Abstract
Background
Syncytins are envelope genes of retroviral origin that have been co-opted by the host to mediate a specialized function in placentation. Two of these genes have already been identified in primates, as well as two distinct, non orthologous genes in rodents.
Results
Here we identified within the rabbit Oryctolagus cuniculus-which belongs to the lagomorpha order- an envelope (env) gene of retroviral origin with the characteristic features of a bona fide syncytin, that we named syncytin-Ory1. An in silico search for full-length env genes with an uninterrupted open reading frame within the rabbit genome first identified two candidate genes that were tested for their specific expression in the placenta by quantitative RT-PCR of RNA isolated from a large set of tissues. This resulted in the identification of an env gene with placenta-specific expression and belonging to a family of endogenous retroelements present at a limited copy number in the rabbit genome. Functional characterization of the identified placenta-expressed env gene after cloning in a CMV-driven expression vector and transient transfection experiments, demonstrated both fusogenic activity in an ex vivo cell-cell fusion assay and infectivity of pseudotypes. The receptor for the rabbit syncytin-Ory1 was found to be the same as that for human syncytin-1, i.e. the previously identified ASCT2 transporter. This was demonstrated by a co-culture fusion assay between hamster A23 cells transduced with an expression vector for ASCT2 and A23 cells transduced with syncytin-Ory1. Finally, in situ hybridization of rabbit placenta sections with a syncytin-Ory1 probe revealed specific expression at the level of the junctional zone between the placental lobe and the maternal decidua, where the invading syncytial fetal tissue contacts the maternal decidua to form the labyrinth, consistent with a role in the formation of the syncytiotrophoblast. The syncytin-Ory1 gene is found in Leporidae but not in Ochotonidae, and should therefore have entered the lagomorpha order 12-30 million years ago.
Conclusion
The identification of a novel syncytin gene within a third order of mammals displaying syncytiotrophoblast formation during placentation strongly supports the notion that on several occasions retroviral infections have resulted in the independent capture of genes that have been positively selected for a convergent physiological role.
Background
Previous studies have identified two pairs of envelope (env) genes of retroviral origin that have been independently captured by their host for a role in placentation. In simians, syncytin-1 [1-3] and syncytin-2 [4,5] entered the primate genome 25 and >40 million years (My) ago, respectively. They retained their coding capacity in all the subsequent branches; syncytin-1 and syncytin-2 display placenta-specific expression, are fusogenic in ex vivo cell-cell fusion assays, and one of them displays immunosuppressive activity [6]. A pair of env genes from endogenous retroviruses (ERVs) were then identified in the mouse, named syncytin-A and -B, which share closely related functional properties although they have a completely distinct origin, showing a divergent sequence and a different genomic location compared to primate syncytins [7]. As found for the latter, syncytin-A and -B have the status of bona fide genes. They have been conserved since their entry into the Muridae genome, approximately 20 million years (My) ago; they display placenta-specific expression, mediate cell-cell fusion in ex vivo assays [7], and one of them is immunosuppressive [6]. Recently we have further unambiguously demonstrated via the generation of syncytin-A knockout mice that these genes are indeed essential for placentation, with a lack of cell-cell fusion observed in vivo at the level of the placenta of the knockout embryos, resulting in impaired maternal-fetal exchanges and death of the embryos at mid-gestation [8]. Therefore, it appears that on some occasions in the course of mammalian evolution, env genes from endogenous retroviruses have been "co-opted" by their host to participate in the formation of the syncytiotrophoblast layer, at the maternal-fetal interface, by mediating the fusion of mononucleated cytotrophoblasts.
A further question that we wanted to answer was whether mammals belonging to orders other than rodents and primates but possessing a placenta with a related architecture, i.e. with a syncytiotrophoblast layer in direct contact with maternal blood at the maternal-fetal interface, have also "captured" retroviral env genes to generate, in a convergent manner, this specific placental structure. Among mammals whose placenta displays such a structural organization at the maternal-fetal interface, i.e. with a haemochorial placenta, the lagomorpha order was selected over the two other orders which also possess a haemochorial placenta -i.e. the Insectivora (hedgehog) and Chiroptera (bats) [9]; this was done because one of its representatives, the rabbit (Oryctolagus cuniculus), has an already sequenced genome, and can be reared and investigated easily at different stages of gestation, and has a placental physiology that has been appropriately described [9-11].
Here, by combining in silico search for env genes within the rabbit genome, RT-PCR assays for their in vivo transcriptional activity in a large panel of tissues including the placenta, cloning of the candidate genes, ex vivo assays for their fusogenicity and, ultimately, in situ hybridization of placenta sections, we identify a new fusogenic and placenta-specific endogenous env gene, displaying all the characteristic features of a bona fide syncytin gene, that we named syncytin-Ory1. Although we demonstrate that the syncytin-Ory1 protein shares the same ASCT2 receptor in common with human syncytin-1, it is divergent from all four syncytins previously identified in rodents and primates and must therefore have been captured independently from a distinct ancestral retrovirus. The occurrence in a third order of Mammals of a new syncytin gene that is specifically expressed at the maternal-fetal interface within the placental junctional zone provides strong support to the notion that ERVs have played a convergent role in the recurrent emergence of syncytiotrophoblast-containing haemochorial placentae in the course of evolution.
