]
- 52.Price JC, Barr EW, Tirupati B, Bollinger JM, Jr, Krebs C. Biochemistry. 2003;42:7497–7508. doi: 10.1021/bi030011f. [DOI] [PubMed] [Google Scholar]
- 53.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Jr, Krebs C. Proc Natl Acad Sci, U S A. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galonic DP, Barr EW, Walsh CT, Bollinger JM, Jr, Krebs C. Nat Chem Biol. 2007;3:113–116. doi: 10.1038/nchembio856. [DOI] [PubMed] [Google Scholar]
- 55.Halfen JA, Mahapatra S, Wilkinson EC, Kaderli S, Young VG, Que L, Zuberbuhler AD, Tolman WB. Science. 1996;271:1397–1400. doi: 10.1126/science.271.5254.1397. [DOI] [PubMed] [Google Scholar]
- 56.Bollinger JM, Jr, Krebs C. Curr Opin Chem Biol. 2007;11:151–158. doi: 10.1016/j.cbpa.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 57.Shan X, Que L., Jr Proc Natl Acad Sci USA. 2005;102:5340–5345. doi: 10.1073/pnas.0409640102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans JP, Ahn K, Klinman JP. J Biol Chem. 2003;278:49691–49698. doi: 10.1074/jbc.M300797200. [DOI] [PubMed] [Google Scholar]
- 59.Chen P, Solomon EI. J Am Chem Soc. 2004;126:4991–5000. doi: 10.1021/ja031564g. [DOI] [PubMed] [Google Scholar]
- 60.Baldwin JE, Bradley M. Chem Rev. 1990;90:1079–1088. [Google Scholar]
- 61.Naber NI, Swan JS, Hamilton GA. Biochemistry. 1986;25:7201–7211. doi: 10.1021/bi00370a065. [DOI] [PubMed] [Google Scholar]
- 62.Rardin RL, Tolman WB, Lippard SJ. New J Chem. 1991;15:417–430. [Google Scholar]
- 63.Nordlund P, Eklund H. J Mol Biol. 1993;232:123–164. doi: 10.1006/jmbi.1993.1374. [DOI] [PubMed] [Google Scholar]
- 64.Logan DT, Su XD, Aberg A, Regnstrom K, Hajdu J, Eklund H, Nordlund P. Structure. 1996;4:1053–1064. doi: 10.1016/s0969-2126(96)00112-8. [DOI] [PubMed] [Google Scholar]
- 65.Rosenzweig AC, Nordlund P, Takahara PM, Frederick CA, Lippard SJ. Chemistry & Biology. 1995;2:409–418. [PubMed] [Google Scholar]
- 66.Voegtli WC, Sommerhalter M, Saleh L, Baldwin J, Bollinger JM, Jr, Rosenzweig AC. J Am Chem Soc. 2003;125:15822–15830. doi: 10.1021/ja0370387. [DOI] [PubMed] [Google Scholar]
- 67.Nordlund P, Eklund H. Curr Opin Struct Biol. 1995;5:758–766. doi: 10.1016/0959-440x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 68.Aravind L, Koonin EV. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 69.Fox BG, Shanklin J, Somerville C, Munck E. Proc Nat Acad Sci USA. 1993;90:2486–2490. doi: 10.1073/pnas.90.6.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- 71.Lindqvist Y, Huang WJ, Schneider G, Shanklin J. EMBO Journal. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- 72.Koehntop KD, Emerson JP, Que L., Jr J Biol Inorg Chem. 2005;10:87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 73.Arner RJ, Prabhu KS, Reddy CC. Biochem Biophys Res Commun. 2004;324:1386–1392. doi: 10.1016/j.bbrc.2004.09.209. [DOI] [PubMed] [Google Scholar]
- 74.Shan X, Que L., Jr J Inorg Biochem. 2006;100:421–433. doi: 10.1016/j.jinorgbio.2006.01.014. [DOI] [PubMed] [Google Scholar]