Results and Discussion
In silico search for retroviral env genes within the rabbit (Oryctolagus cuniculus) genome
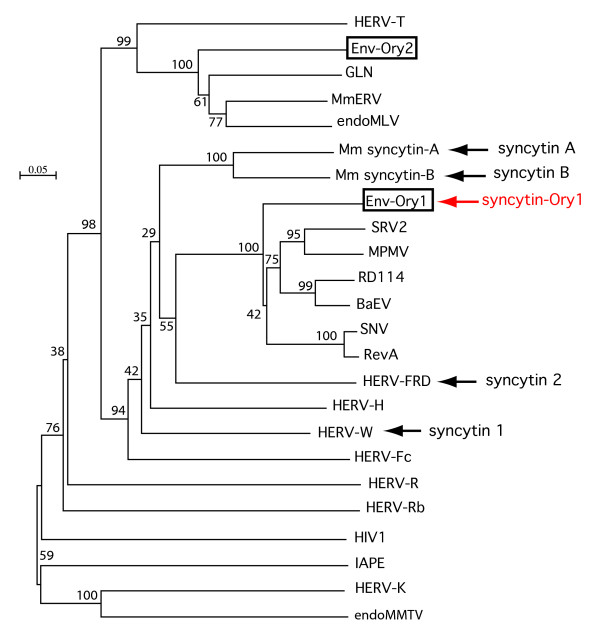
To identify putative env-derived syncytin genes, we made use of the available rabbit genome sequence (low coverage 2× assembly of the Oryctolagus cuniculus genome, Ensembl May 2005 assembly, updated version 49) and of the method that we previously devised to screen the whole human and mouse genomes for such genes [7,12]. Basically, it makes use of the degenerate CKS17u consensus motif, associated with the immunosuppressive domain of retroviral envelope proteins, and is designed to match the majority of env genes of exogenous and endogenous origin [12]. Rabbit sequences from the Ensembl database were screened with this motif using the BIOMOTIF program, and only sequences with open reading frames (ORFs) longer than 1.5 kb were considered. Five ORF-containing sequences were obtained, four of which disclosed >98% nt identity. We named these four sequences Env-Ory1. The fifth sequence that was obtained was unrelated to Env-Ory1. We named this sequence Env-Ory2 (Figure 1). Analysis of the scaffold database identified the former ORF as belonging to a low-copy family of ERVs, most of which had an env gene that was interrupted by stop codons, deletions and/or truncations. Due to the low coverage of the available assembly, it could not be determined whether the identified ORF corresponds to distinct loci or to a single locus - in that case with distinct alleles.
Figure 1.
Retroviral envelope protein-based phylogenic tree with positions of the identified rabbit Env-Ory1- and Env-Ory2, and of the human and mouse syncytins. The tree was determined by the neighbor-joining method using envelope TM subunit sequences (see ref [12]) from murine and human ERVs, and infectious retroviruses. The horizontal branch length and the scale indicate the percentage of amino acid substitutions from the node. Percent bootstrap values obtained from 1,000 replicates are indicated.
Transcription profile and identification of a placenta-specific envelope
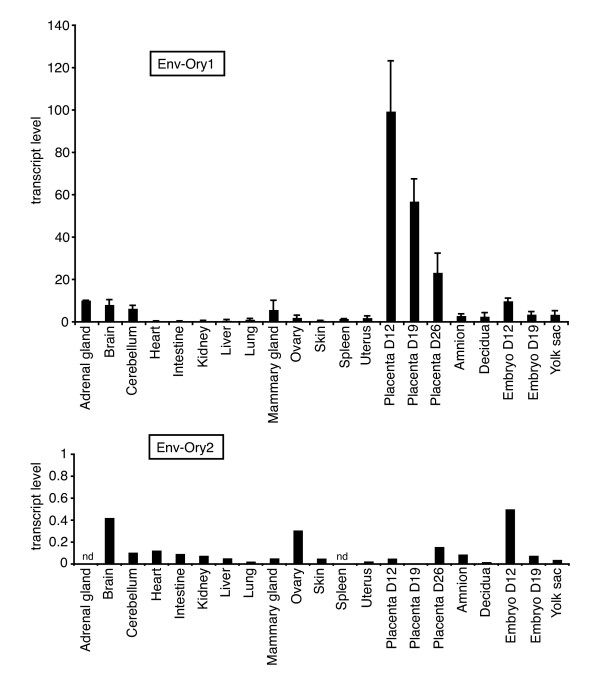
Quantitative RT-PCR analysis of transcript levels for the two identified candidate syncytins was performed using primers specific for each family of elements. As illustrated in Figure 2, the Env-Ory1-encoding gene has the characteristic profile of a bona fide syncytin gene, with high levels of expression in the placenta and very limited expression in other tissues. Expression in the placenta decreases with gestational age, with a four-fold reduction from day 12 to 26 (i.e. 4 days before delivery). The other candidate gene encoding Env-Ory2 is expressed only limitedly (at least 100-fold lower), with no specific expression in the placenta, and was not considered further.
Figure 2.
Real-time quantitative RT-PCR analysis of Env-Ory1 and Env-Ory2 transcripts in rabbit tissues. Transcript levels were normalized relative to the amount of 18S rRNA (arbitrary units). At least 3 samples per organ type were analyzed for Env-Ory1 (from different adult animals for non-fetal tissues; from a given litter for the embryos and placentae); one sample per organ type was analyzed for Env-Ory2.
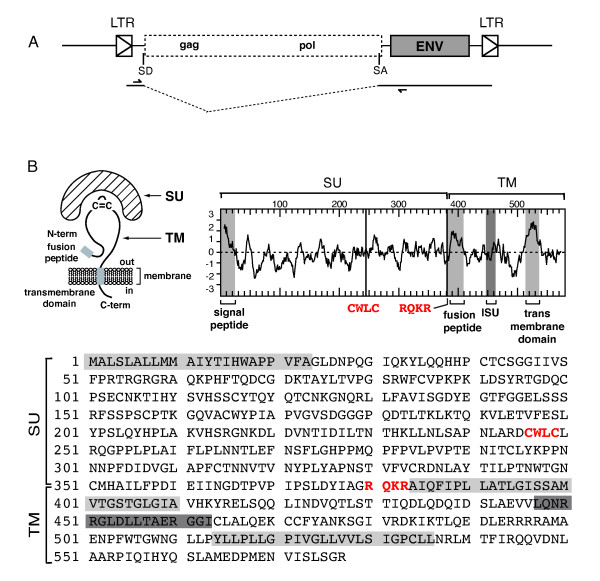
Some of the sequence scaffolds reveal the Env-Ory1-encoding gene to be part of a proviral structure with degenerate but identifiable LTR, gag and pol gene sequences. A phylogenetic tree based on the envelope TM subunit (Figure 1) shows that this envelope protein is related to that of type-D retroviruses such as MPMV, BaEV and RD114, as similarly observed on the basis of a pol-based tree (not shown). A putative donor and acceptor splice site for the generation of a subgenomic env transcript can be identified according to http://www.cbs.dtu.dk/services/NetGene2, as classically observed for retroviruses. Their position and functionality were further ascertained by RT-PCR analysis of Env-Ory1-encoding transcripts in the placenta, using appropriate primers (see Methods and Figure 3).
Figure 3.
Characterization of Env-Ory1. (A) Schematic representation of the Env-Ory1-associated ERV with the LTRs and the splice sites for the sub-genomic env transcript indicated. (B) Schematic structure, hydrophobicity profile and primary sequence of the Env-Ory1 glycoprotein (deposited in GenBank [GenBank:GU196371]). The SU and TM subunits of the envelope protein are delineated, with a canonical furin cleavage site (RQKR; consensus: R/K-N-R/K-R) between the two subunits and the CWLC domain involved in SU-TM interaction indicated in red; the hydrophobic signal peptide and fusion peptide and the transmembrane domain are shaded in light gray, and the putative immunosuppressive domain (ISU) in dark gray.
Analysis of the amino-acid sequence of Env-Ory1 (consensus in Figure 3 of the four sequences in the database, also corresponding to that PCR-amplified, see below) displays the characteristic features of retroviral envelope proteins, with a putative signal peptide at the N-terminus, and a furin cleavage site (RQKR, consensus R/K-X-R/K-R) at amino acid 380 to generate the SU and TM subunits. The hydrophobicity plot of the TM subunit reveals a putative fusion peptide at the TM N-terminus, and a hydrophobic transmembrane domain.
Assay for the fusogenic activity of Env-Ory1
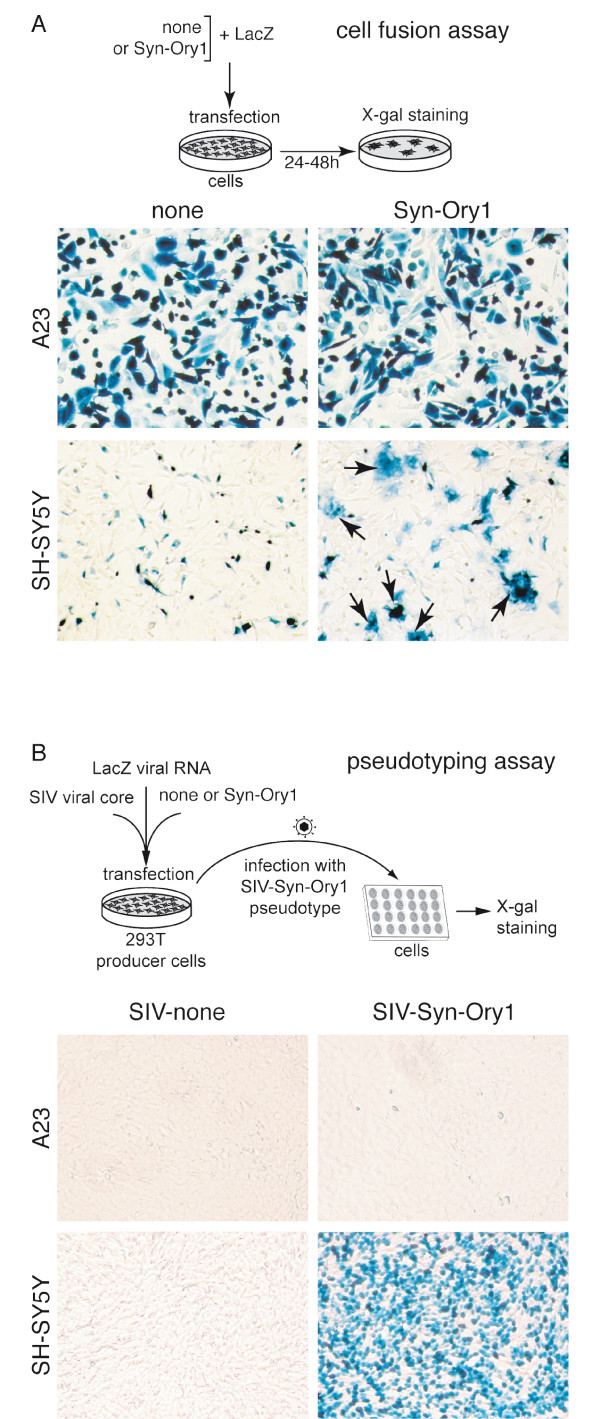
Fusogenic activity of Env-Ory1 was assayed as previously described [4,5,7], by ex vivo assays in cells in culture for both detection of syncytia formation (cell-cell membrane fusion) and generation of infectious pseudotypes (virus-cell membrane fusion). The Env-Ory1-encoding sequence was first PCR-amplified from genomic DNA of Oryctolagus cuniculus and inserted into a CMV promoter-containing expression vector (see Methods). The plasmids were sequenced and those containing a full-length env gene ORF (that were actually >99.7% identical, and therefore most probably corresponding to a single sequence element) were assayed. As illustrated in Figure 4A, transient transfection of human SH-SY5Y neuroblastoma cells with the Env-Ory1-expressing vector triggers cell-cell fusion, as expected for a bona fide syncytin expressed in a cell line carrying its cognate receptor. Cell-cell fusion is not observed with the hamster A23 cells, which were therefore used as a control in the following assays for receptor identification.
Figure 4.
Fusogenic activity of syncytin-Ory1. (A) Assay for cell-cell fusion mediated by syncytin-Ory1. The indicated cell lines were transfected with an expression vector for syncytin-Ory1 or an empty vector (none) together with a LacZ expression vector. Cells were cultured for 1-2 days after transfection, fixed and X-gal-stained. Syncytia (arrows) were detected in syncytin-Ory1-transfected SH-SY5Y cells, with only mononucleated cells visible in the other cases. (B) Assay for cell infection mediated by syncytin-Ory1-pseudotyped virus particles. Pseudotypes were produced by cotransfection of human 293T cells with expression vectors for the SIV core, the syncytin-Ory1 protein (or an empty vector) and a LacZ-containing retroviral transcript. Supernatants were used to infect the indicated target cells, which were X-gal stained 3 days after infection. Abbreviation: Syn-Ory1, syncytin-Ory1.
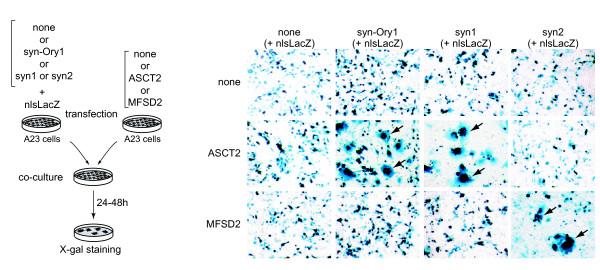
Env-Ory1 can also form infectious pseudotypes, as expected from its retroviral origin. As illustrated in Figure 4B, pseudotypes generated with an SIV core are able to infect SH-SY5Y cells that are positive in the cell-cell fusion assay above, but not A23 cells. The profile of cells positive for infection was found to be similar to that observed for human syncytin-1 (data not shown), thus suggesting that Env-Ory1 could possibly use the same receptor (i.e. the neutral amino acid trasporter ASCT2, [2]). This point was investigated in the experiments illustrated in Figure 5 in which distinct pools of A23 cells transfected with either an ASCT2 or an Env-Ory1 expression vector (supplemented with a β-galactosidase expression vector) were mixed and assayed for cell-cell fusion. As shown in the figure, cell-cell fusion (as revealed by the presence of large LacZ+ syncytia) could be observed with the Env-Ory1 and ASCT2 pair (as well as with the ASCT2/syncytin-1 and MFSD2/syncytin-2 [13] pairs, used as positive controls) but not with any of the other combinations. This strongly suggests that Env-Ory1 uses the ASCT2 receptor, as does human syncytin-1. It is rather unexpected for two independently acquired - and distinct - retroviral envelopes to use the same cellular receptor. ASCT2 seems, however, to be a rather "successful" receptor being also the one used by a series of type-D infectious retroviruses, such as the primate MPMV, feline RD114, and avian SNV viruses [14,15] whose Env actually clusters with Env-Ory1 in phylogenic trees (Figure 1 for the TM domain and data not shown for SU). Finally, database screening indeed reveals the presence of an ASCT2 gene in the rabbit genome (mRNA accession number NM_001082378; 85% amino-acid identity with human ASCT2), and qRT-PCR demonstrates its expression in the rabbit placenta (data not shown). In conclusion, Env-Ory1 can be considered as a bona fide syncytin owing to its fusogenic activity and specific expression in the placenta, and be named Syncytin-Ory1.
Figure 5.
Fusion assay between ASCT2-transduced and syncytin-Ory1-transduced co-cultured cells demonstrates that ASCT2 is the syncytin-Ory1 receptor. Left panel: Cell-cell fusion was assayed upon independent transfections of a set of A23 cells with an empty vector (none) or an expression vector for either the syncytin-Ory1, syncytin-1 or syncytin-2 protein together with an nls-LacZ gene-expression vector, and another set of A23 cells with an expression vector for the syncytin-1 receptor ASCT2, the syncytin-2 receptor MFSD2 [13] or an empty vector (none). One day after transfection, cells were resuspended and pairs of transfected cells from each set were cocultured for 1-2 days, fixed and X-Gal stained. Right panel: Syncytia can be easily detected (arrows) for the syncytin-Ory1/ASCT2, syncytin-1/ASCT2 and syncytin-2/MFSD2 pairs, with only mononucleated cells visible in the other cases. Abbreviations: syn-Ory1, syncytin-Ory1; syn1, syncytin-1; syn2, syncytin-2.
In situ hybridization of placenta sections
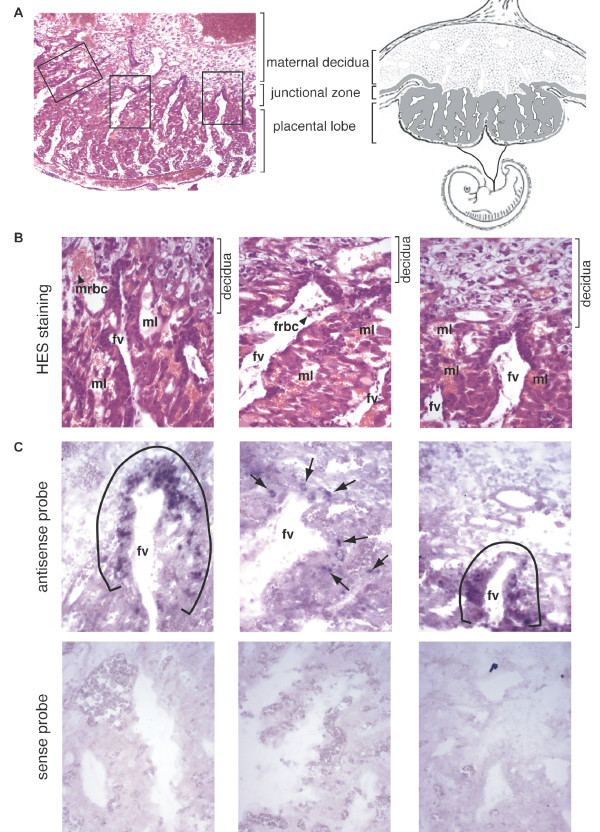
To further assess the physiological relevance of syncytin-Ory1 expression in the placenta, in situ hybridization experiments were performed on paraffin sections of placenta at day 12 of gestation, i.e. the stage showing maximum expression of syncytin-Ory1 by qRT-PCR (Figure 2). Figure 6A shows the representative architecture of rabbit placenta at day 12. Three main zones can be distinguished. The maternal decidua results from the modification of the uterus after implantation, and the placental lobe consists in a labyrinthine structure where fetal-maternal exchanges take place. In the labyrinth, fetal and maternal blood circulations are separated by 2 layers of trophoblasts (haemodichorial placenta). Between the decidua and placental lobe, a junctional zone can be observed where fetal vessels surrounded by invading fetal tissue contact the maternal decidua to form the labyrinth (Figure 6A, B). The invading fetal tissue has been described as a broad syncytial front toward the decidua, backed by and presumably formed from cellular cytotrophoblasts [9]. Thus, as soon as the fetal processes reach the maternal blood spaces, a haemodichorial structure is formed, with both a cellular and a syncytial trophophoblast layer separating maternal and fetal blood spaces (maternal lacuna, ml, and fetal vessels, fv). All of these characterize the definitive labyrinthine placenta [9,11].
Figure 6.
Structure and in situ hybridization for syncytin-Ory1 expression of day 12 rabbit placenta: (A) Schematic representation of a rabbit placenta (right) and haematoxylin and eosin staining of a day 12 placenta section (left) with the 3 main layers of the placenta indicated. (B) Higher magnification of the areas framed in A. Abbreviations: frbc: fetal red blood cell, fv: fetal blood vessel, ml: maternal blood lacuna, mrbc: maternal red blood cell. (C) In situ hybridization on sections of a day 12 rabbit placenta (serial sections of the HES in B) with digoxigenin-labeled syncytin-Ory1 sense (lower panel, negative control) and antisense (upper panel) riboprobes, revealed with an alkaline phosphatase-conjugated anti-digoxigenin antibody. Brackets and arrows highlight the positive labeling of trophoblast cells surrounding the invading fetal vessels in the junctional zone.
A specific digoxigenin-marked antisense probe was synthesized for syncytin-Ory1 transcript detection, as well as the corresponding sense probe as a negative control. As shown in Figure 6C, specific labeling was observed only with the antisense probe (upper panels), but not with the control probe (lower panels). Syncytin-Ory1 expression is restricted to the junctional zone, where its expression (of variable intensity depending on the area, even within the same placental section) is limited to trophoblast cells surrounding the invading fetal vessels (brackets for heavily marked zones and arrows for punctuated domains in Figure 6C). Although we cannot formally discriminate between the syncytial trophoblast and the cellular cytotrophoblasts, the labeling profile is compatible with syncytin-Ory1 being expressed in the cytotrophoblast just before fusion takes place and/or in the newly formed syncytiotrophoblast (no labeling was detected in the placental lobe outside the junctional zone). These observations are consistent with a role for syncytin-Ory1 in the formation of the syncytiotrophoblast.
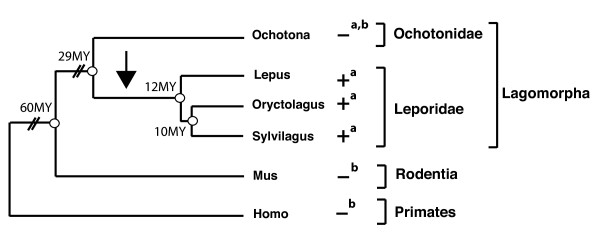
Syncytin-Ory1 sequences are present in Leporidae but not in Ochotonidae
Phylogenetic relationships in the order of Lagomorpha are illustrated in Figure 7 (adapted from [16]). The order includes 2 families, Ochotonidae (pikas) and Leporidae (hares and rabbits). We searched for syncytin-Ory1 genes in lagomorph species belonging to the Ochotonidae family (Ochotona princeps) and the Leporidae family (genus Lepus: Lepus americanus, Lepus europaeus and Lepus starcki; genus Sylvilagus: Sylvilagus brasiliensis, Sylvilagus floridanus). The domestic rabbit, Oryctolagus cuniculus, analyzed in this study, belongs to the genus Oryctolagus within the Leporidae.
Figure 7.
Putative entry date of syncytin-Ory1 during lagomorph evolution. Schematized phylogenetic tree with the evolutionary timeline of four lagomorph genus (adapted from [16]) and the rodent and primate outgroups depicted, with average divergence times indicated for the nodes. The presence of syncytin-Ory1 sequences in each genus, detected either by PCR experiments (a) or database screening (b), is indicated on the right.
Genomic DNA from these different species was PCR-amplified using a pair of primers for the syncytin-Ory1 ORF. A 1900 bp specific amplicon could be obtained from the 3 Lepus species, but neither from the Sylvilagus nor from the Ochotona genus members tested. However, a pair of primers internal to the ORF allowed the amplification of a 1430 bp PCR product from the 2 Sylvilagus species analyzed. Again, no specific amplification could be obtained from the Ochotona princeps genomic DNA. The PCR products obtained from Lepus and Sylvilagus were cloned and sequenced and showed >88% nucleotide identity with the Oryctolagus cuniculus syncytin-Ory1 gene sequence, and therefore most probably correspond to amplification of the same genes.
The absence of a syncytin-Ory1 gene in the Ochotona (as well as in the more ancestrally diverged rodent (mouse) and primate (human) orders) was confirmed by screening (using the BLAST program) the corresponding Ensembl databases.
Conclusively, the identification of syncytin-Ory1 genes in Lepus and Sylvilagus in addition to Oryctolagus indicates that gene capture most probably occurred prior to the divergence of the Lepus and Oryctolagus/Sylvilagus genera ~12 My ago [16]. The absence of a syncytin-Ory1 gene in Ochotona princeps suggests that this integration took place after the Leporidae and Ochotonidae divergence ~30 My ago [16].
Conclusion
Here we have shown that within a placental mammal that has developed a haemochorial placenta with a syncytiotrophoblast layer at the maternal-fetal interface, such as the rabbit, a gene of retroviral origin can be identified which displays all the characteristic features of a bona fide syncytin gene. The identified syncytin-Ory1 has a cell-cell fusion activity and is expressed specifically in the placenta at a location consistent with a direct role in syncytiotrophoblast formation. syncytin-Ory1 can be found in a series of leporidae. Notably, the identified env gene is divergent from any other previously described genes. Thus, for a newly investigated mammalian order, we provide evidence that a retroviral env gene has been captured and has gained the status of a "syncytin" according to a process similar to that previously observed in the two other major orders where haemochorial placentae have emerged: the primates (syncytin-1 and -2) and the rodents (syncytin-A and -B).
It is therefore likely that such gene captures have arisen recurrently during the evolution of placental mammals and that syncytin genes will be found in other placental mammals displaying a syncytiotrophoblast organization. Ongoing investigations carried out on representative animals of the Carnivora order strongly support this hypothesis. An interesting question which remains to be answered is to determine which specific properties of the captured syncytins are responsible for the relative diversity observed in the physiology of mammalian placentation. Among species with hemochorial placentation in particular, most of the species (including human, rabbit and most rodents) have a single layer of syncytiotrophoblast, whereas a few others (Muridae) have a two-layered syncytiotrophoblast. Moreover, it will be of interest to determine whether placental mammals [such as Suidae (pig) or Equidae (horse)] which do not possess a syncytiotrophoblast layer at their maternal-fetal interface, are deprived of syncytins, or whether other functions of retroviral envelope proteins such as their immunosuppressive activity have driven the capture of a non-fusogenic syncytin-like gene. In this case, retroviral envelope proteins would be co-opted solely for an immunological role in relation with maternal-fetal tolerance. Experiments in progress with knock-in mice where syncytin genes have been mutated for immunosuppressive activity without impairment of fusogenicity may also help answer these questions.
Methods
Database screening and sequence analyses
Retroviral env gene sequences were searched by using the Biomotif program http://www.lpta.univ-montp2.fr/users/menes/bioMotif_html_doc/ref_Run.html and the degenerate universal CKS17u consensus motif [12] as a query. We made use of the available rabbit genome sequence (low coverage 2× assembly of the Oryctolagus cuniculus genome, Ensembl RABBIT May 2005 assembly, updated version 49). ORF-containing scaffolds (scaffold number and ORF-coordinates indicated) were: 131951(25124:26887), 15025(429:2192 reverse strand), 90528(17499:19256), 163359(7744:9486 reverse strand) for Env-Ory1, and 1093(1017:3035 reverse strand) for Env-Ory2. Multiple alignments were carried out by using the CLUSTALW program http://bioinfo.hku.hk/services/analyseq/cgi-bin/clustalw_in.pl. Phylogenic trees were constructed from alignments by using the neighbor-joining program within CLUSTALW and were viewed with the NJPLOT program.
The Ochotona princeps (Pika) genome low coverage 1.93× assembly (Ensembl OchPri2.0 June 2007, updated version 53), as well as of the human and mouse assemblies (from the Genome Reference Consortium) were also screened for the presence of the identified rabbit Env-Ory1 ORF sequence, using the BLAST programs at the National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov/BLAST.
Real-time RT-PCR
Env-Ory1 and Env-Ory2 mRNA expression was determined by real-time quantitative RT-PCR. Pregnant New Zealand white rabbit females obtained from INRA (Jouy-en-Josas, France) at various stages of gestation were sacrificed, and dissected organs were stored in liquid nitrogen. Total RNA was extracted from the frozen organs using the RNeasy RNA isolation kit (Qiagen). Reverse transcription was performed with 1 μg of DNase-treated RNA as in [17]. Real-time qPCR was with 5 μl of diluted (1:10) cDNA in a final volume of 25 μl by using SYBR Green PCR Master Mix (or Taqman Universal PCR Master Mix for 18S rRNA detection) (Applied Biosystems). PCR was carried out using an ABI PRISM 7000 sequence detection system. Primer sequences were as follows: 5'-GCTGTTTTTATGCTAACAAGTCC and 5'-GATAAAGGTCATCAGCCTATTGA for Env-Ory1 and 5'-CCTCTAAATGTCATCTTCACCAG and 5'-CTATTGGGACAGCAGTTCTAGTC for Env-Ory2,. The transcript levels were normalized relative to the amount of 18S rRNA (as determined with the primers and Taqman probe from Applied Biosystems). Samples were assayed in duplicate.
Syncytin-Ory1 expression vector
The syncytin-Ory1 expression vector was constructed as follows: syncytin-Ory1 was PCR-amplified from genomic DNA of New Zealand white rabbit using the Accuprime DNA polymerase (Invitrogen) for 30 cycles. XhoI-containing primer sequences were: 5'-ATCACCTCGAGTGCTGGAATTGTTGTCATTGTTG and 5'-ATCACCTCGAGCGTCATTGGCTTACTGCTCATTT. After restriction with XhoI, the PCR product was cloned into the phCMV-G vector (GenBank accession AJ318514, gift F.-L. Cosset) opened with XhoI. Constructs were verified by sequencing.
Cell fusion assay
Cell lines described in [13,18] were grown in DMEM medium supplemented with 10% fetal calf serum (Invitrogen).
For the self-fusion assay, cells seeded at 104 - 5 × 104 cells per well in 24-well plates were transfected by using the Lipofectamine kit (Invitrogen) with 0.2 μg of either the syncytin-Ory1 expressing or an empty vector, supplemented with 0.2 μg of a LacZ-expression vector (pCMV-β, Clontech). Syncytia were visualized by X-Gal staining 24 to 48 h after coculture.
For cell-cell fusion by the coculture assay, A23 cells were seeded at 5 × 105 cells per 60-mm dish. A set of dishes were transfected by using the Lipofectamine LTX kit (Invitrogen) with 5 μg of either an ASCT2 or an MFSD2 expression vector [13] or an empty vector, and another set were transfected with 2.5 μg of either a syncytin-Ory1 or a syncytin-1 or a syncytin-2 expression vector or an empty vector, each cotransfected with 2.5 μg of the nls-LacZ expression vector (R9SA, [19]). One day after transfection, 3.5 × 105 cells from each group of transfected cells were cocultured in 6-well plates. Syncytia were visualized by X-Gal staining 24 to 48 h after coculture.
Pseudotyping assay
SIV virions pseudotyped with syncytin-Ory1 were produced by cotransfecting 8 × 105 293T cells with: 2.25 μg pSIV3+ (encoding SIV retroviral proteins except Env) [20]; 2.25 μg R9SA (a LacZ-marked defective SIV retroviral vector) [19]; and 0.5 μg of syncytin-Ory1 expression vector, using the Lipofectamine LTX transfection kit (Invitrogen). Supernatants from the transfected cells were harvested 48 h after transfection, filtered through 0.45 μm pore-size PVDF membranes, supplemented with Polybrene (4 μg/ml), transferred to target cells seeded in 24-well plates (5 × 104 - 8 × 104 cells per well) the day before infection, followed by spinoculation at 1200 × g for 2 h 30 min at room temperature. X-Gal staining was performed 3 days later.
In situ hybridization
Freshly collected rabbit placentae (at day 12 of gestation) were fixed in 4% paraformaldehyde at 4°C, embedded in paraffin, and serial sections (4 μm) were either stained with haematoxylin and eosin or used for in situ hybridization. A PCR-amplified 1135 bp syncytin-Ory1 fragment (primers: 5'-AGACTGCGGAGATAAAACTGC and 5'-GTGGACCGCGATTCCTAGTC) was cloned into pGEM-T Easy (Promega) for in vitro synthesis of the antisense and sense riboprobes, generated with SP6 RNA polymerase and digoxygenin 11-UTP (Roche Applied Science). Sections were processed, hybridized at 42°C overnight with the riboprobes and incubated further at room temperature for 2 h with alkaline phosphatase-conjugated anti-digoxygenin antibody Fab fragments (Roche Applied Science). Staining was revealed with NBT and BCIP phosphatase alkaline substrates as indicated by the manufacturer (Roche Applied Science).
Search for syncytin-Ory1 in other lagomorphs
Genomic DNAs from Lepus americanus, Lepus europaeus, Ochotona princeps, Sylvilagus brasiliensis and Sylvilagus floridanus were a gift from A. Surridge (Department of Zoology, University of Cambridge, UK). Genomic DNA from Lepus starcki was extracted from tissue given by F. Catzeflis (Laboratoire de Paléontologie, Université de Montpellier 2, France). Genomic DNAs were digested by Not I, which does not cut within the syncytin-Ory1 gene. PCRs were performed on 100 ng of DNA, using Accuprime Taq DNA Polymerase (Invitrogen) for 40 cycles (30 sec at 94°C, 30 sec at 50°C, 2 min at 68°C). The primers used were: 5'-TTCCTGAGGGCTCACTGATTAAC and 5'-GAAGGGGAGAGTCAGTTGTTGGAG (external to the ORF) or 5'-AGACTGCGGAGATAAAACTGC and 5'-gataaaggtcatcagcctattga (internal to the ORF). PCR products were then cloned in pGEM-T Easy vector (Promega) for subsequent sequencing. Primer sequences for splice site determination were: 5'-CTTGGGGTTCGAGCCTGT and 5'-TTGAGCACGGCCACGGCCAC, on each side of the putative splice donor (SD) and acceptor (SA) sequences, respectively (see Figure 3A).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
OH, CV, AD and TH designed research and drafted the manuscript. OH, CV and AD performed research. All authors read and approved the final manuscript.
Contributor Information
Odile Heidmann, Email: oheidmann@igr.fr.
Cécile Vernochet, Email: vernochet@igr.fr.
Anne Dupressoir, Email: dupresso@igr.fr.
Thierry Heidmann, Email: heidmann@igr.fr.
Acknowledgements
The authors wish to acknowledge A. Surridge and F. Catzeflis for the gift of tissues, P. Opolon and O. Bawa for histological analyses, P. Dessen at the Institut Gustave Roussy Bioinformatic Center for help in the computing work and the Institut Gustave Roussy - Service Commun d'Expérimentation Animale and INRA (Jouy en Josas, France) for animal care. We thank Christian Lavialle for comments and critical reading of the manuscript. This work was supported by the CNRS and a fellowship to C.V. from the Fondation pour la Recherche Médicale.
References
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC Jr, McCoy JM. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/JVI.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci USA. 2004;101:1731–1736. doi: 10.1073/pnas.0305763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise S, de Parseval N, Bénit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise S, Ruggieri A, Dewannieux M, Cosset F-L, Heidmann T. Identification of an envelope protein from the FRD family of Human Endogenous Retroviruses (HERV-FRD) conferring infectivity on retroviral particles and functional conservation among simians. J Virol. 2004;78:1050–1054. doi: 10.1128/JVI.78.2.1050-1054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeney M, Renard M, Schlecht-Louf G, Bouallaga I, Heidmann O, Letzelter C, Richaud A, Ducos B, Heidmann T. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci USA. 2007;104:20534–20539. doi: 10.1073/pnas.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Marceau G, Vernochet C, Bénit L, Kanellopoulos C, Sapin V, Heidmann T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci USA. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding P, Burton GJ. Comparative Placentation Structures, Functions and Evolution. Springer; 2008. Haemochorial Placentation: Mouse, Rabbit, Man, Apes, Monkeys; pp. 196–202. [Google Scholar]
- Larsen JF. Electron microscopy of the implantation site in the rabbit. Am J Anat. 1961;109:319–334. doi: 10.1002/aja.1001090307. [DOI] [PubMed] [Google Scholar]
- Larsen JF. Electron Microscopy of the Chorioallantoic Placenta of the Rabbit. I. The Placental labyrinth and the Multinucleated Giant Cells of the Intermediate Zone. J Ultrastructure Research. 1962;7:535–549. doi: 10.1016/S0022-5320(62)90044-8. [DOI] [Google Scholar]
- Benit L, Dessen P, Heidmann T. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J Virol. 2001;75:11709–11719. doi: 10.1128/JVI.75.23.11709-11719.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Priet S, Ribet D, Vernochet C, Bruls T, Lavialle C, Weissenbach J, Heidmann T. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc Natl Acad Sci USA. 2008;105:17532–17537. doi: 10.1073/pnas.0807413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko JE, Battini JL, Gottschalk RJ, Mazo I, Miller AD. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc Natl Acad Sci USA. 1999;96:2129–2134. doi: 10.1073/pnas.96.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor CS, Nouri A, Zhao Y, Takeuchi Y, Kabat D. A sodium-dependent neutral-amino-acid transporter mediates infections of feline and baboon endogenous retroviruses and simian type D retroviruses. J Virol. 1999;73:4470–4474. doi: 10.1128/jvi.73.5.4470-4474.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthee CA, van Vuuren BJ, Bell D, Robinson TJ. A molecular supermatrix of the rabbits and hares (Leporidae) allows for the identification of five intercontinental exchanges during the Miocene. Syst Biol. 2004;53:433–447. doi: 10.1080/10635150490445715. [DOI] [PubMed] [Google Scholar]
- de Parseval N, Lazar V, Bénit L, Casella J, Heidmann T. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise S, de Parseval N, Heidmann T. Functional characterization of two newly identified Human Endogenous Retrovirus coding envelope genes. Retrovirology. 2005;2:19. doi: 10.1186/1742-4690-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre D, Cosset FL. Vectors derived from simian immunodeficiency virus (SIV) Biochimie. 2002;84:1161–1171. doi: 10.1016/S0300-9084(02)00036-6. [DOI] [PubMed] [Google Scholar]
- Negre D, Mangeot PE, Duisit G, Blanchard S, Vidalain PO, Leissner P, Winter AJ, Rabourdin-Combe C, Mehtali M, Moullier P, Darlix JL, Cosset FL. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 2000;7:1613–1623. doi: 10.1038/sj.gt.3301292. [DOI] [PubMed] [Google Scholar]